Drug Information
Drug (ID: DG00109) and It's Reported Resistant Information
| Name |
Doxorubicin
|
||||
|---|---|---|---|---|---|
| Synonyms |
Doxorubicin; 23214-92-8; Doxil; Doxorubicine; Adriablastin; Doxorubicinum; 14-Hydroxydaunomycin; 14-Hydroxydaunorubicine; Doxorubicina; Adriamycin semiquinone; Doxorubicinum [INN-Latin]; Doxorubicine [INN-French]; Doxorubicina [INN-Spanish]; Myocet; FI 106; Doxorubicin [USAN:INN:BAN]; CCRIS 739; NDC 38242-874; HSDB 3070; UNII-80168379AG; NCI-C01514; EINECS 245-495-6; CHEMBL53463; CHEBI:28748; 5,12-Naphthacenedione,; ADM; ADR; ThermoDox; Aerosolized Doxorubicin; Doxorubicin citrate; RDF Rubex; Conjugate of doxorubicin with humanized monoclonal antibody LL1 against CD74; DM2; JT9100000; Adiblastine (hydrochloride salt); Adr iablatina (hydrochloride salt); Adriablastine (hydrochloride salt); Adriablatina (hydrochloride salt); Adriacin (hydrochloride salt); Adriamycin PFS (TN); Adriamycin PFS (hydrochloride salt); Adriamycin RDF (TN); Adriamycin RDF (hydrochloride salt); Adriblastina (TN); Adriblastina (hydrochloride salt); Adriblatina (hydrochloride salt); Caelyx (TN); Conjugate of doxorubicin with monoclonal antibody P4/D10 against GP120; DOX-SL; Doxorubicin hydrochloride (hydrochloride salt); Doxorubicin-hLL1; Doxorubicin-hLL1 conjugate; Farmablastina (hydrochloride salt); Hydroxydaunomycin hydrochlor ide (hydrochloride salt); Hydroxydaunomycin hydrochloride (hydrochloride salt); Hydroxydaunorubicin hydrochloride (hydrochloride salt); Myocet (TN); Rubex (TN); Rubex (hydrochloride salt); TLC D-99; Doxorubicin (USAN/INN); Doxorubicin-P4/D10; Doxorubicin-P4/D10 conjugate; Cantide + adriamycin
Click to Show/Hide
|
||||
| Indication |
In total 3 Indication(s)
|
||||
| Structure |
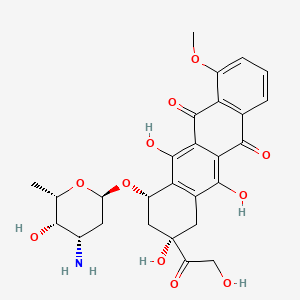
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Clinically Reported Resistance for This Drug
(29 diseases)
[2]
[3]
[4]
[5]
[6]
[7]
[8]
[9]
[10]
[11]
[12]
[8]
[13]
[14]
[15]
[16]
[17]
[18]
[19]
[20]
[21]
[22]
[23]
[24]
[25]
[26]
[27]
[28]
[29]
Disease(s) with Resistance Information Discovered by Cell Line Test for This Drug
(24 diseases)
[30]
[31]
[32]
[1]
[33]
[34]
[35]
[36]
[37]
[38]
[39]
[40]
[41]
[42]
[35]
[43]
[44]
[45]
[46]
[33]
[47]
[48]
[49]
[50]
|
||||
| Target | DNA topoisomerase II (TOP2) |
TOP2A_HUMAN
; TOP2B_HUMAN |
[1] | ||
| TERT messenger RNA (TERT mRNA) | TERT_HUMAN | [1] | |||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C27H29NO11
|
||||
| IsoSMILES |
C[C@H]1[C@H]([C@H](C[C@@H](O1)O[C@H]2C[C@@](CC3=C2C(=C4C(=C3O)C(=O)C5=C(C4=O)C(=CC=C5)OC)O)(C(=O)CO)O)N)O
|
||||
| InChI |
1S/C27H29NO11/c1-10-22(31)13(28)6-17(38-10)39-15-8-27(36,16(30)9-29)7-12-19(15)26(35)21-20(24(12)33)23(32)11-4-3-5-14(37-2)18(11)25(21)34/h3-5,10,13,15,17,22,29,31,33,35-36H,6-9,28H2,1-2H3/t10-,13-,15-,17-,22+,27-/m0/s1
|
||||
| InChIKey |
AOJJSUZBOXZQNB-TZSSRYMLSA-N
|
||||
| PubChem CID | |||||
| ChEBI ID | |||||
| TTD Drug ID | |||||
| VARIDT ID | |||||
| INTEDE ID | |||||
| DrugBank ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-01: Infectious/parasitic diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Putative ABC transporter ATP-binding component (OTRC) | [6] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli BL21 (DE3) | 469008 | ||
| Escherichia coli | 668369 | |||
| Escherichia coli ET12567 (pUZ8002) | 562 | |||
| Streptomyces rimosus M4018 | 1927 | |||
| Streptomyces rimosus SR16 | 1927 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | OtrC is a multidrug resistance protein based on an ATP hydrolysis-dependent active efflux mechanism.OtrC is a multidrug resistance protein based on an ATP hydrolysis-dependent active efflux mechanism. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Multidrug transporter MdfA (MDFA) | [29] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Salmonella enterica infection [ICD-11: 1A09.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Salmonella enterica serovar Typhimurium ATCC 14028s | 588858 | ||
| Experiment for Molecule Alteration |
Quantitative real-time PCR | |||
| Experiment for Drug Resistance |
L agar plate method assay | |||
| Mechanism Description | Overexpression or overproduction of mdfA confers drug resistance. | |||
| Key Molecule: Multidrug resistance protein MdtK (MDTK) | [29] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Salmonella enterica infection [ICD-11: 1A09.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Salmonella enterica serovar Typhimurium ATCC 14028s | 588858 | ||
| Experiment for Molecule Alteration |
Quantitative real-time PCR | |||
| Experiment for Drug Resistance |
L agar plate method assay | |||
| Mechanism Description | Overexpression or overproduction of mdtk confers drug resistance. | |||
ICD-02: Benign/in-situ/malignant neoplasm
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: piR-hsa-39980 | [1] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Neuroblastoma [ICD-11: 2A00.11] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell metastasis | Activation | hsa05205 | ||
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | IMR-32 cells | Abdomen | Homo sapiens (Human) | CVCL_0346 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | piR-39980 is an oncogenic piRNA overexpressed in NB cells which induces the cancer cell growth, enhance metastasis, and inhibit the cellular senescence by targeting JAk3 as well as desensitizes the chemotherapeutic drug. And piR-39980 was found to desensitize the effect of doxorubicin and inhibit drug-induced apoptosis. | |||
| Key Molecule: hsa-mir-125b | [8] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Primitive neuroectodermal tumor [ICD-11: 2A00.08] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| miR125b-p53/BAKT signaling pathway | Activation | hsa05206 | ||
| In Vitro Model | RD-ES cells | Bones | Homo sapiens (Human) | CVCL_2169 |
| Sk-ES cells | Bones | Homo sapiens (Human) | CVCL_0627 | |
| Sk-N-MC cells | Bones | Homo sapiens (Human) | CVCL_0530 | |
| TC-71 cells | Bones | Homo sapiens (Human) | CVCL_2213 | |
| VH-64 cells | Bones | Homo sapiens (Human) | CVCL_9672 | |
| WE-68 cells | Bones | Homo sapiens (Human) | CVCL_9717 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Celltiter-glo luminescent cell viability assay | |||
| Mechanism Description | miR-125b led to the development of chemoresistance by suppressing the expression of p53 and Bak, and repression of miR-125b sensitized EWS cells to apoptosis induced by treatment with various cytotoxic drugs. | |||
|
|
||||
| Key Molecule: Tyrosine-protein kinase JAK3 (JAK3) | [1] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Neuroblastoma [ICD-11: 2A00.11] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell metastasis | Activation | hsa05205 | ||
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | IMR-32 cells | Abdomen | Homo sapiens (Human) | CVCL_0346 |
| Experiment for Molecule Alteration |
Dual-luciferase reporter assay | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | piR-39980 is an oncogenic piRNA overexpressed in NB cells which induces the cancer cell growth, enhance metastasis, and inhibit the cellular senescence by targeting JAk3 as well as desensitizes the chemotherapeutic drug. And piR-39980 was found to desensitize the effect of doxorubicin and inhibit drug-induced apoptosis. | |||
| Key Molecule: Bcl-2 homologous antagonist/killer (BAK1) | [8] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Primitive neuroectodermal tumor [ICD-11: 2A00.08] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| miR125b-p53/BAKT signaling pathway | Activation | hsa05206 | ||
| In Vitro Model | RD-ES cells | Bones | Homo sapiens (Human) | CVCL_2169 |
| Sk-ES cells | Bones | Homo sapiens (Human) | CVCL_0627 | |
| Sk-N-MC cells | Bones | Homo sapiens (Human) | CVCL_0530 | |
| TC-71 cells | Bones | Homo sapiens (Human) | CVCL_2213 | |
| VH-64 cells | Bones | Homo sapiens (Human) | CVCL_9672 | |
| WE-68 cells | Bones | Homo sapiens (Human) | CVCL_9717 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
Celltiter-glo luminescent cell viability assay | |||
| Mechanism Description | miR-125b led to the development of chemoresistance by suppressing the expression of p53 and Bak, and repression of miR-125b sensitized EWS cells to apoptosis induced by treatment with various cytotoxic drugs. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-127 | [51] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Glioma [ICD-11: 2A00.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | AKT signaling pathway | Inhibition | hsa04151 | |
| Cell apoptosis | Activation | hsa04210 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| U87-MG cells | Brain | Homo sapiens (Human) | CVCL_0022 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTS assay; Flow cytometry assay | |||
| Mechanism Description | microRNA-127 silencing significantly affects cell growth and increases the sensitivity to adriamycin. microRNA-127 silencing arrests the cell cycle, potentiates adriamycin-induced apoptosis, and increases cellular Rh-123 uptake. microRNA-127 silencing down-regulates MDR1, MRP1, Runx2, Bcl-2, Survivin and ErbB4 expression while up-regulates p53 expression. microRNA-127 silencing inhibits AkT phosphorylation. | |||
| Key Molecule: hsa-mir-21 | [52] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Glioblastoma [ICD-11: 2A00.02] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | A172 cells | Brain | Homo sapiens (Human) | CVCL_0131 |
| T98G cells | Brain | Homo sapiens (Human) | CVCL_0556 | |
| U87MG cells | Brain | Homo sapiens (Human) | CVCL_GP63 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; TUNEL assay | |||
| Mechanism Description | To validate the possible association of miR-21 with drug resistance of T98G cells, we transfected anti-miR-21 inhibitor into the cells. The expression level of miR-21 was significantly lower in T98G transfected cells (than in the parental control cells). Transfected cells showed a high apoptotic rate compared to control after Dox treatment by TUNEL assay, suggesting that combined Dox and miR-21 inhibitor therapy can sensitize GBM resistant cells to anthracyclines by enhancing apoptosis. | |||
| Key Molecule: hsa-mir-137 | [53] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Neuroblastoma [ICD-11: 2A00.11] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | UkF-NB3 cells | Bone marrow | Homo sapiens (Human) | CVCL_9904 |
| In Vivo Model | Immunodeficient NCr nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Cell titer glo assay assay | |||
| Mechanism Description | Hypermethylation of the miR-137 promoter and negative regulation of miR-137 by CAR contribute in part to reduced miR-137 expression and increased CAR and MDR1 expression in doxorubicin-resistant neuroblastoma cells. | |||
|
|
||||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [51] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Glioma [ICD-11: 2A00.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | AKT signaling pathway | Inhibition | hsa04151 | |
| Cell apoptosis | Activation | hsa04210 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| U87-MG cells | Brain | Homo sapiens (Human) | CVCL_0022 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay; Flow cytometry assay | |||
| Mechanism Description | microRNA-127 silencing significantly affects cell growth and increases the sensitivity to adriamycin. microRNA-127 silencing arrests the cell cycle, potentiates adriamycin-induced apoptosis, and increases cellular Rh-123 uptake. microRNA-127 silencing down-regulates MDR1, MRP1, Runx2, Bcl-2, Survivin and ErbB4 expression while up-regulates p53 expression. microRNA-127 silencing inhibits AkT phosphorylation. | |||
| Key Molecule: Multidrug resistance-associated protein 1 (MRP1) | [51] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Glioma [ICD-11: 2A00.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | AKT signaling pathway | Inhibition | hsa04151 | |
| Cell apoptosis | Activation | hsa04210 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| U87-MG cells | Brain | Homo sapiens (Human) | CVCL_0022 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay; Flow cytometry assay | |||
| Mechanism Description | microRNA-127 silencing significantly affects cell growth and increases the sensitivity to adriamycin. microRNA-127 silencing arrests the cell cycle, potentiates adriamycin-induced apoptosis, and increases cellular Rh-123 uptake. microRNA-127 silencing down-regulates MDR1, MRP1, Runx2, Bcl-2, Survivin and ErbB4 expression while up-regulates p53 expression. microRNA-127 silencing inhibits AkT phosphorylation. | |||
|
|
||||
| Key Molecule: Apoptosis regulator Bcl-2 (BCL2) | [51] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Glioma [ICD-11: 2A00.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | AKT signaling pathway | Inhibition | hsa04151 | |
| Cell apoptosis | Activation | hsa04210 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| U87-MG cells | Brain | Homo sapiens (Human) | CVCL_0022 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay; Flow cytometry assay | |||
| Mechanism Description | microRNA-127 silencing significantly affects cell growth and increases the sensitivity to adriamycin. microRNA-127 silencing arrests the cell cycle, potentiates adriamycin-induced apoptosis, and increases cellular Rh-123 uptake. microRNA-127 silencing down-regulates MDR1, MRP1, Runx2, Bcl-2, Survivin and ErbB4 expression while up-regulates p53 expression. microRNA-127 silencing inhibits AkT phosphorylation. | |||
| Key Molecule: Receptor tyrosine-protein kinase erbB-4 (ERBB4) | [51] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Glioma [ICD-11: 2A00.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | AKT signaling pathway | Inhibition | hsa04151 | |
| Cell apoptosis | Activation | hsa04210 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| U87-MG cells | Brain | Homo sapiens (Human) | CVCL_0022 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay; Flow cytometry assay | |||
| Mechanism Description | microRNA-127 silencing significantly affects cell growth and increases the sensitivity to adriamycin. microRNA-127 silencing arrests the cell cycle, potentiates adriamycin-induced apoptosis, and increases cellular Rh-123 uptake. microRNA-127 silencing down-regulates MDR1, MRP1, Runx2, Bcl-2, Survivin and ErbB4 expression while up-regulates p53 expression. microRNA-127 silencing inhibits AkT phosphorylation. | |||
| Key Molecule: Cellular tumor antigen p53 (TP53) | [51] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Glioma [ICD-11: 2A00.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | AKT signaling pathway | Inhibition | hsa04151 | |
| Cell apoptosis | Activation | hsa04210 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| U87-MG cells | Brain | Homo sapiens (Human) | CVCL_0022 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay; Flow cytometry assay | |||
| Mechanism Description | microRNA-127 silencing significantly affects cell growth and increases the sensitivity to adriamycin. microRNA-127 silencing arrests the cell cycle, potentiates adriamycin-induced apoptosis, and increases cellular Rh-123 uptake. microRNA-127 silencing down-regulates MDR1, MRP1, Runx2, Bcl-2, Survivin and ErbB4 expression while up-regulates p53 expression. microRNA-127 silencing inhibits AkT phosphorylation. | |||
| Key Molecule: Runt-related transcription factor 2 (RUNX2) | [51] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Glioma [ICD-11: 2A00.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | AKT signaling pathway | Inhibition | hsa04151 | |
| Cell apoptosis | Activation | hsa04210 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| U87-MG cells | Brain | Homo sapiens (Human) | CVCL_0022 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay; Flow cytometry assay | |||
| Mechanism Description | microRNA-127 silencing significantly affects cell growth and increases the sensitivity to adriamycin. microRNA-127 silencing arrests the cell cycle, potentiates adriamycin-induced apoptosis, and increases cellular Rh-123 uptake. microRNA-127 silencing down-regulates MDR1, MRP1, Runx2, Bcl-2, Survivin and ErbB4 expression while up-regulates p53 expression. microRNA-127 silencing inhibits AkT phosphorylation. | |||
| Key Molecule: Baculoviral IAP repeat-containing protein 5 (BIRC5) | [51] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Glioma [ICD-11: 2A00.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | AKT signaling pathway | Inhibition | hsa04151 | |
| Cell apoptosis | Activation | hsa04210 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| U87-MG cells | Brain | Homo sapiens (Human) | CVCL_0022 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay; Flow cytometry assay | |||
| Mechanism Description | microRNA-127 silencing significantly affects cell growth and increases the sensitivity to adriamycin. microRNA-127 silencing arrests the cell cycle, potentiates adriamycin-induced apoptosis, and increases cellular Rh-123 uptake. microRNA-127 silencing down-regulates MDR1, MRP1, Runx2, Bcl-2, Survivin and ErbB4 expression while up-regulates p53 expression. microRNA-127 silencing inhibits AkT phosphorylation. | |||
| Key Molecule: Nuclear receptor subfamily 1 group I3 (NR1I3) | [53] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Neuroblastoma [ICD-11: 2A00.11] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | UkF-NB3 cells | Bone marrow | Homo sapiens (Human) | CVCL_9904 |
| In Vivo Model | Immunodeficient NCr nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Chromatin immunoprecipitation assay | |||
| Experiment for Drug Resistance |
Cell titer glo assay assay | |||
| Mechanism Description | Hypermethylation of the miR-137 promoter and negative regulation of miR-137 by CAR contribute in part to reduced miR-137 expression and increased CAR and MDR1 expression in doxorubicin-resistant neuroblastoma cells. | |||
| Key Molecule: Forkhead box protein M1 (FOXM1) | [54] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Rhabdoid tumor [ICD-11: 2A00.0Y] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Tm87-16 cells | Kidney | Homo sapiens (Human) | CVCL_8001 |
| TTC549 cells | Liver | Homo sapiens (Human) | CVCL_8005 | |
| STM91-01 cells | Lung | Homo sapiens (Human) | CVCL_8000 | |
| Experiment for Molecule Alteration |
qRT-PCR; Western blotting assay | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | FOXM1 siRNA and FOXM1 inhibitor (thiostrepton) successfully downregulated the mRNA and protein expression of FOXM1 in vitro and the downregulation of FOXM1 inhibited cell proliferation, drug resistance to doxorubicin, migration, invasion, and caused the cell cycle arrest and apoptosis of MRT cell lines. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: TP53 target 1 (TP53TG1) | [47] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | p53 signaling pathway | Inhibition | hsa04115 | |
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| MkN-45 cells | Gastric | Homo sapiens (Human) | CVCL_0434 | |
| GCIY cells | Gastric | Homo sapiens (Human) | CVCL_1228 | |
| KATO-3 cells | Gastric | Homo sapiens (Human) | CVCL_0371 | |
| MkN-7 cells | Gastric | Homo sapiens (Human) | CVCL_1417 | |
| SNU-1 cells | Gastric | Homo sapiens (Human) | CVCL_0099 | |
| TGBC11TkB cells | Gastric | Homo sapiens (Human) | CVCL_1768 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
MTT assay; TUNEL assay; xCELLigence Real-Time invasion and migration assays | |||
| Mechanism Description | TP53TG1, a p53-induced LncRNA, binds to the multifaceted RNA/RNA binding protein YBX1 to prevent its nuclear localization and thus the YBX1-mediated activation of oncogenes. The epigenetic silencing of TP53TG1 in cancer cells promotes the YBX1-mediated activation of the PI3k/AkT pathway, which then creates further resistance not only to common chemotherapy RNA-damaging agents but also to small drug-targeted inhibitors. | |||
|
|
||||
| Key Molecule: Y-box-binding protein 1 (YBX1) | [47] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | p53 signaling pathway | Inhibition | hsa04115 | |
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| MkN-45 cells | Gastric | Homo sapiens (Human) | CVCL_0434 | |
| GCIY cells | Gastric | Homo sapiens (Human) | CVCL_1228 | |
| KATO-3 cells | Gastric | Homo sapiens (Human) | CVCL_0371 | |
| MkN-7 cells | Gastric | Homo sapiens (Human) | CVCL_1417 | |
| SNU-1 cells | Gastric | Homo sapiens (Human) | CVCL_0099 | |
| TGBC11TkB cells | Gastric | Homo sapiens (Human) | CVCL_1768 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; TUNEL assay; xCELLigence Real-Time invasion and migration assays | |||
| Mechanism Description | TP53TG1, a p53-induced LncRNA, binds to the multifaceted RNA/RNA binding protein YBX1 to prevent its nuclear localization and thus the YBX1-mediated activation of oncogenes. The epigenetic silencing of TP53TG1 in cancer cells promotes the YBX1-mediated activation of the PI3k/AkT pathway, which then creates further resistance not only to common chemotherapy RNA-damaging agents but also to small drug-targeted inhibitors. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-495 | [55] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | A2780 cells | Ovary | Homo sapiens (Human) | CVCL_0134 |
| A2780C cells | Ovary | Homo sapiens (Human) | CVCL_0134 | |
| A2780DX5 cells | Ovary | Homo sapiens (Human) | CVCL_4T98 | |
| SGC7901R cells | Uterus | Homo sapiens (Human) | CVCL_0520 | |
| In Vivo Model | Mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
Annexin-V-FITC apoptosis detection assay; Caspase-3 activity assay; MTT assay; Trypan blue exclusion assay | |||
| Mechanism Description | miR-495 sensitizes MDR cancer cells to the combination of doxorubicin and taxol by inhibiting MDR1 expression, miR-495 was predicted to target ABCB1, which encodes protein MDR1. | |||
|
|
||||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [55] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | A2780 cells | Ovary | Homo sapiens (Human) | CVCL_0134 |
| A2780C cells | Ovary | Homo sapiens (Human) | CVCL_0134 | |
| A2780DX5 cells | Ovary | Homo sapiens (Human) | CVCL_4T98 | |
| SGC7901R cells | Uterus | Homo sapiens (Human) | CVCL_0520 | |
| In Vivo Model | Mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Annexin-V-FITC apoptosis detection assay; Caspase-3 activity assay; MTT assay; Trypan blue exclusion assay | |||
| Mechanism Description | miR-495 sensitizes MDR cancer cells to the combination of doxorubicin and taxol by inhibiting MDR1 expression, miR-495 was predicted to target ABCB1, which encodes protein MDR1. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-21 | [56] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Chronic myeloid leukemia [ICD-11: 2A20.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| PI3K/AKT/mTOR signaling pathway | Activation | hsa04151 | ||
| In Vitro Model | K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 |
| K562/A02 cells | Blood | Homo sapiens (Human) | CVCL_0368 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-21 is associated with inactivation of PTEN, a know tumor suppressor gene, resulting in activation of PI3k/Akt/mTOR signaling pathway, Akt promotes cell survival by inhibiting apoptosis through its ability to phosphorylate/inactivate downstream targets of apoptotic machinery. ADR sensitivity is associated with up-regulation of PTEN resulting from the inhibition of miR-21 expression. | |||
|
|
||||
| Key Molecule: Phosphatase and tensin homolog (PTEN) | [56] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Chronic myeloid leukemia [ICD-11: 2A20.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| PI3K/AKT/mTOR signaling pathway | Activation | hsa04151 | ||
| In Vitro Model | K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 |
| K562/A02 cells | Blood | Homo sapiens (Human) | CVCL_0368 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-21 is associated with inactivation of PTEN, a know tumor suppressor gene, resulting in activation of PI3k/Akt/mTOR signaling pathway, Akt promotes cell survival by inhibiting apoptosis through its ability to phosphorylate/inactivate downstream targets of apoptotic machinery. ADR sensitivity is associated with up-regulation of PTEN resulting from the inhibition of miR-21 expression. | |||
| Key Molecule: Annexin A1 (ANXA1) | [34] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Chronic myelogenous leukemia [ICD-11: 2A20.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The downregulated ANXA1,whose new role in apoptosis and cancer revealed recently,expression contributes considerably to the observed drug resistance in k562/ADR cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-9 | [57] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Chronic myeloid leukemia [ICD-11: 2A20.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR9 regulates the multidrug resistance of chronic myelogenous leukemia by targeting ABCB1. | |||
| Key Molecule: hsa-mir-181c | [58] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Chronic myeloid leukemia [ICD-11: 2A20.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 |
| Ku812 cells | Bone marrow | Homo sapiens (Human) | CVCL_0379 | |
| kCL22 cells | Pleural effusion | Homo sapiens (Human) | CVCL_2091 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | miR-181c directly targeted and inhibited the ST8SIA4 expression, as well as miR-181c was inversely correlated with the levels of ST8SIA4 expression in CML cell lines and samples. Moreover, ST8SIA4 could reverse the effect of miR-181c on drug resistance in k562 and k562/ADR cells in vitro. Upregulation of miR-181c sensitized k562/ADR cells to adriamycin in vivo through directly suppressing ST8SIA4 expression. Further investigation showed that miR-181c mediated the activity of phosphoinositide-3 kinase (PI3k)/AkT signal pathway, and inhibition of PI3k/Akt in k562 cells counteracted miR-181c-mediated MDR phenotype. | |||
|
|
||||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [57] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Chronic myeloid leukemia [ICD-11: 2A20.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR9 regulates the multidrug resistance of chronic myelogenous leukemia by targeting ABCB1. | |||
|
|
||||
| Key Molecule: Sialyltransferase St8Sia IV (SIAT8D) | [58] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Chronic myeloid leukemia [ICD-11: 2A20.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 |
| Ku812 cells | Bone marrow | Homo sapiens (Human) | CVCL_0379 | |
| kCL22 cells | Pleural effusion | Homo sapiens (Human) | CVCL_2091 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | miR-181c directly targeted and inhibited the ST8SIA4 expression, as well as miR-181c was inversely correlated with the levels of ST8SIA4 expression in CML cell lines and samples. Moreover, ST8SIA4 could reverse the effect of miR-181c on drug resistance in k562 and k562/ADR cells in vitro. Upregulation of miR-181c sensitized k562/ADR cells to adriamycin in vivo through directly suppressing ST8SIA4 expression. Further investigation showed that miR-181c mediated the activity of phosphoinositide-3 kinase (PI3k)/AkT signal pathway, and inhibition of PI3k/Akt in k562 cells counteracted miR-181c-mediated MDR phenotype. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-34 | [59] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 |
| HS-5 cells | Bone marrow | Homo sapiens (Human) | CVCL_3720 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | TUG1 epigenetically suppressed miR-34a expression by recruiting EZH2 to the promoter region of miR-34a and increasing H3k27me3 level to confer adriamycin resistance in acute myeloid leukemia. | |||
| Key Molecule: Taurine up-regulated 1 (TUG1) | [59] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 |
| HS-5 cells | Bone marrow | Homo sapiens (Human) | CVCL_3720 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | TUG1 epigenetically suppressed miR-34a expression by recruiting EZH2 to the promoter region of miR-34a and increasing H3k27me3 level to confer adriamycin resistance in acute myeloid leukemia. | |||
| Key Molecule: hsa-miR-520c-3p | [60] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| miR520c-3p/S100A4 signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | THP-1 cells | Blood | Homo sapiens (Human) | CVCL_0006 |
| U937 cells | Blood | Homo sapiens (Human) | CVCL_0007 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | HOXA-AS2 Can enhance S100A4 expression by suppressing miR-520c-3p expression to promote adriamycin resistance in acute myeloid leukemia through the miR-520c-3p /S100A4 pathway. | |||
| Key Molecule: HOXA cluster antisense RNA 2 (HOXA-AS2) | [60] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| miR520c-3p/S100A4 signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | THP-1 cells | Blood | Homo sapiens (Human) | CVCL_0006 |
| U937 cells | Blood | Homo sapiens (Human) | CVCL_0007 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | HOXA-AS2 Can enhance S100A4 expression by suppressing miR-520c-3p expression to promote adriamycin resistance in acute myeloid leukemia through the miR-520c-3p /S100A4 pathway. | |||
| Key Molecule: hsa-miR-153-5p | [61] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | THP-1 cells | Blood | Homo sapiens (Human) | CVCL_0006 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | CircPAN3 mediates drug resistance in acute myeloid leukemia through the miR-153-5p/miR-183-5p-XIAP axis. | |||
| Key Molecule: hsa-miR-183-5p | [61] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | THP-1 cells | Blood | Homo sapiens (Human) | CVCL_0006 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | CircPAN3 mediates drug resistance in acute myeloid leukemia through the miR-153-5p/miR-183-5p-XIAP axis. | |||
| Key Molecule: Long non-protein coding RNA 239 (LINC00239) | [62] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell colony | Activation | hsa05200 | ||
| Cell invasion | Activation | hsa05200 | ||
| Cell migration | Activation | hsa04670 | ||
| Cell proliferation | Activation | hsa05200 | ||
| Cell viability | Activation | hsa05200 | ||
| PI3K/AKT/mTOR signaling pathway | Activation | hsa04151 | ||
| In Vitro Model | KG-1 cells | Bone marrow | Homo sapiens (Human) | CVCL_0374 |
| HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 | |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay; TUNEL assay; Flow cytometry assay | |||
| Mechanism Description | Long non coding RNA linc00239 promotes malignant behaviors and chemoresistance against doxorubicin partially via activation of the PI3k/Akt/mTOR pathway in acute myeloid leukaemia cells. | |||
| Key Molecule: Urothelial cancer associated 1 (UCA1) | [31] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell viability | Activation | hsa05200 | |
| miR125a/hexokinase 2 pathway | Regulation | hsa05206 | ||
| In Vitro Model | HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 |
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Hk2, a target of miR-125a, was positively regulated by uca1 in HL60, and HL60/ADR cells,and UCA1 overexpression significantly attenuated miR-125-mediated inhibition on HIF-1alpha-dependent glycolysis in HL60 and HL60/ADR cells. | |||
| Key Molecule: hsa-mir-125b | [4] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Acute promyelocytic leukemia [ICD-11: 2A60.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 |
| K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 | |
| NB4 cells | Bone marrow | Homo sapiens (Human) | CVCL_0005 | |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | miR-125b could promote leukemic cell proliferation and inhibit cell apoptosis by regulating the expression of tumor suppressor BCL2-antagonist/killer 1 (Bak1). transfection of a miR-125b duplex into AML cells can increase their resistance to therapeutic drugs. | |||
|
|
||||
| Key Molecule: Protein S100-A4 (S100A4) | [60] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| miR520c-3p/S100A4 signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | THP-1 cells | Blood | Homo sapiens (Human) | CVCL_0006 |
| U937 cells | Blood | Homo sapiens (Human) | CVCL_0007 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | HOXA-AS2 Can enhance S100A4 expression by suppressing miR-520c-3p expression to promote adriamycin resistance in acute myeloid leukemia through the miR-520c-3p /S100A4 pathway. | |||
| Key Molecule: E3 ubiquitin-protein ligase XIAP (XIAP) | [61] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | THP-1 cells | Blood | Homo sapiens (Human) | CVCL_0006 |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | CircPAN3 mediates drug resistance in acute myeloid leukemia through the miR-153-5p/miR-183-5p-XIAP axis. | |||
| Key Molecule: RAC serine/threonine-protein kinase (AKT) | [62] | |||
| Molecule Alteration | Phosphorylation | Up-regulation |
||
| Resistant Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell colony | Activation | hsa05200 | ||
| Cell invasion | Activation | hsa05200 | ||
| Cell viability | Activation | hsa05200 | ||
| PI3K/AKT/mTOR signaling pathway | Activation | hsa04151 | ||
| In Vitro Model | KG-1 cells | Bone marrow | Homo sapiens (Human) | CVCL_0374 |
| HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; TUNEL assay; Flow cytometry assay | |||
| Mechanism Description | Long non coding RNA linc00239 promotes malignant behaviors and chemoresistance against doxorubicin partially via activation of the PI3k/Akt/mTOR pathway in acute myeloid leukaemia cells. | |||
| Key Molecule: Hexokinase-2 (HK2) | [31] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell viability | Activation | hsa05200 | |
| miR125a/hexokinase 2 pathway | Regulation | hsa05206 | ||
| In Vitro Model | HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Hk2, a target of miR-125a, was positively regulated by uca1 in HL60, and HL60/ADR cells,and UCA1 overexpression significantly attenuated miR-125-mediated inhibition on HIF-1alpha-dependent glycolysis in HL60 and HL60/ADR cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa_circ_PAN3 | [61] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | THP-1 cells | Blood | Homo sapiens (Human) | CVCL_0006 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | CircPAN3 mediates drug resistance in acute myeloid leukemia through the miR-153-5p/miR-183-5p-XIAP axis. | |||
| Key Molecule: hsa-miR-153-5p | [61] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | THP-1 cells | Blood | Homo sapiens (Human) | CVCL_0006 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | CircPAN3 mediates drug resistance in acute myeloid leukemia through the miR-153-5p/miR-183-5p-XIAP axis. | |||
| Key Molecule: hsa-miR-183-5p | [61] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | THP-1 cells | Blood | Homo sapiens (Human) | CVCL_0006 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | CircPAN3 mediates drug resistance in acute myeloid leukemia through the miR-153-5p/miR-183-5p-XIAP axis. | |||
| Key Molecule: hsa-mir-217 | [63] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 |
| K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 | |
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometric analysis | |||
| Mechanism Description | microRNA 217 inhibits cell proliferation and enhances chemosensitivity to doxorubicin in acute myeloid leukemia by targeting kRAS. | |||
| Key Molecule: Urothelial cancer associated 1 (UCA1) | [31] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell viability | Inhibition | hsa05200 | |
| miR125a/hexokinase 2 pathway | Regulation | hsa05206 | ||
| In Vitro Model | HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Hk2, a target of miR-125a, was positively regulated by uca1 in HL60, and HL60/ADR cells,and UCA1 overexpression significantly attenuated miR-125-mediated inhibition on HIF-1alpha-dependent glycolysis in HL60 and HL60/ADR cells. | |||
| Key Molecule: hsa-mir-181 | [64] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell migration | Inhibition | hsa04670 | ||
| In Vitro Model | HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 |
| K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | The ectopic expression of miR-181b in k562/A02 and HL-60/ADM cells robustly suppressed endogenous HMGB1 and Mcl-1 expression both at mRNA and protein levels. Conversely, knockdown of miR-181b by miR-181b inhibitor markedly increased the expression of both HMGB1 and Mcl-1. Restoration of miR-181b increased the drug sensitivity of AML MDR cells by targeting HMGB1 and Mcl-1. | |||
|
|
||||
| Key Molecule: GTPase KRas (KRAS) | [63] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 |
| K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 | |
| Experiment for Molecule Alteration |
Dual luciferase reporter assay; Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometric analysis | |||
| Mechanism Description | microRNA 217 inhibits cell proliferation and enhances chemosensitivity to doxorubicin in acute myeloid leukemia by targeting kRAS. | |||
| Key Molecule: Hexokinase-2 (HK2) | [31] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell viability | Inhibition | hsa05200 | |
| miR125a/hexokinase 2 pathway | Regulation | hsa05206 | ||
| In Vitro Model | HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Hk2, a target of miR-125a, was positively regulated by uca1 in HL60, and HL60/ADR cells,and UCA1 overexpression significantly attenuated miR-125-mediated inhibition on HIF-1alpha-dependent glycolysis in HL60 and HL60/ADR cells. | |||
| Key Molecule: High mobility group protein B1 (HMGB1) | [64] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell migration | Inhibition | hsa04670 | ||
| In Vitro Model | HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 |
| K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | The ectopic expression of miR-181b in k562/A02 and HL-60/ADM cells robustly suppressed endogenous HMGB1 and Mcl-1 expression both at mRNA and protein levels. Conversely, knockdown of miR-181b by miR-181b inhibitor markedly increased the expression of both HMGB1 and Mcl-1. Restoration of miR-181b increased the drug sensitivity of AML MDR cells by targeting HMGB1 and Mcl-1. | |||
| Key Molecule: Induced myeloid leukemia cell differentiation protein Mcl-1 (MCL1) | [64] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell migration | Inhibition | hsa04670 | ||
| In Vitro Model | HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 |
| K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | The ectopic expression of miR-181b in k562/A02 and HL-60/ADM cells robustly suppressed endogenous HMGB1 and Mcl-1 expression both at mRNA and protein levels. Conversely, knockdown of miR-181b by miR-181b inhibitor markedly increased the expression of both HMGB1 and Mcl-1. Restoration of miR-181b increased the drug sensitivity of AML MDR cells by targeting HMGB1 and Mcl-1. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Bromodomain containing 2 (BRD2) | [2] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Lymphoblastic lymphoma [ICD-11: 2A70.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | RasGRP1/Ras/ERK signaling pathway | Activation | hsa04010 | |
| Cell apoptosis | Inhibition | hsa04210 | ||
| In Vitro Model | Jurkat cells | Pleural effusion | Homo sapiens (Human) | CVCL_0065 |
| SUP-T1 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1714 | |
| MEK-293 cells | Blood vessel | Homo sapiens (Human) | N.A. | |
| In Vivo Model | BALB/C-nu/nu athymic nude mice model | Mus musculus | ||
| Experiment for Molecule Alteration |
sqRT-PCR; Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | The expression of BRD2 was upregulated in drug-resistant adult T-LBL samples. Functional studies of BRD2 further demonstrated the critical role of BRD2. BRD2 induces drug resistance of T-LBL by activating the MEK/ERK/c-Myc signaling pathway. | |||
| Key Molecule: Bromodomain containing 2 (BRD2) | [2] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Lymphoblastic lymphoma [ICD-11: 2A70.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | RasGRP1/Ras/ERK signaling pathway | Activation | hsa04010 | |
| Cell apoptosis | Inhibition | hsa04210 | ||
| In Vitro Model | Jurkat cells | Pleural effusion | Homo sapiens (Human) | CVCL_0065 |
| SUP-T1 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1714 | |
| MEK-293 cells | Blood vessel | Homo sapiens (Human) | N.A. | |
| In Vivo Model | BALB/C-nu/nu athymic nude mice model | Mus musculus | ||
| Experiment for Molecule Alteration |
sqRT-PCR; Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | The expression of BRD2 was upregulated in drug-resistant adult T-LBL samples. Functional studies of BRD2 further demonstrated the critical role of BRD2. BRD2 induces drug resistance of T-LBL by activating the MEK/ERK/c-Myc signaling pathway. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-148b | [11] | |||
| Molecule Alteration | Acetylation | Down-regulation |
||
| Resistant Disease | Diffuse large B-cell lymphoma [ICD-11: 2A81.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell viability | Activation | hsa05200 | |
| HDAC6/miR148b/Ezrin signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | CRL2631 cells | Bone marrow | Homo sapiens (Human) | CVCL_3611 |
| CRL2631/CHOP cells | Bone marrow | Homo sapiens (Human) | CVCL_3611 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | The high level of HDAC6 inhibited miR-148b via maintaining the low acetylation of histones H3 and H4 in the miR-148b promoter, thus rescuing Ezrin expression and promoting CHOP resistance in DLBCL. | |||
|
|
||||
| Key Molecule: hsa-miR-125b-5p | [65] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Diffuse large B-cell lymphoma [ICD-11: 2A81.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | SU-DHL-2 cells | Pleural effusion | Homo sapiens (Human) | CVCL_9550 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | Expression levels of exosomal miR-99a-5p/miR-125b-5p & their correlation with clinicopathological features in DLBCL patients, the expression levels of miR-99a-5p and miR-125b-5p were significantly higher in the chemoresistant group than in the chemosensitive group. | |||
| Key Molecule: hsa-miR-99a-5p | [65] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Diffuse large B-cell lymphoma [ICD-11: 2A81.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | SU-DHL-2 cells | Pleural effusion | Homo sapiens (Human) | CVCL_9550 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | Expression levels of exosomal miR-99a-5p/miR-125b-5p & their correlation with clinicopathological features in DLBCL patients, the expression levels of miR-99a-5p and miR-125b-5p were significantly higher in the chemoresistant group than in the chemosensitive group. | |||
|
|
||||
| Key Molecule: Ezrin (EZR) | [11] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Diffuse large B-cell lymphoma [ICD-11: 2A81.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell viability | Activation | hsa05200 | |
| HDAC6/miR148b/Ezrin signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | CRL2631 cells | Bone marrow | Homo sapiens (Human) | CVCL_3611 |
| CRL2631/CHOP cells | Bone marrow | Homo sapiens (Human) | CVCL_3611 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | The high level of HDAC6 inhibited miR-148b via maintaining the low acetylation of histones H3 and H4 in the miR-148b promoter, thus rescuing Ezrin expression and promoting CHOP resistance in DLBCL. | |||
| Key Molecule: Sirtuin 6 (SIRT6) | [66] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Diffuse large B-cell lymphoma [ICD-11: 2A81.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | MET/PI3K/AKT/mTOR signaling pathway | Activation | hsa04150 | |
| In Vitro Model | Val cells | Bone marrow | Homo sapiens (Human) | CVCL_1819 |
| LY1 cells | Ovary | Homo sapiens (Human) | CVCL_ZU83 | |
| DLBCL cells | Lymph node | Homo sapiens (Human) | N.A. | |
| LY8 cells | Lymph node | Homo sapiens (Human) | CVCL_8803 | |
| LY3 cells | Bone marrow | Homo sapiens (Human) | CVCL_8800 | |
| In Vivo Model | Beige mice xenografts model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Mechanism Description | Sirt6 expression was raised in DLBCL, with its high levels corresponding to poor patient outcomes. Sirt6 was also found to promote tumorigenesis by regulating the PI3K/Akt/mTOR pathway. Targeting Sirt6 exerted anti-lymphoma activity and enhanced chemo-sensitivity. OSS_128167 may prove to be a useful component in further development of novel chemotherapy regimens in DLBCL. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-miR-370-3p | [67] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Diffuse large B-cell lymphoma [ICD-11: 2A81.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | MAPK/BCR/PI signaling pathway | Regulation | hsa04662 | |
| In Vitro Model | SUDHL-4 cells | Peritoneal effusion | Homo sapiens (Human) | CVCL_0539 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CellTiter-Blue Cell Viability assay | |||
| Mechanism Description | miR370-3p, miR381-3p, and miR409-3p miRNAs appear to be the most potent regulators of the MAPk, BCR, and PI signaling system. Overexpression of miR370-3p, miR381-3p, and miR409-3p increases sensitivity to rituximab and doxorubicin. | |||
| Key Molecule: hsa-miR-381-3p | [67] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Diffuse large B-cell lymphoma [ICD-11: 2A81.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | MAPK/BCR/PI signaling pathway | Regulation | hsa04662 | |
| In Vitro Model | SUDHL-4 cells | Peritoneal effusion | Homo sapiens (Human) | CVCL_0539 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CellTiter-Blue Cell Viability assay | |||
| Mechanism Description | miR370-3p, miR381-3p, and miR409-3p miRNAs appear to be the most potent regulators of the MAPk, BCR, and PI signaling system. Overexpression of miR370-3p, miR381-3p, and miR409-3p increases sensitivity to rituximab and doxorubicin. | |||
| Key Molecule: hsa-miR-409-3p | [67] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Diffuse large B-cell lymphoma [ICD-11: 2A81.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | MAPK/BCR/PI signaling pathway | Regulation | hsa04662 | |
| In Vitro Model | SUDHL-4 cells | Peritoneal effusion | Homo sapiens (Human) | CVCL_0539 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CellTiter-Blue Cell Viability assay | |||
| Mechanism Description | miR370-3p, miR381-3p, and miR409-3p miRNAs appear to be the most potent regulators of the MAPk, BCR, and PI signaling system. Overexpression of miR370-3p, miR381-3p, and miR409-3p increases sensitivity to rituximab and doxorubicin. | |||
| Key Molecule: hsa-mir-199a | [68] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Diffuse large B-cell lymphoma [ICD-11: 2A81.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | SUDHL-4 cells | Peritoneal effusion | Homo sapiens (Human) | CVCL_0539 |
| Karpas-422 cells | Peritoneal effusion | Homo sapiens (Human) | CVCL_1325 | |
| RI-1 cells | Peritoneal effusion | Homo sapiens (Human) | CVCL_1885 | |
| U2932 cells | Peritoneal effusion | Homo sapiens (Human) | CVCL_1896 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | High expression of miR-497 or miR-199a was associated with better overall survival (p = 0.042 and p = 0.007). Overexpression of miR-199a and miR-497 led to a statistically significant decrease in viable cells in a dose-dependent fashion after exposure to rituximab and various chemotherapeutics relevant in multi-agent lymphoma therapy. Our data indicate that elevated miR-199a and miR-497 levels are associated with improved survival in aggressive lymphoma patients most likely by modifying drug sensitivity to immunochemotherapy. This functional impairment may serve as a potential novel therapeutic target in future treatment of patients with DLBCL. Overexpression of the individual miRNAs did not result in any difference in cell viability, cell growth or apoptosis. | |||
| Key Molecule: hsa-mir-497 | [68] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Diffuse large B-cell lymphoma [ICD-11: 2A81.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | SUDHL-4 cells | Peritoneal effusion | Homo sapiens (Human) | CVCL_0539 |
| Karpas-422 cells | Peritoneal effusion | Homo sapiens (Human) | CVCL_1325 | |
| RI-1 cells | Peritoneal effusion | Homo sapiens (Human) | CVCL_1885 | |
| U2932 cells | Peritoneal effusion | Homo sapiens (Human) | CVCL_1896 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | High expression of miR-497 or miR-199a was associated with better overall survival (p = 0.042 and p = 0.007). Overexpression of miR-199a and miR-497 led to a statistically significant decrease in viable cells in a dose-dependent fashion after exposure to rituximab and various chemotherapeutics relevant in multi-agent lymphoma therapy. Our data indicate that elevated miR-199a and miR-497 levels are associated with improved survival in aggressive lymphoma patients most likely by modifying drug sensitivity to immunochemotherapy. This functional impairment may serve as a potential novel therapeutic target in future treatment of patients with DLBCL. Overexpression of the individual miRNAs did not result in any difference in cell viability, cell growth or apoptosis. | |||
| Key Molecule: hsa-mir-21 | [69] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Diffuse large B-cell lymphoma [ICD-11: 2A81.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K/AKT signaling pathway | Inhibition | hsa04151 | |
| In Vitro Model | CRL2631 cells | Bone marrow | Homo sapiens (Human) | CVCL_3611 |
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-21 impacts the PI3k/AkT signaling pathway through the regulation of PTEN, thereby affecting cellular sensitivity to the CHOP chemotherapeutic regimen. | |||
|
|
||||
| Key Molecule: Inositol monophosphatase 1 (IMPA1) | [67] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Diffuse large B-cell lymphoma [ICD-11: 2A81.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | MAPK/BCR/PI signaling pathway | Regulation | hsa04662 | |
| In Vitro Model | SUDHL-4 cells | Peritoneal effusion | Homo sapiens (Human) | CVCL_0539 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CellTiter-Blue Cell Viability assay | |||
| Mechanism Description | miR370-3p, miR381-3p, and miR409-3p miRNAs appear to be the most potent regulators of the MAPk, BCR, and PI signaling system. Overexpression of miR370-3p, miR381-3p, and miR409-3p increases sensitivity to rituximab and doxorubicin. | |||
| Key Molecule: Mitogen-activated protein kinase kinase kinase 8 (MAP3K8) | [67] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Diffuse large B-cell lymphoma [ICD-11: 2A81.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | MAPK/BCR/PI signaling pathway | Regulation | hsa04662 | |
| In Vitro Model | SUDHL-4 cells | Peritoneal effusion | Homo sapiens (Human) | CVCL_0539 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CellTiter-Blue Cell Viability assay | |||
| Mechanism Description | miR370-3p, miR381-3p, and miR409-3p miRNAs appear to be the most potent regulators of the MAPk, BCR, and PI signaling system. Overexpression of miR370-3p, miR381-3p, and miR409-3p increases sensitivity to rituximab and doxorubicin. | |||
| Key Molecule: Mitogen-activated protein kinase 1 (MAPK1) | [67] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Diffuse large B-cell lymphoma [ICD-11: 2A81.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | MAPK/BCR/PI signaling pathway | Regulation | hsa04662 | |
| In Vitro Model | SUDHL-4 cells | Peritoneal effusion | Homo sapiens (Human) | CVCL_0539 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CellTiter-Blue Cell Viability assay | |||
| Mechanism Description | miR370-3p, miR381-3p, and miR409-3p miRNAs appear to be the most potent regulators of the MAPk, BCR, and PI signaling system. Overexpression of miR370-3p, miR381-3p, and miR409-3p increases sensitivity to rituximab and doxorubicin. | |||
| Key Molecule: PI3-kinase delta (PIK3CD) | [67] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Diffuse large B-cell lymphoma [ICD-11: 2A81.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | MAPK/BCR/PI signaling pathway | Regulation | hsa04662 | |
| In Vitro Model | SUDHL-4 cells | Peritoneal effusion | Homo sapiens (Human) | CVCL_0539 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CellTiter-Blue Cell Viability assay | |||
| Mechanism Description | miR370-3p, miR381-3p, and miR409-3p miRNAs appear to be the most potent regulators of the MAPk, BCR, and PI signaling system. Overexpression of miR370-3p, miR381-3p, and miR409-3p increases sensitivity to rituximab and doxorubicin. | |||
| Key Molecule: PI3-kinase gamma (PIK3CG) | [67] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Diffuse large B-cell lymphoma [ICD-11: 2A81.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | MAPK/BCR/PI signaling pathway | Regulation | hsa04662 | |
| In Vitro Model | SUDHL-4 cells | Peritoneal effusion | Homo sapiens (Human) | CVCL_0539 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CellTiter-Blue Cell Viability assay | |||
| Mechanism Description | miR370-3p, miR381-3p, and miR409-3p miRNAs appear to be the most potent regulators of the MAPk, BCR, and PI signaling system. Overexpression of miR370-3p, miR381-3p, and miR409-3p increases sensitivity to rituximab and doxorubicin. | |||
| Key Molecule: PI3-kinase regulatory subunit alpha (PIK3R1) | [67] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Diffuse large B-cell lymphoma [ICD-11: 2A81.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | MAPK/BCR/PI signaling pathway | Regulation | hsa04662 | |
| In Vitro Model | SUDHL-4 cells | Peritoneal effusion | Homo sapiens (Human) | CVCL_0539 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CellTiter-Blue Cell Viability assay | |||
| Mechanism Description | miR370-3p, miR381-3p, and miR409-3p miRNAs appear to be the most potent regulators of the MAPk, BCR, and PI signaling system. Overexpression of miR370-3p, miR381-3p, and miR409-3p increases sensitivity to rituximab and doxorubicin. | |||
| Key Molecule: Phosphatase and tensin homolog (PTEN) | [69] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Diffuse large B-cell lymphoma [ICD-11: 2A81.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K/AKT signaling pathway | Inhibition | hsa04151 | |
| In Vitro Model | CRL2631 cells | Bone marrow | Homo sapiens (Human) | CVCL_3611 |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-21 impacts the PI3k/AkT signaling pathway through the regulation of PTEN, thereby affecting cellular sensitivity to the CHOP chemotherapeutic regimen. | |||
| Key Molecule: Sirtuin 6 (SIRT6) | [66] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Diffuse large B-cell lymphoma [ICD-11: 2A81.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | MET/PI3K/AKT/mTOR signaling pathway | Activation | hsa04150 | |
| In Vitro Model | Val cells | Bone marrow | Homo sapiens (Human) | CVCL_1819 |
| LY1 cells | Ovary | Homo sapiens (Human) | CVCL_ZU83 | |
| DLBCL cells | Lymph node | Homo sapiens (Human) | N.A. | |
| LY8 cells | Lymph node | Homo sapiens (Human) | CVCL_8803 | |
| LY3 cells | Bone marrow | Homo sapiens (Human) | CVCL_8800 | |
| In Vivo Model | Beige mice xenografts model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Mechanism Description | Sirt6 expression was raised in DLBCL, with its high levels corresponding to poor patient outcomes. Sirt6 was also found to promote tumorigenesis by regulating the PI3K/Akt/mTOR pathway. Targeting Sirt6 exerted anti-lymphoma activity and enhanced chemo-sensitivity. OSS_128167 may prove to be a useful component in further development of novel chemotherapy regimens in DLBCL. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-155 | [70] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Multiple myeloma [ICD-11: 2A83.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | RPMI8226/Dox cells | Peripheral blood | Homo sapiens (Human) | CVCL_0014 |
| RPMI8226/S cells | Peripheral blood | Homo sapiens (Human) | CVCL_0014 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Targeting inhibition of miR155 expression could restore chemotherapy sensitivity by increasing FOXO3a expression in drug-resistant myeloma cells. | |||
|
|
||||
| Key Molecule: Forkhead box protein O3 (FOXO3) | [70] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Multiple myeloma [ICD-11: 2A83.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | RPMI8226/Dox cells | Peripheral blood | Homo sapiens (Human) | CVCL_0014 |
| RPMI8226/S cells | Peripheral blood | Homo sapiens (Human) | CVCL_0014 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Targeting inhibition of miR155 expression could restore chemotherapy sensitivity by increasing FOXO3a expression in drug-resistant myeloma cells. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-miR-17-92 | [42] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Mantle cell lymphoma [ICD-11: 2A85.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| PI3K/AKT signaling pathway | Activation | hsa04151 | ||
| In Vitro Model | Jeko-1 cells | Blood | Homo sapiens (Human) | CVCL_1865 |
| Granta-519 cells | Blood | Homo sapiens (Human) | CVCL_1818 | |
| Z138c cells | Blood | Homo sapiens (Human) | CVCL_B077 | |
| In Vivo Model | CB-17/SCID nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Xenograft experiments assay | |||
| Mechanism Description | The protein phosphatase PHLPP2, an important negative regulator of the PI3k/AkT pathway, was a direct target of miR-17 92 miRNAs, miRNA-17 92 cluster mediates chemoresistance and enhances tumor growth in mantle cell lymphoma via PI3k/AkT pathway activation. | |||
|
|
||||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [71] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Burkitt lymphoma [ICD-11: 2A85.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HS-Sultan cells | Ascites | Homo sapiens (Human) | CVCL_2516 |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
Trypan blue dye exclusion assay | |||
| Mechanism Description | MDR1 and Survivin upregulation are responsible for resistance to conventional drugs and dasatinib can restore drug sensitivity by reducing MDR1 and Survivin expression in drug-resistant BL cells. Src inhibitors could therefore be a novel treatment strategy for patients with drug resistant BL. | |||
|
|
||||
| Key Molecule: Fructose-bisphosphatase 1 (FBP1) | [20] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | B-cell non-Hodgkin lymphoma [ICD-11: 2A85.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | OCI-LY8 cells | Oral | Homo sapiens (Human) | CVCL_8803 |
| Daudi cells | Peripheral blood | Homo sapiens (Human) | CVCL_0008 | |
| Experiment for Molecule Alteration |
Immunoblot analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Mechanism Description | Cell adhesion mediated drug resistance (CAM DR) remains a major obstacle to the effectiveness of chemotherapeutic treatment of lymphoma. Far upstream element binding protein 1 (FBP1) is a multifunctional protein that is highly expressed in proliferating cells of several solid neoplasms. CAM-DR is considered a major mechanism by which tumor cells escape the cytotoxic effects of therapeutic agents. | |||
|
|
||||
| Key Molecule: PH domain leucine-rich repeat-containing protein phosphatase 2 (PHLPP2) | [42] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Mantle cell lymphoma [ICD-11: 2A85.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| PI3K/AKT signaling pathway | Activation | hsa04151 | ||
| In Vitro Model | Jeko-1 cells | Blood | Homo sapiens (Human) | CVCL_1865 |
| Granta-519 cells | Blood | Homo sapiens (Human) | CVCL_1818 | |
| Z138c cells | Blood | Homo sapiens (Human) | CVCL_B077 | |
| In Vivo Model | CB-17/SCID nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Luciferase assay | |||
| Experiment for Drug Resistance |
Xenograft experiments assay | |||
| Mechanism Description | The protein phosphatase PHLPP2, an important negative regulator of the PI3k/AkT pathway, was a direct target of miR-17 92 miRNAs, miRNA-17 92 cluster mediates chemoresistance and enhances tumor growth in mantle cell lymphoma via PI3k/AkT pathway activation. | |||
| Key Molecule: Baculoviral IAP repeat-containing protein 5 (BIRC5) | [71] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Burkitt lymphoma [ICD-11: 2A85.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HS-Sultan cells | Ascites | Homo sapiens (Human) | CVCL_2516 |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
Trypan blue dye exclusion assay | |||
| Mechanism Description | MDR1 and Survivin upregulation are responsible for resistance to conventional drugs and dasatinib can restore drug sensitivity by reducing MDR1 and Survivin expression in drug-resistant BL cells. Src inhibitors could therefore be a novel treatment strategy for patients with drug resistant BL. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [71] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Burkitt lymphoma [ICD-11: 2A85.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HS-Sultan cells | Ascites | Homo sapiens (Human) | CVCL_2516 |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
Trypan blue dye exclusion assay | |||
| Mechanism Description | MDR1 and Survivin upregulation are responsible for resistance to conventional drugs and dasatinib can restore drug sensitivity by reducing MDR1 and Survivin expression in drug-resistant BL cells. Src inhibitors could therefore be a novel treatment strategy for patients with drug resistant BL. | |||
|
|
||||
| Key Molecule: Fructose-bisphosphatase 1 (FBP1) | [20] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | B-cell non-Hodgkin lymphoma [ICD-11: 2A85.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | OCI-LY8 cells | Oral | Homo sapiens (Human) | CVCL_8803 |
| Daudi cells | Peripheral blood | Homo sapiens (Human) | CVCL_0008 | |
| Experiment for Molecule Alteration |
Immunoblot analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Mechanism Description | Cell adhesion mediated drug resistance (CAM DR) remains a major obstacle to the effectiveness of chemotherapeutic treatment of lymphoma. Far upstream element binding protein 1 (FBP1) is a multifunctional protein that is highly expressed in proliferating cells of several solid neoplasms. CAM-DR is considered a major mechanism by which tumor cells escape the cytotoxic effects of therapeutic agents. | |||
|
|
||||
| Key Molecule: Baculoviral IAP repeat-containing protein 5 (BIRC5) | [71] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Burkitt lymphoma [ICD-11: 2A85.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HS-Sultan cells | Ascites | Homo sapiens (Human) | CVCL_2516 |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
Trypan blue dye exclusion assay | |||
| Mechanism Description | MDR1 and Survivin upregulation are responsible for resistance to conventional drugs and dasatinib can restore drug sensitivity by reducing MDR1 and Survivin expression in drug-resistant BL cells. Src inhibitors could therefore be a novel treatment strategy for patients with drug resistant BL. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-187 | [21] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Peripheral T-cell lymphoma [ICD-11: 2A90.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell proliferation | Activation | hsa05200 | |
| In Vitro Model | MOLT4 cells | Bone marrow | Homo sapiens (Human) | CVCL_0013 |
| HUT78 cells | Lymph | Homo sapiens (Human) | CVCL_0337 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR187 downregulated tumor suppressor gene disabled homolog-2 (Dab2), decreased the interaction of Dab2 with adapter protein Grb2, resulting in Ras activation, phosphorylation/activation of extracellular signal-regulated kinase (ERk) and AkT, and subsequent stabilization of MYC oncoprotein. MiR187-overexpressing cells were resistant to chemotherapeutic agents like doxorubicin, cyclophosphamide, cisplatin and gemcitabine, but sensitive to the proteasome inhibitor bortezomib. | |||
|
|
||||
| Key Molecule: CXC chemokine receptor type 4 (CXCR4) | [23] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | T-cell lymphoma [ICD-11: 2A60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | CXCL12/CXCR4 signaling pathway | Activation | hsa04061 | |
| In Vitro Model | MyLa cells | Embryo | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Western blotting assay | |||
| Experiment for Drug Resistance |
XTT assay | |||
| Mechanism Description | MF-Fs promote migration and chemoresistance of MyLa cells through CXCL12/CXCR4 signaling. Through the entire range of Doxo concentrations, MyLa cells cocultured with MF-Fs were significantly more resistant than MyLa cells cocultured with N-Fs. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: C-X-C motif chemokine ligand 12 (CXCL12) | [23] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Mycosis fungoides [ICD-11: 2B01.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycosis fungoides tissue | N.A. | ||
| Experiment for Molecule Alteration |
Immunohistochemistry assay | |||
| Experiment for Drug Resistance |
XTT assay | |||
| Mechanism Description | MF cultures yielded significantly increased levels of FAPalpha, a CAF marker, and CAF-associated genes and proteins: CXCL12 (ligand of CXCR4 expressed on MF cells), collagen XI, and matrix metalloproteinase 2. Cultured MF fibroblasts showed greater proliferation than normal fibroblasts in ex vivo experiments. A coculture with MyLa cells (MF cell line) increased normal fibroblast growth, reduced the sensitivity of MyLa cells to doxorubicin, and enhanced their migration. Inhibiting the CXCL12/CXCR4 axis increased doxorubicin-induced apoptosis of MyLa cells and reduced MyLa cell motility. | |||
| Key Molecule: CXC chemokine receptor type 4 (CXCR4) | [23] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Mycosis fungoides [ICD-11: 2B01.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycosis fungoides tissue | N.A. | ||
| Experiment for Molecule Alteration |
Immunohistochemistry assay | |||
| Experiment for Drug Resistance |
XTT assay | |||
| Mechanism Description | MF cultures yielded significantly increased levels of FAPalpha, a CAF marker, and CAF-associated genes and proteins: CXCL12 (ligand of CXCR4 expressed on MF cells), collagen XI, and matrix metalloproteinase 2. Cultured MF fibroblasts showed greater proliferation than normal fibroblasts in ex vivo experiments. A coculture with MyLa cells (MF cell line) increased normal fibroblast growth, reduced the sensitivity of MyLa cells to doxorubicin, and enhanced their migration. Inhibiting the CXCL12/CXCR4 axis increased doxorubicin-induced apoptosis of MyLa cells and reduced MyLa cell motility. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-27a | [3] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Leukemia [ICD-11: 2B33.6] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The expression of miR-331-5p and miR-27a was inversely correlated with MDR1 expression. Transfection of exogenous miR-27a or miR-331-5p, or a combination of these two miRNAs, down-regulated MDR1 and increased sensitivity of the k562-resistant cancer cells to DOX. | |||
| Key Molecule: hsa-miR-331-5p | [3] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Leukemia [ICD-11: 2B33.6] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The expression of miR-331-5p and miR-27a was inversely correlated with MDR1 expression. Transfection of exogenous miR-27a or miR-331-5p, or a combination of these two miRNAs, down-regulated MDR1 and increased sensitivity of the k562-resistant cancer cells to DOX. | |||
| Key Molecule: H19, imprinted maternally expressed transcript (H19) | [30] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Leukemia [ICD-11: 2B33.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/AdrVp cells | Breast | Homo sapiens (Human) | CVCL_4Y46 | |
| Experiment for Molecule Alteration |
RT-PCR; Northern blotting analysis | |||
| Experiment for Drug Resistance |
Clonogenic assay | |||
| Mechanism Description | The mRNA of the H19 gene is overexpressed in MCF-7/AdrVp cells relative toparental MCF-7 cells or drug-sensitive MCF-7/AdrVp revertant cells. H19is an imprinted gene with an important role in fetal differentiation, as well as a postulated function as a tumor suppressor gene. Another p95-over-expressing multidrug-resistant cell line, human lung carcinoma NCI-H1688, also displays high levels of 1119 mRNA. | |||
|
|
||||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [3] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Leukemia [ICD-11: 2B33.6] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 |
| Experiment for Molecule Alteration |
Western blotting analysis; Immunofluorescence analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The expression of miR-331-5p and miR-27a was inversely correlated with MDR1 expression. Transfection of exogenous miR-27a or miR-331-5p, or a combination of these two miRNAs, down-regulated MDR1 and increased sensitivity of the k562-resistant cancer cells to DOX. | |||
|
|
||||
| Key Molecule: Bcl-2 homologous antagonist/killer (BAK1) | [4] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Acute promyelocytic leukemia [ICD-11: 2A60.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 |
| K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 | |
| NB4 cells | Bone marrow | Homo sapiens (Human) | CVCL_0005 | |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | miR-125b could promote leukemic cell proliferation and inhibit cell apoptosis by regulating the expression of tumor suppressor BCL2-antagonist/killer 1 (Bak1). transfection of a miR-125b duplex into AML cells can increase their resistance to therapeutic drugs. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Cytochrome P450 family 3 subfamily A member1 (CYP3A4) | [72] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Acute lymphocytic leukemia [ICD-11: 2B33.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | CCRF-CEM cells | Pleural effusion | Homo sapiens (Human) | CVCL_0207 |
| CEM/ADR5000 cells | Bone marrow | Homo sapiens (Human) | CVCL_D544 | |
| Experiment for Molecule Alteration |
CYP450-Glo CYP 3A4 assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | In this study, resveratrol was a significant inhibitor of CYP3A4 enzyme activity with IC50 value 9.32 ( M). Moreover, the CYP3A4 mRNA levels were reduced after treatment with resveratrol 0.03-fold of the control levels with high significance (p < 0.001). | |||
| Key Molecule: Glutathione S-transferase (GST) | [72] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Acute lymphocytic leukemia [ICD-11: 2B33.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | CCRF-CEM cells | Pleural effusion | Homo sapiens (Human) | CVCL_0207 |
| CEM/ADR5000 cells | Bone marrow | Homo sapiens (Human) | CVCL_D544 | |
| Experiment for Molecule Alteration |
Glutathione-S-transferase assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The Glutathione-S-transferases (GSTs) are a multigene family of dimeric proteins which play a central role in the detoxification of electrophilic xenobiotics and catalyze their conjugation with GSH to electrophilic metabolites, thus rendering them more water soluble. GSTs protect cells from cytotoxic and carcinogenic chemicals. GST activity was decreased by resveratrol in a dose dependent manner. IC50 value was 30.73 M. This results were confirmed by RT-PCR data, where the tested samples changed the GST mRNA level by 0.79-fold (p < 0.01) of control level. | |||
|
|
||||
| Key Molecule: hsa-mir-98 | [73] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Leukemia [ICD-11: 2B33.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 |
| K562/A02 cells | Blood | Homo sapiens (Human) | CVCL_0368 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | The targeted upregulated expression of miR98 could decrease the proliferation of leukemia cells and improve the sensitivity to chemotherapeutics by inhibiting E2F1 expression. | |||
| Key Molecule: hsa-miR-485-3p | [74] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Lymphocytic leukemia [ICD-11: 2B33.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | CEM cells | Pleural effusion | Homo sapiens (Human) | N.A. |
| CEM/VM-1-5 cells | Lymph | Homo sapiens (Human) | CVCL_1B35 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-485-3p expression can mediate etoposide sensitivity indirectly by fine-tuning Top2alpha expression through the modification of NF-YB expression. Accordingly, miR-485-3p can be a putative therapeutic target to modulate etoposide resistance in tumor cells. | |||
| Key Molecule: hsa-mir-138 | [75] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Leukemia [ICD-11: 2B33.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 |
| Experiment for Molecule Alteration |
qRT-PCR; Northern blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-138 was found up-regulated in the vincristine-induced multidrug resistance (MDR) leukemia cell line HL-60/VCR as compared with HL-60 cells. Up-regulation of miR-138 could reverse resistance of both P-glycoprotein-related and P-glycoprotein-non-related drugs on HL-60/VCR cells, and promote adriamycin-induced apoptosis, accompanied by increased accumulation and decreased releasing amount of adriamycin. | |||
|
|
||||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [75] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Leukemia [ICD-11: 2B33.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-138 was found up-regulated in the vincristine-induced multidrug resistance (MDR) leukemia cell line HL-60/VCR as compared with HL-60 cells. Up-regulation of miR-138 could reverse resistance of both P-glycoprotein-related and P-glycoprotein-non-related drugs on HL-60/VCR cells, and promote adriamycin-induced apoptosis, accompanied by increased accumulation and decreased releasing amount of adriamycin. | |||
| Key Molecule: ATP-binding cassette sub-family B5 (ABCB5) | [72] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Acute lymphocytic leukemia [ICD-11: 2B33.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | CCRF-CEM cells | Pleural effusion | Homo sapiens (Human) | CVCL_0207 |
| CEM/ADR5000 cells | Bone marrow | Homo sapiens (Human) | CVCL_D544 | |
| Experiment for Molecule Alteration |
Efflux of rhodamine123 assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Resveratrol can restore the sensitivity of Caco-2 and CEM/ADR5000 cell lines to doxorubicin, through enhancing significantly doxorubicin cytotoxicity. ABC-transporter inhibitors, classified according to their action on ABC-transporters proteins into: 1. Function inhibitors, 2. Expression inhibitors, and 3. Functional and expression inhibitors, which have an ideal characters of ABC-transporters inhibitors. Our results indicate that resveratrol falls into the class 3 inhibitors. | |||
|
|
||||
| Key Molecule: Transcription factor E2F1 (E2F1) | [73] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Leukemia [ICD-11: 2B33.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 |
| K562/A02 cells | Blood | Homo sapiens (Human) | CVCL_0368 | |
| Experiment for Molecule Alteration |
RT-PCR; Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | The targeted upregulated expression of miR98 could decrease the proliferation of leukemia cells and improve the sensitivity to chemotherapeutics by inhibiting E2F1 expression. | |||
| Key Molecule: Nuclear transcription factor Y subunit beta (NFYB) | [74] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Lymphocytic leukemia [ICD-11: 2B33.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | CEM cells | Pleural effusion | Homo sapiens (Human) | N.A. |
| CEM/VM-1-5 cells | Lymph | Homo sapiens (Human) | CVCL_1B35 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-485-3p expression can mediate etoposide sensitivity indirectly by fine-tuning Top2alpha expression through the modification of NF-YB expression. Accordingly, miR-485-3p can be a putative therapeutic target to modulate etoposide resistance in tumor cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-125b | [76] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Chondrosarcoma [ICD-11: 2B50.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| Glucose metabolism signaling pathway | Regulation | hsa05230 | ||
| In Vitro Model | CH-2879 cells | Bone | Homo sapiens (Human) | CVCL_9921 |
| OUMS-27 cells | Bone | Homo sapiens (Human) | CVCL_3090 | |
| SW1353 cells | Bone | Homo sapiens (Human) | CVCL_0543 | |
| CS-1 cells | Bone | Homo sapiens (Human) | CVCL_T023 | |
| CSPG cells | Bone | Homo sapiens (Human) | N.A. | |
| JJ012 cells | Bone | Homo sapiens (Human) | CVCL_D605 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-125b was downregulated in chondrosarcoma cells compared with normal human chondrocytes. More importantly, miR-125b was downregulated in doxorubicin resistant cancer cells, with its overexpression enhancing doxorubicin-induced cytotoxicity and apoptosis, subsequently increasing the sensitivity of chondrosarcoma cells to doxorubicin. ErbB2 was a direct target of miR-125b in chondrosarcoma cells. The inhibition of ErbB2 by overexpression of miR-125b led to suppression of glucose metabolism, which rendered chondrosarcoma cells susceptible to doxorubicin. Restoring the expression of ErbB2 and glucose metabolic enzymes recovered doxorubicin resistance in counteracting miR-125b-mediated sensitivity. Taken together, miR-125b plays a critical role in doxorubicin resistance through suppression of ErbB2-induced glucose metabolism, and it may serve as a potential target for overcoming chemoresistance in human chondrosarcoma. | |||
| Key Molecule: hsa-mir-125b | [76] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Chondrosarcoma [ICD-11: 2B50.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell viability | Inhibition | hsa05200 | |
| In Vitro Model | CH-2879 cells | Bone | Homo sapiens (Human) | CVCL_9921 |
| OUMS-27 cells | Bone | Homo sapiens (Human) | CVCL_3090 | |
| SW1353 cells | Bone | Homo sapiens (Human) | CVCL_0543 | |
| CS-1 cells | Bone | Homo sapiens (Human) | CVCL_T023 | |
| CSPG cells | Bone | Homo sapiens (Human) | N.A. | |
| JJ012 cells | Bone | Homo sapiens (Human) | CVCL_D605 | |
| SNM83 cells | Cartilage | Homo sapiens (Human) | N.A. | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-143 enhances the antitumor activity of shikonin by targeting BAG3 and reducing its expression in human glioblastoma stem cell. ErbB2. miR-125 was downregulated in chondrosarcoma cells and doxorubicin resistant cells. Overexpression of miR-125 enhanced the sensitivity of both parental and doxorubicin resistant cells to doxorubicin through direct targeting on the ErbB2-mediated upregulation of glycolysis in chondrosarcoma cells. Moreover, restoration of the expression of ErbB2 and glucose metabolic enzymes in miR-125 pretransfected cells recovered the susceptibility to doxorubicin. | |||
|
|
||||
| Key Molecule: Receptor tyrosine-protein kinase erbB-2 (ERBB2) | [76] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Chondrosarcoma [ICD-11: 2B50.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| Glucose metabolism signaling pathway | Regulation | hsa05230 | ||
| In Vitro Model | CH-2879 cells | Bone | Homo sapiens (Human) | CVCL_9921 |
| OUMS-27 cells | Bone | Homo sapiens (Human) | CVCL_3090 | |
| SW1353 cells | Bone | Homo sapiens (Human) | CVCL_0543 | |
| CS-1 cells | Bone | Homo sapiens (Human) | CVCL_T023 | |
| CSPG cells | Bone | Homo sapiens (Human) | N.A. | |
| JJ012 cells | Bone | Homo sapiens (Human) | CVCL_D605 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-125b was downregulated in chondrosarcoma cells compared with normal human chondrocytes. More importantly, miR-125b was downregulated in doxorubicin resistant cancer cells, with its overexpression enhancing doxorubicin-induced cytotoxicity and apoptosis, subsequently increasing the sensitivity of chondrosarcoma cells to doxorubicin. ErbB2 was a direct target of miR-125b in chondrosarcoma cells. The inhibition of ErbB2 by overexpression of miR-125b led to suppression of glucose metabolism, which rendered chondrosarcoma cells susceptible to doxorubicin. Restoring the expression of ErbB2 and glucose metabolic enzymes recovered doxorubicin resistance in counteracting miR-125b-mediated sensitivity. Taken together, miR-125b plays a critical role in doxorubicin resistance through suppression of ErbB2-induced glucose metabolism, and it may serve as a potential target for overcoming chemoresistance in human chondrosarcoma. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-152 | [77] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| c-Met/PI3K/AKT signaling pathway | Activation | hsa01521 | ||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay; Soft agar assay | |||
| Mechanism Description | LncRNAPVT1 targets miR-152 to enhance chemoresistance of osteosarcoma to doxorubicin through activating c-MET/PI3k/AkT pathway. | |||
| Key Molecule: Pvt1 oncogene (PVT1) | [77] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| c-Met/PI3K/AKT signaling pathway | Activation | hsa01521 | ||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay; Soft agar assay | |||
| Mechanism Description | LncRNAPVT1 targets miR-152 to enhance chemoresistance of osteosarcoma to doxorubicin through activating c-MET/PI3k/AkT pathway. | |||
| Key Molecule: hsa-miR-20a-5p | [78] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | G-292 cells | Bone | Homo sapiens (Human) | CVCL_2909 |
| SJSA-1 cells | Bone | Homo sapiens (Human) | CVCL_1697 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | miR20a-5p modulates multi-drug resistance by repressing SDC2 expression in OS cells. | |||
| Key Molecule: hsa-mir-375 | [79] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Multivariate analysis of overall survival or disease-free survival assay | |||
| Mechanism Description | miR375 overexpression could increase the cisplatin sensitivity of human gastric cancer cells by regulating ERBB2. | |||
| Key Molecule: hsa-miR-3182 | [80] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | ODRUL/miR3182/MMP2 signaling pathway | Regulation | hsa05206 | |
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| SAOS-2 cells | Bone marrow | Homo sapiens (Human) | CVCL_0548 | |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| HOS cells | Bone | Homo sapiens (Human) | CVCL_0312 | |
| 143B cells | Bone | Homo sapiens (Human) | CVCL_2270 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Dual Luciferase Reporter Assay; PCR | |||
| Experiment for Drug Resistance |
Wound Healing Assay; Colony Formation Assay; CCK8 Assay | |||
| Mechanism Description | LncRNA ODRUL Contributes to Osteosarcoma Progression through the miR3182/MMP2 Axis. miR3182 expression and function are inversely correlated with ODRUL expression in vitro and in vivo, miR3182 negatively regulated the mRNA level and the protein level of MMP2 expression. ODRUL could directly interact with miR3182 and upregulate MMP2 expression via its competing endogenous RNA activity on miR3182 at the posttranscriptional level. | |||
| Key Molecule: FOXC2 antisense RNA 1 (FOXC2-AS1) | [80] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | ODRUL/miR3182/MMP2 signaling pathway | Regulation | hsa05206 | |
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| SAOS-2 cells | Bone marrow | Homo sapiens (Human) | CVCL_0548 | |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| HOS cells | Bone | Homo sapiens (Human) | CVCL_0312 | |
| 143B cells | Bone | Homo sapiens (Human) | CVCL_2270 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
PCR | |||
| Experiment for Drug Resistance |
Wound Healing Assay; Colony Formation Assay; CCK8 Assay | |||
| Mechanism Description | LncRNA ODRUL Contributes to Osteosarcoma Progression through the miR3182/MMP2 Axis. miR3182 expression and function are inversely correlated with ODRUL expression in vitro and in vivo, miR3182 negatively regulated the mRNA level and the protein level of MMP2 expression. ODRUL could directly interact with miR3182 and upregulate MMP2 expression via its competing endogenous RNA activity on miR3182 at the posttranscriptional level. | |||
| Key Molecule: hsa-miR-140-5p | [81] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell autophagy | Activation | hsa04140 | |
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| hFOB1.19 cells | Fetal bone | Homo sapiens (Human) | CVCL_3708 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
IC50 assay; Flow cytometric analysis | |||
| Mechanism Description | miR140-5p/HMGN5/autophagy regulatory loop plays a critical role in chemoresistance in osteosarcoma, miR 140-5p regulates osteosarcoma chemoresistance by targeting HMGN5 and autophagy. | |||
| Key Molecule: FOXC2 antisense RNA 1 (FOXC2-AS1) | [24] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| SAOS-2 cells | Bone marrow | Homo sapiens (Human) | CVCL_0548 | |
| HOS cells | Bone | Homo sapiens (Human) | CVCL_0312 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Colony formation assay | |||
| Mechanism Description | Antisense LncRNA FOXC2-AS1 promotes doxorubicin resistance in osteosarcoma by increasing the expression of FOXC2. FOXC2-AS1 contributes to doxorubicin resistance by increasing FOXC2 and further facilitating ABCB1. | |||
| Key Molecule: Delta-like protein 1 (DLL1) | [82] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | ATF2/ATF3/ATF4 signaling pathway | Inhibition | hsa04915 | |
| In Vitro Model | G-292 cells | Bone | Homo sapiens (Human) | CVCL_2909 |
| SJSA-1 cells | Bone | Homo sapiens (Human) | CVCL_1697 | |
| MG63.2 cells | Bone | Homo sapiens (Human) | CVCL_R705 | |
| MNNG/HOS cells | Bone | Homo sapiens (Human) | CVCL_0439 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
IC50 assay; Flow cytometric analysis | |||
| Mechanism Description | miR34a-5p promotes multi-chemoresistance of osteosarcoma through down-regulation of the DLL1 gene. The activity of the ATF2/ATF3/ATF4 pathway was reduced in the miR34a-5p mimic-transfected G-292 cells but increased in the miR34a-5p antagomiRtransfected SJSA-1 cells, hence the ATF2/ATF3/ATF4 pathway was validated to be involved in the OS chemoresistance mediated by miR34a-5p. | |||
| Key Molecule: Small nucleolar RNA host gene 12 (SNHG12) | [83] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell viability | Activation | hsa05200 | ||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| SAOS-2 cells | Bone marrow | Homo sapiens (Human) | CVCL_0548 | |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| HOS cells | Bone | Homo sapiens (Human) | CVCL_0312 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | The up-regulation of MCL1 reversed the sensitivity of doxorubicin induced by miR-320a mimics and knockdown of SNHG12. | |||
| Key Molecule: hsa-mir-320 | [83] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell viability | Activation | hsa05200 | ||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| SAOS-2 cells | Bone marrow | Homo sapiens (Human) | CVCL_0548 | |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| HOS cells | Bone | Homo sapiens (Human) | CVCL_0312 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | The up-regulation of MCL1 reversed the sensitivity of doxorubicin induced by miR-320a mimics and knockdown of SNHG12. | |||
| Key Molecule: hsa-miR-199a-3p | [84] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell viability | Activation | hsa05200 | |
| NF-kappaB signaling pathway | Inhibition | hsa04064 | ||
| In Vitro Model | U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 |
| G-292 cells | Bone | Homo sapiens (Human) | CVCL_2909 | |
| MNNG/HOS cells | Bone | Homo sapiens (Human) | CVCL_0439 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | The Ak4 gene is one of the targets of miR-199a-3p and negatively correlates with the effect of miR-199a-3p on OS drug-resistance. | |||
| Key Molecule: hsa_circ_PVT1 | [85] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell colony | Activation | hsa05200 | |
| Cell viability | Activation | hsa05200 | ||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| SAOS-2 cells | Bone marrow | Homo sapiens (Human) | CVCL_0548 | |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| KHOS cells | Bone | Homo sapiens (Human) | CVCL_2546 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Colony formation assay | |||
| Mechanism Description | CircPVT1 knockdown reduces the expression of classical multidrug resistance related gene-ABCB1 in OS cells. | |||
| Key Molecule: hsa-miR-184 | [86] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | AKT/BCL2 signaling pathway | Regulation | hsa04933 | |
| Cell apoptosis | Activation | hsa04210 | ||
| NF-kappaB signaling pathway | Regulation | hsa04064 | ||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | microRNA-184 modulates doxorubicin resistance in osteosarcoma cells by targeting BCL2L1 and enhancing the level of it. | |||
| Key Molecule: hsa-miR-34a-5p | [87] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Activation | hsa05200 | |
| Cell migration | Activation | hsa04670 | ||
| Cell proliferation | Activation | hsa05200 | ||
| MEF2 signaling pathway | Regulation | hsa04013 | ||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| SAOS-2 cells | Bone marrow | Homo sapiens (Human) | CVCL_0548 | |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| G-292 cells | Bone | Homo sapiens (Human) | CVCL_2909 | |
| SJSA-1 cells | Bone | Homo sapiens (Human) | CVCL_1697 | |
| MG63.2 cells | Bone | Homo sapiens (Human) | CVCL_R705 | |
| MNNG/HOS cells | Bone | Homo sapiens (Human) | CVCL_0439 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The down-regulation of CD117 mediated by miR-34a-5p might be one of the reasons for OS drug resistance. CD117 may also regulate other processes, including cell adhesion, differentiation and migration, which are significant for cancer development and treatment. | |||
| Key Molecule: hsa-mir-30a | [88] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Down-regulation of miR-30a contributed to chemoresistance of osteosarcoma cells through regulating autophagy. Furthermore, to investigate the mechanism of miR-130a in regulating autophagy, bioinformatics analysis was performed. The results showed that the 3'-UTR region of Beclin-1 were the binding sites for miR-30a. Consistently, previous studies demonstrated that Beclin-1 was the directly target of miR-30a. | |||
| Key Molecule: hsa-miR-146b-5p | [89] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| Cell viability | Activation | hsa05200 | ||
| Wnt/Beta-catenin signaling pathway | Regulation | hsa04310 | ||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| hFOB1.19 cells | Fetal bone | Homo sapiens (Human) | CVCL_3708 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | miR-146b-5p was highly expressed in human osteosarcoma tissues and an elevated expression of miR-146b-5p was observed in human osteosarcoma tissues after chemotherapy. Furthermore, it was shown that miR-146b-5p overexpression promoted migration and invasiveness. miR-146b-5p overexpression also increased resistance to chemotherapy. Moreover, knockdown of miR-146b-5p substantially inhibited migration and invasion of osteosarcoma cells as well as rendered them significantly more sensitive to chemotherapy. Results of western blot assay indicated that miR-146b-5p increased MMP-16 protein expression and showed a decrease of ZNRF3 protein. Whereas, IWR-1-endo, an inhibitor of Wnt/beta-catenin, suppressed the decrease in apoptosis of osteosarcoma cells caused by miR-146b-5p overexpression. These results indicated that miR-146b-5p promoted proliferation, migration and invasiveness. | |||
| Key Molecule: hsa-mir-143 | [90] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | SAOS-2 cells | Bone marrow | Homo sapiens (Human) | CVCL_0548 |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| In Vivo Model | NOD-SCID IL2-rgamma -null (NSG) mouse engraftment model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Matrigel colony formation assay; Hoechst33342 staining assay | |||
| Mechanism Description | In chemoresistant SAOS-2 and U2OS osteosarcomas cells, miR-143 levels were significantly downregulated and accompanied by increases in ATG2B, Bcl-2, and/or LC3-II protein levels, high rate of ALDH1+CD133+ cells, and an increase in Matrigel colony formation ability. H2O2 upregulated p53 and miR-143, but downregulated ATG2B, Bcl-2, and LC3-I expression in U2OS cells (wild-type p53) but not in SAOS-2 (p53-null) cells. Forced miR-143 expression significantly reversed chemoresistance as well as downregulation of ATG2B, LC3-I, and Bcl-2 expression in SAOS-2- and U2OS-resistant cells. Forced miR-143 expression significantly inhibited tumor growth in xenograft SAOS-2-Dox and U2OS-Dox animal models. Loss of miR-143 expression is associated with poor prognosis of patients with osteosarcoma underlying chemotherapy. The chemoresistance of osteosarcoma tumor cells to doxorubicin is associated with the downregulation of miR-143 expression, activation of ALDH1+CD133+ cells, activation of autophagy, and inhibition of cell death. | |||
| Key Molecule: hsa-mir-155 | [43] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Activation | hsa05200 | |
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| SAOS-2 cells | Bone marrow | Homo sapiens (Human) | CVCL_0548 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | During treatment with Dox or Cis in osteosarcoma cells, miR-155 expression was strongly induced. The increased miR-155 expression facilitated tumor cell proliferation via upregulating autophagy, thus, facilitated the resistance of osteosarcoma cells to Dox or Cis. | |||
| Key Molecule: hsa-miR-34a-5p | [82], [91] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | ATF2/ATF3/ATF4 signaling pathway | Inhibition | hsa04915 | |
| In Vitro Model | G-292 cells | Bone | Homo sapiens (Human) | CVCL_2909 |
| SJSA-1 cells | Bone | Homo sapiens (Human) | CVCL_1697 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Annexin V-FITC/propidium iodide (PI) staining assay | |||
| Mechanism Description | The miR34a-5p promotes the multi-chemoresistance of osteosarcoma via repression of the AGTR1 gene. And miR34a-5p promotes multi-chemoresistance of osteosarcoma through down-regulation of the DLL1 gene. | |||
|
|
||||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [85] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell colony | Activation | hsa05200 | |
| Cell viability | Activation | hsa05200 | ||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| SAOS-2 cells | Bone marrow | Homo sapiens (Human) | CVCL_0548 | |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| KHOS cells | Bone | Homo sapiens (Human) | CVCL_2546 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Colony formation assay | |||
| Mechanism Description | CircPVT1 knockdown reduces the expression of classical multidrug resistance related gene-ABCB1 in OS cells. | |||
|
|
||||
| Key Molecule: Syndecan-2 (SDC2) | [78] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | G-292 cells | Bone | Homo sapiens (Human) | CVCL_2909 |
| SJSA-1 cells | Bone | Homo sapiens (Human) | CVCL_1697 | |
| Experiment for Molecule Alteration |
Luciferase reporter assay; Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | miR20a-5p modulates multi-drug resistance by repressing SDC2 expression in OS cells. | |||
| Key Molecule: Nucleosome-binding protein 1 (NSBP1) | [81] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell autophagy | Activation | hsa04140 | |
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| hFOB1.19 cells | Fetal bone | Homo sapiens (Human) | CVCL_3708 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Luciferase activity assay; Western blot analysis | |||
| Experiment for Drug Resistance |
IC50 assay; Flow cytometric analysis | |||
| Mechanism Description | miR140-5p/HMGN5/autophagy regulatory loop plays a critical role in chemoresistance in osteosarcoma, miR 140-5p regulates osteosarcoma chemoresistance by targeting HMGN5 and autophagy. | |||
| Key Molecule: Forkhead box protein C2 (FOXC2) | [24] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| SAOS-2 cells | Bone marrow | Homo sapiens (Human) | CVCL_0548 | |
| HOS cells | Bone | Homo sapiens (Human) | CVCL_0312 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Colony formation assay | |||
| Mechanism Description | Antisense LncRNA FOXC2-AS1 promotes doxorubicin resistance in osteosarcoma by increasing the expression of FOXC2. FOXC2-AS1 contributes to doxorubicin resistance by increasing FOXC2 and further facilitating ABCB1. | |||
| Key Molecule: Type-1 angiotensin II receptor (AGTR1) | [91] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | G-292 cells | Bone | Homo sapiens (Human) | CVCL_2909 |
| SJSA-1 cells | Bone | Homo sapiens (Human) | CVCL_1697 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Annexin V-FITC/propidium iodide (PI) staining assay | |||
| Mechanism Description | The miR34a-5p promotes the multi-chemoresistance of osteosarcoma via repression of the AGTR1 gene. | |||
| Key Molecule: Induced myeloid leukemia cell differentiation protein Mcl-1 (MCL1) | [83] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell viability | Activation | hsa05200 | ||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| SAOS-2 cells | Bone marrow | Homo sapiens (Human) | CVCL_0548 | |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| HOS cells | Bone | Homo sapiens (Human) | CVCL_0312 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | The up-regulation of MCL1 reversed the sensitivity of doxorubicin induced by miR-320a mimics and knockdown of SNHG12. | |||
| Key Molecule: Adenylate kinase 4 (AK4) | [84] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell viability | Activation | hsa05200 | |
| NF-kappaB signaling pathway | Inhibition | hsa04064 | ||
| In Vitro Model | U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 |
| G-292 cells | Bone | Homo sapiens (Human) | CVCL_2909 | |
| MNNG/HOS cells | Bone | Homo sapiens (Human) | CVCL_0439 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis; RIP assay; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | The Ak4 gene is one of the targets of miR-199a-3p and negatively correlates with the effect of miR-199a-3p on OS drug-resistance. | |||
| Key Molecule: Bcl-2-like protein 11 (BCL2L11) | [86] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | AKT/BCL2 signaling pathway | Regulation | hsa04933 | |
| Cell apoptosis | Activation | hsa04210 | ||
| NF-kappaB signaling pathway | Regulation | hsa04064 | ||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | microRNA-184 modulates doxorubicin resistance in osteosarcoma cells by targeting BCL2L1 and enhancing the level of it. | |||
| Key Molecule: Mast/stem cell growth factor receptor Kit (KIT) | [87] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Activation | hsa05200 | |
| Cell migration | Activation | hsa04670 | ||
| Cell proliferation | Activation | hsa05200 | ||
| MEF2 signaling pathway | Regulation | hsa04013 | ||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| SAOS-2 cells | Bone marrow | Homo sapiens (Human) | CVCL_0548 | |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| G-292 cells | Bone | Homo sapiens (Human) | CVCL_2909 | |
| SJSA-1 cells | Bone | Homo sapiens (Human) | CVCL_1697 | |
| MG63.2 cells | Bone | Homo sapiens (Human) | CVCL_R705 | |
| MNNG/HOS cells | Bone | Homo sapiens (Human) | CVCL_0439 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The down-regulation of CD117 mediated by miR-34a-5p might be one of the reasons for OS drug resistance. CD117 may also regulate other processes, including cell adhesion, differentiation and migration, which are significant for cancer development and treatment. | |||
| Key Molecule: Beclin-1 (BECN1) | [88] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Down-regulation of miR-30a contributed to chemoresistance of osteosarcoma cells through regulating autophagy. Furthermore, to investigate the mechanism of miR-130a in regulating autophagy, bioinformatics analysis was performed. The results showed that the 3'-UTR region of Beclin-1 were the binding sites for miR-30a. Consistently, previous studies demonstrated that Beclin-1 was the directly target of miR-30a. | |||
| Key Molecule: E3 ubiquitin-protein ligase ZNRF3 (ZNRF3) | [89] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell invasion | Activation | hsa05200 | ||
| Cell migration | Activation | hsa04670 | ||
| Cell proliferation | Activation | hsa05200 | ||
| Wnt/Beta-catenin signaling pathway | Regulation | hsa04310 | ||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| hFOB1.19 cells | Fetal bone | Homo sapiens (Human) | CVCL_3708 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | miR-146b-5p was highly expressed in human osteosarcoma tissues and an elevated expression of miR-146b-5p was observed in human osteosarcoma tissues after chemotherapy. Furthermore, it was shown that miR-146b-5p overexpression promoted migration and invasiveness. miR-146b-5p overexpression also increased resistance to chemotherapy. Moreover, knockdown of miR-146b-5p substantially inhibited migration and invasion of osteosarcoma cells as well as rendered them significantly more sensitive to chemotherapy. Results of western blot assay indicated that miR-146b-5p increased MMP-16 protein expression and showed a decrease of ZNRF3 protein. Whereas, IWR-1-endo, an inhibitor of Wnt/beta-catenin, suppressed the decrease in apoptosis of osteosarcoma cells caused by miR-146b-5p overexpression. These results indicated that miR-146b-5p promoted proliferation, migration and invasiveness. | |||
| Key Molecule: Autophagy-related protein 2 homolog B (ATG2B) | [90] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | SAOS-2 cells | Bone marrow | Homo sapiens (Human) | CVCL_0548 |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Matrigel colony formation assay; Hoechst33342 staining assay | |||
| Mechanism Description | In chemoresistant SAOS-2 and U2OS osteosarcomas cells, miR-143 levels were significantly downregulated and accompanied by increases in ATG2B, Bcl-2, and/or LC3-II protein levels, high rate of ALDH1+CD133+ cells, and an increase in Matrigel colony formation ability. H2O2 upregulated p53 and miR-143, but downregulated ATG2B, Bcl-2, and LC3-I expression in U2OS cells (wild-type p53) but not in SAOS-2 (p53-null) cells. Forced miR-143 expression significantly reversed chemoresistance as well as downregulation of ATG2B, LC3-I, and Bcl-2 expression in SAOS-2- and U2OS-resistant cells. Forced miR-143 expression significantly inhibited tumor growth in xenograft SAOS-2-Dox and U2OS-Dox animal models. Loss of miR-143 expression is associated with poor prognosis of patients with osteosarcoma underlying chemotherapy. The chemoresistance of osteosarcoma tumor cells to doxorubicin is associated with the downregulation of miR-143 expression, activation of ALDH1+CD133+ cells, activation of autophagy, and inhibition of cell death. | |||
| Key Molecule: Apoptosis regulator Bcl-2 (BCL2) | [90] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | SAOS-2 cells | Bone marrow | Homo sapiens (Human) | CVCL_0548 |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Matrigel colony formation assay; Hoechst33342 staining assay | |||
| Mechanism Description | In chemoresistant SAOS-2 and U2OS osteosarcomas cells, miR-143 levels were significantly downregulated and accompanied by increases in ATG2B, Bcl-2, and/or LC3-II protein levels, high rate of ALDH1+CD133+ cells, and an increase in Matrigel colony formation ability. H2O2 upregulated p53 and miR-143, but downregulated ATG2B, Bcl-2, and LC3-I expression in U2OS cells (wild-type p53) but not in SAOS-2 (p53-null) cells. Forced miR-143 expression significantly reversed chemoresistance as well as downregulation of ATG2B, LC3-I, and Bcl-2 expression in SAOS-2- and U2OS-resistant cells. Forced miR-143 expression significantly inhibited tumor growth in xenograft SAOS-2-Dox and U2OS-Dox animal models. Loss of miR-143 expression is associated with poor prognosis of patients with osteosarcoma underlying chemotherapy. The chemoresistance of osteosarcoma tumor cells to doxorubicin is associated with the downregulation of miR-143 expression, activation of ALDH1+CD133+ cells, activation of autophagy, and inhibition of cell death. | |||
| Key Molecule: Microtubule-associated proteins 1A/1B light chain 3A (MAP1LC3A) | [90] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | SAOS-2 cells | Bone marrow | Homo sapiens (Human) | CVCL_0548 |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Matrigel colony formation assay; Hoechst33342 staining assay | |||
| Mechanism Description | In chemoresistant SAOS-2 and U2OS osteosarcomas cells, miR-143 levels were significantly downregulated and accompanied by increases in ATG2B, Bcl-2, and/or LC3-II protein levels, high rate of ALDH1+CD133+ cells, and an increase in Matrigel colony formation ability. H2O2 upregulated p53 and miR-143, but downregulated ATG2B, Bcl-2, and LC3-I expression in U2OS cells (wild-type p53) but not in SAOS-2 (p53-null) cells. Forced miR-143 expression significantly reversed chemoresistance as well as downregulation of ATG2B, LC3-I, and Bcl-2 expression in SAOS-2- and U2OS-resistant cells. Forced miR-143 expression significantly inhibited tumor growth in xenograft SAOS-2-Dox and U2OS-Dox animal models. Loss of miR-143 expression is associated with poor prognosis of patients with osteosarcoma underlying chemotherapy. The chemoresistance of osteosarcoma tumor cells to doxorubicin is associated with the downregulation of miR-143 expression, activation of ALDH1+CD133+ cells, activation of autophagy, and inhibition of cell death. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: FOXF1 adjacent non-coding developmental regulatory RNA (FENDRR) | [92] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| SAOS-2 cells | Bone marrow | Homo sapiens (Human) | CVCL_0548 | |
| HOS cells | Bone | Homo sapiens (Human) | CVCL_0312 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qPCR; Microarray assay | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometric analysis | |||
| Mechanism Description | LncRNA FENDRR sensitizes doxorubicin-resistance of osteosarcoma cells through down-regulating ABCB1 and ABCC1. | |||
| Key Molecule: hsa-mir-100 | [93] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell growth | Inhibition | hsa05200 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; CCK8 assay | |||
| Mechanism Description | Either ZNRF2 overexpression or miR100 depletion increased in vitro OS cell growth and improved cell survival at the presence of Doxorubicin. miR100 bindS to the 3'-UTR of ZNRF2 mRNA to prevent its protein translation, re-expression of miR100 may inhibit OS cell growth and decrease OS cell chemo-resistance. | |||
| Key Molecule: Phosphate cytidylyltransferase 1A, choline (PCYT1A) | [94] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| SAOS-2 cells | Bone marrow | Homo sapiens (Human) | CVCL_0548 | |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
TUNEL Assay; MTT assay; Flow cytometric analysis | |||
| Mechanism Description | LncRNA CTA-miR210 axis plays an important role in reducing OS chemoresistance. LncRNA CTA could be activated by doxorubicin (DOX), and could promote OS cell apoptosis by competitively binding miR210, while inhibit cell autophagy. | |||
| Key Molecule: hsa-mir-210 | [94] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| SAOS-2 cells | Bone marrow | Homo sapiens (Human) | CVCL_0548 | |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR; Dual luciferase reporter assay | |||
| Experiment for Drug Resistance |
TUNEL Assay; MTT assay; Flow cytometric analysis | |||
| Mechanism Description | LncRNA CTA-miR210 axis plays an important role in reducing OS chemoresistance. LncRNA CTA could be activated by doxorubicin (DOX), and could promote OS cell apoptosis by competitively binding miR210, while inhibit cell autophagy. | |||
| Key Molecule: hsa-mir-410 | [95] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 |
| MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 | |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| HFob 1.19 | Bone | Homo sapiens (Human) | CVCL_3708 | |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | microRNA-410 regulates autophagy-related gene ATG16L1 expression and enhances chemosensitivity via autophagy inhibition in osteosarcoma, miR410 directly decreased ATG16L1 expression by targeting its 3'-untranslated region. | |||
| Key Molecule: hsa-mir-124 | [96] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| HCC1937 cells | Breast | Homo sapiens (Human) | CVCL_0290 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-124 may be involved in DNA repair by directly targeting ATMIN and PARP1, suggesting that multiple DNA repair pathways are affected by miR-124 and therefore manipulation of miR-124 level/activity may improve the efficacy of chemotherapies that induce DNA damage. repression of ATMIN (+) the HR repair defect induced by miR-124, and restoration of ATMIN reversed the effect of miR-124 overexpression in breast cancer cells. Therefore, it is intriguing to further speculate which of the multiple roles of ATMIN is specifically affected in breast carcinogenesis. On the other hand, PARP1-mediated processes play a role in oncogenesis, cancer progression, and therapeutic resistance. | |||
| Key Molecule: hsa-miR-199a-3p | [97] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell migration | Inhibition | hsa04670 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 |
| KHOS cells | Bone | Homo sapiens (Human) | CVCL_2546 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | CD44 was overexpressed in metastatic and recurrent osteosarcoma as compared with primary tumors. Higher expression of CD44 was found in both patients with shorter survival and patients who exhibited unfavorable response to chemotherapy before surgical resection. Additionally, the 3'-untranslated region of CD44 mRNA was the direct target of microRNA-199a-3p (miR-199a-3p). Overexpression of miR-199a-3p significantly inhibited CD44 expression in osteosarcoma cells. miR-199a-3p is One of the most dramatically decreased miRs in osteosarcoma cells and tumor tissues as compared with normal osteoblast cells. Transfection of miR-199a-3p significantly increased the drug sensitivity through down-regulation of CD44 in osteosarcoma cells. Taken together, these results suggest that the CD44-miR-199a-3p axis plays an important role in the development of metastasis, recurrence, and drug resistance of osteosarcoma. Developing strategies to target CD44 may improve the clinical outcome of osteosarcoma. | |||
| Key Molecule: hsa-mir-382 | [98] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| MNNG/HOS cells | Bone | Homo sapiens (Human) | CVCL_0439 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Decreased miR-382 was associated with poor survival in OS patients. Overexpression of miR-382 inhibited cell growth and chemoresistance by targeting kLF12 and HIPk3, respectively. In contrast, inhibition of miR-382 or overexpression of target genes stimulated OS cell growth and chemoresistance both in vitro and in vivo. | |||
| Key Molecule: hsa-mir-101 | [99] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 |
| Experiment for Molecule Alteration |
Quantitative GFP-LC3 analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The miR-101 not only decreases the formation of autophagic vesicles but also reduces the expression of LC-3II and Atg 4. This part of the study shows that miR-101 blocks chemotherapy-induced autophagy in OS cells. The sensitivity of OS cells to chemotherapy is increased by miR-101 blocked autophagy. miR-101 blocked the chemotherapy induced autophagy, and the blocked autophagy by miR-101 enhances the sensitivity of the OS cell line U-2 in vitro. | |||
|
|
||||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [92] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| SAOS-2 cells | Bone marrow | Homo sapiens (Human) | CVCL_0548 | |
| HOS cells | Bone | Homo sapiens (Human) | CVCL_0312 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometric analysis | |||
| Mechanism Description | LncRNA FENDRR sensitizes doxorubicin-resistance of osteosarcoma cells through down-regulating ABCB1 and ABCC1. | |||
| Key Molecule: Multidrug resistance-associated protein 1 (MRP1) | [92] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| SAOS-2 cells | Bone marrow | Homo sapiens (Human) | CVCL_0548 | |
| HOS cells | Bone | Homo sapiens (Human) | CVCL_0312 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometric analysis | |||
| Mechanism Description | LncRNA FENDRR sensitizes doxorubicin-resistance of osteosarcoma cells through down-regulating ABCB1 and ABCC1. | |||
|
|
||||
| Key Molecule: E3 ubiquitin-protein ligase ZNRF2 (ZNRF2) | [93] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell growth | Inhibition | hsa05200 | |
| In Vitro Model | U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 |
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
MTT assay; CCK8 assay | |||
| Mechanism Description | Either ZNRF2 overexpression or miR100 depletion increased in vitro OS cell growth and improved cell survival at the presence of Doxorubicin. miR100 bindS to the 3'-UTR of ZNRF2 mRNA to prevent its protein translation, re-expression of miR100 may inhibit OS cell growth and decrease OS cell chemo-resistance. | |||
| Key Molecule: Autophagy-related protein 16-1 (ATG16L1) | [95] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 |
| MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 | |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| HFob 1.19 | Bone | Homo sapiens (Human) | CVCL_3708 | |
| Experiment for Molecule Alteration |
Luciferase reporter assay; Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | microRNA-410 regulates autophagy-related gene ATG16L1 expression and enhances chemosensitivity via autophagy inhibition in osteosarcoma, miR410 directly decreased ATG16L1 expression by targeting its 3'-untranslated region. | |||
| Key Molecule: ATM interactor (ATMIN) | [96] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| HCC1937 cells | Breast | Homo sapiens (Human) | CVCL_0290 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-124 may be involved in DNA repair by directly targeting ATMIN and PARP1, suggesting that multiple DNA repair pathways are affected by miR-124 and therefore manipulation of miR-124 level/activity may improve the efficacy of chemotherapies that induce DNA damage. repression of ATMIN (+) the HR repair defect induced by miR-124, and restoration of ATMIN reversed the effect of miR-124 overexpression in breast cancer cells. Therefore, it is intriguing to further speculate which of the multiple roles of ATMIN is specifically affected in breast carcinogenesis. On the other hand, PARP1-mediated processes play a role in oncogenesis, cancer progression, and therapeutic resistance. | |||
| Key Molecule: Poly[ADP-ribose] synthase 1 (PARP1) | [96] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| HCC1937 cells | Breast | Homo sapiens (Human) | CVCL_0290 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-124 may be involved in DNA repair by directly targeting ATMIN and PARP1, suggesting that multiple DNA repair pathways are affected by miR-124 and therefore manipulation of miR-124 level/activity may improve the efficacy of chemotherapies that induce DNA damage. repression of ATMIN (+) the HR repair defect induced by miR-124, and restoration of ATMIN reversed the effect of miR-124 overexpression in breast cancer cells. Therefore, it is intriguing to further speculate which of the multiple roles of ATMIN is specifically affected in breast carcinogenesis. On the other hand, PARP1-mediated processes play a role in oncogenesis, cancer progression, and therapeutic resistance. | |||
| Key Molecule: Extracellular matrix receptor III (CD44) | [97] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell migration | Inhibition | hsa04670 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 |
| KHOS cells | Bone | Homo sapiens (Human) | CVCL_2546 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | CD44 was overexpressed in metastatic and recurrent osteosarcoma as compared with primary tumors. Higher expression of CD44 was found in both patients with shorter survival and patients who exhibited unfavorable response to chemotherapy before surgical resection. Additionally, the 3'-untranslated region of CD44 mRNA was the direct target of microRNA-199a-3p (miR-199a-3p). Overexpression of miR-199a-3p significantly inhibited CD44 expression in osteosarcoma cells. miR-199a-3p is One of the most dramatically decreased miRs in osteosarcoma cells and tumor tissues as compared with normal osteoblast cells. Transfection of miR-199a-3p significantly increased the drug sensitivity through down-regulation of CD44 in osteosarcoma cells. Taken together, these results suggest that the CD44-miR-199a-3p axis plays an important role in the development of metastasis, recurrence, and drug resistance of osteosarcoma. Developing strategies to target CD44 may improve the clinical outcome of osteosarcoma. | |||
| Key Molecule: Homeodomain-interacting protein kinase 3 (HIPK3) | [98] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| MNNG/HOS cells | Bone | Homo sapiens (Human) | CVCL_0439 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Decreased miR-382 was associated with poor survival in OS patients. Overexpression of miR-382 inhibited cell growth and chemoresistance by targeting kLF12 and HIPk3, respectively. In contrast, inhibition of miR-382 or overexpression of target genes stimulated OS cell growth and chemoresistance both in vitro and in vivo. | |||
| Key Molecule: Krueppel-like factor 12 (KLF12) | [98] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| MNNG/HOS cells | Bone | Homo sapiens (Human) | CVCL_0439 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Decreased miR-382 was associated with poor survival in OS patients. Overexpression of miR-382 inhibited cell growth and chemoresistance by targeting kLF12 and HIPk3, respectively. In contrast, inhibition of miR-382 or overexpression of target genes stimulated OS cell growth and chemoresistance both in vitro and in vivo. | |||
| Key Molecule: Ubiquitin-like modifier-activating enzyme Atg 4 (ATG4) | [99] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The miR-101 not only decreases the formation of autophagic vesicles but also reduces the expression of LC-3II and Atg 4. This part of the study shows that miR-101 blocks chemotherapy-induced autophagy in OS cells. The sensitivity of OS cells to chemotherapy is increased by miR-101 blocked autophagy. miR-101 blocked the chemotherapy induced autophagy, and the blocked autophagy by miR-101 enhances the sensitivity of the OS cell line U-2 in vitro. | |||
| Key Molecule: Microtubule-associated protein 1 light chain3 (LC3) | [99] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The miR-101 not only decreases the formation of autophagic vesicles but also reduces the expression of LC-3II and Atg 4. This part of the study shows that miR-101 blocks chemotherapy-induced autophagy in OS cells. The sensitivity of OS cells to chemotherapy is increased by miR-101 blocked autophagy. miR-101 blocked the chemotherapy induced autophagy, and the blocked autophagy by miR-101 enhances the sensitivity of the OS cell line U-2 in vitro. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-125b | [8] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Ewing sarcoma [ICD-11: 2B52.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| miR125b-p53/BAKT signaling pathway | Activation | hsa05206 | ||
| In Vitro Model | RD-ES cells | Bones | Homo sapiens (Human) | CVCL_2169 |
| Sk-ES cells | Bones | Homo sapiens (Human) | CVCL_0627 | |
| Sk-N-MC cells | Bones | Homo sapiens (Human) | CVCL_0530 | |
| TC-71 cells | Bones | Homo sapiens (Human) | CVCL_2213 | |
| VH-64 cells | Bones | Homo sapiens (Human) | CVCL_9672 | |
| WE-68 cells | Bones | Homo sapiens (Human) | CVCL_9717 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Celltiter-glo luminescent cell viability assay | |||
| Mechanism Description | miR-125b led to the development of chemoresistance by suppressing the expression of p53 and Bak, and repression of miR-125b sensitized EWS cells to apoptosis induced by treatment with various cytotoxic drugs. | |||
|
|
||||
| Key Molecule: Cellular tumor antigen p53 (TP53) | [8] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Ewing sarcoma [ICD-11: 2B52.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| miR125b-p53/BAKT signaling pathway | Activation | hsa05206 | ||
| In Vitro Model | RD-ES cells | Bones | Homo sapiens (Human) | CVCL_2169 |
| Sk-ES cells | Bones | Homo sapiens (Human) | CVCL_0627 | |
| Sk-N-MC cells | Bones | Homo sapiens (Human) | CVCL_0530 | |
| TC-71 cells | Bones | Homo sapiens (Human) | CVCL_2213 | |
| VH-64 cells | Bones | Homo sapiens (Human) | CVCL_9672 | |
| WE-68 cells | Bones | Homo sapiens (Human) | CVCL_9717 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
Celltiter-glo luminescent cell viability assay | |||
| Mechanism Description | miR-125b led to the development of chemoresistance by suppressing the expression of p53 and Bak, and repression of miR-125b sensitized EWS cells to apoptosis induced by treatment with various cytotoxic drugs. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-34 | [100] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Ewing sarcoma [ICD-11: 2B52.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell migration | Inhibition | hsa04670 | ||
| In Vitro Model | Sk-ES-1 cells | Bone | Homo sapiens (Human) | CVCL_0627 |
| Sk-N-MC cells | Bones | Homo sapiens (Human) | CVCL_0530 | |
| TC-71 cells | Bones | Homo sapiens (Human) | CVCL_2213 | |
| IOR/CAR cells | Sarcoma | Homo sapiens (Human) | CVCL_H725 | |
| Experiment for Molecule Alteration |
qRT-PCR; Northern blotting analysis | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | Increased chemo-sensitivity and decreased aggressiveness of EWS cells after enforced expression of miR-34a. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [101] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Embryonal rhabdomyosarcoma [ICD-11: 2B55.1] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | MAST111 cells | N.A. | Homo sapiens (Human) | N.A. |
| MAST139 cells | Embryo | Homo sapiens (Human) | N.A. | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
WST-1 assay | |||
| Mechanism Description | NPE inhibited the activity of ABCB1. Upon 1h combination treatment of MAST139 cells with Vinblastine and 100 ug/ml of NPE , a 40% increase in doxorubicin retention was observed. | |||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [101] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Alveolar rhabdomyosarcoma [ICD-11: 2B55.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | RH4 cells | Embryo | Homo sapiens (Human) | CVCL_C357 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
WST-1 assay | |||
| Mechanism Description | NPE inhibited the activity of ABCB1. Upon 1h combination treatment of MAST139 cells with Vinblastine and 100 ug/ml of NPE , a 40% increase in doxorubicin retention was observed. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Nanog homeobox (NANOG) | [17] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Leiomyosarcoma [ICD-11: 2B58.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Spheroid formation | Activation | hsa04140 | ||
| Cell colony | Activation | hsa05200 | ||
| Cell migration | Activation | hsa04670 | ||
| In Vitro Model | SK-UT-1 cells | Uterus | Homo sapiens (Human) | CVCL_0533 |
| In Vivo Model | BALB/c-nu female mice | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay; Flow cytometry assay; Transwell migration and invasion assay | |||
| Mechanism Description | The expression levels of CSC-related markers in CD133+ subpopulation derived from SK-UT-1 cells, Western blotting was employed to detect the expression levels of CD44, ALDH1, BMI1, and Nanog. Expectedly, researchers found that CD133+subpopulation had higher expression levels of CD44, ALDH1, BMI1, and Nanog compared with those of CD133 subpopulation. Collectively, the above-mentioned results suggested that CD133+ subpopulation derived from SK-UT-1 cells possessed capabilities of resistance to apoptosis after treatment with DXR, as well as stemness feature of cancer stem-like cells. | |||
| Key Molecule: Extracellular matrix receptor III (CD44) | [17] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Leiomyosarcoma [ICD-11: 2B58.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Spheroid formation | Activation | hsa04140 | ||
| Cell colony | Activation | hsa05200 | ||
| Cell migration | Activation | hsa04670 | ||
| In Vitro Model | SK-UT-1 cells | Uterus | Homo sapiens (Human) | CVCL_0533 |
| In Vivo Model | BALB/c-nu female mice | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay; Flow cytometry assay; Transwell migration and invasion assay | |||
| Mechanism Description | The expression levels of CSC-related markers in CD133+ subpopulation derived from SK-UT-1 cells, Western blotting was employed to detect the expression levels of CD44, ALDH1, BMI1, and Nanog. Expectedly, researchers found that CD133+subpopulation had higher expression levels of CD44, ALDH1, BMI1, and Nanog compared with those of CD133 subpopulation. Collectively, the above-mentioned results suggested that CD133+ subpopulation derived from SK-UT-1 cells possessed capabilities of resistance to apoptosis after treatment with DXR, as well as stemness feature of cancer stem-like cells. | |||
| Key Molecule: Polycomb complex protein BMI-1 (BMI1) | [17] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Leiomyosarcoma [ICD-11: 2B58.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Spheroid formation | Activation | hsa04140 | ||
| Cell colony | Activation | hsa05200 | ||
| Cell migration | Activation | hsa04670 | ||
| In Vitro Model | SK-UT-1 cells | Uterus | Homo sapiens (Human) | CVCL_0533 |
| In Vivo Model | BALB/c-nu female mice | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay; Flow cytometry assay; Transwell migration and invasion assay | |||
| Mechanism Description | The expression levels of CSC-related markers in CD133+ subpopulation derived from SK-UT-1 cells, Western blotting was employed to detect the expression levels of CD44, ALDH1, BMI1, and Nanog. Expectedly, researchers found that CD133+subpopulation had higher expression levels of CD44, ALDH1, BMI1, and Nanog compared with those of CD133 subpopulation. Collectively, the above-mentioned results suggested that CD133+ subpopulation derived from SK-UT-1 cells possessed capabilities of resistance to apoptosis after treatment with DXR, as well as stemness feature of cancer stem-like cells. | |||
| Key Molecule: Nanog homeobox (NANOG) | [17] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Leiomyosarcoma [ICD-11: 2B58.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Spheroid formation | Activation | hsa04140 | ||
| Cell colony | Activation | hsa05200 | ||
| Cell migration | Activation | hsa04670 | ||
| In Vitro Model | SK-UT-1 cells | Uterus | Homo sapiens (Human) | CVCL_0533 |
| In Vivo Model | BALB/c-nu female mice | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay; Flow cytometry assay; Transwell migration and invasion assay | |||
| Mechanism Description | The expression levels of CSC-related markers in CD133+ subpopulation derived from SK-UT-1 cells, Western blotting was employed to detect the expression levels of CD44, ALDH1, BMI1, and Nanog. Expectedly, researchers found that CD133+subpopulation had higher expression levels of CD44, ALDH1, BMI1, and Nanog compared with those of CD133 subpopulation. Collectively, the above-mentioned results suggested that CD133+ subpopulation derived from SK-UT-1 cells possessed capabilities of resistance to apoptosis after treatment with DXR, as well as stemness feature of cancer stem-like cells. | |||
| Key Molecule: Extracellular matrix receptor III (CD44) | [17] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Leiomyosarcoma [ICD-11: 2B58.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Spheroid formation | Activation | hsa04140 | ||
| Cell colony | Activation | hsa05200 | ||
| Cell migration | Activation | hsa04670 | ||
| In Vitro Model | SK-UT-1 cells | Uterus | Homo sapiens (Human) | CVCL_0533 |
| In Vivo Model | BALB/c-nu female mice | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay; Flow cytometry assay; Transwell migration and invasion assay | |||
| Mechanism Description | The expression levels of CSC-related markers in CD133+ subpopulation derived from SK-UT-1 cells, Western blotting was employed to detect the expression levels of CD44, ALDH1, BMI1, and Nanog. Expectedly, researchers found that CD133+subpopulation had higher expression levels of CD44, ALDH1, BMI1, and Nanog compared with those of CD133 subpopulation. Collectively, the above-mentioned results suggested that CD133+ subpopulation derived from SK-UT-1 cells possessed capabilities of resistance to apoptosis after treatment with DXR, as well as stemness feature of cancer stem-like cells. | |||
| Key Molecule: Polycomb complex protein BMI-1 (BMI1) | [17] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Leiomyosarcoma [ICD-11: 2B58.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Spheroid formation | Activation | hsa04140 | ||
| Cell colony | Activation | hsa05200 | ||
| Cell migration | Activation | hsa04670 | ||
| In Vitro Model | SK-UT-1 cells | Uterus | Homo sapiens (Human) | CVCL_0533 |
| In Vivo Model | BALB/c-nu female mice | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay; Flow cytometry assay; Transwell migration and invasion assay | |||
| Mechanism Description | The expression levels of CSC-related markers in CD133+ subpopulation derived from SK-UT-1 cells, Western blotting was employed to detect the expression levels of CD44, ALDH1, BMI1, and Nanog. Expectedly, researchers found that CD133+subpopulation had higher expression levels of CD44, ALDH1, BMI1, and Nanog compared with those of CD133 subpopulation. Collectively, the above-mentioned results suggested that CD133+ subpopulation derived from SK-UT-1 cells possessed capabilities of resistance to apoptosis after treatment with DXR, as well as stemness feature of cancer stem-like cells. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-17 | [48] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Synovial sarcoma [ICD-11: 2B5A.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Activation | hsa05200 | |
| p21 | Regulation | |||
| In Vitro Model | HS-SYII cells | Sarcoma | Homo sapiens (Human) | CVCL_8719 |
| SYO-1 cells | Sarcoma | Homo sapiens (Human) | CVCL_7146 | |
| Fuji cells | Sarcoma | Homo sapiens (Human) | CVCL_D880 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Colony formation assay | |||
| Mechanism Description | Overexpression of miR-17 in synovial sarcoma cells, Fuji and HS-SYII, increased colony forming ability in addition to cell growth, but not cell motility and invasion. Tumor volume formed in mice in vivo was significantly increased by miR-17 overexpression with a marked increase of MIB-1 index. According to PicTar and Miranda algorithms, which predicted CDkN1A (p21) as a putative target of miR-17, a luciferase assay was performed and revealed that miR-17 directly targets the 3'-UTR of p21 mRNA. Indeed, p21 protein level was remarkably decreased by miR-17 overexpression in a p53-independent manner. It is noteworthy that miR-17 succeeded in suppressing doxorubicin-evoked higher expression of p21 and conferred the drug resistance. Meanwhile, introduction of anti-miR-17 in Fuji and HS-SYII cells significantly decreased cell growth, consistent with rescued expression of p21. | |||
|
|
||||
| Key Molecule: Ribonuclease P protein subunit p21 (RPP21) | [48] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Synovial sarcoma [ICD-11: 2B5A.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Activation | hsa05200 | |
| p21 | Regulation | |||
| In Vitro Model | HS-SYII cells | Sarcoma | Homo sapiens (Human) | CVCL_8719 |
| SYO-1 cells | Sarcoma | Homo sapiens (Human) | CVCL_7146 | |
| Fuji cells | Sarcoma | Homo sapiens (Human) | CVCL_D880 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Colony formation assay | |||
| Mechanism Description | Overexpression of miR-17 in synovial sarcoma cells, Fuji and HS-SYII, increased colony forming ability in addition to cell growth, but not cell motility and invasion. Tumor volume formed in mice in vivo was significantly increased by miR-17 overexpression with a marked increase of MIB-1 index. According to PicTar and Miranda algorithms, which predicted CDkN1A (p21) as a putative target of miR-17, a luciferase assay was performed and revealed that miR-17 directly targets the 3'-UTR of p21 mRNA. Indeed, p21 protein level was remarkably decreased by miR-17 overexpression in a p53-independent manner. It is noteworthy that miR-17 succeeded in suppressing doxorubicin-evoked higher expression of p21 and conferred the drug resistance. Meanwhile, introduction of anti-miR-17 in Fuji and HS-SYII cells significantly decreased cell growth, consistent with rescued expression of p21. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-221 | [102] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SCC4 cells | Tongue | Homo sapiens (Human) | CVCL_1684 |
| SCC9 cells | Tongue | Homo sapiens (Human) | CVCL_1685 | |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
Annexin V-fluorescein isothiocyanate (FITC)/Hoechst double staining; MTT assay | |||
| Mechanism Description | OSCC cells are resistant to doxorubicin through upregulation of miR221, which in turn downregulates TIMP3. | |||
|
|
||||
| Key Molecule: Metalloproteinase inhibitor 3 (TIMP3) | [102] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SCC4 cells | Tongue | Homo sapiens (Human) | CVCL_1684 |
| SCC9 cells | Tongue | Homo sapiens (Human) | CVCL_1685 | |
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
Annexin V-fluorescein isothiocyanate (FITC)/Hoechst double staining; MTT assay | |||
| Mechanism Description | OSCC cells are resistant to doxorubicin through upregulation of miR221, which in turn downregulates TIMP3. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-214 | [12] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | ECA-109 cells | Esophagus | Homo sapiens (Human) | CVCL_6898 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | High expression of miR-483 and miR-214 might predict less chemotherapy effect. Down-regulation of miR-483 and miR-214 could confer sensitivity of both P-glycoprotein-related and P-glycoprotein-nonrelated drugs to esophageal cancer cells, and it might induce increased accumulation of adriamycin (ADR) and decreased amount of ADR released. | |||
| Key Molecule: hsa-mir-483 | [12] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | ECA-109 cells | Esophagus | Homo sapiens (Human) | CVCL_6898 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | High expression of miR-483 and miR-214 might predict less chemotherapy effect. Down-regulation of miR-483 and miR-214 could confer sensitivity of both P-glycoprotein-related and P-glycoprotein-nonrelated drugs to esophageal cancer cells, and it might induce increased accumulation of adriamycin (ADR) and decreased amount of ADR released. | |||
|
|
||||
| Key Molecule: ATP-binding cassette sub-family G2 (ABCG2) | [37] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Esophageal cancer [ICD-11: 2B70.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell viability | Activation | hsa05200 | ||
| In Vitro Model | ECA-109 cells | Esophagus | Homo sapiens (Human) | CVCL_6898 |
| Experiment for Molecule Alteration |
Flow cytometry assay | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Extracellular vesicles released by drug-resistant cells were proved that they could upregulate the expression of ABCG2 in esophageal cancer cells and thus regulate the drug resistance of esophageal cancer cells, which was related to the linc-VLDLR carried by EVs. | |||
|
|
||||
| Key Molecule: Very low density lipoprotein receptor (VLDLR) | [37] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Esophageal cancer [ICD-11: 2B70.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell viability | Activation | hsa05200 | ||
| In Vitro Model | ECA-109 cells | Esophagus | Homo sapiens (Human) | CVCL_6898 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Extracellular vesicles released by drug-resistant cells were proved that they could upregulate the expression of ABCG2 in esophageal cancer cells and thus regulate the drug resistance of esophageal cancer cells, which was related to the linc-VLDLR carried by EVs. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-200c | [103] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Esophageal cancer [ICD-11: 2B70.1] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Endoscopy; Computed tomography assay; Positron emission tomography assay | |||
| Mechanism Description | Serum miR-200c levels are useful for predicting the response to chemotherapy (cisplatin, 5-fluorouracil, and Adriamycin (ACF) or cisplatin, 5-fluorouracil, and docetaxel (DCF) ) in patients with esophageal cancer who underwent preoperative chemotherapy followed by surgery. | |||
| Key Molecule: hsa-mir-223 | [104] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Esophageal adenocarcinoma [ICD-11: 2B70.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell migration | Activation | hsa04670 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | OE33 cellss | Esophagus | Homo sapiens (Human) | CVCL_0471 |
| HEEpiC cells | Esophagus | Homo sapiens (Human) | N.A. | |
| JHesoAD1 cells | Esophagus | Homo sapiens (Human) | CVCL_8098 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | The DNA damage repair protein poly(ADP-ribose) polymerase 1 (PARP1) is a bona fide target of miR-223, miR-223 up-regulation is also associated with reduced PARP1 transcripts, and an increased sensitivity to cis-diamminedichloroplatinum (II) (Cisplatin), Doxorubicin and Mitomycin C. | |||
| Key Molecule: hsa-mir-296 | [105] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell growth | Inhibition | hsa05200 | ||
| In Vitro Model | ECA-109 cells | Esophagus | Homo sapiens (Human) | CVCL_6898 |
| Experiment for Molecule Alteration |
RT-PCR; Northern blotting analysis | |||
| Experiment for Drug Resistance |
WST-1 assay | |||
| Mechanism Description | Down-regulation of miR-296 could confer sensitivity of both P-glycoprotein-related and P-glycoprotein-nonrelated drugs on esophageal cancer cells, and might promote ADR-induced apoptosis, accompanied by increased accumulation and decreased releasing amount of ADR. Down-regulation of miR-296 could significantly decrease the expression of P-glycoprotein, Bcl-2, and the transcription of MDR1, but up-regulate the expression of Bax. | |||
| Key Molecule: hsa-mir-27a | [106] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | ECA-109 cells | Esophagus | Homo sapiens (Human) | CVCL_6898 |
| TE13 cells | Esophageal | Homo sapiens (Human) | CVCL_4463 | |
| Experiment for Molecule Alteration |
qRT-PCR; Northern blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Down-regulation of miR-27a significantly decreased expression of MDR1, but did not alter the expression of MRP, miR-27a could possibly mediate drug resistance, at least in part through regulation of MDR1 and apoptosis. | |||
|
|
||||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [105], [106] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell growth | Inhibition | hsa05200 | ||
| In Vitro Model | ECA-109 cells | Esophagus | Homo sapiens (Human) | CVCL_6898 |
| TE13 cells | Esophageal | Homo sapiens (Human) | CVCL_4463 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Down-regulation of miR-296 could confer sensitivity of both P-glycoprotein-related and P-glycoprotein-nonrelated drugs on esophageal cancer cells, and might promote ADR-induced apoptosis, accompanied by increased accumulation and decreased releasing amount of ADR. Down-regulation of miR-296 could significantly decrease the expression of P-glycoprotein, Bcl-2, and the transcription of MDR1, but up-regulate the expression of Bax. And down-regulation of miR-27a significantly decreased expression of MDR1, but did not alter the expression of MRP, miR-27a could possibly mediate drug resistance, at least in part through regulation of MDR1 and apoptosis. | |||
|
|
||||
| Key Molecule: Poly[ADP-ribose] synthase 1 (PARP1) | [104] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Esophageal adenocarcinoma [ICD-11: 2B70.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell migration | Activation | hsa04670 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | OE33 cellss | Esophagus | Homo sapiens (Human) | CVCL_0471 |
| HEEpiC cells | Esophagus | Homo sapiens (Human) | N.A. | |
| JHesoAD1 cells | Esophagus | Homo sapiens (Human) | CVCL_8098 | |
| Experiment for Molecule Alteration |
Luciferase reporter assay | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | The DNA damage repair protein poly(ADP-ribose) polymerase 1 (PARP1) is a bona fide target of miR-223, miR-223 up-regulation is also associated with reduced PARP1 transcripts, and an increased sensitivity to cis-diamminedichloroplatinum (II) (Cisplatin), Doxorubicin and Mitomycin C. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-1 | [107] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | miR-1 reverses multidrug resistance in gastric cancer cells via downregulation of sorcin through promoting the accumulation of intracellular drugs and apoptosis of cells. | |||
| Key Molecule: hsa-miR-633 | [108] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell invasion | Activation | hsa05200 | ||
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | miR-633 regulates chemotherapy resistance through downregulating FADD in gastric tumor cells. | |||
| Key Molecule: hsa-mir-27b | [109] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| SGC-7901/DDP cells | Gastric | Homo sapiens (Human) | CVCL_0520 | |
| SGC-7901/FU cells | Gastric | Homo sapiens (Human) | CVCL_0520 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Annexin V-FITC Apoptosis assay | |||
| Mechanism Description | LncRNA urothelial carcinoma associated 1 (UCA1) increases multi-drug resistance of gastric cancer via downregulating miR27b. | |||
| Key Molecule: Urothelial cancer associated 1 (UCA1) | [109] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| SGC-7901/DDP cells | Gastric | Homo sapiens (Human) | CVCL_0520 | |
| SGC-7901/FU cells | Gastric | Homo sapiens (Human) | CVCL_0520 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Annexin V-FITC Apoptosis assay | |||
| Mechanism Description | LncRNA urothelial carcinoma associated 1 (UCA1) increases multi-drug resistance of gastric cancer via downregulating miR27b. | |||
| Key Molecule: hsa-mir-494 | [110] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell colony | Activation | hsa05200 | |
| Cell viability | Activation | hsa05200 | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| AGS cells | Gastric | Homo sapiens (Human) | CVCL_0139 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | microRNA 494 increases chemosensitivity to doxorubicin in gastric cancer cells by targeting phosphodiesterases 4D. | |||
| Key Molecule: hsa-mir-20a | [111] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell viability | Activation | hsa05200 | ||
| MAPK/ERK signaling pathway | Inhibition | hsa04010 | ||
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | The restoration of miR-20a expression significantly reduced LRIG1-induced GC cell chemosensitivity. | |||
| Key Molecule: hsa-miR-135a-5p | [112] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| miR135a-5p/AP2alpha /BCL2 signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | BGC-823 cells | Gastric | Homo sapiens (Human) | CVCL_3360 |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | AP-2alpha contains a putative miRNA-135a-5p target, which was confirmed as a direct target using the 3'-UTR luciferase reporter system. Additionally, an increase and decrease of miRNA-135a-5p inhi bited or impaired adriamycin-induced apoptosis in BGC-823 cells (p<0.05, compared with the group without gene intervention), respectively. Luciferase reporter experiments confirmed that AP-2alpha bound to the BCL-2 promoter and affected its transcription. Therefore, miRNA-135a-5p increased BCL-2 via AP-2alpha and consequently (+) cell resistance to apoptosis. | |||
| Key Molecule: hsa-mir-106a | [113] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| TGF-beta signaling pathway | Regulation | hsa04350 | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-106a, elevated in multidrug-resistant GC cell lines, suppressed the sensitivity of GC cells to chemo-therapeutic drugs by accelerating drug efflux and reducing apoptosis. Moreover, we validated RUNX3 as a target of miR-106a in GC cells, indicating that miR-106a might modulate MDR by regulating RUNX3 in GC. | |||
| Key Molecule: hsa-mir-19a | [38] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| PTEN/AKT signaling pathway | Inhibition | hsa05235 | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| SGC7901/VCR cells | Gastric | Homo sapiens (Human) | CVCL_VU58 | |
| SGC7901/ADR cells | Gastric | Homo sapiens (Human) | CVCL_VU57 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-19a/b are upregulated in multidrug-resistant gastric cancer cell line, miR-19a/b suppress the sensitivity of gastric cancer cells to anticancer drugs, miR-19a/b accelerate the efflux of ADR through P-gp upregulation. | |||
| Key Molecule: hsa-mir-19b | [38] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| PTEN/AKT signaling pathway | Inhibition | hsa05235 | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| SGC7901/VCR cells | Gastric | Homo sapiens (Human) | CVCL_VU58 | |
| SGC7901/ADR cells | Gastric | Homo sapiens (Human) | CVCL_VU57 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-19a/b are upregulated in multidrug-resistant gastric cancer cell line, miR-19a/b suppress the sensitivity of gastric cancer cells to anticancer drugs, miR-19a/b accelerate the efflux of ADR through P-gp upregulation. | |||
|
|
||||
| Key Molecule: Sorcin (SRI) | [107] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | miR-1 reverses multidrug resistance in gastric cancer cells via downregulation of sorcin through promoting the accumulation of intracellular drugs and apoptosis of cells. | |||
| Key Molecule: FAS-associated death domain protein (FADD) | [108] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell invasion | Activation | hsa05200 | ||
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | miR-633 regulates chemotherapy resistance through downregulating FADD in gastric tumor cells. | |||
| Key Molecule: Phosphodiesterase 4D (PDE4D) | [110] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell colony | Activation | hsa05200 | |
| Cell viability | Activation | hsa05200 | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| AGS cells | Gastric | Homo sapiens (Human) | CVCL_0139 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | microRNA 494 increases chemosensitivity to doxorubicin in gastric cancer cells by targeting phosphodiesterases 4D. | |||
| Key Molecule: Leucine-rich repeats and immunoglobulin-like domains protein 1 (LRIG1) | [111] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell viability | Activation | hsa05200 | ||
| MAPK/ERK signaling pathway | Inhibition | hsa04010 | ||
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | The restoration of miR-20a expression significantly reduced LRIG1-induced GC cell chemosensitivity. | |||
| Key Molecule: Transcription factor AP2 alpha (TFAP2A) | [112] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| miR135a-5p/AP2alpha /BCL2 signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | BGC-823 cells | Gastric | Homo sapiens (Human) | CVCL_3360 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | AP-2alpha contains a putative miRNA-135a-5p target, which was confirmed as a direct target using the 3'-UTR luciferase reporter system. Additionally, an increase and decrease of miRNA-135a-5p inhi bited or impaired adriamycin-induced apoptosis in BGC-823 cells (p<0.05, compared with the group without gene intervention), respectively. Luciferase reporter experiments confirmed that AP-2alpha bound to the BCL-2 promoter and affected its transcription. Therefore, miRNA-135a-5p increased BCL-2 via AP-2alpha and consequently (+) cell resistance to apoptosis. | |||
| Key Molecule: Runt-related transcription factor 3 (RUNX3) | [113] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| TGF-beta signaling pathway | Regulation | hsa04350 | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-106a, elevated in multidrug-resistant GC cell lines, suppressed the sensitivity of GC cells to chemo-therapeutic drugs by accelerating drug efflux and reducing apoptosis. Moreover, we validated RUNX3 as a target of miR-106a in GC cells, indicating that miR-106a might modulate MDR by regulating RUNX3 in GC. | |||
| Key Molecule: Phosphatase and tensin homolog (PTEN) | [38] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| PTEN/AKT signaling pathway | Inhibition | hsa05235 | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| SGC7901/VCR cells | Gastric | Homo sapiens (Human) | CVCL_VU58 | |
| SGC7901/ADR cells | Gastric | Homo sapiens (Human) | CVCL_VU57 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-19a/b are upregulated in multidrug-resistant gastric cancer cell line, miR-19a/b suppress the sensitivity of gastric cancer cells to anticancer drugs, miR-19a/b accelerate the efflux of ADR through P-gp upregulation. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Long non-protein coding RNA (D63785) | [114] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| miR422a/MEF2D signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | MGC-803 cells | Gastric | Homo sapiens (Human) | CVCL_5334 |
| SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 | |
| BGC823 cells | Gastric | Homo sapiens (Human) | CVCL_3360 | |
| GES-1 cells | Gastric | Homo sapiens (Human) | CVCL_EQ22 | |
| NCI-N87 cells | Gastric | Homo sapiens (Human) | CVCL_1603 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | LncRNA-D6378 mediates gastric cancer cell resistance to apoptosis by modulating the miR-422a-MEF2D signaling pathway and Silencing of Lncr-D63785 enhanced the sensitivity of gastric cancer cells to chemotherapy by inducing apoptosis. | |||
| Key Molecule: hsa-miR-422a | [114] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| miR422a/MEF2D signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | MGC-803 cells | Gastric | Homo sapiens (Human) | CVCL_5334 |
| SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 | |
| BGC823 cells | Gastric | Homo sapiens (Human) | CVCL_3360 | |
| GES-1 cells | Gastric | Homo sapiens (Human) | CVCL_EQ22 | |
| NCI-N87 cells | Gastric | Homo sapiens (Human) | CVCL_1603 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | miR-422a has an enhancer activity in DOX-mediated chemosensitivity and cell death. | |||
| Key Molecule: hsa-mir-16 | [115] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| SGC7901/AR cells | Gastric | Homo sapiens (Human) | CVCL_VU57 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Transwell invasion assay; CCK8 assay | |||
| Mechanism Description | The expression of miR16-1 was positively related with the chemosensitivity of GC to adriamycin, and miR16-1 could targeted silence FUBP1 to advance the chemosensitivity to adriamycin in GC. | |||
| Key Molecule: Nuclear paraspeckle assembly transcript 1 (NEAT1) | [116] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| GES-1 cells | Gastric | Homo sapiens (Human) | CVCL_EQ22 | |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Transwell Invasion assay; Annexin V-FITC apoptosis detection assay | |||
| Mechanism Description | NEAT1 silence in SGC7901 cells could inhibit proliferation and invasion ability, and promote cell apoptosis significantly. NEAT1 knockdown Inhibited Chemotherapy Resistance to Adriamycin in GC Adriamycin-Resistant Cells. | |||
| Key Molecule: Hepatocellular carcinoma up-regulated long non-coding RNA (HULC) | [117] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | MGC-803 cells | Gastric | Homo sapiens (Human) | CVCL_5334 |
| GES-1 cells | Gastric | Homo sapiens (Human) | CVCL_EQ22 | |
| MkN-45 cells | Gastric | Homo sapiens (Human) | CVCL_0434 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometric analysis | |||
| Mechanism Description | Silencing LncRNA HULC could enhance chemotherapy induced apoptosis in GC cells. | |||
| Key Molecule: hsa-mir-126 | [118] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| SGC7901/VCR cells | Gastric | Homo sapiens (Human) | CVCL_VU58 | |
| SGC7901/ADR cells | Gastric | Homo sapiens (Human) | CVCL_VU57 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Caspase3/7 activity assay | |||
| Mechanism Description | microRNA-126 increases chemosensitivity in drug-resistant gastric cancer cells by targeting EZH2. | |||
| Key Molecule: hsa-mir-495 | [119] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| mTOR signaling pathway | Inhibition | hsa04150 | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | The miR-495 exerts promotive effects on GC chemosensitivity via inactivation of the mTOR signaling pathway by suppressing ERBB2. | |||
| Key Molecule: hsa-miR-21-5p | [120] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | Suppression of miR-21-5p expression sensitizes SGC7901/DOX cells to DOX via upregulating PTEN and TIMP3. | |||
| Key Molecule: Protein lin-28 homolog A (CSDD1) | [121] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| Lin28/miR107 pathway | Regulation | hsa05206 | ||
| In Vitro Model | MkN-45 cells | Gastric | Homo sapiens (Human) | CVCL_0434 |
| MkN28 cells | Gastric | Homo sapiens (Human) | CVCL_1416 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Lin28 could inhibit the expression of miR-107, thereby up-regulating C-myc, P-gp and down-regulating Cyclin D1, subsequently result in chemo-resistance of gastric cancer cells. The Lin28/miR-107 pathway might be served as one of many signaling pathways that is associated with gastric cancer chemo-resistance. | |||
| Key Molecule: Protein lin-28 homolog B (CSDD2) | [121] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| Lin28/miR107 pathway | Regulation | hsa05206 | ||
| In Vitro Model | MkN-45 cells | Gastric | Homo sapiens (Human) | CVCL_0434 |
| MkN28 cells | Gastric | Homo sapiens (Human) | CVCL_1416 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Lin28 could inhibit the expression of miR-107, thereby up-regulating C-myc, P-gp and down-regulating Cyclin D1, subsequently result in chemo-resistance of gastric cancer cells. The Lin28/miR-107 pathway might be served as one of many signaling pathways that is associated with gastric cancer chemo-resistance. | |||
| Key Molecule: hsa-miR-107 | [121] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| Lin28/miR107 pathway | Regulation | hsa05206 | ||
| In Vitro Model | MkN-45 cells | Gastric | Homo sapiens (Human) | CVCL_0434 |
| MkN28 cells | Gastric | Homo sapiens (Human) | CVCL_1416 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Lin28 could inhibit the expression of miR-107, thereby up-regulating C-myc, P-gp and down-regulating Cyclin D1, subsequently result in chemo-resistance of gastric cancer cells. The Lin28/miR-107 pathway might be served as one of many signaling pathways that is associated with gastric cancer chemo-resistance. | |||
| Key Molecule: hsa-mir-218 | [122] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-218 may inhibit efflux of ADM and oxaliplatin by down-regulating P-gp expression. | |||
| Key Molecule: hsa-miR-129-5p | [123] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The over-expressed miR-129-5p reduced the chemo-resistance of SGC7901/VCR and SGC7901/ADR cells, while down-regulation of miR-129-5p had an opposite effect. Furthermore, three members of multi-drug resistance (MDR) related ABC transporters (ABCB1, ABCC5 and ABCG1) were found to be direct targets of miR-129-5p using bioinformatics analysis and report gene assays. The present study indicated that hyper-methylation of miR-129-5p CpG island might play important roles in the development of gastric cancer chemo-resistance by targeting MDR related ABC transporters and might be used as a potential therapeutic target in preventing the chemo-resistance of gastric cancer. | |||
| Key Molecule: Cyclin D binding myb like transcription factor 1 (DMTF1) | [124] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay assay | |||
| Mechanism Description | MRUL depletion enhances the chemosensitivity of stomach cancer cells via inhibiting ABCB1 expression and increasing cell apoptosis. | |||
| Key Molecule: hsa-mir-185 | [125] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | BGC-823 cells | Gastric | Homo sapiens (Human) | CVCL_3360 |
| MGC-803 cells | Gastric | Homo sapiens (Human) | CVCL_5334 | |
| AGS cells | Gastric | Homo sapiens (Human) | CVCL_0139 | |
| GES-1 cells | Gastric | Homo sapiens (Human) | CVCL_EQ22 | |
| NCI-N87 cells | Gastric | Homo sapiens (Human) | CVCL_1603 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Trypan blue exclusion assay; Tunel assay | |||
| Mechanism Description | Restoration of miR-185 alone can inhibit gastric cancer tumor growth. Moreover, combination therapy using enforced miR-185 expression and lower dose chemotherapeutic drugs had an effective therapeutic activity against large established tumors, with decreased host toxicity. miR-185 increases the chemosensitivity of gastric cancer cells in vitro and in vivo. It exerts tumor-suppressing function through negatively regulating ARC. Besides, miR-185 upregulation in response to cisplatin or doxorubicin treatment in gastric cancer cells is dependent on RUNX3 transcriptional activity. | |||
| Key Molecule: hsa-miR-520h | [126] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MkN-45 cells | Gastric | Homo sapiens (Human) | CVCL_0434 |
| MkN28 cells | Gastric | Homo sapiens (Human) | CVCL_1416 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | miR-520h is up-regulated by doxorubicin to target HDAC1 and sensitizes gastric cancer cells to doxorubicin, doxorubicin down-regulates HDAC1 expression to aggravate DNA-doxorubicin interaction by inducing the expression of HDAC1-targeting miR-520h. | |||
| Key Molecule: hsa-miR-508-5p | [127] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| SGC7901/VCR cells | Gastric | Homo sapiens (Human) | CVCL_VU58 | |
| SGC7901/ADR cells | Gastric | Homo sapiens (Human) | CVCL_VU57 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The overexpression of miR-508-5p was sufficient to reverse cancer cell resistance to multiple chemotherapeutics in vitro and sensitize tumours to chemotherapy in vivo. Further studies showed that miR-508-5p could directly target the 3'-untranslated regions of ABCB1 and Zinc ribbon domain-containing 1 (ZNRD1), and suppress their expression at the mRNA and protein levels. Meanwhile, the suppression of ZNRD1 led to a decrease in ABCB1. | |||
| Key Molecule: hsa-mir-27a | [128] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| Tumorigenesis | Inhibition | hsa05200 | ||
| In Vitro Model | MkN-45 cells | Gastric | Homo sapiens (Human) | CVCL_0434 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Down-regulation of miR-27a could also confer sensitivity of drugs on gastric cancer cells, and might increase accumulation and decrease releasing amount of adriamycin in gastric cancer cells. Down-regulation of miR-27a could significantly decrease the expression of P-glycoprotein and the transcriptional activity of cyclin D1, and up-regulate the expression of p21. | |||
| Key Molecule: hsa-mir-181 | [129] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Gastric adenocarcinoma [ICD-11: 2B72.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| SGC7901/VCR cells | Gastric | Homo sapiens (Human) | CVCL_VU58 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The antiapoptotic protein BCL2 is upregulated, whereas miR-181b is downregulated in both SGC7901/VCR and A549/CDDP cells, compared with SGC7901 and A549 cells, respectively. Enforced miR-181b expression reduced BCL2 protein level and sensitized SGC7901/VCR and A549/CDDP cells to VCR-induced and CDDP-induced apoptosis, respectively. | |||
| Key Molecule: hsa-mir-34 | [130] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | p53 signaling pathway | Inhibition | hsa04115 | |
| In Vitro Model | AGS cells | Gastric | Homo sapiens (Human) | CVCL_0139 |
| NCI-N87 cells | Gastric | Homo sapiens (Human) | CVCL_1603 | |
| MkN-45 cells | Gastric | Homo sapiens (Human) | CVCL_0434 | |
| KATO-3 cells | Gastric | Homo sapiens (Human) | CVCL_0371 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Human gastric cancer kato III cells with miR-34 restoration reduced the expression of target genes Bcl-2, Notch, and HMGA2. MicroRNA miR-34 was recently found to be a direct target of p53, functioning downstream of the p53 pathway as a tumor suppressor, miR-34 impaired cell growth, accumulated the cells in G1 phase, increased caspase-3 activation, and, more significantly, inhibited tumorsphere formation and growth. | |||
| Key Molecule: hsa-mir-15b | [131] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Mitochondrial signaling pathway | Activation | hsa04217 | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| SGC7901/VCR cells | Gastric | Homo sapiens (Human) | CVCL_VU58 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-15b and miR-16, among the downregulated miRNAs in SGC7901/VCR cells, were demonstrated to play a role in the development of MDR in gastric cancer cells by targeting the antiapoptotic gene BCL2. | |||
| Key Molecule: hsa-mir-16 | [131] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Mitochondrial signaling pathway | Activation | hsa04217 | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| SGC7901/VCR cells | Gastric | Homo sapiens (Human) | CVCL_VU58 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-15b and miR-16, among the downregulated miRNAs in SGC7901/VCR cells, were demonstrated to play a role in the development of MDR in gastric cancer cells by targeting the antiapoptotic gene BCL2. | |||
|
|
||||
| Key Molecule: ATP-binding cassette sub-family B5 (ABCB5) | [122] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-218 may inhibit efflux of ADM and oxaliplatin by down-regulating P-gp expression. | |||
| Key Molecule: ATP-binding cassette sub-family C5 (ABCC5) | [123] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The over-expressed miR-129-5p reduced the chemo-resistance of SGC7901/VCR and SGC7901/ADR cells, while down-regulation of miR-129-5p had an opposite effect. Furthermore, three members of multi-drug resistance (MDR) related ABC transporters (ABCB1, ABCC5 and ABCG1) were found to be direct targets of miR-129-5p using bioinformatics analysis and report gene assays. The present study indicated that hyper-methylation of miR-129-5p CpG island might play important roles in the development of gastric cancer chemo-resistance by targeting MDR related ABC transporters and might be used as a potential therapeutic target in preventing the chemo-resistance of gastric cancer. | |||
| Key Molecule: ATP-binding cassette sub-family G1 (ABCG1) | [123] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The over-expressed miR-129-5p reduced the chemo-resistance of SGC7901/VCR and SGC7901/ADR cells, while down-regulation of miR-129-5p had an opposite effect. Furthermore, three members of multi-drug resistance (MDR) related ABC transporters (ABCB1, ABCC5 and ABCG1) were found to be direct targets of miR-129-5p using bioinformatics analysis and report gene assays. The present study indicated that hyper-methylation of miR-129-5p CpG island might play important roles in the development of gastric cancer chemo-resistance by targeting MDR related ABC transporters and might be used as a potential therapeutic target in preventing the chemo-resistance of gastric cancer. | |||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [127] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| SGC7901/VCR cells | Gastric | Homo sapiens (Human) | CVCL_VU58 | |
| SGC7901/ADR cells | Gastric | Homo sapiens (Human) | CVCL_VU57 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The overexpression of miR-508-5p was sufficient to reverse cancer cell resistance to multiple chemotherapeutics in vitro and sensitize tumours to chemotherapy in vivo. Further studies showed that miR-508-5p could directly target the 3'-untranslated regions of ABCB1 and Zinc ribbon domain-containing 1 (ZNRD1), and suppress their expression at the mRNA and protein levels. Meanwhile, the suppression of ZNRD1 led to a decrease in ABCB1. | |||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [123], [124] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay assay | |||
| Mechanism Description | MRUL depletion enhances the chemosensitivity of stomach cancer cells via inhibiting ABCB1 expression and increasing cell apoptosis. And The over-expressed miR-129-5p reduced the chemo-resistance of SGC7901/VCR and SGC7901/ADR cells, while down-regulation of miR-129-5p had an opposite effect. Furthermore, three members of multi-drug resistance (MDR) related ABC transporters (ABCB1, ABCC5 and ABCG1) were found to be direct targets of miR-129-5p using bioinformatics analysis and report gene assays. The present study indicated that hyper-methylation of miR-129-5p CpG island might play important roles in the development of gastric cancer chemo-resistance by targeting MDR related ABC transporters and might be used as a potential therapeutic target in preventing the chemo-resistance of gastric cancer. | |||
|
|
||||
| Key Molecule: Myocyte-specific enhancer factor 2D (MEF2D) | [114] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| miR422a/MEF2D signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | MGC-803 cells | Gastric | Homo sapiens (Human) | CVCL_5334 |
| SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 | |
| BGC823 cells | Gastric | Homo sapiens (Human) | CVCL_3360 | |
| GES-1 cells | Gastric | Homo sapiens (Human) | CVCL_EQ22 | |
| NCI-N87 cells | Gastric | Homo sapiens (Human) | CVCL_1603 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | MEF2D expression markedly inhibited BGC823 cell proliferation and migration and invasion and enhanced the sensitivity of gastric cancer cells to chemotherapy. | |||
| Key Molecule: Far upstream element-binding protein 1 (FUBP1) | [115] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| SGC7901/AR cells | Gastric | Homo sapiens (Human) | CVCL_VU57 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Transwell invasion assay; CCK8 assay | |||
| Mechanism Description | The expression of miR16-1 was positively related with the chemosensitivity of GC to adriamycin, and miR16-1 could targeted silence FUBP1 to advance the chemosensitivity to adriamycin in GC. | |||
| Key Molecule: Histone-lysine N-methyltransferase EZH2 (EZH2) | [118] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| SGC7901/VCR cells | Gastric | Homo sapiens (Human) | CVCL_VU58 | |
| SGC7901/ADR cells | Gastric | Homo sapiens (Human) | CVCL_VU57 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Caspase3/7 activity assay | |||
| Mechanism Description | microRNA-126 increases chemosensitivity in drug-resistant gastric cancer cells by targeting EZH2. | |||
| Key Molecule: Receptor tyrosine-protein kinase erbB-2 (ERBB2) | [119] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| mTOR signaling pathway | Inhibition | hsa04150 | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | The miR-495 exerts promotive effects on GC chemosensitivity via inactivation of the mTOR signaling pathway by suppressing ERBB2. | |||
| Key Molecule: Phosphatase and tensin homolog (PTEN) | [120] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | Suppression of miR-21-5p expression sensitizes SGC7901/DOX cells to DOX via upregulating PTEN and TIMP3. | |||
| Key Molecule: Metalloproteinase inhibitor 3 (TIMP3) | [120] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | Suppression of miR-21-5p expression sensitizes SGC7901/DOX cells to DOX via upregulating PTEN and TIMP3. | |||
| Key Molecule: Apoptosis regulator Bcl-2 (BCL2) | [122] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Higher miR-218 levels increased the level of Bax and reduced the level of Bcl-2 and miR-218 inhibits multidrug resistance (MDR) of gastric cancer cells by targeting Hedgehog/smoothened. | |||
| Key Molecule: Smoothened homolog (SMO) | [122] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Higher miR-218 levels increased the level of Bax and reduced the level of Bcl-2 and miR-218 inhibits multidrug resistance (MDR) of gastric cancer cells by targeting Hedgehog/smoothened. | |||
| Key Molecule: Activity-regulated cytoskeleton-associated protein (ARC) | [125] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | BGC-823 cells | Gastric | Homo sapiens (Human) | CVCL_3360 |
| MGC-803 cells | Gastric | Homo sapiens (Human) | CVCL_5334 | |
| AGS cells | Gastric | Homo sapiens (Human) | CVCL_0139 | |
| GES-1 cells | Gastric | Homo sapiens (Human) | CVCL_EQ22 | |
| NCI-N87 cells | Gastric | Homo sapiens (Human) | CVCL_1603 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Trypan blue exclusion assay; Tunel assay | |||
| Mechanism Description | Restoration of miR-185 alone can inhibit gastric cancer tumor growth. Moreover, combination therapy using enforced miR-185 expression and lower dose chemotherapeutic drugs had an effective therapeutic activity against large established tumors, with decreased host toxicity. miR-185 increases the chemosensitivity of gastric cancer cells in vitro and in vivo. It exerts tumor-suppressing function through negatively regulating ARC. Besides, miR-185 upregulation in response to cisplatin or doxorubicin treatment in gastric cancer cells is dependent on RUNX3 transcriptional activity. | |||
| Key Molecule: Histone deacetylase 1 (HDAC1) | [126] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MkN-45 cells | Gastric | Homo sapiens (Human) | CVCL_0434 |
| MkN28 cells | Gastric | Homo sapiens (Human) | CVCL_1416 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | miR-520h is up-regulated by doxorubicin to target HDAC1 and sensitizes gastric cancer cells to doxorubicin, doxorubicin down-regulates HDAC1 expression to aggravate DNA-doxorubicin interaction by inducing the expression of HDAC1-targeting miR-520h. | |||
| Key Molecule: DNA-directed RNA polymerase I subunit RPA12 (RPA12) | [127] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| SGC7901/VCR cells | Gastric | Homo sapiens (Human) | CVCL_VU58 | |
| SGC7901/ADR cells | Gastric | Homo sapiens (Human) | CVCL_VU57 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The overexpression of miR-508-5p was sufficient to reverse cancer cell resistance to multiple chemotherapeutics in vitro and sensitize tumours to chemotherapy in vivo. Further studies showed that miR-508-5p could directly target the 3'-untranslated regions of ABCB1 and Zinc ribbon domain-containing 1 (ZNRD1), and suppress their expression at the mRNA and protein levels. Meanwhile, the suppression of ZNRD1 led to a decrease in ABCB1. | |||
| Key Molecule: G1/S-specific cyclin-D1 (CCND1) | [128] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| Tumorigenesis | Inhibition | hsa05200 | ||
| In Vitro Model | MkN-45 cells | Gastric | Homo sapiens (Human) | CVCL_0434 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Down-regulation of miR-27a could also confer sensitivity of drugs on gastric cancer cells, and might increase accumulation and decrease releasing amount of adriamycin in gastric cancer cells. Down-regulation of miR-27a could significantly decrease the expression of P-glycoprotein and the transcriptional activity of cyclin D1, and up-regulate the expression of p21. | |||
| Key Molecule: Apoptosis regulator Bcl-2 (BCL2) | [129] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Gastric adenocarcinoma [ICD-11: 2B72.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| SGC7901/VCR cells | Gastric | Homo sapiens (Human) | CVCL_VU58 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The antiapoptotic protein BCL2 is upregulated, whereas miR-181b is downregulated in both SGC7901/VCR and A549/CDDP cells, compared with SGC7901 and A549 cells, respectively. Enforced miR-181b expression reduced BCL2 protein level and sensitized SGC7901/VCR and A549/CDDP cells to VCR-induced and CDDP-induced apoptosis, respectively. | |||
| Key Molecule: Apoptosis regulator Bcl-2 (BCL2) | [130] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | p53 signaling pathway | Inhibition | hsa04115 | |
| In Vitro Model | AGS cells | Gastric | Homo sapiens (Human) | CVCL_0139 |
| NCI-N87 cells | Gastric | Homo sapiens (Human) | CVCL_1603 | |
| MkN-45 cells | Gastric | Homo sapiens (Human) | CVCL_0434 | |
| KATO-3 cells | Gastric | Homo sapiens (Human) | CVCL_0371 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Human gastric cancer kato III cells with miR-34 restoration reduced the expression of target genes Bcl-2, Notch, and HMGA2. MicroRNA miR-34 was recently found to be a direct target of p53, functioning downstream of the p53 pathway as a tumor suppressor, miR-34 impaired cell growth, accumulated the cells in G1 phase, increased caspase-3 activation, and, more significantly, inhibited tumorsphere formation and growth. | |||
| Key Molecule: High mobility group protein HMGI-C (HMGA2) | [130] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | p53 signaling pathway | Inhibition | hsa04115 | |
| In Vitro Model | AGS cells | Gastric | Homo sapiens (Human) | CVCL_0139 |
| NCI-N87 cells | Gastric | Homo sapiens (Human) | CVCL_1603 | |
| MkN-45 cells | Gastric | Homo sapiens (Human) | CVCL_0434 | |
| KATO-3 cells | Gastric | Homo sapiens (Human) | CVCL_0371 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Human gastric cancer kato III cells with miR-34 restoration reduced the expression of target genes Bcl-2, Notch, and HMGA2. MicroRNA miR-34 was recently found to be a direct target of p53, functioning downstream of the p53 pathway as a tumor suppressor, miR-34 impaired cell growth, accumulated the cells in G1 phase, increased caspase-3 activation, and, more significantly, inhibited tumorsphere formation and growth. | |||
| Key Molecule: Neurogenic locus notch homolog protein (NOTCH) | [130] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | p53 signaling pathway | Inhibition | hsa04115 | |
| In Vitro Model | AGS cells | Gastric | Homo sapiens (Human) | CVCL_0139 |
| NCI-N87 cells | Gastric | Homo sapiens (Human) | CVCL_1603 | |
| MkN-45 cells | Gastric | Homo sapiens (Human) | CVCL_0434 | |
| KATO-3 cells | Gastric | Homo sapiens (Human) | CVCL_0371 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Human gastric cancer kato III cells with miR-34 restoration reduced the expression of target genes Bcl-2, Notch, and HMGA2. MicroRNA miR-34 was recently found to be a direct target of p53, functioning downstream of the p53 pathway as a tumor suppressor, miR-34 impaired cell growth, accumulated the cells in G1 phase, increased caspase-3 activation, and, more significantly, inhibited tumorsphere formation and growth. | |||
| Key Molecule: Apoptosis regulator Bcl-2 (BCL2) | [131] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Mitochondrial signaling pathway | Activation | hsa04217 | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| SGC7901/VCR cells | Gastric | Homo sapiens (Human) | CVCL_VU58 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-15b and miR-16, among the downregulated miRNAs in SGC7901/VCR cells, were demonstrated to play a role in the development of MDR in gastric cancer cells by targeting the antiapoptotic gene BCL2. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-424 | [35] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Colon cancer [ICD-11: 2B90.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |
| U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 | |
| HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 | |
| PARP cells | Skin | Homo sapiens (Human) | N.A. | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Hypoxia induces miR-424 expression and that miR-424 in turn suppresses the level of PDCD4 protein, a tumor suppressor that is involved in apoptosis, by targeting its 3' untranslated region. Functionally, miR-424 overexpression decreases the sensitivity of cancer cells (HCT116 and A375) to doxorubicin (Dox) and etoposide. In contrast, the inhibition of miR-424 (+) apoptosis and increased the sensitivity of cancer cells to Dox. In a xenograft tumor model, miR-424 overexpression promoted tumor growth following Dox treatment, suggesting that miR-424 promotes tumor cell resistance to Dox. Furthermore, miR-424 levels are inversely correlated with PDCD4 expression in clinical breast cancer samples. | |||
|
|
||||
| Key Molecule: Programmed cell death protein 4 (PDCD4) | [35] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Colon cancer [ICD-11: 2B90.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |
| U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 | |
| HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 | |
| PARP cells | Skin | Homo sapiens (Human) | N.A. | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Hypoxia induces miR-424 expression and that miR-424 in turn suppresses the level of PDCD4 protein, a tumor suppressor that is involved in apoptosis, by targeting its 3' untranslated region. Functionally, miR-424 overexpression decreases the sensitivity of cancer cells (HCT116 and A375) to doxorubicin (Dox) and etoposide. In contrast, the inhibition of miR-424 (+) apoptosis and increased the sensitivity of cancer cells to Dox. In a xenograft tumor model, miR-424 overexpression promoted tumor growth following Dox treatment, suggesting that miR-424 promotes tumor cell resistance to Dox. Furthermore, miR-424 levels are inversely correlated with PDCD4 expression in clinical breast cancer samples. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-522 | [132] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Colon cancer [ICD-11: 2B90.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | HT29 Cells | Colon | Homo sapiens (Human) | CVCL_A8EZ |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | When miR 522 was overexpressed in the HT29/DOX cells, the protein expression levels of ABCB5 were downregulated. Furthermore, knockdown of ABCB5 significantly increased the growth inhibition rate of the HT29/DOX cells, compared with the control group. These results suggested that miR 522 may affect the sensitivity of colon cancer cell lines to DOX treatment by targeting ABCB5. | |||
| Key Molecule: hsa-mir-137 | [53] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Colon cancer [ICD-11: 2B90.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | LS174 cells | Colon | Homo sapiens (Human) | CVCL_YJ85 |
| In Vivo Model | Immunodeficient NCr nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Cell titer glo assay assay | |||
| Mechanism Description | Hypermethylation of the miR-137 promoter and negative regulation of miR-137 by CAR contribute in part to reduced miR-137 expression and increased CAR and MDR1 expression in doxorubicin-resistant neuroblastoma cells. | |||
| Key Molecule: hsa-mir-195 | [133] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Colon cancer [ICD-11: 2B90.1] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | HT29 Cells | Colon | Homo sapiens (Human) | CVCL_A8EZ |
| LOVO cells | Colon | Homo sapiens (Human) | CVCL_0399 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Suppression of miR-195 leads to elevation of its direct target gene BCL2L2 expression therefore makes the human colon cancer cells more resistant to Dox. | |||
|
|
||||
| Key Molecule: ATP-binding cassette sub-family B5 (ABCB5) | [132] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Colon cancer [ICD-11: 2B90.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | HT29 Cells | Colon | Homo sapiens (Human) | CVCL_A8EZ |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | When miR 522 was overexpressed in the HT29/DOX cells, the protein expression levels of ABCB5 were downregulated. Furthermore, knockdown of ABCB5 significantly increased the growth inhibition rate of the HT29/DOX cells, compared with the control group. These results suggested that miR 522 may affect the sensitivity of colon cancer cell lines to DOX treatment by targeting ABCB5. | |||
|
|
||||
| Key Molecule: Nuclear receptor subfamily 1 group I3 (NR1I3) | [53] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Colon cancer [ICD-11: 2B90.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | LS174 cells | Colon | Homo sapiens (Human) | CVCL_YJ85 |
| In Vivo Model | Immunodeficient NCr nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Chromatin immunoprecipitation assay | |||
| Experiment for Drug Resistance |
Cell titer glo assay assay | |||
| Mechanism Description | Hypermethylation of the miR-137 promoter and negative regulation of miR-137 by CAR contribute in part to reduced miR-137 expression and increased CAR and MDR1 expression in doxorubicin-resistant neuroblastoma cells. | |||
| Key Molecule: Bcl-2-like protein 2 (BCL2L2) | [133] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Colon cancer [ICD-11: 2B90.1] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | HT29 Cells | Colon | Homo sapiens (Human) | CVCL_A8EZ |
| LOVO cells | Colon | Homo sapiens (Human) | CVCL_0399 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Suppression of miR-195 leads to elevation of its direct target gene BCL2L2 expression therefore makes the human colon cancer cells more resistant to Dox. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Glycogen synthase kinase-3 beta (GSK3B) | [36] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Wnt/Beta-catenin signaling pathway | Regulation | hsa04310 | |
| In Vitro Model | SW480 cells | Colon | Homo sapiens (Human) | CVCL_0546 |
| SW480/ADM cells | Colon | Homo sapiens (Human) | CVCL_0546 | |
| Experiment for Molecule Alteration |
Dual luciferase gene reporter assay | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay; EdU staining | |||
| Mechanism Description | miR224 up-regulation is associated with ADM resistance of CRC cells. Suppression of miR224 expression up-regulated GSk-3beta expression, inhibited Wnt/beta-catenin signal pathway activity and Survivin expression, as well as reduced ADM resistance of CRC SW480 cells. | |||
| Key Molecule: hsa-mir-224 | [36] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Wnt/Beta-catenin signaling pathway | Regulation | hsa04310 | |
| In Vitro Model | SW480 cells | Colon | Homo sapiens (Human) | CVCL_0546 |
| SW480/ADM cells | Colon | Homo sapiens (Human) | CVCL_0546 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay; EdU staining | |||
| Mechanism Description | miR224 up-regulation is associated with ADM resistance of CRC cells. Suppression of miR224 expression up-regulated GSk-3beta expression, inhibited Wnt/beta-catenin signal pathway activity and Survivin expression, as well as reduced ADM resistance of CRC SW480 cells. | |||
|
|
||||
| Key Molecule: SLC25A25 antisense RNA 1 (SLC25A25-AS1) | [10] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell migration | Activation | hsa04670 | |
| Cell proliferation | Activation | hsa05200 | ||
| ERK/p38 signaling pathway | Inhibition | hsa04210 | ||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| HT-29 cells | Colon | Homo sapiens (Human) | CVCL_0320 | |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | SLC25A25-AS1 overexpression significantly inhibited proliferation and colony formation in colorectal cancer cell lines, and downregulation of SLC25A25-AS1 obviously (+) chemoresistance and promoted EMT process in vitro associated with Erk and p38 signaling pathway activation. Therefore, SLC25A25-AS1 was determined to play a tumor suppressive role in CRC. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Cytochrome P450 family 3 subfamily A member1 (CYP3A4) | [72] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | CaCo2 cells | Colon | Homo sapiens (Human) | CVCL_0025 |
| Experiment for Molecule Alteration |
CYP450-Glo CYP 3A4 assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | In this study, resveratrol was a significant inhibitor of CYP3A4 enzyme activity with IC50 value 9.32 ( M). Moreover, the CYP3A4 mRNA levels were reduced after treatment with resveratrol 0.03-fold of the control levels with high significance (p < 0.001). | |||
| Key Molecule: Glutathione S-transferase (GST) | [72] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | CaCo2 cells | Colon | Homo sapiens (Human) | CVCL_0025 |
| Experiment for Molecule Alteration |
Glutathione-S-transferase assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The Glutathione-S-transferases (GSTs) are a multigene family of dimeric proteins which play a central role in the detoxification of electrophilic xenobiotics and catalyze their conjugation with GSH to electrophilic metabolites, thus rendering them more water soluble. GSTs protect cells from cytotoxic and carcinogenic chemicals. GST activity was decreased by resveratrol in a dose dependent manner. IC50 value was 30.73 M. This results were confirmed by RT-PCR data, where the tested samples changed the GST mRNA level by 0.79-fold (p < 0.01) of control level. | |||
|
|
||||
| Key Molecule: ATP-binding cassette sub-family B5 (ABCB5) | [134] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Colorectal carcinoma [ICD-11: 2B91.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | HCT8 cells | Colon | Homo sapiens (Human) | CVCL_2478 |
| CT26 cells | Colon | Mus musculus (Mouse) | CVCL_7254 | |
| Salmonella enterica serovar Typhimurium SL1344 | 216597 | |||
| Salmonella enterica serovar Typhimurium SL1344 detaSipA | 216597 | |||
| Salmonella enterica serovar Typhimurium SL1344 detaSipB | 216597 | |||
| Salmonella enterica serovar Typhimurium SL1344 detaSipC | 216597 | |||
| Salmonella enterica serovar Typhimurium SL1344 detaSopB | 216597 | |||
| In Vivo Model | BALB/c mice xenograft model | Mus musculus | ||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | Mimicking the ability of Salmonella to reverse multidrug resistance, we constructed a gold nanoparticle system packaged with a SipA corona, and found this bacterial mimic not only accumulates in tumours but also reduces P-gp at a SipA dose significantly lower than free SipA. Moreover, the Salmonella nanoparticle mimic suppresses tumour growth with a concomitant reduction in P-gp when used with an existing chemotherapeutic drug (that is, doxorubicin). | |||
| Key Molecule: ATP-binding cassette sub-family B5 (ABCB5) | [135] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Colorectal carcinoma [ICD-11: 2B91.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | CaCo2 cells | Colon | Homo sapiens (Human) | CVCL_0025 |
| HCT-8 cells | Colon | Homo sapiens (Human) | CVCL_2478 | |
| NIH-G185 cells | Ovary | Homo sapiens (Human) | CVCL_L991 | |
| NIH 3T3 cells | Colon | Homo sapiens (Human) | CVCL_0594 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | G185 cells were 27-135 fold more resistant to the cytotoxic drugs doxorubicin, vinblastine, colchicine and paclitaxel than the parental NIH 3T3 cells. Co-administration of TPGS enhanced the cytotoxicity of doxorubicin, vinblastine, paclitaxel, and colchicine in the G185 cells to levels comparable to the parental. | |||
| Key Molecule: ATP-binding cassette sub-family B5 (ABCB5) | [72] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | CaCo2 cells | Colon | Homo sapiens (Human) | CVCL_0025 |
| Experiment for Molecule Alteration |
Efflux of rhodamine123 assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Resveratrol can restore the sensitivity of Caco-2 and CEM/ADR5000 cell lines to doxorubicin, through enhancing significantly doxorubicin cytotoxicity. ABC-transporter inhibitors, classified according to their action on ABC-transporters proteins into: 1. Function inhibitors, 2. Expression inhibitors, and 3. Functional and expression inhibitors, which have an ideal characters of ABC-transporters inhibitors. Our results indicate that resveratrol falls into the class 3 inhibitors. | |||
|
|
||||
| Key Molecule: F-box/WD repeat-containing protein 7 (FBXW7) | [136] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell viability | Activation | hsa05200 | |
| miR223/FBXW7 signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | SW480 cells | Colon | Homo sapiens (Human) | CVCL_0546 |
| SW620 cells | Colon | Homo sapiens (Human) | CVCL_0547 | |
| LOVO cells | Colon | Homo sapiens (Human) | CVCL_0399 | |
| HT-29 cells | Colon | Homo sapiens (Human) | CVCL_0320 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Overexpression of miR-223 decreased FBXW7 expression and the sensitivity of CRC cells to doxorubicin, while suppression of miR-223 had the opposite effect. | |||
| Key Molecule: hsa-mir-223 | [136] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell viability | Activation | hsa05200 | |
| miR223/FBXW7 signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | SW480 cells | Colon | Homo sapiens (Human) | CVCL_0546 |
| SW620 cells | Colon | Homo sapiens (Human) | CVCL_0547 | |
| LOVO cells | Colon | Homo sapiens (Human) | CVCL_0399 | |
| HT-29 cells | Colon | Homo sapiens (Human) | CVCL_0320 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Overexpression of miR-223 decreased FBXW7 expression and the sensitivity of CRC cells to doxorubicin, while suppression of miR-223 had the opposite effect. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-miR-1291 | [137] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Pancreatic carcinoma [ICD-11: 2C10.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell growth | Inhibition | hsa05200 | |
| In Vitro Model | H69 cells | Lung | Homo sapiens (Human) | CVCL_8121 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Hsa-miR-1291-directed downregulation of ABCC1 led to a greater intracellular drug accumulation and sensitized the cells to doxorubicin. | |||
|
|
||||
| Key Molecule: Multidrug resistance-associated protein 1 (MRP1) | [137] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Pancreatic carcinoma [ICD-11: 2C10.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell growth | Inhibition | hsa05200 | |
| In Vitro Model | H69 cells | Lung | Homo sapiens (Human) | CVCL_8121 |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Hsa-miR-1291-directed downregulation of ABCC1 led to a greater intracellular drug accumulation and sensitized the cells to doxorubicin. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-33a | [138] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell viability | Activation | hsa05200 | ||
| In Vitro Model | BEL-7402 cells | Liver | Homo sapiens (Human) | CVCL_5492 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Downregulated LncRNA CRNDE could up-regulate miR-33a expression and inhibit HMGA2 expression, thus it could significantly promote apoptosis of liver cancer drug-resistant cells on different chemotherapeutic drugs (ADM, DDP, 5-FU)and inhibit its proliferation, migration, invasion and drug resistance. | |||
| Key Molecule: Colorectal neoplasia differentially expressed (CRNDE) | [138] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | BEL-7402 cells | Liver | Homo sapiens (Human) | CVCL_5492 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Downregulated LncRNA CRNDE could up-regulate miR-33a expression and inhibit HMGA2 expression, thus it could significantly promote apoptosis of liver cancer drug-resistant cells on different chemotherapeutic drugs (ADM, DDP, 5-FU)and inhibit its proliferation, migration, invasion and drug resistance. | |||
| Key Molecule: hsa-miR-589-5p | [139] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | STAT3 signaling pathway | Activation | hsa04550 | |
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| QGY-7703 cells | Liver | Homo sapiens (Human) | CVCL_6715 | |
| SMMC7721 cells | Uterus | Homo sapiens (Human) | CVCL_0534 | |
| 97H cells | Liver | Homo sapiens (Human) | N.A. | |
| PLC cells | Liver | Homo sapiens (Human) | CVCL_0485 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Flow cytometric analysis; Spheroid formation assay | |||
| Mechanism Description | miR589-5p promotes the cancer stem cell characteristics and chemoresistance via targeting multiple negative regulators of STAT3 signaling pathway, including SOCS2, SOCS5, PTPN1 and PTPN11, leading to constitutive activation of STAT3 signaling. | |||
| Key Molecule: Ribosomal protein L13a pseudogene 20 (RPL13AP20) | [18] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| GSKIP/GSK-3beta signaling pathway | Activation | hsa04550 | ||
| Tumorigenesis | Activation | hsa05206 | ||
| Wnt/Beta-catenin signaling pathway | Activation | hsa04310 | ||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| L02 cells | Liver | Homo sapiens (Human) | CVCL_6926 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qPCR; Microarray assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | HANR bind to GSkIP for regulating the phosphorylation of GSk3beta in HCC, knock-down of HANR markedly retarded cell proliferation, suppressed HCC xenograft/orthotopic tumor growth, induced apoptosis and enhanced chemosensitivity to doxorubicin. GSkIP is the direct target of HANR to influence GSk3beta phosphorylation, HANR is physically associated with GSkIP to regulate the GSkIP/GSk3beta pathway. | |||
| Key Molecule: LncRNA regulator of Akt signaling associated with HCC and RCC (LNCARSR) | [140] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K/AKT signaling pathway | Activation | hsa04151 | |
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| SMMC7721 cells | Uterus | Homo sapiens (Human) | CVCL_0534 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
TUNEL assays | |||
| Mechanism Description | lncARSR physically associates with PTEN mRNA, promotes PTEN mRNA degradation, decreases PTEN expression, and activates PI3k/Akt pathway. Upregulated lncARSR promotes doxorubicin resistance in HCC via modulating PTEN-PI3k/Akt pathway. | |||
| Key Molecule: hsa-mir-146a | [141] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HCC Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The HCC Huh-7 cell line was treated with adramycin (ADM), cisplatin (DDP), carboplatin (CBP), mitomycin C (MMC) or vincristine (VCR) at increasing concentrations to develop drug-resistant sublines. Among these 51 upregulated and downregulated miRNAs, 12 miRNAs were upregulated and 13 miRNAs were downregulated in Huh-7/VCR. Upregulation of miR-27b, miR-181a, miR-146b-5p, miR-181d and miR-146a expression was verified using real-time RT-PCR in the parental and the five drug-resistant cell lines. | |||
| Key Molecule: hsa-miR-146b-5p | [141] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HCC Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The HCC Huh-7 cell line was treated with adramycin (ADM), cisplatin (DDP), carboplatin (CBP), mitomycin C (MMC) or vincristine (VCR) at increasing concentrations to develop drug-resistant sublines. Among these 51 upregulated and downregulated miRNAs, 12 miRNAs were upregulated and 13 miRNAs were downregulated in Huh-7/VCR. Upregulation of miR-27b, miR-181a, miR-146b-5p, miR-181d and miR-146a expression was verified using real-time RT-PCR in the parental and the five drug-resistant cell lines. | |||
| Key Molecule: hsa-mir-181a | [141] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HCC Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The HCC Huh-7 cell line was treated with adramycin (ADM), cisplatin (DDP), carboplatin (CBP), mitomycin C (MMC) or vincristine (VCR) at increasing concentrations to develop drug-resistant sublines. Among these 51 upregulated and downregulated miRNAs, 12 miRNAs were upregulated and 13 miRNAs were downregulated in Huh-7/VCR. Upregulation of miR-27b, miR-181a, miR-146b-5p, miR-181d and miR-146a expression was verified using real-time RT-PCR in the parental and the five drug-resistant cell lines. | |||
| Key Molecule: hsa-mir-181d | [141] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HCC Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The HCC Huh-7 cell line was treated with adramycin (ADM), cisplatin (DDP), carboplatin (CBP), mitomycin C (MMC) or vincristine (VCR) at increasing concentrations to develop drug-resistant sublines. Among these 51 upregulated and downregulated miRNAs, 12 miRNAs were upregulated and 13 miRNAs were downregulated in Huh-7/VCR. Upregulation of miR-27b, miR-181a, miR-146b-5p, miR-181d and miR-146a expression was verified using real-time RT-PCR in the parental and the five drug-resistant cell lines. | |||
| Key Molecule: hsa-mir-27b | [141] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HCC Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The HCC Huh-7 cell line was treated with adramycin (ADM), cisplatin (DDP), carboplatin (CBP), mitomycin C (MMC) or vincristine (VCR) at increasing concentrations to develop drug-resistant sublines. Among these 51 upregulated and downregulated miRNAs, 12 miRNAs were upregulated and 13 miRNAs were downregulated in Huh-7/VCR. Upregulation of miR-27b, miR-181a, miR-146b-5p, miR-181d and miR-146a expression was verified using real-time RT-PCR in the parental and the five drug-resistant cell lines. | |||
| Key Molecule: hsa-mir-181 | [142] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Activation | hsa05200 | |
| Cell migration | Activation | hsa04670 | ||
| TGF-beta signaling pathway | Activation | hsa04350 | ||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | TIMP3 is a tumor suppressor and a validated miR-181 target. Ectopic expression and depletion of miR-181b showed that miR-181b enhanced MMP2 and MMP9 activity and promoted growth, clonogenic survival, migration and invasion of HCC cells that could be reversed by modulating TIMP3 level. | |||
| Key Molecule: hsa-let-7a | [33] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Caspase-3 is the key executioner caspase in apoptosis. Ectopic expression of let-7adecreased the luciferase activity of a reporter constructcontaining the 30untranslated region of caspase-3. Enforced let-7aexpression increased the resistance in A431 cells andHepG2 cells to apoptosis induced by therapeutic drugs suchas interferon-gamma, doxorubicin and paclitaxel. | |||
| Key Molecule: H19, imprinted maternally expressed transcript (H19) | [40] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Antisense H19 oligonucleotides transfection induced a marked increase in the percentage of MDR1 promoter methylation and decrease in MDR1 expression in R-HepG2 cells. Thus, the H19 gene is believed to induce P-glycoprotein expression and MDR1-associated drug resistance at least in liver cancer cells through regulation of MDR1 promoter methylation. | |||
|
|
||||
| Key Molecule: ATP-binding cassette sub-family G2 (ABCG2) | [143] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Hepatocellular cancer [ICD-11: 2C12.4] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| PLC/PRF-5 cells | Liver | Homo sapiens (Human) | CVCL_0485 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay; Flow cytometry assay | |||
| Mechanism Description | LincRNA-VLDLR (linc-VLDLR) was significantly up-regulated in malignant hepatocytes. Exposure of HCC cells to diverse anti-cancer agents such as sorafenib, camptothecin, and doxorubicin increased linc-VLDLR expression in cells as well as within EVs released from these cells. Incubation with EVs reduced chemotherapy-induced cell death and also increased linc-VLDLR expression in recipient cells. RNAi-mediated knockdown of linc-VLDLR decreased cell viability and abrogated cell cycle progression. Moreover, knockdown of VLDLR reduced expression of ABCG2 (ATP-binding cassette, sub-family G member 2), whereas over-expression of this protein reduced the effects of VLDLR knockdown on sorafenib-induced cell death. Here, linc-VLDLR is identified as an extracellular vesicle enriched LncRNA that contributes to cellular stress responses. | |||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [40] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
Northern blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Antisense H19 oligonucleotides transfection induced a marked increase in the percentage of MDR1 promoter methylation and decrease in MDR1 expression in R-HepG2 cells. Thus, the H19 gene is believed to induce P-glycoprotein expression and MDR1-associated drug resistance at least in liver cancer cells through regulation of MDR1 promoter methylation. | |||
|
|
||||
| Key Molecule: Very low density lipoprotein receptor (VLDLR) | [143] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Hepatocellular cancer [ICD-11: 2C12.4] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| PLC/PRF-5 cells | Liver | Homo sapiens (Human) | CVCL_0485 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTS assay; Flow cytometry assay | |||
| Mechanism Description | LincRNA-VLDLR (linc-VLDLR) was significantly up-regulated in malignant hepatocytes. Exposure of HCC cells to diverse anti-cancer agents such as sorafenib, camptothecin, and doxorubicin increased linc-VLDLR expression in cells as well as within EVs released from these cells. Incubation with EVs reduced chemotherapy-induced cell death and also increased linc-VLDLR expression in recipient cells. RNAi-mediated knockdown of linc-VLDLR decreased cell viability and abrogated cell cycle progression. Moreover, knockdown of VLDLR reduced expression of ABCG2 (ATP-binding cassette, sub-family G member 2), whereas over-expression of this protein reduced the effects of VLDLR knockdown on sorafenib-induced cell death. Here, linc-VLDLR is identified as an extracellular vesicle enriched LncRNA that contributes to cellular stress responses. | |||
|
|
||||
| Key Molecule: High mobility group protein HMGI-C (HMGA2) | [138] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | BEL-7402 cells | Liver | Homo sapiens (Human) | CVCL_5492 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR; Luciferase activity assay | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Downregulated LncRNA CRNDE could up-regulate miR-33a expression and inhibit HMGA2 expression, thus it could significantly promote apoptosis of liver cancer drug-resistant cells on different chemotherapeutic drugs (ADM, DDP, 5-FU)and inhibit its proliferation, migration, invasion and drug resistance. | |||
| Key Molecule: Protein-tyrosine phosphatase 1B (PTPN1) | [139] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | STAT3 signaling pathway | Activation | hsa04550 | |
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| QGY-7703 cells | Liver | Homo sapiens (Human) | CVCL_6715 | |
| SMMC7721 cells | Uterus | Homo sapiens (Human) | CVCL_0534 | |
| 97H cells | Liver | Homo sapiens (Human) | N.A. | |
| PLC cells | Liver | Homo sapiens (Human) | CVCL_0485 | |
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
Flow cytometric analysis; Spheroid formation assay | |||
| Mechanism Description | miR589-5p promotes the cancer stem cell characteristics and chemoresistance via targeting multiple negative regulators of STAT3 signaling pathway, including SOCS2, SOCS5, PTPN1 and PTPN11, leading to constitutive activation of STAT3 signaling. | |||
| Key Molecule: Tyrosine-protein phosphatase non-receptor type 11 (PTPN11) | [139] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | STAT3 signaling pathway | Activation | hsa04550 | |
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| QGY-7703 cells | Liver | Homo sapiens (Human) | CVCL_6715 | |
| SMMC7721 cells | Uterus | Homo sapiens (Human) | CVCL_0534 | |
| 97H cells | Liver | Homo sapiens (Human) | N.A. | |
| PLC cells | Liver | Homo sapiens (Human) | CVCL_0485 | |
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
Flow cytometric analysis; Spheroid formation assay | |||
| Mechanism Description | miR589-5p promotes the cancer stem cell characteristics and chemoresistance via targeting multiple negative regulators of STAT3 signaling pathway, including SOCS2, SOCS5, PTPN1 and PTPN11, leading to constitutive activation of STAT3 signaling. | |||
| Key Molecule: Suppressor of cytokine signaling 2 (SOCS2) | [139] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | STAT3 signaling pathway | Activation | hsa04550 | |
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| QGY-7703 cells | Liver | Homo sapiens (Human) | CVCL_6715 | |
| SMMC7721 cells | Uterus | Homo sapiens (Human) | CVCL_0534 | |
| 97H cells | Liver | Homo sapiens (Human) | N.A. | |
| PLC cells | Liver | Homo sapiens (Human) | CVCL_0485 | |
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
Flow cytometric analysis; Spheroid formation assay | |||
| Mechanism Description | miR589-5p promotes the cancer stem cell characteristics and chemoresistance via targeting multiple negative regulators of STAT3 signaling pathway, including SOCS2, SOCS5, PTPN1 and PTPN11, leading to constitutive activation of STAT3 signaling. | |||
| Key Molecule: Suppressor of cytokine signaling 5 (SOCS5) | [139] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | STAT3 signaling pathway | Activation | hsa04550 | |
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| QGY-7703 cells | Liver | Homo sapiens (Human) | CVCL_6715 | |
| SMMC7721 cells | Uterus | Homo sapiens (Human) | CVCL_0534 | |
| 97H cells | Liver | Homo sapiens (Human) | N.A. | |
| PLC cells | Liver | Homo sapiens (Human) | CVCL_0485 | |
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
Flow cytometric analysis; Spheroid formation assay | |||
| Mechanism Description | miR589-5p promotes the cancer stem cell characteristics and chemoresistance via targeting multiple negative regulators of STAT3 signaling pathway, including SOCS2, SOCS5, PTPN1 and PTPN11, leading to constitutive activation of STAT3 signaling. | |||
| Key Molecule: Glycogen synthase kinase-3 beta (GSK3B) | [18] | |||
| Molecule Alteration | Phosphorylation | Down-regulation |
||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| GSKIP/GSK-3beta signaling pathway | Activation | hsa04550 | ||
| Wnt/Beta-catenin signaling pathway | Activation | hsa04310 | ||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| L02 cells | Liver | Homo sapiens (Human) | CVCL_6926 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Immunohistochemical staining | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | HANR bind to GSkIP for regulating the phosphorylation of GSk3beta in HCC, knock-down of HANR markedly retarded cell proliferation, suppressed HCC xenograft/orthotopic tumor growth, induced apoptosis and enhanced chemosensitivity to doxorubicin. GSkIP is the direct target of HANR to influence GSk3beta phosphorylation, HANR is physically associated with GSkIP to regulate the GSkIP/GSk3beta pathway. | |||
| Key Molecule: Phosphatase and tensin homolog (PTEN) | [140] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K/AKT signaling pathway | Activation | hsa04151 | |
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| SMMC7721 cells | Uterus | Homo sapiens (Human) | CVCL_0534 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
TUNEL assays | |||
| Mechanism Description | lncARSR physically associates with PTEN mRNA, promotes PTEN mRNA degradation, decreases PTEN expression, and activates PI3k/Akt pathway. Upregulated lncARSR promotes doxorubicin resistance in HCC via modulating PTEN-PI3k/Akt pathway. | |||
| Key Molecule: Metalloproteinase inhibitor 3 (TIMP3) | [142] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Activation | hsa05200 | |
| Cell migration | Activation | hsa04670 | ||
| TGF-beta signaling pathway | Activation | hsa04350 | ||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | TIMP3 is a tumor suppressor and a validated miR-181 target. Ectopic expression and depletion of miR-181b showed that miR-181b enhanced MMP2 and MMP9 activity and promoted growth, clonogenic survival, migration and invasion of HCC cells that could be reversed by modulating TIMP3 level. | |||
| Key Molecule: Caspase-3 (CASP3) | [33] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Caspase-3 is the key executioner caspase in apoptosis. Ectopic expression of let-7adecreased the luciferase activity of a reporter constructcontaining the 30untranslated region of caspase-3. Enforced let-7aexpression increased the resistance in A431 cells andHepG2 cells to apoptosis induced by therapeutic drugs suchas interferon-gamma, doxorubicin and paclitaxel. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Cytochrome P450 family 1 subfamily B member1 (CYP1B1) | [144] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | miR27b/CCNG1/p53 signaling pathway | Regulation | hsa05206 | |
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| SNU182 cells | Liver | Homo sapiens (Human) | CVCL_0090 | |
| SNU-739 cells | Liver | Homo sapiens (Human) | CVCL_5088 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | |||
| Mechanism Description | miR-27b synergizes with anticancer drugs througth enhancing anticancer drug-induced cell death which due to p53 activation and CYP1B1 suppression. | |||
| Key Molecule: Cytochrome P450 family 3 subfamily A member1 (CYP3A4) | [72] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
CYP450-Glo CYP 3A4 assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | In this study, resveratrol was a significant inhibitor of CYP3A4 enzyme activity with IC50 value 9.32 ( M). Moreover, the CYP3A4 mRNA levels were reduced after treatment with resveratrol 0.03-fold of the control levels with high significance (p < 0.001). | |||
| Key Molecule: Glutathione S-transferase (GST) | [72] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
Glutathione-S-transferase assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The Glutathione-S-transferases (GSTs) are a multigene family of dimeric proteins which play a central role in the detoxification of electrophilic xenobiotics and catalyze their conjugation with GSH to electrophilic metabolites, thus rendering them more water soluble. GSTs protect cells from cytotoxic and carcinogenic chemicals. GST activity was decreased by resveratrol in a dose dependent manner. IC50 value was 30.73 M. This results were confirmed by RT-PCR data, where the tested samples changed the GST mRNA level by 0.79-fold (p < 0.01) of control level. | |||
| Key Molecule: Cytochrome P450 family 3 subfamily A member1 (CYP3A4) | [145] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
CYP450-Glo TM CYP 3A4 assay, RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Synergistic interaction between the MDR mechanisms include ABCT proteins (P-gp, BCRP, and MDR1) and metabolic enzymes of phase I of metabolism mainly CYP3A4, phase II of metabolism mainly GST was observed. In this study, FUC alone and in combination with DOX inhibited the enzyme activities of CYP3A4 and GST and down regulated their genes. We interpret this effect as a consequence of a down-regulation of pregnane X receptor (PXR) gene. FUC overcame MDR by significantly suppressing PXR mediated pathways that regulated the expression of CYP3A and ABCB1 genes in HepG-2 cells. | |||
| Key Molecule: Glutathione S-transferase (GST) | [145] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
GST colorimetric assay, RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Synergistic interaction between the MDR mechanisms include ABCT proteins (P-gp, BCRP, and MDR1) and metabolic enzymes of phase I of metabolism mainly CYP3A4, phase II of metabolism mainly GST was observed. In this study, FUC alone and in combination with DOX inhibited the enzyme activities of CYP3A4 and GST and down regulated their genes. We interpret this effect as a consequence of a down-regulation of pregnane X receptor (PXR) gene. FUC overcame MDR by significantly suppressing PXR mediated pathways that regulated the expression of CYP3A and ABCB1 genes in HepG-2 cells. | |||
|
|
||||
| Key Molecule: hsa-mir-122 | [146] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Flow Cytometric Analysis | |||
| Mechanism Description | Transfection of miR122 mimics into cultured HepG2 cells induces cell-cycle arrest and sensitizes these cells to doxorubicin by modulating the expression of multidrug resistance genes, ABCB1 and ABCF2. | |||
| Key Molecule: hsa-mir-26a | [147] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell migration | Inhibition | hsa04670 | ||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | miR26a/b can promote apoptosis and sensitize HCC to chemotherapy via suppressing the expression of autophagy initiator ULk. | |||
| Key Molecule: hsa-mir-26b | [147] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell autophagy | Inhibition | hsa04140 | ||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | miR26a/b can promote apoptosis and sensitize HCC to chemotherapy via suppressing the expression of autophagy initiator ULk. | |||
| Key Molecule: hsa-mir-379 | [148] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | IGF1/IGF1R signaling pathway | Inhibition | hsa05200 | |
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Propidium Iodide (PI) Staining | |||
| Mechanism Description | IGF1 is a hub gene in HCC and is involved in the p53 signaling pathway regulation. miR379 can sensitize HCC cells to chemotherapeutic reagents via targeting IGF1R and suppressing its expression, and suppressing the IGF1/IGF1R signaling pathway. | |||
| Key Molecule: hsa-mir-383 | [149] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| SNU449 cells | Liver | Homo sapiens (Human) | CVCL_0454 | |
| SNU387 cells | Liver | Homo sapiens (Human) | CVCL_0250 | |
| In Vivo Model | BALB/c nude mice model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | miR-383 inhibited Dox resistance in HCC cells by downregulating EIF5A2. | |||
| Key Molecule: Long non-protein coding RNA 607 (LINC00607) | [150] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| NF-kappaB p65/p53 signaling pathway | Regulation | hsa04064 | ||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| HCCLM3 cells | Liver | Homo sapiens (Human) | CVCL_6832 | |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| SMMC7721 cells | Uterus | Homo sapiens (Human) | CVCL_0534 | |
| PLC cells | Liver | Homo sapiens (Human) | CVCL_0485 | |
| MHCC97-H cells | Liver | Homo sapiens (Human) | CVCL_4972 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Real-time RT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | LncRNA 00607 overexpression leads to decreased HCC cell proliferation in vitro and in vivo, enhanced apoptosis and chemotherapeutic drug sensitivity, inhibiting the p65 transcription by binding to the p65 promoter region, therefore contributing to increased p53 levels in HCC. | |||
| Key Molecule: hsa-miR-590-5p | [151] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| Hippo signaling pathway | Regulation | hsa04392 | ||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| In Vivo Model | NU/NU nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | miR-590-5p suppresses hepatocellular carcinoma chemoresistance by downregulating YAP1 expression. | |||
| Key Molecule: hsa-miR-760 | [152] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell viability | Inhibition | hsa05200 | ||
| Notch1/HES1-PTEN/AKT signaling pathway | Regulation | hsa04330 | ||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| SMMC7721 cells | Uterus | Homo sapiens (Human) | CVCL_0534 | |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Caspase-3 Activity kit assay | |||
| Mechanism Description | miR-760 inhibits Dox-resistance in HCC cells through inhibiting Notch1 and promoting PTEN expression. | |||
| Key Molecule: hsa-mir-122 | [153] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| miR122/PKM2 signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| PLC cells | Liver | Homo sapiens (Human) | CVCL_0485 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Overexpression of miR-122 in Huh7/R cells reversed the doxorubicin-resistance through the inhibition of PkM2, inducing the apoptosis in doxorubicin-resistant cancer cells. | |||
| Key Molecule: hsa-mir-101 | [154] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| PLC cells | Liver | Homo sapiens (Human) | CVCL_0485 | |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-101 was downregulated in HCC cell lines, while its overexpression (+) the sensitivity of HepG2 cells to the chemotherapeutic agent DOX by facilitating apoptosis. Of note, Mcl-1 was confirmed as a functional target of miR-101 in HCC, demonstrating that miR-101 may enhance the sensitivity of cancer cells by downregulating Mcl-1 expression. | |||
| Key Molecule: WT1 antisense RNA (WT1-AS) | [155] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| JAKT2/STAT3/MAPK signaling pathway | Inhibition | hsa04659 | ||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| L02 cells | Liver | Homo sapiens (Human) | CVCL_6926 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
EDU assay; Flow cytometry assay | |||
| Mechanism Description | WT1-AS promotes cell apoptosis in hepatocellular carcinoma through down-regulating of WT1.WT1-AS expression correlated negatively with WT1 expression in HCC tumor tissue. kaplan-Meier curve analysis revealed that WT1-AS expression is a reliable indicator of HCC prognosis. The downregulation of WT1 expression by WT1-AS promoted cell apoptosis by suppressing the JAk/STAT3 signaling pathway. Bioinformatics analysis showed that WT1-AS downregulates WT1 by binding to the TATA region of the WT1 promotor. WT1-AS was also able to reverse WT1-mediated resistance to Dox based chemotherapy in HCC cells. | |||
| Key Molecule: WT1 antisense RNA (WT1-AS) | [155] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| JAKT2/STAT3/MAPK signaling pathway | Inhibition | hsa04659 | ||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| L02 cells | Liver | Homo sapiens (Human) | CVCL_6926 | |
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
EDU assay; Flow cytometry assay | |||
| Mechanism Description | WT1-AS promotes cell apoptosis in hepatocellular carcinoma through down-regulating of WT1.WT1-AS expression correlated negatively with WT1 expression in HCC tumor tissue. kaplan-Meier curve analysis revealed that WT1-AS expression is a reliable indicator of HCC prognosis. The downregulation of WT1 expression by WT1-AS promoted cell apoptosis by suppressing the JAk/STAT3 signaling pathway. Bioinformatics analysis showed that WT1-AS downregulates WT1 by binding to the TATA region of the WT1 promotor. WT1-AS was also able to reverse WT1-mediated resistance to Dox based chemotherapy in HCC cells. | |||
| Key Molecule: hsa-mir-27b | [144] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | miR27b/CCNG1/p53 signaling pathway | Regulation | hsa05206 | |
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| SNU182 cells | Liver | Homo sapiens (Human) | CVCL_0090 | |
| SNU-739 cells | Liver | Homo sapiens (Human) | CVCL_5088 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | |||
| Mechanism Description | miR-27b synergizes with anticancer drugs througth enhancing anticancer drug-induced cell death which due to p53 activation and CYP1B1 suppression. | |||
| Key Molecule: hsa-mir-503 | [156] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
MTS assay; Flow cytometry assay | |||
| Mechanism Description | The expression of a number drug resistance related proteins, including multidrug resistance 1, multi drug resistance associated protein 1, DNA excision repair protein ERCC 1, survivin and B cell lymphoma 2, was significantly downregulated by miR 503 overexpression. | |||
| Key Molecule: hsa-mir-26b | [157] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| NF-kappaB signaling pathway | Inhibition | hsa04064 | ||
| In Vitro Model | QGY-7703 cells | Liver | Homo sapiens (Human) | CVCL_6715 |
| MHCC97-H cells | Liver | Homo sapiens (Human) | CVCL_4972 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-26b suppresses NF-kB signaling and thereby sensitized HCC cells to the doxorubicin-induced apoptosis by inhibiting the expression of TAk1 and TAB3. | |||
| Key Molecule: hsa-mir-221 | [158] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-221 can activate the p53/mdm2 axis by inhibiting MDM2 and, in turn, p53 activation contributes to miR-221 enhanced expression. Moreover, by modulating the p53 axis, miR-221 impacts cell-cycle progression and apoptotic response to doxorubicin in hepatocellular carcinoma-derived cell lines. | |||
| Key Molecule: hsa-mir-101 | [159] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| SNU182 cells | Liver | Homo sapiens (Human) | CVCL_0090 | |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
WST-1 assay | |||
| Mechanism Description | miR-101-mediated EZH2 silencing sensitized hepatoblastoma cells to 5-FU- and doxorubicin-induced apoptosis, whereas antagomiR-mediated downregulation of endogenous miR-101 reversed the pro-apoptotic effect. | |||
| Key Molecule: Long non-protein coding RNA (BX537613) | [160] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | LncRNA-BX537613 knockdown could sensitize MCF-7/ADR cell to adriamycin again. | |||
| Key Molecule: hsa-mir-137 | [53] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| In Vivo Model | Immunodeficient NCr nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Cell titer glo assay assay | |||
| Mechanism Description | Hypermethylation of the miR-137 promoter and negative regulation of miR-137 by CAR contribute in part to reduced miR-137 expression and increased CAR and MDR1 expression in doxorubicin-resistant neuroblastoma cells. | |||
| Key Molecule: hsa-mir-223 | [161] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| BEL-7402 cells | Liver | Homo sapiens (Human) | CVCL_5492 | |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| SMMC7721 cells | Uterus | Homo sapiens (Human) | CVCL_0534 | |
| Skhep1 cells | Liver | Homo sapiens (Human) | CVCL_0525 | |
| HCC3 cells | Liver | Homo sapiens (Human) | CVCL_0C57 | |
| LM-6 cells | Liver | Homo sapiens (Human) | CVCL_7680 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
WST-1 assay | |||
| Mechanism Description | miR-223 targeted ABCB1 3'UTR directly, and miR-223 down-regulated ABCB1 at both mRNA and protein levels. The over-expression of miR-223 increased the HCC cellsensitivity to anticancer drugs, and the inhibition of miR-223 had the opposite effect. Importantly, the over-expression or silencingof ABCB1 can rescue the cell response to the anticancer drugs mediated by miR-223 over-expression or inhibition. | |||
| Key Molecule: hsa-mir-122 | [162] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| PLC/PRF/5 cells | Liver | Homo sapiens (Human) | CVCL_0485 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Overexpression of miR-122 could modulate the sensitivity of the HCC cells to chemotherapeutic drugs through downregulating MDR related genes MDR-1, GST-Pi, and MRP, antiapoptotic gene Bcl-w and cell cycle related gene cyclin B1. | |||
| Key Molecule: hsa-miR-199a-3p | [163] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| PLC/PRF/5 cells | Liver | Homo sapiens (Human) | CVCL_0485 | |
| HLE cells | Liver | Homo sapiens (Human) | CVCL_1281 | |
| HLF cells | Liver | Homo sapiens (Human) | CVCL_2947 | |
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
WST-1 assay | |||
| Mechanism Description | There is an inverse correlation between the expression of miR-199a-3p and CD44 protein. Transfection of miR-199a-3p into SNU449 cells reduced in vitro invasion and sensitized the cells to doxorubicin. Inhibition of CD44 in CD44+ HCC cell lines using antisense oligonucleotides increased apoptosis, enhanced chemosensitivity, reduced tumorigensis and invasion. | |||
| Key Molecule: hsa-miR-199a-3p | [164] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Hepatocellular cancer [ICD-11: 2C12.4] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| mTOR signaling pathway | Inhibition | hsa04150 | ||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| SNU475 cells | Liver | Homo sapiens (Human) | CVCL_0497 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Cell invasion assay | |||
| Mechanism Description | The mTOR pathway is activated by multiple extracellular signals, such as growth factors, nutrients, amino acids, hormones, and mitogens leading to the phosphorylation of the translational regulator, phospho-p70S6 kinase, which, in turn, regulates cell proliferation, regulates protein synthesis, and allows progression from the G1 to the S phase of the cell cycle. There is an inverse correlation linking miR-199a-3p and mTOR. miR-199a-3p restoration blocks the G1-S transition of the cell cycle, impairs invasion capability, and sensitizes HCC cells to doxorubicin challenge. | |||
| Key Molecule: hsa-mir-122 | [165] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Hepatocellular cancer [ICD-11: 2C12.4] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| p53 signaling pathway | Activation | hsa04115 | ||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Annexin V/propidium iodide detection kit | |||
| Mechanism Description | miR-122 enforced expression, as well as cyclin G1 silencing, leads to increased p53 protein stability and transcriptional activity and reduced invasion capability of HCC-derived cell lines, miR-122, through down-regulation of cyclin G1, can trigger apoptosis and increase sensitivity of HCC-derived cells to doxorubicin. | |||
|
|
||||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [146] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Flow Cytometric Analysis | |||
| Mechanism Description | Transfection of miR122 mimics into cultured HepG2 cells induces cell-cycle arrest and sensitizes these cells to doxorubicin by modulating the expression of multidrug resistance genes, ABCB1 and ABCF2. | |||
| Key Molecule: ATP-binding cassette sub-family F2 (ABCF2) | [146] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Flow Cytometric Analysis | |||
| Mechanism Description | Transfection of miR122 mimics into cultured HepG2 cells induces cell-cycle arrest and sensitizes these cells to doxorubicin by modulating the expression of multidrug resistance genes, ABCB1 and ABCF2. | |||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [156] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay; Flow cytometry assay | |||
| Mechanism Description | The expression of a number drug resistance related proteins, including multidrug resistance 1, multi drug resistance associated protein 1, DNA excision repair protein ERCC 1, survivin and B cell lymphoma 2, was significantly downregulated by miR 503 overexpression. | |||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [161] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| BEL-7402 cells | Liver | Homo sapiens (Human) | CVCL_5492 | |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| SMMC7721 cells | Uterus | Homo sapiens (Human) | CVCL_0534 | |
| Skhep1 cells | Liver | Homo sapiens (Human) | CVCL_0525 | |
| HCC3 cells | Liver | Homo sapiens (Human) | CVCL_0C57 | |
| LM-6 cells | Liver | Homo sapiens (Human) | CVCL_7680 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
WST-1 assay | |||
| Mechanism Description | miR-223 targeted ABCB1 3'UTR directly, and miR-223 down-regulated ABCB1 at both mRNA and protein levels. The over-expression of miR-223 increased the HCC cellsensitivity to anticancer drugs, and the inhibition of miR-223 had the opposite effect. Importantly, the over-expression or silencingof ABCB1 can rescue the cell response to the anticancer drugs mediated by miR-223 over-expression or inhibition. | |||
| Key Molecule: ATP-binding cassette sub-family G2 (ABCG2) | [145] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Synergistic interaction between the MDR mechanisms include ABCT proteins (P-gp, BCRP, and MDR1) and metabolic enzymes of phase I of metabolism mainly CYP3A4, phase II of metabolism mainly GST was observed. In this study, FUC alone and in combination with DOX inhibited the enzyme activities of CYP3A4 and GST and down regulated their genes. We interpret this effect as a consequence of a down-regulation of pregnane X receptor (PXR) gene. FUC overcame MDR by significantly suppressing PXR mediated pathways that regulated the expression of CYP3A and ABCB1 genes in HepG-2 cells. | |||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [145] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Synergistic interaction between the MDR mechanisms include ABCT proteins (P-gp, BCRP, and MDR1) and metabolic enzymes of phase I of metabolism mainly CYP3A4, phase II of metabolism mainly GST was observed. In this study, FUC alone and in combination with DOX inhibited the enzyme activities of CYP3A4 and GST and down regulated their genes. We interpret this effect as a consequence of a down-regulation of pregnane X receptor (PXR) gene. FUC overcame MDR by significantly suppressing PXR mediated pathways that regulated the expression of CYP3A and ABCB1 genes in HepG-2 cells. | |||
| Key Molecule: ATP-binding cassette sub-family B5 (ABCB5) | [72] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
Efflux of rhodamine123 assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Resveratrol can restore the sensitivity of Caco-2 and CEM/ADR5000 cell lines to doxorubicin, through enhancing significantly doxorubicin cytotoxicity. ABC-transporter inhibitors, classified according to their action on ABC-transporters proteins into: 1. Function inhibitors, 2. Expression inhibitors, and 3. Functional and expression inhibitors, which have an ideal characters of ABC-transporters inhibitors. Our results indicate that resveratrol falls into the class 3 inhibitors. | |||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [166] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | High glucose upregulated the level of MDR-1, which can be expected the intracellular accumulation of anticancer drugs. Interestingly, reduced accumulation of doxorubicin was recorded in cells cultured in high glucose media. Curcumin-mediated inhibition of MDR-1 expression can be suggested as critical event leading to retention of anticancer drug in cellular interior. | |||
| Key Molecule: ATP-binding cassette sub-family B5 (ABCB5) | [145] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Synergistic interaction between the MDR mechanisms include ABCT proteins (P-gp, BCRP, and MDR1) and metabolic enzymes of phase I of metabolism mainly CYP3A4, phase II of metabolism mainly GST was observed. In this study, FUC alone and in combination with DOX inhibited the enzyme activities of CYP3A4 and GST and down regulated their genes. We interpret this effect as a consequence of a down-regulation of pregnane X receptor (PXR) gene. FUC overcame MDR by significantly suppressing PXR mediated pathways that regulated the expression of CYP3A and ABCB1 genes in HepG-2 cells. | |||
|
|
||||
| Key Molecule: Serine/threonine-protein kinase ULK1 (ULK1) | [147] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell autophagy | Inhibition | hsa04140 | ||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | miR26a/b can promote apoptosis and sensitize HCC to chemotherapy via suppressing the expression of autophagy initiator ULk. | |||
| Key Molecule: Insulin-like growth factor 1 receptor (IGF1R) | [148] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | IGF1/IGF1R signaling pathway | Inhibition | hsa05200 | |
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| Experiment for Molecule Alteration |
Dual luciferase assay; Western blot analysis | |||
| Experiment for Drug Resistance |
Propidium Iodide (PI) Staining | |||
| Mechanism Description | IGF1 is a hub gene in HCC and is involved in the p53 signaling pathway regulation. miR379 can sensitize HCC cells to chemotherapeutic reagents via targeting IGF1R and suppressing its expression, and suppressing the IGF1/IGF1R signaling pathway. | |||
| Key Molecule: Eukaryotic translation initiation factor 5A-2 (EIF5A2) | [149] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| SNU449 cells | Liver | Homo sapiens (Human) | CVCL_0454 | |
| SNU387 cells | Liver | Homo sapiens (Human) | CVCL_0250 | |
| In Vivo Model | BALB/c nude mice model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | miR-383 inhibited Dox resistance in HCC cells by downregulating EIF5A2. | |||
| Key Molecule: Cellular tumor antigen p53 (TP53) | [150] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| NF-kappaB p65/p53 signaling pathway | Regulation | hsa04064 | ||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| HCCLM3 cells | Liver | Homo sapiens (Human) | CVCL_6832 | |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| SMMC7721 cells | Uterus | Homo sapiens (Human) | CVCL_0534 | |
| PLC cells | Liver | Homo sapiens (Human) | CVCL_0485 | |
| MHCC97-H cells | Liver | Homo sapiens (Human) | CVCL_4972 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | LncRNA 00607 overexpression leads to decreased HCC cell proliferation in vitro and in vivo, enhanced apoptosis and chemotherapeutic drug sensitivity, inhibiting the p65 transcription by binding to the p65 promoter region, therefore contributing to increased p53 levels in HCC. | |||
| Key Molecule: Transcriptional coactivator YAP1 (YAP1) | [151] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| Hippo signaling pathway | Regulation | hsa04392 | ||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| In Vivo Model | NU/NU nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | miR-590-5p suppresses hepatocellular carcinoma chemoresistance by downregulating YAP1 expression. | |||
| Key Molecule: Neurogenic locus notch homolog protein 1 (NOTCH1) | [152] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell colony | Inhibition | hsa05200 | ||
| Cell viability | Inhibition | hsa05200 | ||
| Notch1/HES1-PTEN/AKT signaling pathway | Regulation | hsa04330 | ||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| SMMC7721 cells | Uterus | Homo sapiens (Human) | CVCL_0534 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Caspase-3 Activity kit assay | |||
| Mechanism Description | miR-760 inhibits Dox-resistance in HCC cells through inhibiting Notch1 and promoting PTEN expression. | |||
| Key Molecule: Pyruvate kinase M2 (PKM) | [153] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| miR122/PKM2 signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| PLC cells | Liver | Homo sapiens (Human) | CVCL_0485 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Overexpression of miR-122 in Huh7/R cells reversed the doxorubicin-resistance through the inhibition of PkM2, inducing the apoptosis in doxorubicin-resistant cancer cells. | |||
| Key Molecule: Induced myeloid leukemia cell differentiation protein Mcl-1 (MCL1) | [154] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| PLC cells | Liver | Homo sapiens (Human) | CVCL_0485 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-101 was downregulated in HCC cell lines, while its overexpression (+) the sensitivity of HepG2 cells to the chemotherapeutic agent DOX by facilitating apoptosis. Of note, Mcl-1 was confirmed as a functional target of miR-101 in HCC, demonstrating that miR-101 may enhance the sensitivity of cancer cells by downregulating Mcl-1 expression. | |||
| Key Molecule: Cyclin-G1 (CCNG1) | [144] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| miR27b/CCNG1/p53 signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| SNU182 cells | Liver | Homo sapiens (Human) | CVCL_0090 | |
| SNU-739 cells | Liver | Homo sapiens (Human) | CVCL_5088 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | |||
| Mechanism Description | miR-27b synergizes with anticancer drugs througth enhancing anticancer drug-induced cell death which due to p53 activation and CYP1B1 suppression. | |||
| Key Molecule: TAK1-binding protein 3 (TAB3) | [157] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| NF-kappaB signaling pathway | Inhibition | hsa04064 | ||
| In Vitro Model | QGY-7703 cells | Liver | Homo sapiens (Human) | CVCL_6715 |
| MHCC97-H cells | Liver | Homo sapiens (Human) | CVCL_4972 | |
| Experiment for Molecule Alteration |
Immunoblotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-26b suppresses NF-kB signaling and thereby sensitized HCC cells to the doxorubicin-induced apoptosis by inhibiting the expression of TAk1 and TAB3. | |||
| Key Molecule: Nuclear receptor subfamily 2 group C2 (NR2C2) | [157] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| NF-kappaB signaling pathway | Inhibition | hsa04064 | ||
| In Vitro Model | QGY-7703 cells | Liver | Homo sapiens (Human) | CVCL_6715 |
| MHCC97-H cells | Liver | Homo sapiens (Human) | CVCL_4972 | |
| Experiment for Molecule Alteration |
Immunoblotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-26b suppresses NF-kB signaling and thereby sensitized HCC cells to the doxorubicin-induced apoptosis by inhibiting the expression of TAk1 and TAB3. | |||
| Key Molecule: E3 ubiquitin-protein ligase Mdm2 (MDM2) | [158] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-221 can activate the p53/mdm2 axis by inhibiting MDM2 and, in turn, p53 activation contributes to miR-221 enhanced expression. Moreover, by modulating the p53 axis, miR-221 impacts cell-cycle progression and apoptotic response to doxorubicin in hepatocellular carcinoma-derived cell lines. | |||
| Key Molecule: Histone-lysine N-methyltransferase EZH2 (EZH2) | [159] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| SNU182 cells | Liver | Homo sapiens (Human) | CVCL_0090 | |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
WST-1 assay | |||
| Mechanism Description | miR-101-mediated EZH2 silencing sensitized hepatoblastoma cells to 5-FU- and doxorubicin-induced apoptosis, whereas antagomiR-mediated downregulation of endogenous miR-101 reversed the pro-apoptotic effect. | |||
| Key Molecule: Nuclear receptor subfamily 1 group I3 (NR1I3) | [53] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| In Vivo Model | Immunodeficient NCr nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Chromatin immunoprecipitation assay | |||
| Experiment for Drug Resistance |
Cell titer glo assay assay | |||
| Mechanism Description | Hypermethylation of the miR-137 promoter and negative regulation of miR-137 by CAR contribute in part to reduced miR-137 expression and increased CAR and MDR1 expression in doxorubicin-resistant neuroblastoma cells. | |||
| Key Molecule: Extracellular matrix receptor III (CD44) | [163] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | |
| Cell migration | Inhibition | hsa04670 | ||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| PLC/PRF/5 cells | Liver | Homo sapiens (Human) | CVCL_0485 | |
| HLE cells | Liver | Homo sapiens (Human) | CVCL_1281 | |
| HLF cells | Liver | Homo sapiens (Human) | CVCL_2947 | |
| Experiment for Molecule Alteration |
Luciferase assay | |||
| Experiment for Drug Resistance |
WST-1 assay | |||
| Mechanism Description | There is an inverse correlation between the expression of miR-199a-3p and CD44 protein. Transfection of miR-199a-3p into SNU449 cells reduced in vitro invasion and sensitized the cells to doxorubicin. Inhibition of CD44 in CD44+ HCC cell lines using antisense oligonucleotides increased apoptosis, enhanced chemosensitivity, reduced tumorigensis and invasion. | |||
| Key Molecule: Serine/threonine-protein kinase mTOR (mTOR) | [164] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Hepatocellular cancer [ICD-11: 2C12.4] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| mTOR signaling pathway | Inhibition | hsa04150 | ||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| SNU475 cells | Liver | Homo sapiens (Human) | CVCL_0497 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
Cell invasion assay | |||
| Mechanism Description | The mTOR pathway is activated by multiple extracellular signals, such as growth factors, nutrients, amino acids, hormones, and mitogens leading to the phosphorylation of the translational regulator, phospho-p70S6 kinase, which, in turn, regulates cell proliferation, regulates protein synthesis, and allows progression from the G1 to the S phase of the cell cycle. There is an inverse correlation linking miR-199a-3p and mTOR. miR-199a-3p restoration blocks the G1-S transition of the cell cycle, impairs invasion capability, and sensitizes HCC cells to doxorubicin challenge. | |||
| Key Molecule: Cyclin-G1 (CCNG1) | [165] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Hepatocellular cancer [ICD-11: 2C12.4] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| p53 signaling pathway | Activation | hsa04115 | ||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
Annexin V/propidium iodide detection kit | |||
| Mechanism Description | miR-122 enforced expression, as well as cyclin G1 silencing, leads to increased p53 protein stability and transcriptional activity and reduced invasion capability of HCC-derived cell lines, miR-122, through down-regulation of cyclin G1, can trigger apoptosis and increase sensitivity of HCC-derived cells to doxorubicin. | |||
| Key Molecule: Fatty acid synthase (FASN) | [166] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Curcumin mediated the amputation of chemoresistance by repressing the hyperglycolytic behavior of malignant cells via modulated expression of metabolic enzymes (HkII, PFk1, GAPDH, PkM2, LDH, SDH, IDH, and FASN), transporters (GLUT-1, MCT-1, and MCT-4), and their regulators. Along altered constitution of extracellular milieu, these molecular changes culminated into improved drug accumulation, chromatin condensation, and induction of cell death. | |||
| Key Molecule: Solute carrier family 2 member 1 (SLC2A1) | [166] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Curcumin mediated the amputation of chemoresistance by repressing the hyperglycolytic behavior of malignant cells via modulated expression of metabolic enzymes (HkII, PFk1, GAPDH, PkM2, LDH, SDH, IDH, and FASN), transporters (GLUT-1, MCT-1, and MCT-4), and their regulators. Along altered constitution of extracellular milieu, these molecular changes culminated into improved drug accumulation, chromatin condensation, and induction of cell death. | |||
| Key Molecule: Isocitrate dehydrogenase NAD 3 alpha (IDH3A) | [166] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Curcumin mediated the amputation of chemoresistance by repressing the hyperglycolytic behavior of malignant cells via modulated expression of metabolic enzymes (HkII, PFk1, GAPDH, PkM2, LDH, SDH, IDH, and FASN), transporters (GLUT-1, MCT-1, and MCT-4), and their regulators. Along altered constitution of extracellular milieu, these molecular changes culminated into improved drug accumulation, chromatin condensation, and induction of cell death. | |||
| Key Molecule: Solute carrier family 16 member 1 (SLC16A1) | [166] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Curcumin mediated the amputation of chemoresistance by repressing the hyperglycolytic behavior of malignant cells via modulated expression of metabolic enzymes (HkII, PFk1, GAPDH, PkM2, LDH, SDH, IDH, and FASN), transporters (GLUT-1, MCT-1, and MCT-4), and their regulators. Along altered constitution of extracellular milieu, these molecular changes culminated into improved drug accumulation, chromatin condensation, and induction of cell death. | |||
| Key Molecule: Solute carrier family 16 member 3 (SLC16A3) | [166] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Curcumin mediated the amputation of chemoresistance by repressing the hyperglycolytic behavior of malignant cells via modulated expression of metabolic enzymes (HkII, PFk1, GAPDH, PkM2, LDH, SDH, IDH, and FASN), transporters (GLUT-1, MCT-1, and MCT-4), and their regulators. Along altered constitution of extracellular milieu, these molecular changes culminated into improved drug accumulation, chromatin condensation, and induction of cell death. | |||
| Key Molecule: Succinate dehydrogenase [ubiquinone] iron-sulfur subunit (SDHB) | [166] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Curcumin was able to induce SDH expression and repress the IDH3a in HepG2 cells both in a normal or elevated level of glucose. Such changes in SDH and IDH3a levels can bring a reduction in the succinate accumulation and hindering the succinate-HIF-1alpha axis. The augmented expression of HIF-1alpha in high glucose conditions was resisted by curcumin. HIF-1alpha is known for metabolic regulation in malignant cells, their hyperglycolytic behavior, and the onset of chemoresistance. HIF-1 exerts protumor effects through the upregulated expression of enzymes and transporters favoring the hyperglycolytic and therapy-resistant phenotype. | |||
| Key Molecule: Succinate dehydrogenase [ubiquinone] iron-sulfur subunit (SDHB) | [166] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Curcumin mediated the amputation of chemoresistance by repressing the hyperglycolytic behavior of malignant cells via modulated expression of metabolic enzymes (HkII, PFk1, GAPDH, PkM2, LDH, SDH, IDH, and FASN), transporters (GLUT-1, MCT-1, and MCT-4), and their regulators. Along altered constitution of extracellular milieu, these molecular changes culminated into improved drug accumulation, chromatin condensation, and induction of cell death. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Long non-protein coding RNA (GBCDRlnc1) | [13] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Gallbladder cancer [ICD-11: 2C13.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell autophagy | Activation | hsa04140 | |
| GBCD/Rlnc1/PGK1 signaling pathway | Activation | hsa04066 | ||
| In Vitro Model | GBC-SD cells | Gallbladder | Homo sapiens (Human) | CVCL_6903 |
| NOZ cells | Gallbladder | Homo sapiens (Human) | CVCL_3079 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | LncRNA GBCDRlnc1 induces autophagy and drug-resistance of gallbladder cancer cells by interacting with PGk1 and preventing its degradation, which eventually upregulates ATG5-ATG12 expression. | |||
|
|
||||
| Key Molecule: Phosphoglycerate kinase 1 (PGK1) | [13] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Gallbladder cancer [ICD-11: 2C13.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell autophagy | Activation | hsa04140 | |
| GBCD/Rlnc1/PGK1 signaling pathway | Activation | hsa04066 | ||
| In Vitro Model | GBC-SD cells | Gallbladder | Homo sapiens (Human) | CVCL_6903 |
| NOZ cells | Gallbladder | Homo sapiens (Human) | CVCL_3079 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RNA immunoprecipitation; Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | LncRNA GBCDRlnc1 induces autophagy and drug-resistance of gallbladder cancer cells by interacting with PGk1 and preventing its degradation, which eventually upregulates ATG5-ATG12 expression. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-125a | [167] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Laryngeal cancer [ICD-11: 2C23.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HEp-2 cells | Skin | Homo sapiens (Human) | CVCL_1906 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Annexin V-FITC apoptosis assay | |||
| Mechanism Description | Inhibition of HAX-1 by miR125a reverses cisplatin resistance in laryngeal cancer stem cells. Overexpression of miR125a increases the sensitivity of Hep-2-CSCs to cisplatin by inhibiting HAX-1. | |||
|
|
||||
| Key Molecule: HCLS1-associated protein X-1 (HAX1) | [167] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Laryngeal cancer [ICD-11: 2C23.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HEp-2 cells | Skin | Homo sapiens (Human) | CVCL_1906 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
MTT assay; Annexin V-FITC apoptosis assay | |||
| Mechanism Description | Inhibition of HAX-1 by miR125a reverses cisplatin resistance in laryngeal cancer stem cells. Overexpression of miR125a increases the sensitivity of Hep-2-CSCs to cisplatin by inhibiting HAX-1. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) | [168] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | |
| SPC-A1 cells | Lung | Homo sapiens (Human) | CVCL_6955 | |
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; CCK8 assay; Colony formation assay; Flow cytometry assay | |||
| Mechanism Description | MALAT1 could alter chemo-resistance (Cisplatin, Adriamycin, Gefitinib and Paclitaxel) of NSCLC cells by targeting miR-197-3p and regulating p120-ctn expression, which might assist in improvement of chemo-therapies for NSCLC. | |||
| Key Molecule: hsa-miR-197-3p | [168] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | |
| SPC-A1 cells | Lung | Homo sapiens (Human) | CVCL_6955 | |
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; CCK8 assay; Colony formation assay; Flow cytometry assay | |||
| Mechanism Description | MALAT1 could alter chemo-resistance (Cisplatin, Adriamycin, Gefitinib and Paclitaxel) of NSCLC cells by targeting miR-197-3p and regulating p120-ctn expression, which might assist in improvement of chemo-therapies for NSCLC. | |||
| Key Molecule: HOX transcript antisense RNA (HOTAIR) | [169] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Lung small cell carcinoma [ICD-11: 2C25.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | NCI-H446 cells | Lung | Homo sapiens (Human) | CVCL_1562 |
| NCI-H69 cells | Lung | Homo sapiens (Human) | CVCL_1579 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | H3kH3k27me3 induces multidrug resistance in small cell lung cancer by affecting HOXA1 DNA methylation via regulation of the LncRNA HOTAIR. | |||
| Key Molecule: hsa-mir-216a | [170] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Lung small cell carcinoma [ICD-11: 2C25.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | 16HBE cells | Lung | Homo sapiens (Human) | CVCL_0112 |
| H446 cells | Lung | Homo sapiens (Human) | CVCL_1562 | |
| H69 cells | Lung | Homo sapiens (Human) | CVCL_8121 | |
| H69AR cells | Lung | Homo sapiens (Human) | CVCL_3513 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Long non-coding RNA HOTTIP promotes BCL-2 expression and induces chemoresistance in small cell lung cancer by sponging miR216a. HOTTIP functioned as an oncogene in SCLC progression by binding miR216a and abrogating its tumor-suppressive function in this setting. HOTTIP also increased the expression of anti-apoptotic factor BCL-2, a target gene of miR216a, and jointly enhanced chemoresistance of SCLC by regulating BCL-2. | |||
| Key Molecule: hsa-mir-100 | [19] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Lung small cell carcinoma [ICD-11: 2C25.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | NCI-H69 cells | Lung | Homo sapiens (Human) | CVCL_1579 |
| NCI-H69AR cells | Lung | Homo sapiens (Human) | CVCL_3513 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Forced expression of HOXA1 in immortalised human mammary epithelial cells results in oncogenic transformation and tumour formation in vivo. HOXA1 expression was inversely correlated with miR-100. HOXA1-mediated SCLC chemoresistance is under the regulation of miR-100. HOXA1 may be a prognostic predictor and potential therapeutic target in human SCLC. | |||
| Key Molecule: hsa-mir-128 | [41] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K/AKT signaling pathway | Activation | hsa04151 | |
| In Vitro Model | H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 |
| A459 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |
| H1299 clone 23 cells | Lung | Homo sapiens (Human) | N.A. | |
| H1299 clone 41 cells | Lung | Homo sapiens (Human) | N.A. | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | The expression of the transcriptional repressor E2F5, a target of miR-128-2, strongly decreases after miR-128-2 exogenous expression. This leads to the abrogation of E2F5 repressive activity on p21waf1 promoter and, consequently, to the transcriptional induction of p21waf1. The newly synthesized p21waf1 protein is mainly localized into the cytoplasmic compartment, where it exerts an anti-apoptotic function in response to anticancer drug treatments. | |||
|
|
||||
| Key Molecule: HOXA distal transcript antisense RNA (HOTTIP) | [170] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Lung small cell carcinoma [ICD-11: 2C25.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | 16HBE cells | Lung | Homo sapiens (Human) | CVCL_0112 |
| H446 cells | Lung | Homo sapiens (Human) | CVCL_1562 | |
| H69 cells | Lung | Homo sapiens (Human) | CVCL_8121 | |
| H69AR cells | Lung | Homo sapiens (Human) | CVCL_3513 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | HOTTIP acts as sponge of miR216a and enhanced the expression of its another target gene, anti-apoptotic gene BCL-2. | |||
| Key Molecule: Ephrin type-A receptor 3 (EPHA3) | [171] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Lung small cell carcinoma [ICD-11: 2C25.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | NCI-H446 cells | Lung | Homo sapiens (Human) | CVCL_1562 |
| NCI-H69 cells | Lung | Homo sapiens (Human) | CVCL_1579 | |
| H69/AR cells | Lung | Homo sapiens (Human) | CVCL_3513 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Immunohistochemical staining; Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Cell scratch-wound healing assay; Flow cytometry assay | |||
| Mechanism Description | miR495 promotes the chemoresistance of SCLC through the epithelial-mesenchymal transition via Etk/BMX. Ectopic expression of Etk/BMX obviously rescued the miR495 elevation elevation-induced inhibition of drug resistance. | |||
| Key Molecule: Cytoplasmic tyrosine-protein kinase BMX (BMX) | [171] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Lung small cell carcinoma [ICD-11: 2C25.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | NCI-H446 cells | Lung | Homo sapiens (Human) | CVCL_1562 |
| NCI-H69 cells | Lung | Homo sapiens (Human) | CVCL_1579 | |
| H69/AR cells | Lung | Homo sapiens (Human) | CVCL_3513 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Immunohistochemical staining; Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Cell scratch-wound healing assay; Flow cytometry assay | |||
| Mechanism Description | miR495 promotes the chemoresistance of SCLC through the epithelial-mesenchymal transition via Etk/BMX. Ectopic expression of Etk/BMX obviously rescued the miR495 elevation elevation-induced inhibition of drug resistance. | |||
| Key Molecule: hsa-mir-495 | [171] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Lung small cell carcinoma [ICD-11: 2C25.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | NCI-H446 cells | Lung | Homo sapiens (Human) | CVCL_1562 |
| NCI-H69 cells | Lung | Homo sapiens (Human) | CVCL_1579 | |
| H69/AR cells | Lung | Homo sapiens (Human) | CVCL_3513 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Cell scratch-wound healing assay; Flow cytometry assay | |||
| Mechanism Description | miR495 promotes the chemoresistance of SCLC through the epithelial-mesenchymal transition via Etk/BMX. Ectopic expression of Etk/BMX obviously rescued the miR495 elevation elevation-induced inhibition of drug resistance. | |||
| Key Molecule: ATPase H+ transporting V0 subunit d1 (ATP6V0D1) | [172] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Non-small cell lung cancer isolates | Lung | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Immunofluorescence assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The drug resistance of cancer cells is likely to be related to the changes in pH gradient between the extracellular environment and the cytoplasm.Vacuolar-H+ -ATPase(V-ATPase) plays a major role in the regulation of cellular pH conditions.The expression of V-ATPase was shown to be related to the pathological type and grade of the cancer and might be associated with the chemotherapy drug resistance in NSCLC. | |||
| Key Molecule: ATPase H+ transporting V0 subunit d1 (ATP6V0D1) | [172] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Lung squamous cell carcinoma [ICD-11: 2C25.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Non-small cell lung cancer isolates | Lung | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Immunofluorescence assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The drug resistance of cancer cells is likely to be related to the changes in pH gradient between the extracellular environment and the cytoplasm.Vacuolar-H+ -ATPase(V-ATPase) plays a major role in the regulation of cellular pH conditions.The expression of V-ATPase was shown to be related to the pathological type and grade of the cancer and might be associated with the chemotherapy drug resistance in NSCLC. | |||
| Key Molecule: ATPase H+ transporting V0 subunit d1 (ATP6V0D1) | [172] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Non-small cell lung cancer isolates | Lung | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Immunofluorescence assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The drug resistance of cancer cells is likely to be related to the changes in pH gradient between the extracellular environment and the cytoplasm.Vacuolar-H+ -ATPase(V-ATPase) plays a major role in the regulation of cellular pH conditions.The expression of V-ATPase was shown to be related to the pathological type and grade of the cancer and might be associated with the chemotherapy drug resistance in NSCLC. | |||
|
|
||||
| Key Molecule: Catenin delta-1 (CTNND1) | [168] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | |
| SPC-A1 cells | Lung | Homo sapiens (Human) | CVCL_6955 | |
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; CCK8 assay; Colony formation assay; Flow cytometry assay | |||
| Mechanism Description | MALAT1 could alter chemo-resistance (Cisplatin, Adriamycin, Gefitinib and Paclitaxel) of NSCLC cells by targeting miR-197-3p and regulating p120-ctn expression, which might assist in improvement of chemo-therapies for NSCLC. | |||
| Key Molecule: H3 lysine 27 trimethylation (H3K27) | [169] | |||
| Molecule Alteration | Methylation | Up-regulation |
||
| Resistant Disease | Lung small cell carcinoma [ICD-11: 2C25.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | NCI-H446 cells | Lung | Homo sapiens (Human) | CVCL_1562 |
| NCI-H69 cells | Lung | Homo sapiens (Human) | CVCL_1579 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | H3kH3k27me3 induces multidrug resistance in small cell lung cancer by affecting HOXA1 DNA methylation via regulation of the LncRNA HOTAIR. | |||
| Key Molecule: Homeobox protein Hox-A13 (HOXA13) | [170] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Lung small cell carcinoma [ICD-11: 2C25.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | 16HBE cells | Lung | Homo sapiens (Human) | CVCL_0112 |
| H446 cells | Lung | Homo sapiens (Human) | CVCL_1562 | |
| H69 cells | Lung | Homo sapiens (Human) | CVCL_8121 | |
| H69AR cells | Lung | Homo sapiens (Human) | CVCL_3513 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | HOTTIP acts at least partly by controlling HOXA13 in SCLC poor prognostic and chemoresistance progression. | |||
| Key Molecule: Homeobox protein Hox-A1 (HOXA1) | [19] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Lung small cell carcinoma [ICD-11: 2C25.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | NCI-H69 cells | Lung | Homo sapiens (Human) | CVCL_1579 |
| NCI-H69AR cells | Lung | Homo sapiens (Human) | CVCL_3513 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Forced expression of HOXA1 in immortalised human mammary epithelial cells results in oncogenic transformation and tumour formation in vivo. HOXA1 expression was inversely correlated with miR-100. HOXA1-mediated SCLC chemoresistance is under the regulation of miR-100. HOXA1 may be a prognostic predictor and potential therapeutic target in human SCLC. | |||
| Key Molecule: Protein LYRIC (MTDH) | [41] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K/AKT signaling pathway | Activation | hsa04151 | |
| In Vitro Model | H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 |
| A459 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |
| H1299 clone 23 cells | Lung | Homo sapiens (Human) | N.A. | |
| H1299 clone 41 cells | Lung | Homo sapiens (Human) | N.A. | |
| Experiment for Molecule Alteration |
Luciferase assay | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | The expression of the transcriptional repressor E2F5, a target of miR-128-2, strongly decreases after miR-128-2 exogenous expression. This leads to the abrogation of E2F5 repressive activity on p21waf1 promoter and, consequently, to the transcriptional induction of p21waf1. The newly synthesized p21waf1 protein is mainly localized into the cytoplasmic compartment, where it exerts an anti-apoptotic function in response to anticancer drug treatments. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Glutathione S-transferase P (GSTP1) | [173] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | AKT/ERK signaling pathway | Inhibition | hsa04010 | |
| Cell apoptosis | Activation | hsa04210 | ||
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Suppression of miR-155 in this cell line considerably reversed doxorubicin resistance, and doxorubicin-induced apoptosis and cell cycle arrest were recovered. Furthermore, reverse transcription-polymerase chain reaction and western blot analysis revealed that miR-155 suppression downregulated the expression of multidrug resistance protein 1, multidrug resistance-associated protein 1, breast cancer resistance protein, glutathione S-transferase-Pi, Survivin and B-cell lymphoma 2, and upregulated the expression of caspase-3 and caspase-8. In addition, it was found that miR-155 suppression inhibited the activation of AkT and extracellular signal-regulated kinase. The transcriptional activity of nuclear factor-kB and activator protein-1 was also downregulated. | |||
|
|
||||
| Key Molecule: Apoptosis regulator Bcl-2 (BCL2) | [170] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Lung small cell carcinoma [ICD-11: 2C25.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | 16HBE cells | Lung | Homo sapiens (Human) | CVCL_0112 |
| H446 cells | Lung | Homo sapiens (Human) | CVCL_1562 | |
| H69 cells | Lung | Homo sapiens (Human) | CVCL_8121 | |
| H69AR cells | Lung | Homo sapiens (Human) | CVCL_3513 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Luciferase reporter assay; RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Long non-coding RNA HOTTIP promotes BCL-2 expression and induces chemoresistance in small cell lung cancer by sponging miR216a. HOTTIP functioned as an oncogene in SCLC progression by binding miR216a and abrogating its tumor-suppressive function in this setting. HOTTIP also increased the expression of anti-apoptotic factor BCL-2, a target gene of miR216a, and jointly enhanced chemoresistance of SCLC by regulating BCL-2. | |||
| Key Molecule: hsa-miR-142-3p | [174] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K/AKT/mTOR signaling pathway | Activation | hsa04151 | |
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| 16HBE cells | Lung | Homo sapiens (Human) | CVCL_0112 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Caspase-3 and TUNEL staining assay; MTT assay | |||
| Mechanism Description | miR142-3p regulates starvation-induced autophagy of NSCLC cells by directly downregulating HMGB1 and subsequently activating the PI3k/Akt/mTOR pathway. miR142-3p overexpression inhibited anticancer drug-induced autophagy and increased chemo-sensitivity of NSCLC in vitro and in vivo. | |||
| Key Molecule: hsa-mir-335 | [175] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Lung small cell carcinoma [ICD-11: 2C25.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | NF-kappaB signaling pathway | Inhibition | hsa04064 | |
| In Vitro Model | H69 cells | Lung | Homo sapiens (Human) | CVCL_8121 |
| H69AR cells | Lung | Homo sapiens (Human) | CVCL_3513 | |
| H446 cells | Lung | Homo sapiens (Human) | CVCL_1562 | |
| H446/DDP cells | Lung | Homo sapiens (Human) | CVCL_RT21 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Annexin V-PE Apoptosis assay; Flow cytometry assay; Wound healing assay; Colony formation assay | |||
| Mechanism Description | Overexpression of miR335 sensitized human SCLC cells to chemotherapy and radiotherapy, promoted cell apoptosis and inhibited cell migration ability of human SCLC in vitro, and inhibited tumor growth in vivo. Overexpression of miR335 decreased the expression of PARP-1 mRNA and protein, and NF-kB protein levels were correspondingly downregulated, thus regulating the chemo-radiosensitivity of SCLC. | |||
| Key Molecule: hsa-mir-138 | [176] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| ERK signaling pathway | Regulation | hsa04210 | ||
| RhoC/FAKT/Src/ROCK2 signaling pathways | Regulation | hsa05200 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| NCI-H23 cells | Lung | Homo sapiens (Human) | CVCL_1547 | |
| In Vivo Model | CB-17 SCID Mus musculus(Mouse) Orthotopic breast tumor model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-138 sensitizes NSCLC cells to ADM through regulation of EMT regulator ZEB2. Ectopic expression of miR-138 decreased ZEB2 expression and inhibited the luciferase activity in chemoresistant tumor cells, suggesting that miR-138 could regulate EMT, at least partly, through targeting ZEB2 in NSCLC cells. | |||
| Key Molecule: hsa-mir-155 | [173] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | AKT/ERK signaling pathway | Inhibition | hsa04010 | |
| Cell apoptosis | Activation | hsa04210 | ||
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Suppression of miR-155 in this cell line considerably reversed doxorubicin resistance, and doxorubicin-induced apoptosis and cell cycle arrest were recovered. Furthermore, reverse transcription-polymerase chain reaction and western blot analysis revealed that miR-155 suppression downregulated the expression of multidrug resistance protein 1, multidrug resistance-associated protein 1, breast cancer resistance protein, glutathione S-transferase-Pi, Survivin and B-cell lymphoma 2, and upregulated the expression of caspase-3 and caspase-8. In addition, it was found that miR-155 suppression inhibited the activation of AkT and extracellular signal-regulated kinase. The transcriptional activity of nuclear factor-kB and activator protein-1 was also downregulated. | |||
| Key Molecule: hsa-miR-299-3p | [177] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | NCI-H69 cells | Lung | Homo sapiens (Human) | CVCL_1579 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | miR-299-3p in doxorubicin-sensitive lung cancer was decreased less than that in doxorubicin-resistant lung cancer samples, which directly regulated the expression of ABCE1. Over-expression of miR-299-3p was significantly inhibited the cell proliferation and increased cell apoptosis in H69/ADR lung cancer cells, and also promoted cell inhibitory rate. Over-expression of miR-299-3p promotes the sensibility of lung cancer to doxorubicin. | |||
| Key Molecule: hsa-mir-494 | [178] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Lung small cell carcinoma [ICD-11: 2C25.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | BCL2 signaling pathway | Activation | hsa04210 | |
| Cell apoptosis | Activation | hsa04210 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | H69 cells | Lung | Homo sapiens (Human) | CVCL_8121 |
| H446 cells | Lung | Homo sapiens (Human) | CVCL_1562 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | Knockdown of SCGN led to significantly increasing of chemosensitivity, which is similar to those induced by miR-494 mimics, and ectopic expression of SCGN could rescue the suppressive effect of miR-494. | |||
| Key Molecule: hsa-miR-1291 | [137] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Lung small cell carcinoma [ICD-11: 2C25.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell growth | Inhibition | hsa05200 | |
| In Vitro Model | PANC-1 cells | Pancreas | Homo sapiens (Human) | CVCL_0480 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Hsa-miR-1291-directed downregulation of ABCC1 led to a greater intracellular drug accumulation and sensitized the cells to doxorubicin. | |||
| Key Molecule: hsa-mir-126 | [179] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K/AKT signaling pathway | Inhibition | hsa04151 | |
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | VEGF activates the downstream PI3k/Akt signaling pathway, which is a critical regulator of cellular growth, differentiation, and metabolism. miR-126 could overcome the resistance of NSCLC cells to antineoplastic drugs through inhibition of a VEGF-PI3k/Akt signaling pathway that resulted in the down-regulation of MRP1. | |||
| Key Molecule: hsa-mir-181 | [129] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| A549/CDDP cells | Lung | Homo sapiens (Human) | CVCL_0023 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The antiapoptotic protein BCL2 is upregulated, whereas miR-181b is downregulated in both SGC7901/VCR and A549/CDDP cells, compared with SGC7901 and A549 cells, respectively. Enforced miR-181b expression reduced BCL2 protein level and sensitized SGC7901/VCR and A549/CDDP cells to VCR-induced and CDDP-induced apoptosis, respectively. | |||
|
|
||||
| Key Molecule: ATP-binding cassette sub-family G2 (ABCG2) | [173] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | AKT/ERK signaling pathway | Inhibition | hsa04010 | |
| Cell apoptosis | Activation | hsa04210 | ||
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR; Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Suppression of miR-155 in this cell line considerably reversed doxorubicin resistance, and doxorubicin-induced apoptosis and cell cycle arrest were recovered. Furthermore, reverse transcription-polymerase chain reaction and western blot analysis revealed that miR-155 suppression downregulated the expression of multidrug resistance protein 1, multidrug resistance-associated protein 1, breast cancer resistance protein, glutathione S-transferase-Pi, Survivin and B-cell lymphoma 2, and upregulated the expression of caspase-3 and caspase-8. In addition, it was found that miR-155 suppression inhibited the activation of AkT and extracellular signal-regulated kinase. The transcriptional activity of nuclear factor-kB and activator protein-1 was also downregulated. | |||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [173] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | AKT/ERK signaling pathway | Inhibition | hsa04010 | |
| Cell apoptosis | Activation | hsa04210 | ||
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Suppression of miR-155 in this cell line considerably reversed doxorubicin resistance, and doxorubicin-induced apoptosis and cell cycle arrest were recovered. Furthermore, reverse transcription-polymerase chain reaction and western blot analysis revealed that miR-155 suppression downregulated the expression of multidrug resistance protein 1, multidrug resistance-associated protein 1, breast cancer resistance protein, glutathione S-transferase-Pi, Survivin and B-cell lymphoma 2, and upregulated the expression of caspase-3 and caspase-8. In addition, it was found that miR-155 suppression inhibited the activation of AkT and extracellular signal-regulated kinase. The transcriptional activity of nuclear factor-kB and activator protein-1 was also downregulated. | |||
| Key Molecule: Multidrug resistance-associated protein 1 (MRP1) | [173] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | AKT/ERK signaling pathway | Inhibition | hsa04010 | |
| Cell apoptosis | Activation | hsa04210 | ||
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Suppression of miR-155 in this cell line considerably reversed doxorubicin resistance, and doxorubicin-induced apoptosis and cell cycle arrest were recovered. Furthermore, reverse transcription-polymerase chain reaction and western blot analysis revealed that miR-155 suppression downregulated the expression of multidrug resistance protein 1, multidrug resistance-associated protein 1, breast cancer resistance protein, glutathione S-transferase-Pi, Survivin and B-cell lymphoma 2, and upregulated the expression of caspase-3 and caspase-8. In addition, it was found that miR-155 suppression inhibited the activation of AkT and extracellular signal-regulated kinase. The transcriptional activity of nuclear factor-kB and activator protein-1 was also downregulated. | |||
| Key Molecule: ATP-binding cassette sub-family E1 (ABCE1) | [177] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | NCI-H69 cells | Lung | Homo sapiens (Human) | CVCL_1579 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | miR-299-3p in doxorubicin-sensitive lung cancer was decreased less than that in doxorubicin-resistant lung cancer samples, which directly regulated the expression of ABCE1. Over-expression of miR-299-3p was significantly inhibited the cell proliferation and increased cell apoptosis in H69/ADR lung cancer cells, and also promoted cell inhibitory rate. Over-expression of miR-299-3p promotes the sensibility of lung cancer to doxorubicin. | |||
| Key Molecule: Multidrug resistance-associated protein 1 (MRP1) | [137] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Lung small cell carcinoma [ICD-11: 2C25.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell growth | Inhibition | hsa05200 | |
| In Vitro Model | PANC-1 cells | Pancreas | Homo sapiens (Human) | CVCL_0480 |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Hsa-miR-1291-directed downregulation of ABCC1 led to a greater intracellular drug accumulation and sensitized the cells to doxorubicin. | |||
|
|
||||
| Key Molecule: High mobility group protein B1 (HMGB1) | [174] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K/AKT/mTOR signaling pathway | Activation | hsa04151 | |
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| 16HBE cells | Lung | Homo sapiens (Human) | CVCL_0112 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
Caspase-3 and TUNEL staining assay; MTT assay | |||
| Mechanism Description | miR142-3p regulates starvation-induced autophagy of NSCLC cells by directly downregulating HMGB1 and subsequently activating the PI3k/Akt/mTOR pathway. miR142-3p overexpression inhibited anticancer drug-induced autophagy and increased chemo-sensitivity of NSCLC in vitro and in vivo. | |||
| Key Molecule: Poly[ADP-ribose] synthase 1 (PARP1) | [175] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Lung small cell carcinoma [ICD-11: 2C25.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | NF-kappaB signaling pathway | Inhibition | hsa04064 | |
| In Vitro Model | H69 cells | Lung | Homo sapiens (Human) | CVCL_8121 |
| H69AR cells | Lung | Homo sapiens (Human) | CVCL_3513 | |
| H446 cells | Lung | Homo sapiens (Human) | CVCL_1562 | |
| H446/DDP cells | Lung | Homo sapiens (Human) | CVCL_RT21 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR; Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Annexin V-PE Apoptosis assay; Flow cytometry assay; Wound healing assay; Colony formation assay | |||
| Mechanism Description | Overexpression of miR335 sensitized human SCLC cells to chemotherapy and radiotherapy, promoted cell apoptosis and inhibited cell migration ability of human SCLC in vitro, and inhibited tumor growth in vivo. Overexpression of miR335 decreased the expression of PARP-1 mRNA and protein, and NF-kB protein levels were correspondingly downregulated, thus regulating the chemo-radiosensitivity of SCLC. | |||
| Key Molecule: Zinc finger E-box-binding homeobox 2 (ZEB2) | [176] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell migration | Inhibition | hsa04670 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| ERK signaling pathway | Regulation | hsa04210 | ||
| RhoC/FAKT/Src/ROCK2 signaling pathways | Regulation | hsa05200 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| NCI-H23 cells | Lung | Homo sapiens (Human) | CVCL_1547 | |
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase activity assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-138 sensitizes NSCLC cells to ADM through regulation of EMT regulator ZEB2. Ectopic expression of miR-138 decreased ZEB2 expression and inhibited the luciferase activity in chemoresistant tumor cells, suggesting that miR-138 could regulate EMT, at least partly, through targeting ZEB2 in NSCLC cells. | |||
| Key Molecule: Caspase-3 (CASP3) | [173] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | AKT/ERK signaling pathway | Inhibition | hsa04010 | |
| Cell apoptosis | Activation | hsa04210 | ||
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Suppression of miR-155 in this cell line considerably reversed doxorubicin resistance, and doxorubicin-induced apoptosis and cell cycle arrest were recovered. Furthermore, reverse transcription-polymerase chain reaction and western blot analysis revealed that miR-155 suppression downregulated the expression of multidrug resistance protein 1, multidrug resistance-associated protein 1, breast cancer resistance protein, glutathione S-transferase-Pi, Survivin and B-cell lymphoma 2, and upregulated the expression of caspase-3 and caspase-8. In addition, it was found that miR-155 suppression inhibited the activation of AkT and extracellular signal-regulated kinase. The transcriptional activity of nuclear factor-kB and activator protein-1 was also downregulated. | |||
| Key Molecule: Caspase-8 (CASP8) | [173] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | AKT/ERK signaling pathway | Inhibition | hsa04010 | |
| Cell apoptosis | Activation | hsa04210 | ||
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Suppression of miR-155 in this cell line considerably reversed doxorubicin resistance, and doxorubicin-induced apoptosis and cell cycle arrest were recovered. Furthermore, reverse transcription-polymerase chain reaction and western blot analysis revealed that miR-155 suppression downregulated the expression of multidrug resistance protein 1, multidrug resistance-associated protein 1, breast cancer resistance protein, glutathione S-transferase-Pi, Survivin and B-cell lymphoma 2, and upregulated the expression of caspase-3 and caspase-8. In addition, it was found that miR-155 suppression inhibited the activation of AkT and extracellular signal-regulated kinase. The transcriptional activity of nuclear factor-kB and activator protein-1 was also downregulated. | |||
| Key Molecule: Secretagogin (SCGN) | [178] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Lung small cell carcinoma [ICD-11: 2C25.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | BCL2 signaling pathway | Activation | hsa04210 | |
| Cell apoptosis | Activation | hsa04210 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | H69 cells | Lung | Homo sapiens (Human) | CVCL_8121 |
| H446 cells | Lung | Homo sapiens (Human) | CVCL_1562 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | Knockdown of SCGN led to significantly increasing of chemosensitivity, which is similar to those induced by miR-494 mimics, and ectopic expression of SCGN could rescue the suppressive effect of miR-494. | |||
| Key Molecule: Vascular endothelial growth factor A (VEGFA) | [179] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K/AKT signaling pathway | Inhibition | hsa04151 | |
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | VEGF activates the downstream PI3k/Akt signaling pathway, which is a critical regulator of cellular growth, differentiation, and metabolism. miR-126 could overcome the resistance of NSCLC cells to antineoplastic drugs through inhibition of a VEGF-PI3k/Akt signaling pathway that resulted in the down-regulation of MRP1. | |||
| Key Molecule: Apoptosis regulator Bcl-2 (BCL2) | [129] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| A549/CDDP cells | Lung | Homo sapiens (Human) | CVCL_0023 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The antiapoptotic protein BCL2 is upregulated, whereas miR-181b is downregulated in both SGC7901/VCR and A549/CDDP cells, compared with SGC7901 and A549 cells, respectively. Enforced miR-181b expression reduced BCL2 protein level and sensitized SGC7901/VCR and A549/CDDP cells to VCR-induced and CDDP-induced apoptosis, respectively. | |||
| Key Molecule: Claudin-2 (CLDN2) | [180] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Nrf2 signaling pathway | Inhibition | hsa05208 | |
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Mechanism Description | CLDN2 knockdown attenuated the expression of Nrf2 and the Nrf2-targeted genes. The DXR-induced toxicity was enhanced by CLDN2 knockdown, which was inhibited by an Nrf2 activator sulforaphane. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-301 | [22] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Melanoma [ICD-11: 2C30.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | AKT/FAKT signaling pathway | Activation | hsa04151 | |
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 |
| SkMEL1 cells | Skin | Homo sapiens (Human) | CVCL_0068 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Colony formation assay; Annexin V-fluorescein isothiocyanate (FITC) apoptosis analysis; Wound scratch healing or transwell invasion assay | |||
| Mechanism Description | PTEN can interact with AkT and FAk and inhibit their activity through their dephosphorylation, Akt and FAk signaling pathways are involved in miR301a/PTEN-promoting malignant phenotypes in MM cells, miR301a promotes MM progression via activation of Akt and FAk signaling pathways by down regulating PTEN. | |||
| Key Molecule: hsa-mir-424 | [35] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Melanoma [ICD-11: 2C30.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |
| U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 | |
| HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 | |
| PARP cells | Skin | Homo sapiens (Human) | N.A. | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Hypoxia induces miR-424 expression and that miR-424 in turn suppresses the level of PDCD4 protein, a tumor suppressor that is involved in apoptosis, by targeting its 3' untranslated region. Functionally, miR-424 overexpression decreases the sensitivity of cancer cells (HCT116 and A375) to doxorubicin (Dox) and etoposide. In contrast, the inhibition of miR-424 (+) apoptosis and increased the sensitivity of cancer cells to Dox. In a xenograft tumor model, miR-424 overexpression promoted tumor growth following Dox treatment, suggesting that miR-424 promotes tumor cell resistance to Dox. Furthermore, miR-424 levels are inversely correlated with PDCD4 expression in clinical breast cancer samples. | |||
|
|
||||
| Key Molecule: Phosphatase and tensin homolog (PTEN) | [22] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Melanoma [ICD-11: 2C30.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | AKT/FAKT signaling pathway | Activation | hsa04151 | |
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 |
| SkMEL1 cells | Skin | Homo sapiens (Human) | CVCL_0068 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Colony formation assay; Annexin V-fluorescein isothiocyanate (FITC) apoptosis analysis; Wound scratch healing or transwell invasion assay | |||
| Mechanism Description | PTEN can interact with AkT and FAk and inhibit their activity through their dephosphorylation, Akt and FAk signaling pathways are involved in miR301a/PTEN-promoting malignant phenotypes in MM cells, miR301a promotes MM progression via activation of Akt and FAk signaling pathways by down regulating PTEN. | |||
| Key Molecule: Programmed cell death protein 4 (PDCD4) | [35] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Melanoma [ICD-11: 2C30.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |
| U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 | |
| HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 | |
| PARP cells | Skin | Homo sapiens (Human) | N.A. | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Hypoxia induces miR-424 expression and that miR-424 in turn suppresses the level of PDCD4 protein, a tumor suppressor that is involved in apoptosis, by targeting its 3' untranslated region. Functionally, miR-424 overexpression decreases the sensitivity of cancer cells (HCT116 and A375) to doxorubicin (Dox) and etoposide. In contrast, the inhibition of miR-424 (+) apoptosis and increased the sensitivity of cancer cells to Dox. In a xenograft tumor model, miR-424 overexpression promoted tumor growth following Dox treatment, suggesting that miR-424 promotes tumor cell resistance to Dox. Furthermore, miR-424 levels are inversely correlated with PDCD4 expression in clinical breast cancer samples. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-let-7a | [33] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Cutaneous squamous cell carcinoma [ICD-11: 2C31.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | A431 cells | Skin | Homo sapiens (Human) | CVCL_0037 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Caspase-3 is the key executioner caspase in apoptosis. Ectopic expression of let-7adecreased the luciferase activity of a reporter constructcontaining the 30untranslated region of caspase-3. Enforced let-7aexpression increased the resistance in A431 cells andHepG2 cells to apoptosis induced by therapeutic drugs suchas interferon-gamma, doxorubicin and paclitaxel. | |||
|
|
||||
| Key Molecule: Caspase-3 (CASP3) | [33] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Cutaneous squamous cell carcinoma [ICD-11: 2C31.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | A431 cells | Skin | Homo sapiens (Human) | CVCL_0037 |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Caspase-3 is the key executioner caspase in apoptosis. Ectopic expression of let-7adecreased the luciferase activity of a reporter constructcontaining the 30untranslated region of caspase-3. Enforced let-7aexpression increased the resistance in A431 cells andHepG2 cells to apoptosis induced by therapeutic drugs suchas interferon-gamma, doxorubicin and paclitaxel. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: ATP-binding cassette sub-family G2 (ABCG2) | [181] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Sarcoma [ICD-11: 2C35.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SW-872 cells | Skin | Homo sapiens (Human) | CVCL_1730 |
| SW-1353 cells | Brain | Homo sapiens (Human) | CVCL_0543 | |
| TE-671 cells | Peripheral blood | Homo sapiens (Human) | CVCL_1756 | |
| SW-684 cells | Skin | Homo sapiens (Human) | CVCL_1726 | |
| SW-982 cells | Testicular | Homo sapiens (Human) | CVCL_1734 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | By investigating of important regulators of stem cell biology, real-time RT-PCR data showed an increased expression of c-Myc, beta-catenin, and SOX-2 in the ALDH1high population and a significant higher level of ABCG2. Statistical analysis of data demonstrated that ALDH1high cells of SW-982 and SW-1353 showed higher resistance to commonly used chemotherapeutic agents like doxorubicin, epirubicin, and cisplatin than ALDH1low cells. This study demonstrates that in different sarcoma cell lines, high ALDH1 activity can be used to identify a subpopulation of cells characterized by a significantly higher proliferation rate, increased colony forming, increased expression of ABC transporter genes and stemness markers compared to control cells. In addition, enhanced drug resistance was demonstrated. | |||
|
|
||||
| Key Molecule: Aldehyde dehydrogenase 1 family member A1 (ALDH1A1) | [181] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Sarcoma [ICD-11: 2C35.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SW-872 cells | Skin | Homo sapiens (Human) | CVCL_1730 |
| SW-1353 cells | Brain | Homo sapiens (Human) | CVCL_0543 | |
| TE-671 cells | Peripheral blood | Homo sapiens (Human) | CVCL_1756 | |
| SW-684 cells | Skin | Homo sapiens (Human) | CVCL_1726 | |
| SW-982 cells | Testicular | Homo sapiens (Human) | CVCL_1734 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | By investigating of important regulators of stem cell biology, real-time RT-PCR data showed an increased expression of c-Myc, beta-catenin, and SOX-2 in the ALDH1high population and a significant higher level of ABCG2. Statistical analysis of data demonstrated that ALDH1high cells of SW-982 and SW-1353 showed higher resistance to commonly used chemotherapeutic agents like doxorubicin, epirubicin, and cisplatin than ALDH1low cells. This study demonstrates that in different sarcoma cell lines, high ALDH1 activity can be used to identify a subpopulation of cells characterized by a significantly higher proliferation rate, increased colony forming, increased expression of ABC transporter genes and stemness markers compared to control cells. In addition, enhanced drug resistance was demonstrated. | |||
| Key Molecule: Myc proto-oncogene protein (MYC) | [181] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Sarcoma [ICD-11: 2C35.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SW-872 cells | Skin | Homo sapiens (Human) | CVCL_1730 |
| SW-1353 cells | Brain | Homo sapiens (Human) | CVCL_0543 | |
| TE-671 cells | Peripheral blood | Homo sapiens (Human) | CVCL_1756 | |
| SW-684 cells | Skin | Homo sapiens (Human) | CVCL_1726 | |
| SW-982 cells | Testicular | Homo sapiens (Human) | CVCL_1734 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | By investigating of important regulators of stem cell biology, real-time RT-PCR data showed an increased expression of c-Myc, beta-catenin, and SOX-2 in the ALDH1high population and a significant higher level of ABCG2. Statistical analysis of data demonstrated that ALDH1high cells of SW-982 and SW-1353 showed higher resistance to commonly used chemotherapeutic agents like doxorubicin, epirubicin, and cisplatin than ALDH1low cells. This study demonstrates that in different sarcoma cell lines, high ALDH1 activity can be used to identify a subpopulation of cells characterized by a significantly higher proliferation rate, increased colony forming, increased expression of ABC transporter genes and stemness markers compared to control cells. In addition, enhanced drug resistance was demonstrated. | |||
| Key Molecule: Transcription factor SOX-2 (SOX2) | [181] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Sarcoma [ICD-11: 2C35.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SW-872 cells | Skin | Homo sapiens (Human) | CVCL_1730 |
| SW-1353 cells | Brain | Homo sapiens (Human) | CVCL_0543 | |
| TE-671 cells | Peripheral blood | Homo sapiens (Human) | CVCL_1756 | |
| SW-684 cells | Skin | Homo sapiens (Human) | CVCL_1726 | |
| SW-982 cells | Testicular | Homo sapiens (Human) | CVCL_1734 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | By investigating of important regulators of stem cell biology, real-time RT-PCR data showed an increased expression of c-Myc, beta-catenin, and SOX-2 in the ALDH1high population and a significant higher level of ABCG2. Statistical analysis of data demonstrated that ALDH1high cells of SW-982 and SW-1353 showed higher resistance to commonly used chemotherapeutic agents like doxorubicin, epirubicin, and cisplatin than ALDH1low cells. This study demonstrates that in different sarcoma cell lines, high ALDH1 activity can be used to identify a subpopulation of cells characterized by a significantly higher proliferation rate, increased colony forming, increased expression of ABC transporter genes and stemness markers compared to control cells. In addition, enhanced drug resistance was demonstrated. | |||
| Key Molecule: Catenin beta interacting protein 1 (CTNNBIP1) | [181] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Sarcoma [ICD-11: 2C35.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SW-872 cells | Skin | Homo sapiens (Human) | CVCL_1730 |
| SW-1353 cells | Brain | Homo sapiens (Human) | CVCL_0543 | |
| TE-671 cells | Peripheral blood | Homo sapiens (Human) | CVCL_1756 | |
| SW-684 cells | Skin | Homo sapiens (Human) | CVCL_1726 | |
| SW-982 cells | Testicular | Homo sapiens (Human) | CVCL_1734 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | By investigating of important regulators of stem cell biology, real-time RT-PCR data showed an increased expression of c-Myc, beta-catenin, and SOX-2 in the ALDH1high population and a significant higher level of ABCG2. Statistical analysis of data demonstrated that ALDH1high cells of SW-982 and SW-1353 showed higher resistance to commonly used chemotherapeutic agents like doxorubicin, epirubicin, and cisplatin than ALDH1low cells. This study demonstrates that in different sarcoma cell lines, high ALDH1 activity can be used to identify a subpopulation of cells characterized by a significantly higher proliferation rate, increased colony forming, increased expression of ABC transporter genes and stemness markers compared to control cells. In addition, enhanced drug resistance was demonstrated. | |||
| Key Molecule: Tyrosine-protein kinase Yes (YES) | [46] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Vascular sarcoma [ICD-11: 2C35.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | COSB cells | Nasopharynx | Canis lupus familiaris (Dog) (Canis familiaris) | CVCL_5I33 |
| DD-1 cells | Synovium | Canis lupus familiaris (Dog) | CVCL_0C94 | |
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | For this study, we demonstrate that both hemangiosarcoma and angiosarcoma cells with high expression of CSF-1R are more drug resistant than their CSF-1R low-expressing counterparts, indicating a shared mechanism for the observed treatment failures and subsequent drug resistance. Our data also suggest that part of this resistance may be achieved through drug sequestration within cellular lysosomes. | |||
| Key Molecule: Tyrosine-protein kinase Yes (YES) | [46] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Vascular sarcoma [ICD-11: 2C35.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | AS5 cells | Blood vessel | Homo sapiens (Human) | N.A. |
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | For this study, we demonstrate that both hemangiosarcoma and angiosarcoma cells with high expression of CSF-1R are more drug resistant than their CSF-1R low-expressing counterparts, indicating a shared mechanism for the observed treatment failures and subsequent drug resistance. Our data also suggest that part of this resistance may be achieved through drug sequestration within cellular lysosomes. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: HOX transcript antisense RNA (HOTAIR) | [182] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Sarcoma [ICD-11: 2C35.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Response evaluation criteria in solid tumors assay | |||
| Mechanism Description | High level expression of both of MTDH/AEG1 and HOTAIR in the primary tumor correlated with a likelihood to metastasize. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-222 | [183] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Bim signaling pathway | Activation | hsa05206 | |
| Cell apoptosis | Activation | hsa04210 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | microRNA-222 promotes drug resistance to doxorubicin in breast cancer via regulation of miR-222/bim pathway. | |||
| Key Molecule: LncRNA in non-homologous end joining pathway 1 (LINP1) | [184] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell invasion | Activation | hsa05200 | ||
| Cell migration | Activation | hsa04670 | ||
| Epithelial mesenchymal transition signaling pathway | Activation | hsa01521 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Long noncoding RNA LINP1 acts as an oncogene and promotes chemoresistance against 5-fluoroutacil and doxorubicin by inhibiting chemotherapeutics-induced apoptosis in breast cancer. | |||
| Key Molecule: H19, imprinted maternally expressed transcript (H19) | [185] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cullin4A/MDR1 signaling pathway | Regulation | hsa05206 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Quantitative real-time RT-PCR | |||
| Experiment for Drug Resistance |
CellTiter AQueous One Solution Cell Proliferation Assay | |||
| Mechanism Description | LncRNA H19 is a major mediator of doxorubicin chemoresistance in breast cancer cells through a cullin4A-MDR1 pathway. H19 overexpression was contributed to cancer cell resistance to anthracyclines and paclitaxel as knockdown of H19 LncRNA by a specific H19 shRNA in Dox-resistant cells significantly improved the cell sensitivity to anthracyclines and paclitaxel. | |||
| Key Molecule: hsa-mir-130b | [186] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| PI3K/AKT signaling pathway | Activation | hsa04151 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Colony Formation assay; FITC Annexin V Apoptosis assay | |||
| Mechanism Description | microRNA-130b targets PTEN to mediate drug resistance and proliferation of breast cancer cells via the PI3k/Akt signaling pathway. PTEN acted as a tumor inhibitor gene by specifically reversely regulating the PI3k/Akt pathway, miR130b may activate PI3k/Akt signaling by silencing PTEN. | |||
| Key Molecule: hsa-miR-3609 | [187] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell viability | Activation | hsa05200 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| In Vivo Model | BALB/c mouse model | Mus musculus | ||
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Transfection of a miR-3609 mimic markedly suppressed PD-L1 protein expression in MDA-MB-231 and MDA-MB-468 cells in a dose-dependent manner and increased the sensitivity of MCF7/ADR cells to adriamycin, whereas transfection of a miR-3609 inhibitor enhanced PD-L1 protein expression in HBL-100 and MCF-7 cells. | |||
| Key Molecule: hsa-mir-381 | [188] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell viability | Activation | hsa05200 | ||
| MAPK signaling pathway | Activation | hsa04010 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | miR-381 inactivated MAPk signaling by down-regulating FYN, thereby promoting the chemosensitization of breast cancer cells to DOX. | |||
| Key Molecule: Cancer susceptibility 9 (CASC9) | [189] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell invasion | Activation | hsa05200 | ||
| Cell migration | Activation | hsa04670 | ||
| Cell viability | Activation | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | CASC9 is upregulated in breast cancer tissues and breast cancer drug-resistant cell lines and overexpressed CASC9 may significantly increase the protein expression of EZH2. | |||
| Key Molecule: hsa-mir-132 | [190] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell viability | Activation | hsa05200 | ||
| PTEN/AKT/NF-kappaB signaling pathway | Regulation | hsa05235 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | AkT/NF-kB pathway contributes to the miR-132/-212-mediated drug resistance phenotype in breast cancer cells, which is likely regulated by suppressing PTEN expression at the molecular level. | |||
| Key Molecule: hsa-mir-212 | [190] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell viability | Activation | hsa05200 | ||
| PTEN/AKT/NF-kappaB signaling pathway | Regulation | hsa05235 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | AkT/NF-kB pathway contributes to the miR-132/-212-mediated drug resistance phenotype in breast cancer cells, which is likely regulated by suppressing PTEN expression at the molecular level. | |||
| Key Molecule: hsa-mir-574 | [191] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell colony | Inhibition | hsa05200 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | microRNA-574 enhances doxorubicin resistance through down-regulating SMAD4 in breast cancer cells. | |||
| Key Molecule: hsa-mir-222 | [192] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | miR-222 induced Adr-resistance at least in part via suppressing p27kip1 expression and altering its subcellular localization, and miR-222 inhibitors could reverse Adr-resistance of breast cancer cells. | |||
| Key Molecule: hsa-mir-181 | [193] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell migration | Activation | hsa04670 | ||
| Cell proliferation | Activation | hsa05200 | ||
| miR181b/Bim/MMP/caspase signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |
| MDA-MB-435 cells | Breast | Homo sapiens (Human) | CVCL_0417 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The miR-181b level was significantly upregulated in patient serum and breast cancer cell lines compared with that in normal controls. The results of in vitro 3H thymidine incorporation and Transwell migration assay indicated that miR-181b overexpression markedly promoted the proliferation and metastasis of breast cancer cells. These data suggest that miR-181b is a tumor promoter in breast cancer. Furthermore, miR-181b expression was found to be upregulated in doxorubicin (DOX)-resistant T-47D cells (T-47D-R) compared with that in the parental T-47D cells, and upregulation of miR-181b expression decreased the anticancer effect of DOX in the T-47D cells. Mechanistic studies demonstrated that the Bim gene, an essential initiator of apoptosis, was inhibited by miR-181b overexpression. knockdown of miR-181b by its specific inhibitors significantly re-sensitized the T-47D-R cells to the cytotoxicity of DOX. miR-181b inhibitors increased the level of Bim in the T-47D-R cells, resulting in the loss of mitochondrial membrane potential (MMP) and the activation of caspases caused by DOX. | |||
| Key Molecule: hsa-let-7e | [194] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Compared to the breast cancer tissues from chemotherapy responders, 10 miRNAs were identified to be dysregulated in the chemoresistant breast cancer tissues. Three of these miRNAs were up-regulated (miR-141, miR-200c, and miR-31), and 7 were down-regulated (let-7e, miR-576-3p, miR-125b-1, miR-370, miR-145, miR-765, and miR-760). | |||
| Key Molecule: hsa-mir-125b | [194] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Compared to the breast cancer tissues from chemotherapy responders, 10 miRNAs were identified to be dysregulated in the chemoresistant breast cancer tissues. Three of these miRNAs were up-regulated (miR-141, miR-200c, and miR-31), and 7 were down-regulated (let-7e, miR-576-3p, miR-125b-1, miR-370, miR-145, miR-765, and miR-760). | |||
| Key Molecule: hsa-mir-141 | [194] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| In Vivo Model | Mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Compared to the breast cancer tissues from chemotherapy responders, 10 miRNAs were identified to be dysregulated in the chemoresistant breast cancer tissues. Three of these miRNAs were up-regulated (miR-141, miR-200c, and miR-31), and 7 were down-regulated (let-7e, miR-576-3p, miR-125b-1, miR-370, miR-145, miR-765, and miR-760). | |||
| Key Molecule: hsa-mir-145 | [194] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Compared to the breast cancer tissues from chemotherapy responders, 10 miRNAs were identified to be dysregulated in the chemoresistant breast cancer tissues. Three of these miRNAs were up-regulated (miR-141, miR-200c, and miR-31), and 7 were down-regulated (let-7e, miR-576-3p, miR-125b-1, miR-370, miR-145, miR-765, and miR-760). | |||
| Key Molecule: hsa-mir-200c | [194] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Compared to the breast cancer tissues from chemotherapy responders, 10 miRNAs were identified to be dysregulated in the chemoresistant breast cancer tissues. Three of these miRNAs were up-regulated (miR-141, miR-200c, and miR-31), and 7 were down-regulated (let-7e, miR-576-3p, miR-125b-1, miR-370, miR-145, miR-765, and miR-760). | |||
| Key Molecule: hsa-mir-31 | [194] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Compared to the breast cancer tissues from chemotherapy responders, 10 miRNAs were identified to be dysregulated in the chemoresistant breast cancer tissues. Three of these miRNAs were up-regulated (miR-141, miR-200c, and miR-31), and 7 were down-regulated (let-7e, miR-576-3p, miR-125b-1, miR-370, miR-145, miR-765, and miR-760). | |||
| Key Molecule: hsa-mir-370 | [194] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Compared to the breast cancer tissues from chemotherapy responders, 10 miRNAs were identified to be dysregulated in the chemoresistant breast cancer tissues. Three of these miRNAs were up-regulated (miR-141, miR-200c, and miR-31), and 7 were down-regulated (let-7e, miR-576-3p, miR-125b-1, miR-370, miR-145, miR-765, and miR-760). | |||
| Key Molecule: hsa-miR-576-3p | [194] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Compared to the breast cancer tissues from chemotherapy responders, 10 miRNAs were identified to be dysregulated in the chemoresistant breast cancer tissues. Three of these miRNAs were up-regulated (miR-141, miR-200c, and miR-31), and 7 were down-regulated (let-7e, miR-576-3p, miR-125b-1, miR-370, miR-145, miR-765, and miR-760). | |||
| Key Molecule: hsa-miR-765 | [194] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Compared to the breast cancer tissues from chemotherapy responders, 10 miRNAs were identified to be dysregulated in the chemoresistant breast cancer tissues. Three of these miRNAs were up-regulated (miR-141, miR-200c, and miR-31), and 7 were down-regulated (let-7e, miR-576-3p, miR-125b-1, miR-370, miR-145, miR-765, and miR-760). | |||
| Key Molecule: hsa-mir-149 | [195] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| HS signaling pathway | Inhibition | hsa00534 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR; RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-149 modulated chemoresistance through targeting the expression of GlcNAc N-deacetylase/N-sulfotransferase-1 (NDST1). With downregulated miR-149, NDST1 expression was increased in chemoresistant MCF-7/ADM cells versus control MCF-7 wild-type cells. The increased NDST1 then activated a heparan sulfate-related pathway involving activation of heparanase. Finally, expression of miR-149 and NDST1 was confirmed in clinical chemoresistant samples of breast cancers receiving anthracycline/taxane-based chemotherapies. The high expression of NDST1 was also an unfavorable predictor for distant relapse-free survival in Her2 and basal breast cancers. | |||
| Key Molecule: hsa-mir-452 | [196] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | miR-452 could modulate the sensitivity of breast cancer cells to ADR, maybe in part by regulating the expression of IGF-1R. | |||
| Key Molecule: hsa-mir-30c | [197] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | p38/MAPK signaling pathway | Inhibition | hsa04010 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Down-regulation of miR-30c correlated with overexpression of YWHAZ in breast doxorubicin-resistant cells, Overexpression of miR-30c sensitized MCF-7/ADR cells to doxorubicin, miR-30c suppressed expression of the YWHAZ gene, YWHAZ was a key signal molecule in doxorubicin resistance by reducing activation of the p38MAPk signal pathway in MCF-7/ADR cells, miR-30c regulated doxorubicin resistance in vivo. | |||
| Key Molecule: hsa-mir-181a | [198] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | miR-181a played an important role in chemosensitivity to adriamycin in MCF-7 and MCF-7/ADR cells via targeting Bcl-2. | |||
| Key Molecule: hsa-mir-222 | [199] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | PTEN plays major roles in suppressing cancer and embryonic development, cell migration and apoptosis, miR-222 and -29a could regulate the expression of PTEN, maybe through which the two miRNAs conferred Adr and Doc resistance in MCF-7 cells. | |||
| Key Molecule: hsa-mir-29a | [199] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | PTEN plays major roles in suppressing cancer and embryonic development, cell migration and apoptosis, miR-222 and -29a could regulate the expression of PTEN, maybe through which the two miRNAs conferred Adr and Doc resistance in MCF-7 cells. | |||
| Key Molecule: hsa-mir-128a | [200] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | miR-128 regulates sensitivity to drugs and apoptosis in breast cancer cells, pro-apoptotic protein bax is negatively post-transcriptionally regulated by miR-128. Bax overexpression could lead to a generalised enhancement of the apoptotic response to death stimuli, miR-128 was significantly associated with a drug fast in breast cancer cells by resisting the activation of the apoptosis pathway. | |||
| Key Molecule: Polycomb complex protein BMI-1 (BMI1) | [201] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| BT474 cells | Breast | Homo sapiens (Human) | CVCL_0179 | |
| Experiment for Molecule Alteration |
Immunoblotting analysis | |||
| Experiment for Drug Resistance |
Celltiter glo assay | |||
| Mechanism Description | TrkB is signaling via PLCGamma, the MAP kinases or PI3k and Akt. Phosphorylation of Akt plays a pivotal role in cell survival and anti-apoptotic signaling by phosphorylating and thereby inhibiting pro-apoptotic factors like Bad or Caspase 9. Bmi1 may mediate resistance via other pathways than p53, for instance via the PI3k/Akt pathway. TrkB and Bmi1 Protein Expression is Hampered by Overexpression of miR-200c in MDA-MB 436 Cells, and cause thr resistance to Doxorubicin. | |||
| Key Molecule: hsa-mir-137 | [202] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Elevated miR-137 expression could sensitize breast cancer cells to chemotherapeutic agents (like Vincristine) through modulating the expression of P-gp by targeting YB-1. | |||
| Key Molecule: hsa-miR-298 | [203] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell viability | Inhibition | hsa05200 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| Experiment for Molecule Alteration |
Northern blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Overexpression of miR-298 down-regulated P-gp expression, increasing nuclear accumulation of doxorubicin and cytotoxicity in doxorubicin-resistant breast cancer cells. | |||
| Key Molecule: hsa-mir-221 | [204] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Patient treatment and assessment assay | |||
| Mechanism Description | The expression level of miR-221 was significantly associated with hormone receptor (HR) status. Patients with higher plasma miR-221 levels tended to be HR-negative. plasma miR-221 may be a predictive biomarker for sensitivity to NAC in breast cancer patients. | |||
| Key Molecule: hsa-mir-128a | [205] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Reduction of miR-128 in BT-ICs leads to overexpression of Bmi-1 and ABCC5, two independent targets of miR-128. Ectopic expression of miR-128 decreases cell viability and increases apoptosis and DNA damage in the presence of doxorubicin, hence sensitizes BT-ICs to chemotherapy. | |||
| Key Molecule: hsa-mir-21 | [206] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-21 regulates ADR resistance of breast cancer cells, at least in part, by targeting the tumor suppressor gene PTEN. | |||
| Key Molecule: hsa-mir-155 | [9] | |||
| Molecule Alteration | Expressiom | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| TGF-beta/Smad signaling pathway | Regulation | hsa04350 | ||
| In Vitro Model | Breast cancer cell lines | Colon | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
qRT-PCR; Northern blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Loss of FOXO3a is often linked to a decline in apoptotic activity and increased chemoresistance in cancer cells. miR-155 directly interacts with 3'-UTR of FOXO3a and blocks FOXO3a translation. knockdown of miR-155 renders cells to apoptosis and enhances chemosensitivity. | |||
| Key Molecule: hsa-let-7a | [33] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Caspase-3 is the key executioner caspase in apoptosis. Ectopic expression of let-7adecreased the luciferase activity of a reporter constructcontaining the 30untranslated region of caspase-3. Enforced let-7aexpression increased the resistance in A431 cells andHepG2 cells to apoptosis induced by therapeutic drugs suchas interferon-gamma, doxorubicin and paclitaxel. | |||
| Key Molecule: hsa-mir-222 | [207], [208] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | PTEN/AKT/FOXO1 signaling pathway | Inhibition | hsa05235 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| MCF-7/S cells | Breast | Homo sapiens (Human) | CVCL_0031 | |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Annexin-V-APC apoptosis assay; Flow cytometry assay | |||
| Mechanism Description | miR222 promotes drug-resistance of breast cancer cells to adriamycin via modulation of PTEN/Akt/FOXO1 pathway, inhibition of miR222 resulted in increased PTEN expression and decreased p-Akt expression. The expression of miR222 is negatively correlated with FOXO1 expression in breast cancer cells. Inhibition of miR222 could reverse the ADR-resistance and improve the prognosis of breast cancer patients. | |||
| Key Molecule: hsa-miR-760 | [194], [209] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | TGF-beta/Wnt/G protein signaling pathway | Regulation | hsa04010 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Compared to the breast cancer tissues from chemotherapy responders, 10 miRNAs were identified to be dysregulated in the chemoresistant breast cancer tissues. Three of these miRNAs were up-regulated (miR-141, miR-200c, and miR-31), and 7 were down-regulated (let-7e, miR-576-3p, miR-125b-1, miR-370, miR-145, miR-765, and miR-760). And miR-760 downregulation can enhance glioblastoma cells resistance to temozolomide through TGF-beta signaling pathway. | |||
| Key Molecule: H19, imprinted maternally expressed transcript (H19) | [30] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/AdrVp cells | Breast | Homo sapiens (Human) | CVCL_4Y46 | |
| Experiment for Molecule Alteration |
RT-PCR; Northern blotting analysis | |||
| Experiment for Drug Resistance |
Clonogenic assay | |||
| Mechanism Description | The mRNA of the H19 gene is overexpressed in MCF-7/AdrVp cells relative toparental MCF-7 cells or drug-sensitive MCF-7/AdrVp revertant cells. H19is an imprinted gene with an important role in fetal differentiation, as well as a postulated function as a tumor suppressor gene. Another p95-over-expressing multidrug-resistant cell line, human lung carcinoma NCI-H1688, also displays high levels of 1119 mRNA. | |||
|
|
||||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [185] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter AQueous One Solution Cell Proliferation Assay | |||
| Mechanism Description | H19 LncRNA plays a leading role in breast cancer chemoresistance, mediated mainly through a H19-CUL4A-ABCB1/MDR1 pathway. H19 overexpression was contributed to cancer cell resistance to anthracyclines and paclitaxel. | |||
| Key Molecule: ATP-binding cassette sub-family A1 (ABCA1) | [209] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | TGF-beta/Wnt/G protein signaling pathway | Regulation | hsa04010 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | ABCA1 might be the target gene of miR-760, and lead to resistance to chemotherapeutic agents predominantly arises from decreased drug intracellular concentrations due to increased efflux. | |||
| Key Molecule: ATP-binding cassette sub-family B5 (ABCB5) | [203] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell viability | Inhibition | hsa05200 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Overexpression of miR-298 down-regulated P-gp expression, increasing nuclear accumulation of doxorubicin and cytotoxicity in doxorubicin-resistant breast cancer cells. | |||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [210] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Epithelial mesenchymal transition signaling pathway | Activation | hsa01521 | |
| p53 signaling pathway | Regulation | hsa04115 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| Experiment for Molecule Alteration |
Flow cytometry assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Up-regulation of miR-200c with transfection of miR-200c mimics in breast cancer cells could enhance the chemosensitivity to epirubicin and reduce expression of multidrug resistance 1 mRNA and P-glycoprotein. | |||
| Key Molecule: ATP-binding cassette sub-family C5 (ABCC5) | [205] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Reduction of miR-128 in BT-ICs leads to overexpression of Bmi-1 and ABCC5, two independent targets of miR-128. Ectopic expression of miR-128 decreases cell viability and increases apoptosis and DNA damage in the presence of doxorubicin, hence sensitizes BT-ICs to chemotherapy. | |||
| Key Molecule: ATP-binding cassette sub-family G2 (ABCG2) | [211] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/AdrVp cells | Breast | Homo sapiens (Human) | CVCL_4Y46 | |
| MCF-7/AdrVpPR cells | Breast | Homo sapiens (Human) | CVCL_4Y46 | |
| Experiment for Drug Resistance |
Flow cytometric assay | |||
| Mechanism Description | Enforced expression of the full-length BCRP cDNA in MCF-7 breast cancer cells confers resistance to mitoxantrone, doxorubicin, and daunorubicin, reduces daunorubicin accumulation and retention, and causes an ATP-dependent enhancement of the efflux of rhodamine 123 in the cloned transfected. | |||
|
|
||||
| Key Molecule: hsa-mir-155 | [212] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| Experiment for Molecule Alteration |
PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Exosome-mediated breast cancer chemoresistance via miR155 transfeRNA Increased miR155 expression increases miR155 content of exosomes, leading to EMT-associated chemoresistance. | |||
| Key Molecule: hsa-mir-93 | [213] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell motility | Activation | hsa04510 | |
| Cell proliferation | Activation | hsa05200 | ||
| Self-renewal signaling pathway | Activation | hsa04550 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometric analysis | |||
| Mechanism Description | miR93 contributes to inducing EMT and drug resistance of breast cancer cells by targeting PTEN. | |||
| Key Molecule: Phosphatase and tensin homolog (PTEN) | [213] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Activation | hsa05200 | |
| Epithelial mesenchymal transition signaling pathway | Activation | hsa01521 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| Experiment for Molecule Alteration |
Dual-luciferase reporter assay; qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometric analysis | |||
| Mechanism Description | miR93 contributes to inducing EMT and drug resistance of breast cancer cells by targeting PTEN. | |||
| Key Molecule: Rho-related GTP-binding protein RhoB (RHOB) | [209] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | TGF-beta/Wnt/G protein signaling pathway | Regulation | hsa04010 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-760 enchance the expression of RHOB and RHOB overexpression was suggested to be associated angiogenesis and tumor progression in breast cancer,which can cause resistance in the cancer cells. | |||
| Key Molecule: Zinc finger E-box-binding homeobox 1 (ZEB1) | [214] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PTEN/AKT signaling pathway | Activation | hsa05235 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The loss of miRNA-200c correlates with the acquired resistance of breast cancer cells to ADR,the loss of miRNA-200c correlated with decreased levels of E-cadherin and PTEN, and increased levels of ZEB1 and phospho-Akt (p-Akt) in ADR-resistant breast cancer cells (MCF-7/ADR cells). miRNA-200c inhibited Akt signaling through its effects on E-cadherin and PTEN, resulting in the inhibition of ADR resistance in breast cancer cells. | |||
| Key Molecule: hsa-mir-200c | [201], [210], [214] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | DNA damage repair signaling pathway | Activation | hsa03410 | |
| PTEN/AKT signaling pathway | Activation | hsa05235 | ||
| miR151a-3p/XRCC4 signaling pathway | Regulation | hsa05206 | ||
| p53 signaling pathway | Regulation | hsa04115 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| BT474 cells | Breast | Homo sapiens (Human) | CVCL_0179 | |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Celltiter glo assay | |||
| Mechanism Description | The loss of miRNA-200c correlates with the acquired resistance of breast cancer cells to ADR, the loss of miRNA-200c correlated with decreased levels of E-cadherin and PTEN, and increased levels of ZEB1 and phospho-Akt (p-Akt) in ADR-resistant breast cancer cells (MCF-7/ADR cells). miRNA-200c inhibited Akt signaling through its effects on E-cadherin and PTEN, resulting in the inhibition of ADR resistance in breast cancer cells. | |||
|
|
||||
| Key Molecule: Bcl-2-like protein 11 (BCL2L11) | [183] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Bim signaling pathway | Activation | hsa05206 | |
| Cell migration | Activation | hsa04670 | ||
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | microRNA-222 promotes drug resistance to doxorubicin in breast cancer via regulation of miR-222/bim pathway. | |||
| Key Molecule: Apoptosis regulator BAX (BAX) | [184] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| Experiment for Molecule Alteration |
Western blot analysis; TUNEL assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Long noncoding RNA LINP1 acts as an oncogene and promotes chemoresistance against 5-fluoroutacil and doxorubicin by inhibiting chemotherapeutics-induced apoptosis (apoptosis-related proteins such as caspase-8, caspase-9 and Bax proteins) in breast cancer. | |||
| Key Molecule: Caspase-8 (CASP8) | [184] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell invasion | Activation | hsa05200 | ||
| Cell migration | Activation | hsa04670 | ||
| Epithelial mesenchymal transition signaling pathway | Activation | hsa01521 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| Experiment for Molecule Alteration |
Western blot analysis; TUNEL assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Long noncoding RNA LINP1 acts as an oncogene and promotes chemoresistance against 5-fluoroutacil and doxorubicin by inhibiting chemotherapeutics-induced apoptosis (apoptosis-related proteins such as caspase-8, caspase-9 and Bax proteins) in breast cancer. | |||
| Key Molecule: Caspase-9 (CASP9) | [184] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell invasion | Activation | hsa05200 | ||
| Cell migration | Activation | hsa04670 | ||
| Epithelial mesenchymal transition signaling pathway | Activation | hsa01521 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| Experiment for Molecule Alteration |
Western blot analysis; TUNEL assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Long noncoding RNA LINP1 acts as an oncogene and promotes chemoresistance against 5-fluoroutacil and doxorubicin by inhibiting chemotherapeutics-induced apoptosis (apoptosis-related proteins such as caspase-8, caspase-9 and Bax proteins) in breast cancer. | |||
| Key Molecule: Cullin-4A (CUL4A) | [185] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter AQueous One Solution Cell Proliferation Assay | |||
| Mechanism Description | H19 LncRNA plays a leading role in breast cancer chemoresistance, mediated mainly through a H19-CUL4A-ABCB1/MDR1 pathway. H19 overexpression was contributed to cancer cell resistance to anthracyclines and paclitaxel. | |||
| Key Molecule: Phosphatase and tensin homolog (PTEN) | [186] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| PI3K/AKT signaling pathway | Activation | hsa04151 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Colony Formation assay; FITC Annexin V Apoptosis assay | |||
| Mechanism Description | microRNA-130b targets PTEN to mediate drug resistance and proliferation of breast cancer cells via the PI3k/Akt signaling pathway. PTEN acted as a tumor inhibitor gene by specifically reversely regulating the PI3k/Akt pathway, miR130b may activate PI3k/Akt signaling by silencing PTEN. | |||
| Key Molecule: Forkhead box protein O1 (FOXO1) | [208] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PTEN/AKT/FOXO1 signaling pathway | Inhibition | hsa05235 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| MCF-7/S cells | Breast | Homo sapiens (Human) | CVCL_0031 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Annexin-V-APC apoptosis assay | |||
| Mechanism Description | miR222 promotes drug-resistance of breast cancer cells to adriamycin via modulation of PTEN/Akt/FOXO1 pathway, inhibition of miR222 resulted in increased PTEN expression and decreased p-Akt expression. The expression of miR222 is negatively correlated with FOXO1 expression in breast cancer cells. Inhibition of miR222 could reverse the ADR-resistance and improve the prognosis of breast cancer patients. | |||
| Key Molecule: Programmed cell death 1 ligand 1 (PD-L1) | [187] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell viability | Activation | hsa05200 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| In Vivo Model | BALB/c mouse model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Transfection of a miR-3609 mimic markedly suppressed PD-L1 protein expression in MDA-MB-231 and MDA-MB-468 cells in a dose-dependent manner and increased the sensitivity of MCF7/ADR cells to adriamycin, whereas transfection of a miR-3609 inhibitor enhanced PD-L1 protein expression in HBL-100 and MCF-7 cells. | |||
| Key Molecule: Tyrosine-protein kinase Fyn (FYN) | [188] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell viability | Activation | hsa05200 | ||
| MAPK signaling pathway | Activation | hsa04010 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | miR-381 inactivated MAPk signaling by down-regulating FYN, thereby promoting the chemosensitization of breast cancer cells to DOX. | |||
| Key Molecule: Histone-lysine N-methyltransferase EZH2 (EZH2) | [189] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell invasion | Activation | hsa05200 | ||
| Cell migration | Activation | hsa04670 | ||
| Cell viability | Activation | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | CASC9 is upregulated in breast cancer tissues and breast cancer drug-resistant cell lines and overexpressed CASC9 may significantly increase the protein expression of EZH2. | |||
| Key Molecule: Phosphatase and tensin homolog (PTEN) | [190] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell viability | Activation | hsa05200 | ||
| PTEN/AKT/NF-kappaB signaling pathway | Regulation | hsa05235 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | AkT/NF-kB pathway contributes to the miR-132/-212-mediated drug resistance phenotype in breast cancer cells, which is likely regulated by suppressing PTEN expression at the molecular level. | |||
| Key Molecule: Mothers against decapentaplegic homolog 4 (SMAD4) | [191] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | microRNA-574 enhances doxorubicin resistance through down-regulating SMAD4 in breast cancer cells. | |||
| Key Molecule: Cyclin-dependent kinase inhibitor 1B (CDKN1B) | [192] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
RT-qPCR and Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | miR-222 induced Adr-resistance at least in part via suppressing p27kip1 expression and altering its subcellular localization, and miR-222 inhibitors could reverse Adr-resistance of breast cancer cells. | |||
| Key Molecule: Bcl-2-like protein 11 (BCL2L11) | [193] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell migration | Activation | hsa04670 | ||
| Cell proliferation | Activation | hsa05200 | ||
| miR181b/Bim/MMP/caspase signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |
| MDA-MB-435 cells | Breast | Homo sapiens (Human) | CVCL_0417 | |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The miR-181b level was significantly upregulated in patient serum and breast cancer cell lines compared with that in normal controls. The results of in vitro 3H thymidine incorporation and Transwell migration assay indicated that miR-181b overexpression markedly promoted the proliferation and metastasis of breast cancer cells. These data suggest that miR-181b is a tumor promoter in breast cancer. Furthermore, miR-181b expression was found to be upregulated in doxorubicin (DOX)-resistant T-47D cells (T-47D-R) compared with that in the parental T-47D cells, and upregulation of miR-181b expression decreased the anticancer effect of DOX in the T-47D cells. Mechanistic studies demonstrated that the Bim gene, an essential initiator of apoptosis, was inhibited by miR-181b overexpression. knockdown of miR-181b by its specific inhibitors significantly re-sensitized the T-47D-R cells to the cytotoxicity of DOX. miR-181b inhibitors increased the level of Bim in the T-47D-R cells, resulting in the loss of mitochondrial membrane potential (MMP) and the activation of caspases caused by DOX. | |||
| Key Molecule: Bifunctional heparan sulfate N-deacetylase/sulfotransferase 1 (NDST1) | [195] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| HS signaling pathway | Inhibition | hsa00534 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-149 modulated chemoresistance through targeting the expression of GlcNAc N-deacetylase/N-sulfotransferase-1 (NDST1). With downregulated miR-149, NDST1 expression was increased in chemoresistant MCF-7/ADM cells versus control MCF-7 wild-type cells. The increased NDST1 then activated a heparan sulfate-related pathway involving activation of heparanase. Finally, expression of miR-149 and NDST1 was confirmed in clinical chemoresistant samples of breast cancers receiving anthracycline/taxane-based chemotherapies. The high expression of NDST1 was also an unfavorable predictor for distant relapse-free survival in Her2 and basal breast cancers. | |||
| Key Molecule: Insulin-like growth factor 1 receptor (IGF1R) | [196] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | miR-452 could modulate the sensitivity of breast cancer cells to ADR, maybe in part by regulating the expression of IGF-1R. | |||
| Key Molecule: Protein zeta/delta 14-3-3 (YWHAZ) | [197] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | p38/MAPK signaling pathway | Inhibition | hsa04010 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Down-regulation of miR-30c correlated with overexpression of YWHAZ in breast doxorubicin-resistant cells, Overexpression of miR-30c sensitized MCF-7/ADR cells to doxorubicin, miR-30c suppressed expression of the YWHAZ gene, YWHAZ was a key signal molecule in doxorubicin resistance by reducing activation of the p38MAPk signal pathway in MCF-7/ADR cells, miR-30c regulated doxorubicin resistance in vivo. | |||
| Key Molecule: Apoptosis regulator Bcl-2 (BCL2) | [198] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | miR-181a played an important role in chemosensitivity to adriamycin in MCF-7 and MCF-7/ADR cells via targeting Bcl-2. | |||
| Key Molecule: Phosphatase and tensin homolog (PTEN) | [199] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | PTEN plays major roles in suppressing cancer and embryonic development, cell migration and apoptosis, miR-222 and -29a could regulate the expression of PTEN, maybe through which the two miRNAs conferred Adr and Doc resistance in MCF-7 cells. | |||
| Key Molecule: Apoptosis regulator BAX (BAX) | [200] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| Experiment for Molecule Alteration |
luciferase report assay | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | miR-128 regulates sensitivity to drugs and apoptosis in breast cancer cells, pro-apoptotic protein bax is negatively post-transcriptionally regulated by miR-128. Bax overexpression could lead to a generalised enhancement of the apoptotic response to death stimuli, miR-128 was significantly associated with a drug fast in breast cancer cells by resisting the activation of the apoptosis pathway. | |||
| Key Molecule: BDNF/NT-3 growth factors receptor (NTRK2) | [201] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| BT474 cells | Breast | Homo sapiens (Human) | CVCL_0179 | |
| Experiment for Molecule Alteration |
Immunoblotting analysis | |||
| Experiment for Drug Resistance |
Celltiter glo assay | |||
| Mechanism Description | TrkB is signaling via PLCGamma, the MAP kinases or PI3k and Akt. Phosphorylation of Akt plays a pivotal role in cell survival and anti-apoptotic signaling by phosphorylating and thereby inhibiting pro-apoptotic factors like Bad or Caspase 9. Bmi1 may mediate resistance via other pathways than p53, for instance via the PI3k/Akt pathway. TrkB and Bmi1 Protein Expression is Hampered by Overexpression of miR-200c in MDA-MB 436 Cells, and cause thr resistance to Doxorubicin. | |||
| Key Molecule: Y-box-binding protein 1 (YBX1) | [202] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Elevated miR-137 expression could sensitize breast cancer cells to chemotherapeutic agents (like Vincristine) through modulating the expression of P-gp by targeting YB-1. | |||
| Key Molecule: Polycomb complex protein BMI-1 (BMI1) | [205] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Reduction of miR-128 in BT-ICs leads to overexpression of Bmi-1 and ABCC5, two independent targets of miR-128. Ectopic expression of miR-128 decreases cell viability and increases apoptosis and DNA damage in the presence of doxorubicin, hence sensitizes BT-ICs to chemotherapy. | |||
| Key Molecule: Phosphatase and tensin homolog (PTEN) | [206] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-21 regulates ADR resistance of breast cancer cells, at least in part, by targeting the tumor suppressor gene PTEN. | |||
| Key Molecule: Forkhead box protein O3 (FOXO3) | [9] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| TGF-beta/Smad signaling pathway | Regulation | hsa04350 | ||
| In Vitro Model | Breast cancer cell lines | Colon | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Loss of FOXO3a is often linked to a decline in apoptotic activity and increased chemoresistance in cancer cells. miR-155 directly interacts with 3'-UTR of FOXO3a and blocks FOXO3a translation. knockdown of miR-155 renders cells to apoptosis and enhances chemosensitivity. | |||
| Key Molecule: Caspase-3 (CASP3) | [33] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Caspase-3 is the key executioner caspase in apoptosis. Ectopic expression of let-7adecreased the luciferase activity of a reporter constructcontaining the 30untranslated region of caspase-3. Enforced let-7aexpression increased the resistance in A431 cells andHepG2 cells to apoptosis induced by therapeutic drugs suchas interferon-gamma, doxorubicin and paclitaxel. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Cytochrome P450 family 3 subfamily A member1 (CYP3A4) | [145] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
CYP450-Glo TM CYP 3A4 assay, RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Synergistic interaction between the MDR mechanisms include ABCT proteins (P-gp, BCRP, and MDR1) and metabolic enzymes of phase I of metabolism mainly CYP3A4, phase II of metabolism mainly GST was observed. In this study, FUC alone and in combination with DOX inhibited the enzyme activities of CYP3A4 and GST and down regulated their genes. We interpret this effect as a consequence of a down-regulation of pregnane X receptor (PXR) gene. FUC overcame MDR by significantly suppressing PXR mediated pathways that regulated the expression of CYP3A and ABCB1 genes in HepG-2 cells. | |||
| Key Molecule: Glutathione S-transferase (GST) | [145] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
GST colorimetric assay, RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Synergistic interaction between the MDR mechanisms include ABCT proteins (P-gp, BCRP, and MDR1) and metabolic enzymes of phase I of metabolism mainly CYP3A4, phase II of metabolism mainly GST was observed. In this study, FUC alone and in combination with DOX inhibited the enzyme activities of CYP3A4 and GST and down regulated their genes. We interpret this effect as a consequence of a down-regulation of pregnane X receptor (PXR) gene. FUC overcame MDR by significantly suppressing PXR mediated pathways that regulated the expression of CYP3A and ABCB1 genes in HepG-2 cells. | |||
|
|
||||
| Key Molecule: hsa-mir-124 | [215] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | STAT3/HIF-1 signaling pathway | Inhibition | hsa04066 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-124 sensitizes DOX-resistant BCSCs to DOX by modulating the STAT3 and HIF-1 signaling pathways. | |||
| Key Molecule: HOX transcript antisense RNA (HOTAIR) | [216] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K/AKT/mTOR signaling pathway | Inhibition | hsa04151 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Colony formation assay | |||
| Mechanism Description | Knockdown of LncRNA-HOTAIR weakened the resistance of breast cancer cells to DOX via PI3k/AkT/mTOR signaling, suggesting that LncRNA-HOTAIR may be a novel intervention target to reverse DOX-resistance in breast cancer. | |||
| Key Molecule: HOX transcript antisense RNA (HOTAIR) | [216] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| PI3K/AKT/mTOR signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Colony formation assay | |||
| Mechanism Description | The protein levels of MDR1, MRP1 and ABCB1 were significantly decreased in DOXR-MCF-7 siR-HOTAIR1 cells compared with the siR-NC DOXR-MCF-7 cells and HOTAIR silencing reduces the sensitivity of drug resistance in drug-resistant MCF-7 cells. | |||
| Key Molecule: HOX transcript antisense RNA (HOTAIR) | [216] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| PI3K/AKT/mTOR signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Colony formation assay | |||
| Mechanism Description | The protein levels of MDR1, MRP1 and ABCB1 were significantly decreased in DOXR-MCF-7 siR-HOTAIR1 cells compared with the siR-NC DOXR-MCF-7 cells and HOTAIR silencing reduces the sensitivity of drug resistance in drug-resistant MCF-7 cells. | |||
| Key Molecule: Long non-protein coding RNA 968 (LINC00968) | [217] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Wnt2/Beta-catenin signaling pathway | Inhibition | hsa04310 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| KPL-4 cells | Breast | Homo sapiens (Human) | CVCL_5310 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR,Northern blot | |||
| Experiment for Drug Resistance |
CCK8 assay; Colony formation assay | |||
| Mechanism Description | Long non-coding RNA LINC00968 attenuates drug (Doxorubicin; Vincristine; Taxol) resistance of breast cancer cells through inhibiting the Wnt2/beta-catenin signaling pathway by regulating WNT2. | |||
| Key Molecule: hsa-mir-324 | [218] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell migration | Activation | hsa04670 | ||
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | PIWI-interacting RNA-36712 restrains breast cancer progression and chemoresistance by interaction with SEPW1 pseudogene SEPW1P RNA. | |||
| Key Molecule: hsa-mir-7 | [218] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell migration | Activation | hsa04670 | ||
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | PIWI-interacting RNA-36712 restrains breast cancer progression and chemoresistance by interaction with SEPW1 pseudogene SEPW1P RNA. | |||
| Key Molecule: piR-hsa-36712 | [218] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell migration | Activation | hsa04670 | ||
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | PIWI-interacting RNA-36712 restrains breast cancer progression and chemoresistance by interaction with SEPW1 pseudogene SEPW1P RNA. | |||
| Key Molecule: hsa-mir-770 | [219] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR770, a potential tumor suppressor, could specifically target and down-regulate STMN1, thus inhibit the metastasis and chemo-resistance of TNBC cells. | |||
| Key Molecule: hsa-miR-1268b | [220] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometric analysis; Transwell invasion assay | |||
| Mechanism Description | miR1268b confers chemosensitivity in breast cancer by targeting ERBB2-mediated PI3k-AkT pathway. miR1268b could repress the PI3k-AkT signaling pathway by targeting ERBB2 and inhibit the anti-apoptosis protein Bcl2. | |||
| Key Molecule: hsa-mir-503 | [221] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometric analysis | |||
| Mechanism Description | Down-regulation of eIF4G by microRNA-503 enhances drug sensitivity of MCF-7/ADR cells through suppressing the expression of ABC transport proteins. | |||
| Key Molecule: hsa-miR-129-5p | [222] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADM cells | Breast | Homo sapiens (Human) | CVCL_0031 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | There is a reciprocal regulation between miR129-5p and SOX4 via the SOX4/EZH2 complex mediated H3k27me3 modification in breast cancer cells. miR129-5p is an important miRNA modulating EMT and MDR in breast cancer cells. | |||
| Key Molecule: hsa-mir-223 | [223] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |
| MDA-MB-435 cells | Breast | Homo sapiens (Human) | CVCL_0417 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; FITC-Annexin V and PI staining assay; Flow cytometry assay | |||
| Mechanism Description | microRNA-223 increases the sensitivity of triple-negative breast cancer stem cells to TRAIL-induced apoptosis by targeting HAX-1. | |||
| Key Molecule: hsa-miR-124-3p | [224] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | |
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay; Transwell assay; Scratch assay | |||
| Mechanism Description | The combination of downregulation of ABCC4 with overexpression of miR-124-3p significantly increased sensitivity to ADR in MCF-7-ADR cells. | |||
| Key Molecule: hsa-miR-135b-5p | [225] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell viability | Inhibition | hsa05200 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | miR-135b-5p enhances doxorubicin-sensitivity of breast cancer cells through targeting anterior gradient 2. | |||
| Key Molecule: hsa-mir-181c | [226] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell colony | Inhibition | hsa05200 | ||
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell viability | Inhibition | hsa05200 | ||
| p53 signaling pathway | Activation | hsa04115 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | The microRNA miR-181c enhances chemosensitivity and reduces chemoresistance in breast cancer cells via down-regulating osteopontin. | |||
| Key Molecule: hsa-mir-199a | [227] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| miR199a/MRP1 signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
XTT assay; Flow cytometry assay; Caspase 9 activity assay | |||
| Mechanism Description | Linc00518 downregulation reduced MDR by upregulating miR-199a which downregulates MRP1 in breast cancer. | |||
| Key Molecule: Long non-protein coding RNA 518 (LINC00518) | [227] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| miR199a/MRP1 signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
XTT assay; Flow cytometry assay; Caspase 9 activity assay | |||
| Mechanism Description | Linc00518 downregulation reduced MDR by upregulating miR-199a which downregulates MRP1 in breast cancer. | |||
| Key Molecule: hsa-miR-129-5p | [228] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8; Colony formation assay; wound healing; Transwell invasion; Flow cytometry assay | |||
| Mechanism Description | miR-129-5p suppresses Adriamycin resistance in breast cancer by directly targeting SOX2. | |||
| Key Molecule: hsa-mir-489 | [229] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-489 acts as a therapeutic sensitizer in breast cancer cells by inhibiting doxorubicin-induced cytoprotective autophagy and directly targeting LAPTM4B. | |||
| Key Molecule: hsa-miR-485-5p | [230] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Transwell assay | |||
| Mechanism Description | miR-485-5p suppresses breast cancer progression and enhances chemosensitivity through down-regulation of survivin expression. | |||
| Key Molecule: hsa-mir-125b | [231] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| miR125b/HAX1 signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Enforced expression of miR-125b resensitizes MCF-7/R cells to DOX via downregulation of HAX-1. | |||
| Key Molecule: hsa-mir-29a | [232] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| PTEN/AKT/GSk3Beta signaling pathway | Activation | hsa05235 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTS assay; Flow cytometry assay | |||
| Mechanism Description | Down-regulation of miR-29a expression in MCF-7/ADR cells increased PTEN expression levels, resulting in decreased phospho-Akt (p-Akt) and phospho-GSk3beta (p-GSk3beta) expression. Conversely, upregulation of miR-29a expression in MCF-7/S cells is associated with decreasing PTEN expression and increasing p-Akt and p-GSk3beta expression. PTEN and GSk3beta are targeted by miR-29a, and miR-29a may contribute to ADR resistance through inhibition of the PTEN/AkT/GSk3beta pathway in breast cancer cells. | |||
| Key Molecule: hsa-mir-145 | [233] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |
| MDA-kb2 cells | Breast | Homo sapiens (Human) | CVCL_6421 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-145 suppressed MRP1 expression by directly targeting MRP1 3'-untranslated regions. Overexpression of miR-145 sensitized breast cancer cells to doxorubicin in vitro and (+) to doxorubicin chemotherapy in vivo through inducing intracellular doxorubicin accumulation via inhibiting MRP1. | |||
| Key Molecule: hsa-mir-375 | [234] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | MCF-7/PTX cells | Breast | Homo sapiens (Human) | CVCL_4V97 |
| Experiment for Molecule Alteration |
qRT-PCR; MSP assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-375 is downregulated in MCF-7/ADM and MCF-7/PTX cells, and its downregulation is a result of promoter methylation. miR-375 can directly target 3'UTR of YBX1 and, thereby, decrease its expression, which might be an important mechanism of MDR in breast cancer cells. miR-375 is downregulated in MCF-7/ADM and MCF-7/PTX cells, and its downregulation is a result of promoter methylation. miR-375 can directly target 3'UTR of YBX1 and thereby decrease its expression, which might be an important mechanism of MDR in breast cancer cells. | |||
| Key Molecule: hsa-mir-214 | [235] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| p53 signaling pathway | Activation | hsa04115 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| MDA-MB-157 cells | Breast | Homo sapiens (Human) | CVCL_0618 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
WST-8 cell viability assay; CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | miR-214 promotes apoptosis and sensitizes breast cancer cells to doxorubicin by targeting the RFWD2-p53 cascade. | |||
| Key Molecule: hsa-mir-205 | [236] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| PI3K/AKT signaling pathway | Regulation | hsa04151 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Drug resistance clonogenic assay | |||
| Mechanism Description | miR-205 enhances chemosensitivity of breast cancer cells to TAC chemotherapy by suppressing both VEGFA and FGF2, leading to evasion of apoptosis. | |||
| Key Molecule: hsa-mir-489 | [237] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| PI3K/AKT signaling pathway | Regulation | hsa04151 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| MCF-7/ADM cells | Breast | Homo sapiens (Human) | CVCL_0031 | |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | miR-489 inhibited proliferation and induced apoptosis in breast cancer cells. miR-489 suppressed cell migration and invasion of breast cancer cells in vitro. miR-489 increased the chemosensitivity of breast cancer and impaired tumour growth and invasion capabilities in vivo. PIk3CA, AkT, CREB1 and BCL2 act downstream of SPIN1 and were found to be decreased by miR-489. | |||
| Key Molecule: hsa-mir-302a | [238] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| ERK signaling pathway | Regulation | hsa04210 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | MAP/ERk kinase kinase 1 (MEkk1) as a direct and functional target of miR-302. miR-302 showed combinatorial effects on MkEE1 repression and MEkk1-mediated ERk pathway. The suppression of P-gp by miR-302 was reversed by MEkk1 overexpression. miR-302 cooperatively sensitizes breast cancer cells to adriamycin via suppressing P-glycoprotein by targeting MEkk1 of ERk pathway. | |||
| Key Molecule: hsa-mir-302b | [238] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| ERK signaling pathway | Regulation | hsa04210 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | MAP/ERk kinase kinase 1 (MEkk1) as a direct and functional target of miR-302. miR-302 showed combinatorial effects on MkEE1 repression and MEkk1-mediated ERk pathway. The suppression of P-gp by miR-302 was reversed by MEkk1 overexpression. miR-302 cooperatively sensitizes breast cancer cells to adriamycin via suppressing P-glycoprotein by targeting MEkk1 of ERk pathway. | |||
| Key Molecule: hsa-mir-302c | [238] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| ERK signaling pathway | Regulation | hsa04210 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | MAP/ERk kinase kinase 1 (MEkk1) as a direct and functional target of miR-302. miR-302 showed combinatorial effects on MkEE1 repression and MEkk1-mediated ERk pathway. The suppression of P-gp by miR-302 was reversed by MEkk1 overexpression. miR-302 cooperatively sensitizes breast cancer cells to adriamycin via suppressing P-glycoprotein by targeting MEkk1 of ERk pathway. | |||
| Key Molecule: hsa-mir-302d | [238] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| ERK signaling pathway | Regulation | hsa04210 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | MAP/ERk kinase kinase 1 (MEkk1) as a direct and functional target of miR-302. miR-302 showed combinatorial effects on MkEE1 repression and MEkk1-mediated ERk pathway. The suppression of P-gp by miR-302 was reversed by MEkk1 overexpression. miR-302 cooperatively sensitizes breast cancer cells to adriamycin via suppressing P-glycoprotein by targeting MEkk1 of ERk pathway. | |||
| Key Molecule: hsa-miR-224-3p | [239] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Increased fucosylation has a pivotal role in multidrug resistance of breast cancer cells through miR-224-3p targeting FUT4. | |||
| Key Molecule: hsa-mir-193b | [240] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| miR193b/MCL1 apoptosis signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | MCL-1 was significantly overexpressed in MCF-7/DOXR cells, suggesting that the MCL-1 might be essential for doxorubicin resistance in breast cancer. Further results showed that MCL-1 was directly regulated by miR-193b, which is in accordance with the prior finding in melanoma. There was a negative correlation between the expression levels of miR-193b and MCL-1 in MCF-7/DOXR cells. Doxorubicin-induced apoptosis was inhibited in MCF-7/DOXR cells cotransfected with MCL-1 expression vector and miR-193b mimic, indicating that MCL-1 plays a pivotal role in mediating miR-193b-modulated doxorubicin resistance in human breast cancer. | |||
| Key Molecule: hsa-mir-124 | [96] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| HCC1937 cells | Breast | Homo sapiens (Human) | CVCL_0290 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-124 may be involved in DNA repair by directly targeting ATMIN and PARP1, suggesting that multiple DNA repair pathways are affected by miR-124 and therefore manipulation of miR-124 level/activity may improve the efficacy of chemotherapies that induce DNA damage. repression of ATMIN (+) the HR repair defect induced by miR-124, and restoration of ATMIN reversed the effect of miR-124 overexpression in breast cancer cells. Therefore, it is intriguing to further speculate which of the multiple roles of ATMIN is specifically affected in breast carcinogenesis. On the other hand, PARP1-mediated processes play a role in oncogenesis, cancer progression, and therapeutic resistance. | |||
| Key Molecule: hsa-let-7a | [241] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| Redox signaling pathway | Regulation | hsa01100 | ||
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| WM239 cells | Breast | Homo sapiens (Human) | CVCL_6795 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Let-7a represses proliferation and clonogenic capacity of MDA-MB-231 cells. Let-7a down-regulates key anabolic enzymes in MDA-MB-231 cells. Let-7a regulates energy metabolism and mitochondrial ROS in MDA-MB-231 cells. Let-7a regulates mitochondrial ROS in WM239 melanoma cells. Let-7a sensitizes breast cancer and melanoma cells to doxorubicin. | |||
| Key Molecule: hsa-mir-34 | [242] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | miR-34a negatively regulates the expression of Notch1 at mRNA and protein levels, and overexpression of Mir-34A can increase the drug sensitivity of breast cancer cells to ADR. | |||
| Key Molecule: hsa-mir-34 | [243] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| miR34a/Notch1 signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
XTT assay; Flow cytometry assay | |||
| Mechanism Description | Primary and mature miR34a were suppressed by treatment with p53 RNAi or the dominant-negative p53 mutant in MCF7 cells. Ectopic miR34a expression reduced cancer stem cell properties and increased sensitivity to doxorubicin treatment by directly targeting NOTCH1. Furthermore, tumors from nude mice treated with miR34a were significantly smaller compared with those of mice treated with control lentivirus. | |||
| Key Molecule: hsa-mir-21 | [244] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| c-Jun signaling pathway | Inhibition | hsa04210 | ||
| In Vitro Model | MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 |
| Experiment for Molecule Alteration |
PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | HA/CD44 activates c-Jun signaling which, in turn, stimulates miR-21 expression and function. These events lead to the production of an anti-apoptosis protein, Bcl-2 and upregulation of survival proteins (IAPs) and Doxorubicin chemoresistance in MDA-MB-468 cells. cells. Inhibition of c-Jun signaling or silencing miR-21 expression/function not only results in Bcl-2 downregulation, but also causes a reduction of survival protein expression and enhances chemosensitivity to Doxorubicin. | |||
| Key Molecule: hsa-mir-223 | [245] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell migration | Inhibition | hsa04670 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Cell Death assay | |||
| Mechanism Description | miR-223 Impairs Tumor Cell Migration and Invasion, miR-223 Expression Enhances Cell Death in Anoikis Conditions or in Presence of Chemotherapeutic Drugs (Doxorubicin and Paclitaxel), miR-223 Affects Signal Transduction Pathways Involved in Cell Death and Directly Targets STAT5A, Down-modulation of STAT5A Accounts for miR-223 Biological Effects. | |||
| Key Molecule: Long non-protein coding RNA (BX537613) | [160] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | LncRNA-BX537613 knockdown could sensitize MCF-7/ADR cell to adriamycin again. | |||
| Key Molecule: hsa-mir-133a | [246] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell viability | Inhibition | hsa05200 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/DOX cells | Breast | Homo sapiens (Human) | CVCL_0031 | |
| CVCL_4V97 | Breast | Homo sapiens (Human) | N.A. | |
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Targeted downregulation of overexpressed FTL protein by microRNA miR-133a increases the sensitivity of drug-resistant cells to doxorubicin and cisplatin. | |||
| Key Molecule: hsa-mir-195 | [247] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| PI3K/PTEN/AKT signaling pathway | Regulation | hsa05235 | ||
| RAS/RAF/MEK/ERK signaling pathway | Regulation | hsa04010 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| HBL-100 cells | Breast | Homo sapiens (Human) | CVCL_4362 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Induction of miR-195 expression promoted tumor cell apoptosis and inhibited breast cancer cell viability, but induced the sensitivity of breast cancer cells to Adriamycin treatment and was associated with inhibition of Raf-1 expression in breast cancer cells. | |||
| Key Molecule: hsa-mir-31 | [248] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Chemoresistance | Inhibition | hsa05207 | ||
| NF-kappaB signaling pathway | Inhibition | hsa04064 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Celltiter glo assay | |||
| Mechanism Description | We identified protein kinase C epsilon (PkC encoded by the PRkCE gene) as a novel direct target of miR-31 and show that down-regulation of PkC results in impaired NF-kB signaling, enhanced apoptosis, and increased sensitivity of MCF10A breast epithelial and MDA-MB-231 triple-negative breast cancer cells toward ionizing radiation as well as treatment with chemotherapeutics. | |||
| Key Molecule: hsa-mir-34 | [249] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Notch signaling pathway | Inhibition | hsa04330 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-34a expression modulated breast cancer cells response to ADR by targeting Notch1 and Notch signaling pathway. | |||
| Key Molecule: hsa-miR-326 | [250] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell proliferation | Activation | hsa05200 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
RT-PCR; qRT-PCR | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | The elevated levels of miR-326 in the mimics-transfected VP-16-resistant cell line, MCF-7/VP, downregulated MRP-1 expression and sensitized these cells to VP-16 and doxorubicin. | |||
| Key Molecule: hsa-mir-451 | [251] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/DOX cells | Breast | Homo sapiens (Human) | CVCL_0031 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Celltiter-blue cell viability assay | |||
| Mechanism Description | Expression of miR-451 is inversely correlated with mdr1 expression in breast cancer drug-resistant cells. Furthermore, the enforced increase of miR-451 levels in the MCF-7/DOX cells down-regulates expression of mdr1 and increases sensitivity of the MCF-7-resistant cancer cells to DOX | |||
|
|
||||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [216] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| PI3K/AKT/mTOR signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Colony formation assay | |||
| Mechanism Description | The protein levels of MDR1, MRP1 and ABCB1 were significantly decreased in DOXR-MCF-7 siR-HOTAIR1 cells compared with the siR-NC DOXR-MCF-7 cells and HOTAIR silencing reduces the sensitivity of drug resistance in drug-resistant MCF-7 cells. | |||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [216] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| PI3K/AKT/mTOR signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Colony formation assay | |||
| Mechanism Description | The protein levels of MDR1, MRP1 and ABCB1 were significantly decreased in DOXR-MCF-7 siR-HOTAIR1 cells compared with the siR-NC DOXR-MCF-7 cells and HOTAIR silencing reduces the sensitivity of drug resistance in drug-resistant MCF-7 cells. | |||
| Key Molecule: Multidrug resistance-associated protein 1 (MRP1) | [216] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| PI3K/AKT/mTOR signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Colony formation assay | |||
| Mechanism Description | The protein levels of MDR1, MRP1 and ABCB1 were significantly decreased in DOXR-MCF-7 siR-HOTAIR1 cells compared with the siR-NC DOXR-MCF-7 cells and HOTAIR silencing reduces the sensitivity of drug resistance in drug-resistant MCF-7 cells. | |||
| Key Molecule: ATP-binding cassette sub-family C4 (ABCC4) | [224] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell metastasis | Inhibition | hsa05205 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay; Transwell assay; Scratch assay | |||
| Mechanism Description | The combination of downregulation of ABCC4 with overexpression of miR-124-3p significantly increased sensitivity to ADR in MCF-7-ADR cells. | |||
| Key Molecule: Multidrug resistance-associated protein 1 (MRP1) | [227] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| miR199a/MRP1 signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
XTT assay; Flow cytometry assay; Caspase 9 activity assay | |||
| Mechanism Description | Linc00518 downregulation reduced MDR by upregulating miR-199a which downregulates MRP1 in breast cancer. | |||
| Key Molecule: Multidrug resistance-associated protein 1 (MRP1) | [233] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |
| MDA-kb2 cells | Breast | Homo sapiens (Human) | CVCL_6421 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-145 suppressed MRP1 expression by directly targeting MRP1 3'-untranslated regions. Overexpression of miR-145 sensitized breast cancer cells to doxorubicin in vitro and (+) to doxorubicin chemotherapy in vivo through inducing intracellular doxorubicin accumulation via inhibiting MRP1. | |||
| Key Molecule: Multidrug resistance-associated protein 1 (MRP1) | [250] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell proliferation | Activation | hsa05200 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | The elevated levels of miR-326 in the mimics-transfected VP-16-resistant cell line, MCF-7/VP, downregulated MRP-1 expression and sensitized these cells to VP-16 and doxorubicin. | |||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [251] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/DOX cells | Breast | Homo sapiens (Human) | CVCL_0031 | |
| Experiment for Molecule Alteration |
Western blotting analysis; Immunofluorescence analysis | |||
| Experiment for Drug Resistance |
Celltiter-blue cell viability assay | |||
| Mechanism Description | Expression of miR-451 is inversely correlated with mdr1 expression in breast cancer drug-resistant cells. Furthermore, the enforced increase of miR-451 levels in the MCF-7/DOX cells down-regulates expression of mdr1 and increases sensitivity of the MCF-7-resistant cancer cells to DOX | |||
| Key Molecule: ATP-binding cassette sub-family B5 (ABCB5) | [134] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| CT26 cells | Colon | Mus musculus (Mouse) | CVCL_7254 | |
| Salmonella enterica serovar Typhimurium SL1344 | 216597 | |||
| Salmonella enterica serovar Typhimurium SL1344 detaSipA | 216597 | |||
| Salmonella enterica serovar Typhimurium SL1344 detaSipB | 216597 | |||
| Salmonella enterica serovar Typhimurium SL1344 detaSipC | 216597 | |||
| Salmonella enterica serovar Typhimurium SL1344 detaSopB | 216597 | |||
| In Vivo Model | BALB/c nude mice xenograft model | Mus musculus | ||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | Mimicking the ability of Salmonella to reverse multidrug resistance, we constructed a gold nanoparticle system packaged with a SipA corona, and found this bacterial mimic not only accumulates in tumours but also reduces P-gp at a SipA dose significantly lower than free SipA. Moreover, the Salmonella nanoparticle mimic suppresses tumour growth with a concomitant reduction in P-gp when used with an existing chemotherapeutic drug (that is, doxorubicin). | |||
| Key Molecule: ATP-binding cassette sub-family B5 (ABCB5) | [252] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7/DX1 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Sf9 cells | Ovary | Homo sapiens (Human) | CVCL_0549 | |
| Experiment for Molecule Alteration |
ATPase assay | |||
| Experiment for Drug Resistance |
Flow cytometric assay | |||
| Mechanism Description | Gal-2 was found to inhibit the efflux of the fluorescent P-gp substrate rhodamine 123 in cancer cells that over express P-gp with an IC50 value of approximately 0.8 M. In addition, Gal-2 was found to inhibit the efflux of therapeutic substrates of P-gp, such as doxorubicin, daunomycin and verapamil with IC50 values ranging from 0.5 uM - 2 uM. | |||
| Key Molecule: ATP-binding cassette sub-family G2 (ABCG2) | [145] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Synergistic interaction between the MDR mechanisms include ABCT proteins (P-gp, BCRP, and MDR1) and metabolic enzymes of phase I of metabolism mainly CYP3A4, phase II of metabolism mainly GST was observed. In this study, FUC alone and in combination with DOX inhibited the enzyme activities of CYP3A4 and GST and down regulated their genes. We interpret this effect as a consequence of a down-regulation of pregnane X receptor (PXR) gene. FUC overcame MDR by significantly suppressing PXR mediated pathways that regulated the expression of CYP3A and ABCB1 genes in HepG-2 cells. | |||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [145] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Synergistic interaction between the MDR mechanisms include ABCT proteins (P-gp, BCRP, and MDR1) and metabolic enzymes of phase I of metabolism mainly CYP3A4, phase II of metabolism mainly GST was observed. In this study, FUC alone and in combination with DOX inhibited the enzyme activities of CYP3A4 and GST and down regulated their genes. We interpret this effect as a consequence of a down-regulation of pregnane X receptor (PXR) gene. FUC overcame MDR by significantly suppressing PXR mediated pathways that regulated the expression of CYP3A and ABCB1 genes in HepG-2 cells. | |||
| Key Molecule: ATP-binding cassette sub-family B5 (ABCB5) | [145] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Synergistic interaction between the MDR mechanisms include ABCT proteins (P-gp, BCRP, and MDR1) and metabolic enzymes of phase I of metabolism mainly CYP3A4, phase II of metabolism mainly GST was observed. In this study, FUC alone and in combination with DOX inhibited the enzyme activities of CYP3A4 and GST and down regulated their genes. We interpret this effect as a consequence of a down-regulation of pregnane X receptor (PXR) gene. FUC overcame MDR by significantly suppressing PXR mediated pathways that regulated the expression of CYP3A and ABCB1 genes in HepG-2 cells. | |||
|
|
||||
| Key Molecule: hsa-miR-760 | [253] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Epithelial mesenchymal transition signaling pathway | Inhibition | hsa01521 | |
| In Vitro Model | MCF-7/DOX cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | miR760 mediates chemoresistance through inhibition of epithelial mesenchymal transition in breast cancer cells, overexpression of miR760 suppressed the expression of Nanog, a transcriptional factor involved in chemoresistance, and resulted in the reversal of EMT in breast cancer cells. | |||
| Key Molecule: Noggin (NOG) | [253] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Epithelial mesenchymal transition signaling pathway | Inhibition | hsa01521 | |
| In Vitro Model | MCF-7/DOX cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | miR760 mediates chemoresistance through inhibition of epithelial mesenchymal transition in breast cancer cells, overexpression of miR760 suppressed the expression of Nanog,a transcriptional factor involved in chemoresistance, and resulted in the reversal of EMT in breast cancer cells. | |||
| Key Molecule: Cadherin-2 (CDH2) | [254] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Overexpression of miR-708-3p dramatically inhibits breast cancer cell lung metastasis and the expression of ZEB1, CDH2 and vimentin was significantly decreased in miR-708-3p-overexpressing cells at both the mRNA and protein levels compared to that in vector control cells. | |||
| Key Molecule: Vimentin (VIM) | [254] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Overexpression of miR-708-3p dramatically inhibits breast cancer cell lung metastasis and the expression of ZEB1, CDH2 and vimentin was significantly decreased in miR-708-3p-overexpressing cells at both the mRNA and protein levels compared to that in vector control cells. | |||
| Key Molecule: Zinc finger E-box-binding homeobox 1 (ZEB1) | [254] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Overexpression of miR-708-3p dramatically inhibits breast cancer cell lung metastasis and the expression of ZEB1, CDH2 and vimentin was significantly decreased in miR-708-3p-overexpressing cells at both the mRNA and protein levels compared to that in vector control cells. | |||
| Key Molecule: hsa-miR-708-3p | [254] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Overexpression of miR-708-3p dramatically inhibits breast cancer cell lung metastasis and the expression of ZEB1, CDH2 and vimentin was significantly decreased in miR-708-3p-overexpressing cells at both the mRNA and protein levels compared to that in vector control cells. | |||
| Key Molecule: C-terminal-binding protein 1 (CTBP1) | [255] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| miR644a/CTBP1/p53 signaling pathway | Activation | hsa05206 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| BT474 cells | Breast | Homo sapiens (Human) | CVCL_0179 | |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |
| MCF-12A cells | Breast | Homo sapiens (Human) | CVCL_3744 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
RTCA proliferation and migration assay; Promega assay | |||
| Mechanism Description | miR-644a directly targets transcriptional co-repressor CTBP1 and thereby upregulates p53 levels and the miR-644a/CTBP1/p53 axis suppresses drug resistance by simultaneous inhibition of cell survival and epithelial-mesenchymal transition in breast cancer. | |||
| Key Molecule: hsa-miR-644a | [255] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| miR644a/CTBP1/p53 signaling pathway | Activation | hsa05206 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| BT474 cells | Breast | Homo sapiens (Human) | CVCL_0179 | |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |
| MCF-12A cells | Breast | Homo sapiens (Human) | CVCL_3744 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
RTCA proliferation and migration assay; Promega assay | |||
| Mechanism Description | miR-644a directly targets transcriptional co-repressor CTBP1 and thereby upregulates p53 levels and the miR-644a/CTBP1/p53 axis suppresses drug resistance by simultaneous inhibition of cell survival and epithelial-mesenchymal transition in breast cancer. | |||
| Key Molecule: hsa-mir-489 | [256] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell migration | Inhibition | hsa04670 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-489 was significantly suppressed in MCF-7/ADM cells compared with chemosensitive parental control MCF-7/WT cells. Forced-expression of miR-489 reversed chemoresistance. Furthermore, Smad3 was identified as the target of miR-489 and is highly expressed in MCF-7/ADM cells. Forced expression of miR-489 both inhibited Smad3 expression and Smad3 related EMT properties. Finally, the interactions between Smad3, miR-489 and EMT were confirmed in chemoresistant tumor xenografts and clinical samples, indicating their potential implication for treatment of chemoresistance. | |||
| Key Molecule: Mothers against decapentaplegic homolog 3 (SMAD3) | [256] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell migration | Inhibition | hsa04670 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-489 was significantly suppressed in MCF-7/ADM cells compared with chemosensitive parental control MCF-7/WT cells. Forced-expression of miR-489 reversed chemoresistance. Furthermore, Smad3 was identified as the target of miR-489 and is highly expressed in MCF-7/ADM cells. Forced expression of miR-489 both inhibited Smad3 expression and Smad3 related EMT properties. Finally, the interactions between Smad3, miR-489 and EMT were confirmed in chemoresistant tumor xenografts and clinical samples, indicating their potential implication for treatment of chemoresistance. | |||
| Key Molecule: Interleukin-11 (IL11) | [257] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 | |
| G418 cells | Breast | Homo sapiens (Human) | N.A. | |
| In Vivo Model | NOD/SCID nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | miR-30c plays a pivotal role in Paclitaxel and Doxorubicin chemo-resistance by a direct targeting of TWF1, which encodes an actin-binding protein and promotes EMT. | |||
| Key Molecule: hsa-mir-30c | [257] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 | |
| G418 cells | Breast | Homo sapiens (Human) | N.A. | |
| In Vivo Model | NOD/SCID nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | miR-30c plays a pivotal role in Paclitaxel and Doxorubicin chemo-resistance by a direct targeting of TWF1, which encodes an actin-binding protein and promotes EMT. | |||
| Key Molecule: hsa-mir-200b | [258] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | |
| p53 signaling pathway | Activation | hsa04115 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| BT549 cells | Breast | Homo sapiens (Human) | CVCL_1092 | |
| HCC70 cells | Breast | Homo sapiens (Human) | CVCL_1270 | |
| Hs-578T cells | Breast | Homo sapiens (Human) | CVCL_0332 | |
| MDA-MB-361 cells | Breast | Homo sapiens (Human) | CVCL_0620 | |
| CAMA-1 cells | Breast | Homo sapiens (Human) | CVCL_1115 | |
| MCF-10-2A cells | Breast | Homo sapiens (Human) | CVCL_3743 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Celltiter-blue cell viability assay | |||
| Mechanism Description | The up-regulation of the miR-200b and miR-200c diminishes EMT by directly targeting the transcriptional repressor ZEB1 leading to up-regulation of E-cadherin. Restoration of E-cadherin expression increases the sensitivity of cancer cells to chemotherapeutic agents. Disruption of ZEB1-histone deacetylase repressor complexes and down-regulation of histone deacetylase, in particular SIRT1, positively affect the p53 apoptotic pathway leading to the increased sensitivity of breast cancer cells to chemotherapy and radiotherapy. | |||
| Key Molecule: hsa-mir-200c | [258] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | |
| p53 signaling pathway | Activation | hsa04115 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| BT549 cells | Breast | Homo sapiens (Human) | CVCL_1092 | |
| HCC70 cells | Breast | Homo sapiens (Human) | CVCL_1270 | |
| Hs-578T cells | Breast | Homo sapiens (Human) | CVCL_0332 | |
| MDA-MB-361 cells | Breast | Homo sapiens (Human) | CVCL_0620 | |
| CAMA-1 cells | Breast | Homo sapiens (Human) | CVCL_1115 | |
| MCF-10-2A cells | Breast | Homo sapiens (Human) | CVCL_3743 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Celltiter-blue cell viability assay | |||
| Mechanism Description | The up-regulation of the miR-200b and miR-200c diminishes EMT by directly targeting the transcriptional repressor ZEB1 leading to up-regulation of E-cadherin. Restoration of E-cadherin expression increases the sensitivity of cancer cells to chemotherapeutic agents. Disruption of ZEB1-histone deacetylase repressor complexes and down-regulation of histone deacetylase, in particular SIRT1, positively affect the p53 apoptotic pathway leading to the increased sensitivity of breast cancer cells to chemotherapy and radiotherapy. | |||
| Key Molecule: Zinc finger E-box-binding homeobox 1 (ZEB1) | [258] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | |
| p53 signaling pathway | Activation | hsa04115 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| BT549 cells | Breast | Homo sapiens (Human) | CVCL_1092 | |
| HCC70 cells | Breast | Homo sapiens (Human) | CVCL_1270 | |
| Hs-578T cells | Breast | Homo sapiens (Human) | CVCL_0332 | |
| MDA-MB-361 cells | Breast | Homo sapiens (Human) | CVCL_0620 | |
| CAMA-1 cells | Breast | Homo sapiens (Human) | CVCL_1115 | |
| MCF-10-2A cells | Breast | Homo sapiens (Human) | CVCL_3743 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
Celltiter-blue cell viability assay | |||
| Mechanism Description | The up-regulation of the miR-200b and miR-200c diminishes EMT by directly targeting the transcriptional repressor ZEB1 leading to up-regulation of E-cadherin. Restoration of E-cadherin expression increases the sensitivity of cancer cells to chemotherapeutic agents. Disruption of ZEB1-histone deacetylase repressor complexes and down-regulation of histone deacetylase, in particular SIRT1, positively affect the p53 apoptotic pathway leading to the increased sensitivity of breast cancer cells to chemotherapy and radiotherapy. | |||
| Key Molecule: Zinc finger E-box-binding homeobox 2 (ZEB2) | [258] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | |
| p53 signaling pathway | Activation | hsa04115 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| BT549 cells | Breast | Homo sapiens (Human) | CVCL_1092 | |
| HCC70 cells | Breast | Homo sapiens (Human) | CVCL_1270 | |
| Hs-578T cells | Breast | Homo sapiens (Human) | CVCL_0332 | |
| MDA-MB-361 cells | Breast | Homo sapiens (Human) | CVCL_0620 | |
| CAMA-1 cells | Breast | Homo sapiens (Human) | CVCL_1115 | |
| MCF-10-2A cells | Breast | Homo sapiens (Human) | CVCL_3743 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
Celltiter-blue cell viability assay | |||
| Mechanism Description | The up-regulation of the miR-200b and miR-200c diminishes EMT by directly targeting the transcriptional repressor ZEB1 leading to up-regulation of E-cadherin. Restoration of E-cadherin expression increases the sensitivity of cancer cells to chemotherapeutic agents. Disruption of ZEB1-histone deacetylase repressor complexes and down-regulation of histone deacetylase, in particular SIRT1, positively affect the p53 apoptotic pathway leading to the increased sensitivity of breast cancer cells to chemotherapy and radiotherapy. | |||
|
|
||||
| Key Molecule: Signal transducer activator transcription 3 (STAT3) | [215] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | STAT3/HIF-1 signaling pathway | Inhibition | hsa04066 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-124 sensitizes DOX-resistant BCSCs to DOX by modulating the STAT3 and HIF-1 signaling pathways. | |||
| Key Molecule: Hairy/enhancer-of-split related with YRPW motif protein 1 (HEY1) | [217] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Wnt2/Beta-catenin signaling pathway | Inhibition | hsa04310 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| KPL-4 cells | Breast | Homo sapiens (Human) | CVCL_5310 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RIP assay; ChIP assay; Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Colony formation assay | |||
| Mechanism Description | Long non-coding RNA LINC00968 attenuates drug (Doxorubicin; Vincristine; Taxol) resistance of breast cancer cells through inhibiting the Wnt2/beta-catenin signaling pathway by regulating WNT2. | |||
| Key Molecule: Selenoprotein W (SEPW1) | [218] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell migration | Activation | hsa04670 | ||
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Luciferase reporter assay | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | PIWI-interacting RNA-36712 restrains breast cancer progression and chemoresistance by interaction with SEPW1 pseudogene SEPW1P RNA. | |||
| Key Molecule: Stathmin (STMN1) | [219] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| Experiment for Molecule Alteration |
qRT-PCR; Western blot analysis; Dua-luciferase reporter assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR770, a potential tumor suppressor, could specifically target and down-regulate STMN1, thus inhibit the metastasis and chemo-resistance of TNBC cells. | |||
| Key Molecule: Receptor tyrosine-protein kinase erbB-2 (ERBB2) | [220] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| Experiment for Molecule Alteration |
Western blot analysis; Dual luciferase reporter assay | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometric analysis; Transwell invasion assay | |||
| Mechanism Description | miR1268b confers chemosensitivity in breast cancer by targeting ERBB2-mediated PI3k-AkT pathway. miR1268b could repress the PI3k-AkT signaling pathway by targeting ERBB2 and inhibit the anti-apoptosis protein Bcl2. | |||
| Key Molecule: Eukaryotic translation initiation factor 4 gamma 1 (EIF4G1) | [221] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometric analysis | |||
| Mechanism Description | Down-regulation of eIF4G by microRNA-503 enhances drug sensitivity of MCF-7/ADR cells through suppressing the expression of ABC transport proteins. | |||
| Key Molecule: Transcription factor SOX-4 (SOX4) | [222] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADM cells | Breast | Homo sapiens (Human) | CVCL_0031 | |
| Experiment for Molecule Alteration |
IP assay; ChIP assay; Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | There is a reciprocal regulation between miR129-5p and SOX4 via the SOX4/EZH2 complex mediated H3k27me3 modification in breast cancer cells. miR129-5p is an important miRNA modulating EMT and MDR in breast cancer cells. | |||
| Key Molecule: HCLS1-associated protein X-1 (HAX1) | [223] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |
| MDA-MB-435 cells | Breast | Homo sapiens (Human) | CVCL_0417 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; FITC-Annexin V and PI staining assay; Flow cytometry assay | |||
| Mechanism Description | microRNA-223 increases the sensitivity of triple-negative breast cancer stem cells to TRAIL-induced apoptosis by targeting HAX-1. | |||
| Key Molecule: Anterior gradient protein 2 homolog (AGR2) | [225] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell viability | Inhibition | hsa05200 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | miR-135b-5p enhances doxorubicin-sensitivity of breast cancer cells through targeting anterior gradient 2. | |||
| Key Molecule: Osteopontin (OPN) | [226] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell colony | Inhibition | hsa05200 | ||
| Cell invasion | Inhibition | hsa05200 | ||
| Cell viability | Inhibition | hsa05200 | ||
| p53 signaling pathway | Activation | hsa04115 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | The microRNA miR-181c enhances chemosensitivity and reduces chemoresistance in breast cancer cells via down-regulating osteopontin. | |||
| Key Molecule: Transcription factor SOX-2 (SOX2) | [228] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell colony | Inhibition | hsa05200 | ||
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis; RIP assay; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
CCK8; Colony formation assay; wound healing; Transwell invasion; Flow cytometry assay | |||
| Mechanism Description | miR-129-5p suppresses Adriamycin resistance in breast cancer by directly targeting SOX2. | |||
| Key Molecule: LAMTM4B (Unclear) | [229] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis; RIP assay; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-489 acts as a therapeutic sensitizer in breast cancer cells by inhibiting doxorubicin-induced cytoprotective autophagy and directly targeting LAPTM4B. | |||
| Key Molecule: Serine/threonine-protein kinase ULK1 (ULK1) | [229] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis; RIP assay; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-489 acts as a therapeutic sensitizer in breast cancer cells by inhibiting doxorubicin-induced cytoprotective autophagy and directly targeting LAPTM4B. | |||
| Key Molecule: Baculoviral IAP repeat-containing protein 5 (BIRC5) | [230] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Transwell assay | |||
| Mechanism Description | miR-485-5p suppresses breast cancer progression and enhances chemosensitivity through down-regulation of survivin expression. | |||
| Key Molecule: HCLS1-associated protein X-1 (HAX1) | [231] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| miR125b/HAX1 signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Enforced expression of miR-125b resensitizes MCF-7/R cells to DOX via downregulation of HAX-1. | |||
| Key Molecule: Glycogen synthase kinase-3 beta (GSK3B) | [232] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| PTEN/AKT/GSk3Beta signaling pathway | Activation | hsa05235 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay; Flow cytometry assay | |||
| Mechanism Description | Down-regulation of miR-29a expression in MCF-7/ADR cells increased PTEN expression levels, resulting in decreased phospho-Akt (p-Akt) and phospho-GSk3beta (p-GSk3beta) expression. Conversely, upregulation of miR-29a expression in MCF-7/S cells is associated with decreasing PTEN expression and increasing p-Akt and p-GSk3beta expression. PTEN and GSk3beta are targeted by miR-29a, and miR-29a may contribute to ADR resistance through inhibition of the PTEN/AkT/GSk3beta pathway in breast cancer cells. | |||
| Key Molecule: Phosphatase and tensin homolog (PTEN) | [232] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| PTEN/AKT/GSk3Beta signaling pathway | Activation | hsa05235 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay; Flow cytometry assay | |||
| Mechanism Description | Down-regulation of miR-29a expression in MCF-7/ADR cells increased PTEN expression levels, resulting in decreased phospho-Akt (p-Akt) and phospho-GSk3beta (p-GSk3beta) expression. Conversely, upregulation of miR-29a expression in MCF-7/S cells is associated with decreasing PTEN expression and increasing p-Akt and p-GSk3beta expression. PTEN and GSk3beta are targeted by miR-29a, and miR-29a may contribute to ADR resistance through inhibition of the PTEN/AkT/GSk3beta pathway in breast cancer cells. | |||
| Key Molecule: Y-box-binding protein 1 (YBX1) | [234] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | MCF-7/PTX cells | Breast | Homo sapiens (Human) | CVCL_4V97 |
| Experiment for Molecule Alteration |
Dual luciferase assay; Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-375 is downregulated in MCF-7/ADM and MCF-7/PTX cells, and its downregulation is a result of promoter methylation. miR-375 can directly target 3'UTR of YBX1 and, thereby, decrease its expression, which might be an important mechanism of MDR in breast cancer cells. miR-375 is downregulated in MCF-7/ADM and MCF-7/PTX cells, and its downregulation is a result of promoter methylation. miR-375 can directly target 3'UTR of YBX1 and thereby decrease its expression, which might be an important mechanism of MDR in breast cancer cells. | |||
| Key Molecule: E3 ubiquitin-protein ligase COP1 (COP1) | [235] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| p53 signaling pathway | Activation | hsa04115 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| MDA-MB-157 cells | Breast | Homo sapiens (Human) | CVCL_0618 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
WST-8 Cell viability assay; CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | miR-214 promotes apoptosis and sensitizes breast cancer cells to doxorubicin by targeting the RFWD2-p53 cascade. | |||
| Key Molecule: Fibroblast growth factor 2 (FGF1) | [236] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| PI3K/AKT signaling pathway | Regulation | hsa04151 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Drug resistance clonogenic assay | |||
| Mechanism Description | miR-205 enhances chemosensitivity of breast cancer cells to TAC chemotherapy by suppressing both VEGFA and FGF2, leading to evasion of apoptosis. | |||
| Key Molecule: Vascular endothelial growth factor A (VEGFA) | [236] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| PI3K/AKT signaling pathway | Regulation | hsa04151 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Drug resistance clonogenic assay | |||
| Mechanism Description | miR-205 enhances chemosensitivity of breast cancer cells to TAC chemotherapy by suppressing both VEGFA and FGF2, leading to evasion of apoptosis. | |||
| Key Molecule: Spindlin-1 (SFPQ) | [237] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| PI3K/AKT signaling pathway | Regulation | hsa04151 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| MCF-7/ADM cells | Breast | Homo sapiens (Human) | CVCL_0031 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | miR-489 inhibited proliferation and induced apoptosis in breast cancer cells. miR-489 suppressed cell migration and invasion of breast cancer cells in vitro. miR-489 increased the chemosensitivity of breast cancer and impaired tumour growth and invasion capabilities in vivo. PIk3CA, AkT, CREB1 and BCL2 act downstream of SPIN1 and were found to be decreased by miR-489. | |||
| Key Molecule: Mitogen-activated protein kinase kinase kinase 1 (MAP3K1) | [238] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| ERK signaling pathway | Regulation | hsa04210 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | MAP/ERk kinase kinase 1 (MEkk1) as a direct and functional target of miR-302. miR-302 showed combinatorial effects on MkEE1 repression and MEkk1-mediated ERk pathway. The suppression of P-gp by miR-302 was reversed by MEkk1 overexpression. miR-302 cooperatively sensitizes breast cancer cells to adriamycin via suppressing P-glycoprotein by targeting MEkk1 of ERk pathway. | |||
| Key Molecule: Alpha-(1,3)-fucosyltransferase 4 (FUT4) | [239] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Increased fucosylation has a pivotal role in multidrug resistance of breast cancer cells through miR-224-3p targeting FUT4. | |||
| Key Molecule: Induced myeloid leukemia cell differentiation protein Mcl-1 (MCL1) | [240] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| miR193b/MCL1 apoptosis signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | MCL-1 was significantly overexpressed in MCF-7/DOXR cells, suggesting that the MCL-1 might be essential for doxorubicin resistance in breast cancer. Further results showed that MCL-1 was directly regulated by miR-193b, which is in accordance with the prior finding in melanoma. There was a negative correlation between the expression levels of miR-193b and MCL-1 in MCF-7/DOXR cells. Doxorubicin-induced apoptosis was inhibited in MCF-7/DOXR cells cotransfected with MCL-1 expression vector and miR-193b mimic, indicating that MCL-1 plays a pivotal role in mediating miR-193b-modulated doxorubicin resistance in human breast cancer. | |||
| Key Molecule: ATM interactor (ATMIN) | [96] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| HCC1937 cells | Breast | Homo sapiens (Human) | CVCL_0290 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-124 may be involved in DNA repair by directly targeting ATMIN and PARP1, suggesting that multiple DNA repair pathways are affected by miR-124 and therefore manipulation of miR-124 level/activity may improve the efficacy of chemotherapies that induce DNA damage. repression of ATMIN (+) the HR repair defect induced by miR-124, and restoration of ATMIN reversed the effect of miR-124 overexpression in breast cancer cells. Therefore, it is intriguing to further speculate which of the multiple roles of ATMIN is specifically affected in breast carcinogenesis. On the other hand, PARP1-mediated processes play a role in oncogenesis, cancer progression, and therapeutic resistance. | |||
| Key Molecule: Poly[ADP-ribose] synthase 1 (PARP1) | [96] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| HCC1937 cells | Breast | Homo sapiens (Human) | CVCL_0290 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-124 may be involved in DNA repair by directly targeting ATMIN and PARP1, suggesting that multiple DNA repair pathways are affected by miR-124 and therefore manipulation of miR-124 level/activity may improve the efficacy of chemotherapies that induce DNA damage. repression of ATMIN (+) the HR repair defect induced by miR-124, and restoration of ATMIN reversed the effect of miR-124 overexpression in breast cancer cells. Therefore, it is intriguing to further speculate which of the multiple roles of ATMIN is specifically affected in breast carcinogenesis. On the other hand, PARP1-mediated processes play a role in oncogenesis, cancer progression, and therapeutic resistance. | |||
| Key Molecule: Neurogenic locus notch homolog protein 1 (NOTCH1) | [242] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | miR-34a negatively regulates the expression of Notch1 at mRNA and protein levels, and overexpression of Mir-34A can increase the drug sensitivity of breast cancer cells to ADR. | |||
| Key Molecule: Neurogenic locus notch homolog protein 1 (NOTCH1) | [243] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| miR34a/Notch1 signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
XTT assay; Flow cytometry assay | |||
| Mechanism Description | Primary and mature miR34a were suppressed by treatment with p53 RNAi or the dominant-negative p53 mutant in MCF7 cells. Ectopic miR34a expression reduced cancer stem cell properties and increased sensitivity to doxorubicin treatment by directly targeting NOTCH1. Furthermore, tumors from nude mice treated with miR34a were significantly smaller compared with those of mice treated with control lentivirus. | |||
| Key Molecule: Apoptosis regulator Bcl-2 (BCL2) | [244] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| c-Jun signaling pathway | Inhibition | hsa04210 | ||
| In Vitro Model | MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 |
| Experiment for Molecule Alteration |
Immunoblotting techniques assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | HA/CD44 activates c-Jun signaling which, in turn, stimulates miR-21 expression and function. These events lead to the production of an anti-apoptosis protein, Bcl-2 and upregulation of survival proteins (IAPs) and Doxorubicin chemoresistance in MDA-MB-468 cells. cells. Inhibition of c-Jun signaling or silencing miR-21 expression/function not only results in Bcl-2 downregulation, but also causes a reduction of survival protein expression and enhances chemosensitivity to Doxorubicin. | |||
| Key Molecule: Signal transducer activator transcription 5A (STAT5A) | [245] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell migration | Inhibition | hsa04670 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| Experiment for Molecule Alteration |
Luciferase assay | |||
| Experiment for Drug Resistance |
Cell Death assay | |||
| Mechanism Description | miR-223 Impairs Tumor Cell Migration and Invasion, miR-223 Expression Enhances Cell Death in Anoikis Conditions or in Presence of Chemotherapeutic Drugs (Doxorubicin and Paclitaxel), miR-223 Affects Signal Transduction Pathways Involved in Cell Death and Directly Targets STAT5A, Down-modulation of STAT5A Accounts for miR-223 Biological Effects. | |||
| Key Molecule: Ferritin light chain (FTL) | [246] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell viability | Inhibition | hsa05200 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/DOX cells | Breast | Homo sapiens (Human) | CVCL_0031 | |
| CVCL_4V97 | Breast | Homo sapiens (Human) | N.A. | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Targeted downregulation of overexpressed FTL protein by microRNA miR-133a increases the sensitivity of drug-resistant cells to doxorubicin and cisplatin. | |||
| Key Molecule: RAF proto-oncogene serine/threonine-protein kinase (RAF1) | [247] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| PI3K/PTEN/AKT signaling pathway | Regulation | hsa05235 | ||
| RAS/RAF/MEK/ERK signaling pathway | Regulation | hsa04010 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| HBL-100 cells | Breast | Homo sapiens (Human) | CVCL_4362 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Induction of miR-195 expression promoted tumor cell apoptosis and inhibited breast cancer cell viability, but induced the sensitivity of breast cancer cells to Adriamycin treatment and was associated with inhibition of Raf-1 expression in breast cancer cells. | |||
| Key Molecule: Twinfilin-1 (TWF1) | [257] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 | |
| G418 cells | Breast | Homo sapiens (Human) | N.A. | |
| In Vivo Model | NOD/SCID nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | miR-30c plays a pivotal role in Paclitaxel and Doxorubicin chemo-resistance by a direct targeting of TWF1, which encodes an actin-binding protein and promotes EMT. | |||
| Key Molecule: Neurogenic locus notch homolog protein 1 (NOTCH1) | [249] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Notch signaling pathway | Inhibition | hsa04330 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-34a expression modulated breast cancer cells response to ADR by targeting Notch1 and Notch signaling pathway. | |||
| Key Molecule: Cadherin-1 (CDH1) | [258] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | |
| p53 signaling pathway | Activation | hsa04115 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| BT549 cells | Breast | Homo sapiens (Human) | CVCL_1092 | |
| HCC70 cells | Breast | Homo sapiens (Human) | CVCL_1270 | |
| Hs-578T cells | Breast | Homo sapiens (Human) | CVCL_0332 | |
| MDA-MB-361 cells | Breast | Homo sapiens (Human) | CVCL_0620 | |
| CAMA-1 cells | Breast | Homo sapiens (Human) | CVCL_1115 | |
| MCF-10-2A cells | Breast | Homo sapiens (Human) | CVCL_3743 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
Celltiter-blue cell viability assay | |||
| Mechanism Description | The up-regulation of the miR-200b and miR-200c diminishes EMT by directly targeting the transcriptional repressor ZEB1 leading to up-regulation of E-cadherin. Restoration of E-cadherin expression increases the sensitivity of cancer cells to chemotherapeutic agents. Disruption of ZEB1-histone deacetylase repressor complexes and down-regulation of histone deacetylase, in particular SIRT1, positively affect the p53 apoptotic pathway leading to the increased sensitivity of breast cancer cells to chemotherapy and radiotherapy. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Protein mono-ADP-ribosyltransferase PARP8 (PARP8) | [259] | |||
| Molecule Alteration | Missense mutation | p.P81T |
||
| Resistant Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | AXLK signaling pathway | Activation | hsa01521 | |
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Circulating-free DNA assay; Whole exome sequencing assay | |||
| Mechanism Description | Quantification of allele fractions in plasma identified increased representation of mutant alleles in association with emergence of therapy resistance. | |||
| Key Molecule: Low-density lipoprotein receptor-related protein 1B (LRP1B) | [25] | |||
| Molecule Alteration | Structural variation | Copy number loss |
||
| Resistant Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | KURAMOCHI cells | Ovary | Homo sapiens (Human) | CVCL_1345 |
| IGROV1 cells | Ovary | Homo sapiens (Human) | CVCL_1304 | |
| JHOS3 cells | Ovary | Homo sapiens (Human) | CVCL_4648 | |
| OVCAR4 cells | Ovary | Homo sapiens (Human) | CVCL_1627 | |
| Experiment for Molecule Alteration |
High-resolution single-nucleotide polymorphism array assay; Single-cell sequencing assay | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | Functional studies showed that reducing LRP1B expression was sufficient to reduce the sensitivity of HGSC cell lines to liposomal doxorubicin, but not to doxorubicin, whereas LRP1B overexpression was sufficient to increase sensitivity to liposomal doxorubicin. Together, our findings underscore the large degree of variation in DNA copy number in spatially and temporally separated tumors in HGSC patients, and they define LRP1B as a potential contributor to the emergence of chemotherapy resistance in these patients. | |||
| Key Molecule: Low-density lipoprotein receptor-related protein 1B (LRP1B) | [25] | |||
| Molecule Alteration | Structural variation | Copy number loss |
||
| Resistant Disease | Ovarian serous carcinoma [ICD-11: 2C73.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | KURAMOCHI cells | Ovary | Homo sapiens (Human) | CVCL_1345 |
| IGROV1 cells | Ovary | Homo sapiens (Human) | CVCL_1304 | |
| JHOS3 cells | Ovary | Homo sapiens (Human) | CVCL_4648 | |
| OVCAR4 cells | Ovary | Homo sapiens (Human) | CVCL_1627 | |
| Experiment for Molecule Alteration |
High-resolution single-nucleotide polymorphism array assay; Single-cell sequencing assay | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | Functional studies showed that reducing LRP1B expression was sufficient to reduce the sensitivity of HGSC cell lines to liposomal doxorubicin, but not to doxorubicin, whereas LRP1B overexpression was sufficient to increase sensitivity to liposomal doxorubicin. Together, our findings underscore the large degree of variation in DNA copy number in spatially and temporally separated tumors in HGSC patients, and they define LRP1B as a potential contributor to the emergence of chemotherapy resistance in these patients. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Cytochrome P450 family 3 subfamily A member1 (CYP3A4) | [145] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SkOV3 cells | Ovary | Homo sapiens (Human) | CVCL_0532 |
| Experiment for Molecule Alteration |
CYP450-Glo TM CYP 3A4 assay, RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Synergistic interaction between the MDR mechanisms include ABCT proteins (P-gp, BCRP, and MDR1) and metabolic enzymes of phase I of metabolism mainly CYP3A4, phase II of metabolism mainly GST was observed. In this study, FUC alone and in combination with DOX inhibited the enzyme activities of CYP3A4 and GST and down regulated their genes. We interpret this effect as a consequence of a down-regulation of pregnane X receptor (PXR) gene. FUC overcame MDR by significantly suppressing PXR mediated pathways that regulated the expression of CYP3A and ABCB1 genes in HepG-2 cells. | |||
| Key Molecule: Glutathione S-transferase (GST) | [145] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SkOV3 cells | Ovary | Homo sapiens (Human) | CVCL_0532 |
| Experiment for Molecule Alteration |
GST colorimetric assay, RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Synergistic interaction between the MDR mechanisms include ABCT proteins (P-gp, BCRP, and MDR1) and metabolic enzymes of phase I of metabolism mainly CYP3A4, phase II of metabolism mainly GST was observed. In this study, FUC alone and in combination with DOX inhibited the enzyme activities of CYP3A4 and GST and down regulated their genes. We interpret this effect as a consequence of a down-regulation of pregnane X receptor (PXR) gene. FUC overcame MDR by significantly suppressing PXR mediated pathways that regulated the expression of CYP3A and ABCB1 genes in HepG-2 cells. | |||
|
|
||||
| Key Molecule: hsa-miR-634 | [260] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| MAPK/RAS signaling pathway | Regulation | hsa04010 | ||
| In Vitro Model | A2780 cells | Ovary | Homo sapiens (Human) | CVCL_0134 |
| T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |
| HCT8 cells | Colon | Homo sapiens (Human) | CVCL_2478 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-634 is an important player in cisplatin-resistance. First of all, miR-634 was the only miR miR-634 overexpression in ovarian cancer cell lines and patient samples negatively regulates important cell-cycle genes (CCND1) and Ras-MAPk pathway components (GRB2, ERk2, RSk1 and RSk2). Inhibition of the Ras-MAPk pathway resulted in increased sensitivity to cisplatin, suggesting that the miR-634-mediated repression of this pathway is responsible for the effect of miR-634 on cisplatin resistance. | |||
| Key Molecule: hsa-mir-199a | [261] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell migration | Activation | hsa04670 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | CD44+/CD117+ ovarian CICs cells | Ovary | Homo sapiens (Human) | N.A. |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | CD44 plays an important role in cellular adhesion, lymphocyte activation/migration, tumorigenesis, and the formation of metastases, endogenous mature miR-199a may prevent the growth of human ovarian CICs via decreasing the expression of CD44. | |||
|
|
||||
| Key Molecule: ATP-binding cassette sub-family G2 (ABCG2) | [145] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SkOV3 cells | Ovary | Homo sapiens (Human) | CVCL_0532 |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Synergistic interaction between the MDR mechanisms include ABCT proteins (P-gp, BCRP, and MDR1) and metabolic enzymes of phase I of metabolism mainly CYP3A4, phase II of metabolism mainly GST was observed. In this study, FUC alone and in combination with DOX inhibited the enzyme activities of CYP3A4 and GST and down regulated their genes. We interpret this effect as a consequence of a down-regulation of pregnane X receptor (PXR) gene. FUC overcame MDR by significantly suppressing PXR mediated pathways that regulated the expression of CYP3A and ABCB1 genes in HepG-2 cells. | |||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [145] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SkOV3 cells | Ovary | Homo sapiens (Human) | CVCL_0532 |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Synergistic interaction between the MDR mechanisms include ABCT proteins (P-gp, BCRP, and MDR1) and metabolic enzymes of phase I of metabolism mainly CYP3A4, phase II of metabolism mainly GST was observed. In this study, FUC alone and in combination with DOX inhibited the enzyme activities of CYP3A4 and GST and down regulated their genes. We interpret this effect as a consequence of a down-regulation of pregnane X receptor (PXR) gene. FUC overcame MDR by significantly suppressing PXR mediated pathways that regulated the expression of CYP3A and ABCB1 genes in HepG-2 cells. | |||
| Key Molecule: ATP-binding cassette sub-family B5 (ABCB5) | [145] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SkOV3 cells | Ovary | Homo sapiens (Human) | CVCL_0532 |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Synergistic interaction between the MDR mechanisms include ABCT proteins (P-gp, BCRP, and MDR1) and metabolic enzymes of phase I of metabolism mainly CYP3A4, phase II of metabolism mainly GST was observed. In this study, FUC alone and in combination with DOX inhibited the enzyme activities of CYP3A4 and GST and down regulated their genes. We interpret this effect as a consequence of a down-regulation of pregnane X receptor (PXR) gene. FUC overcame MDR by significantly suppressing PXR mediated pathways that regulated the expression of CYP3A and ABCB1 genes in HepG-2 cells. | |||
|
|
||||
| Key Molecule: G1/S-specific cyclin-D1 (CCND1) | [260] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| MAPK/RAS signaling pathway | Regulation | hsa04010 | ||
| In Vitro Model | A2780 cells | Ovary | Homo sapiens (Human) | CVCL_0134 |
| T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |
| HCT8 cells | Colon | Homo sapiens (Human) | CVCL_2478 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-634 is an important player in cisplatin-resistance. First of all, miR-634 was the only miR miR-634 overexpression in ovarian cancer cell lines and patient samples negatively regulates important cell-cycle genes (CCND1) and Ras-MAPk pathway components (GRB2, ERk2, RSk1 and RSk2). Inhibition of the Ras-MAPk pathway resulted in increased sensitivity to cisplatin, suggesting that the miR-634-mediated repression of this pathway is responsible for the effect of miR-634 on cisplatin resistance. | |||
| Key Molecule: Mitogen-activated protein kinase 1 (MAPK1) | [260] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| MAPK/RAS signaling pathway | Regulation | hsa04010 | ||
| In Vitro Model | A2780 cells | Ovary | Homo sapiens (Human) | CVCL_0134 |
| T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |
| HCT8 cells | Colon | Homo sapiens (Human) | CVCL_2478 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-634 is an important player in cisplatin-resistance. First of all, miR-634 was the only miR miR-634 overexpression in ovarian cancer cell lines and patient samples negatively regulates important cell-cycle genes (CCND1) and Ras-MAPk pathway components (GRB2, ERk2, RSk1 and RSk2). Inhibition of the Ras-MAPk pathway resulted in increased sensitivity to cisplatin, suggesting that the miR-634-mediated repression of this pathway is responsible for the effect of miR-634 on cisplatin resistance. | |||
| Key Molecule: Growth factor receptor-bound protein 2 (GRB2) | [260] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| MAPK/RAS signaling pathway | Regulation | hsa04010 | ||
| In Vitro Model | A2780 cells | Ovary | Homo sapiens (Human) | CVCL_0134 |
| T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |
| HCT8 cells | Colon | Homo sapiens (Human) | CVCL_2478 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-634 is an important player in cisplatin-resistance. First of all, miR-634 was the only miR miR-634 overexpression in ovarian cancer cell lines and patient samples negatively regulates important cell-cycle genes (CCND1) and Ras-MAPk pathway components (GRB2, ERk2, RSk1 and RSk2). Inhibition of the Ras-MAPk pathway resulted in increased sensitivity to cisplatin, suggesting that the miR-634-mediated repression of this pathway is responsible for the effect of miR-634 on cisplatin resistance. | |||
| Key Molecule: Ribosomal protein S6 kinase alpha-3 (RPS6KA3) | [260] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| MAPK/RAS signaling pathway | Regulation | hsa04010 | ||
| In Vitro Model | A2780 cells | Ovary | Homo sapiens (Human) | CVCL_0134 |
| T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |
| HCT8 cells | Colon | Homo sapiens (Human) | CVCL_2478 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-634 is an important player in cisplatin-resistance. First of all, miR-634 was the only miR miR-634 overexpression in ovarian cancer cell lines and patient samples negatively regulates important cell-cycle genes (CCND1) and Ras-MAPk pathway components (GRB2, ERk2, RSk1 and RSk2). Inhibition of the Ras-MAPk pathway resulted in increased sensitivity to cisplatin, suggesting that the miR-634-mediated repression of this pathway is responsible for the effect of miR-634 on cisplatin resistance. | |||
| Key Molecule: Extracellular matrix receptor III (CD44) | [261] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell migration | Activation | hsa04670 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | CD44+/CD117+ ovarian CICs cells | Ovary | Homo sapiens (Human) | N.A. |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | CD44 plays an important role in cellular adhesion, lymphocyte activation/migration, tumorigenesis, and the formation of metastases, endogenous mature miR-199a may prevent the growth of human ovarian CICs via decreasing the expression of CD44. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Cytochrome P450 family 3 subfamily A member1 (CYP3A4) | [72] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Cervical carcinoma [ICD-11: 2C77.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 |
| Experiment for Molecule Alteration |
CYP450-Glo CYP 3A4 assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | In this study, resveratrol was a significant inhibitor of CYP3A4 enzyme activity with IC50 value 9.32 ( M). Moreover, the CYP3A4 mRNA levels were reduced after treatment with resveratrol 0.03-fold of the control levels with high significance (p < 0.001). | |||
| Key Molecule: Glutathione S-transferase (GST) | [72] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Cervical carcinoma [ICD-11: 2C77.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 |
| Experiment for Molecule Alteration |
Glutathione-S-transferase assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The Glutathione-S-transferases (GSTs) are a multigene family of dimeric proteins which play a central role in the detoxification of electrophilic xenobiotics and catalyze their conjugation with GSH to electrophilic metabolites, thus rendering them more water soluble. GSTs protect cells from cytotoxic and carcinogenic chemicals. GST activity was decreased by resveratrol in a dose dependent manner. IC50 value was 30.73 M. This results were confirmed by RT-PCR data, where the tested samples changed the GST mRNA level by 0.79-fold (p < 0.01) of control level. | |||
|
|
||||
| Key Molecule: hsa-mir-135a | [262] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Cervical cancer [ICD-11: 2C77.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Promega viability kit assay | |||
| Mechanism Description | Down-regulated of FAk increased sensitivity of HeLa cancer cells to Fluorouracil chemotherapy, targeting and down-regulation of FAk expression by miR-135 and miR-138, overexpressed miR-138 and miR-135 had increased sensitivity to chemotherapy. | |||
| Key Molecule: hsa-mir-138 | [262] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Cervical cancer [ICD-11: 2C77.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Promega viability kit assay | |||
| Mechanism Description | Down-regulated of FAk increased sensitivity of HeLa cancer cells to Fluorouracil chemotherapy, targeting and down-regulation of FAk expression by miR-135 and miR-138, overexpressed miR-138 and miR-135 had increased sensitivity to chemotherapy. | |||
|
|
||||
| Key Molecule: ATP-binding cassette sub-family B5 (ABCB5) | [72] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Cervical carcinoma [ICD-11: 2C77.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 |
| Experiment for Molecule Alteration |
Efflux of rhodamine123 assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Resveratrol can restore the sensitivity of Caco-2 and CEM/ADR5000 cell lines to doxorubicin, through enhancing significantly doxorubicin cytotoxicity. ABC-transporter inhibitors, classified according to their action on ABC-transporters proteins into: 1. Function inhibitors, 2. Expression inhibitors, and 3. Functional and expression inhibitors, which have an ideal characters of ABC-transporters inhibitors. Our results indicate that resveratrol falls into the class 3 inhibitors. | |||
|
|
||||
| Key Molecule: Focal adhesion kinase 1 (FAK1) | [262] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Cervical cancer [ICD-11: 2C77.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
Promega viability kit assay | |||
| Mechanism Description | Down-regulated of FAk increased sensitivity of HeLa cancer cells to Fluorouracil chemotherapy, targeting and down-regulation of FAk expression by miR-135 and miR-138, overexpressed miR-138 and miR-135 had increased sensitivity to chemotherapy. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Reticulocalbin 1 (RCN1) | [50] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Uterine cancer [ICD-11: 2C78.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MES-SA cells | Uterus | Homo sapiens (Human) | CVCL_1404 |
| Experiment for Molecule Alteration |
Western blotting assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | RCN1 silencing has a significant role on doxorubicin-induced apoptosis in resistant cells, MES-SA/DxR-2 uM and MES-SA/DxR-8 uM, implying RCN1 knockdown has an auxiliary effect to increase resistant cell death during doxorubicin treatment. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: LOXL1 antisense RNA 1 (LOXL1-AS1) | [263] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Prostate cancer [ICD-11: 2C82.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell migration | Activation | hsa04670 | ||
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | DU-145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | LncRNA LOXL1-AS1/miR-let-7a-5p/EGFR-related pathway regulates the doxorubicin resistance of prostate cancer DU-145 cells. | |||
| Key Molecule: hsa-let-7a-5p | [263] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Prostate cancer [ICD-11: 2C82.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell migration | Activation | hsa04670 | ||
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | DU-145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | LncRNA LOXL1-AS1/miR-let-7a-5p/EGFR-related pathway regulates the doxorubicin resistance of prostate cancer DU-145 cells. | |||
| Key Molecule: PCGEM1 prostate-specific transcript (PCGEM1) | [45] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Prostate cancer [ICD-11: 2C82.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | LNCaP cells | Prostate | Homo sapiens (Human) | CVCL_0395 |
| Experiment for Molecule Alteration |
Northern blotting analysis | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | PCGEM1overexpression in LNCaP cell culturemodel results in the inhibition of apoptosis induced by doxorubicin (DOX). Induction of p53 and p21Waf1/Cip1by DOX were delayed in LNCaP cells stably overexpressing PCGEM1(LNCaP-PCGEM1cells) compared tocontrol LNCaP cells. | |||
|
|
||||
| Key Molecule: ATP-binding cassette sub-family C2 (ABCC2) | [264] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Prostate cancer [ICD-11: 2C82.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | PC3 cells | Prostate | Homo sapiens (Human) | CVCL_0035 |
| 22RV1 cells | Prostate | Homo sapiens (Human) | CVCL_1045 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | However, higher concentrations of probenecid (500 uM) failed to demonstrate a chemosensitizing effect. Consistent with this lower chemosensitizing efficacy in higher-concentration probenecid treatment, we observed that the expression of ABCG2, a drug-efflux transporter, increased in a dose-dependent manner following probenecid treatment. Thus, probenecid could enhance the chemosensitivity of 3D-cultured prostate cancer cells, but not at higher concentr. | |||
| Key Molecule: Multidrug resistance-associated protein 1 (MRP1) | [264] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Prostate cancer [ICD-11: 2C82.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | PC3 cells | Prostate | Homo sapiens (Human) | CVCL_0035 |
| 22RV1 cells | Prostate | Homo sapiens (Human) | CVCL_1045 | |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | However, higher concentrations of probenecid (500 uM) failed to demonstrate a chemosensitizing effect. Consistent with this lower chemosensitizing efficacy in higher-concentration probenecid treatment, we observed that the expression of ABCG2, a drug-efflux transporter, increased in a dose-dependent manner following probenecid treatment. Thus, probenecid could enhance the chemosensitivity of 3D-cultured prostate cancer cells, but not at higher concentr. | |||
|
|
||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [263] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Prostate cancer [ICD-11: 2C82.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell migration | Activation | hsa04670 | ||
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | DU-145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | LncRNA LOXL1-AS1/miR-let-7a-5p/EGFR-related pathway regulates the doxorubicin resistance of prostate cancer DU-145 cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: ATP-binding cassette sub-family B5 (ABCB5) | [265] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Prostate cancer [ICD-11: 2C82.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | DU-145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 |
| DU-145Nox1 cells | Prostate | Homo sapiens (Human) | CVCL_0105 | |
| Experiment for Molecule Alteration |
Immunohistochemistry assay | |||
| Experiment for Drug Resistance |
Annexin V staining assay | |||
| Mechanism Description | In DU-145Nox1 tumor spheroids, expression of HIF-1alpha as well as P-gp was significantly decreased as compared to DU-145 spheroids, which resulted in an increased retention of the anticancer agent doxorubicin. Pretreatment with the free radical scavengers vitamin E and vitamin C increased the expression of P-gp as well as HIF-1alpha in Nox-1-overexpressing cells, whereas no effect of free radical scavengers was observed on mdr-1 mRNA expression. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-451 | [39] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Renal cell carcinoma [ICD-11: 2C90.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | ACHN cells | Pleural effusion | Homo sapiens (Human) | CVCL_1067 |
| GRC-1 cells | Kidney | Homo sapiens (Human) | N.A. | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Annexin V-FITC Apoptosis Detection assay; MTT assay | |||
| Mechanism Description | microRNA-451 regulates chemoresistance in renal cell carcinoma by targeting ATF-2 gene. | |||
| Key Molecule: SET and MYND domain containing 2 (SMYD2) | [15] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Kidney cancer [ICD-11: 2C90.1] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 |
| HK-2 cells | Kidney | Homo sapiens (Human) | CVCL_0302 | |
| In Vivo Model | Balb/c athymic nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blotting assay | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | SMYD2 is a histone methyltransferase.The estimated IC50 values of cisplatin, doxorubicin, or 5-FU (but not docetaxel) for AZ505-treated RCC cells were significantly lower than those for the control cells, indicating that the SMYD2 inhibition enhanced the drug sensitivity in renal cancer cells. | |||
|
|
||||
| Key Molecule: Cyclic AMP-dependent transcription factor ATF-2 (ATF2) | [39] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Renal cell carcinoma [ICD-11: 2C90.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | ACHN cells | Pleural effusion | Homo sapiens (Human) | CVCL_1067 |
| GRC-1 cells | Kidney | Homo sapiens (Human) | N.A. | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Annexin V-FITC Apoptosis Detection assay; MTT assay | |||
| Mechanism Description | microRNA-451 regulates chemoresistance in renal cell carcinoma by targeting ATF-2 gene. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Cytochrome P450 family 1 subfamily B member1 (CYP1B1) | [144] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Kidney cancer [ICD-11: 2C90.1] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | miR27b/CCNG1/p53 signaling pathway | Regulation | hsa05206 | |
| In Vitro Model | 769-P cells | Kidney | Homo sapiens (Human) | CVCL_1050 |
| 786-O cells | Kidney | Homo sapiens (Human) | CVCL_1051 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | |||
| Mechanism Description | miR-27b synergizes with anticancer drugs througth enhancing anticancer drug-induced cell death which due to p53 activation and CYP1B1 suppression. | |||
|
|
||||
| Key Molecule: hsa-mir-708 | [266] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Kidney cancer [ICD-11: 2C90.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| TRAIL-mediated signaling pathway | Regulation | hsa04210 | ||
| In Vitro Model | Caki cells | Kidney | Homo sapiens (Human) | CVCL_0234 |
| Experiment for Molecule Alteration |
RT-PCR; RT-PCR | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | C-FLIPL expression was upregulated while miR-708 expression was downregulated in RCC tissues compared to normal tissues. miR-708 functioned as a pro-apoptotic miRNA via specific downregulation of c-FLIPL expression but did not have any effect on the expression of c-FLIPs, which can also increase the drug sensitivity of renal cancer cells. | |||
| Key Molecule: hsa-mir-27b | [144] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Kidney cancer [ICD-11: 2C90.1] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | miR27b/CCNG1/p53 signaling pathway | Regulation | hsa05206 | |
| In Vitro Model | 769-P cells | Kidney | Homo sapiens (Human) | CVCL_1050 |
| 786-O cells | Kidney | Homo sapiens (Human) | CVCL_1051 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | |||
| Mechanism Description | miR-27b synergizes with anticancer drugs througth enhancing anticancer drug-induced cell death which due to p53 activation and CYP1B1 suppression. | |||
|
|
||||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [267] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Renal cell carcinoma [ICD-11: 2C90.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Flp-In-293/Mock cells | Kidney | Homo sapiens (Human) | CVCL_U421 |
| Flp-In-293/ABCB1 cells | Kidney | Homo sapiens (Human) | CVCL_U421 | |
| Experiment for Molecule Alteration |
ATPase assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Through calcein assays, we found that epimagnolin A inhibited the ABCB1-mediated export of calcein. This result suggests that epimagnolin A behaved as inhibitor or substrate for ABCB1. In ATPase assays, epimagnolin A stimulated ABCB1-dependent ATPase activity. This result indicates that epimagnolin A was recognised as a substrate by ABCB1, since ABCB1 utilises energy derived from ATP hydrolysis for substrate transport. Furthermore, in MTT assays we found that the cytotoxicity of daunorubicin, doxorubicin, vinblastine, and vincristine was enhanced by epimagnolin A in a manner comparable to verapamil, a typical substrate for ABCB1. | |||
|
|
||||
| Key Molecule: CASP8 and FADD-like apoptosis regulator (CFLAR) | [266] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Kidney cancer [ICD-11: 2C90.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| TRAIL-mediated signaling pathway | Regulation | hsa04210 | ||
| In Vitro Model | Caki cells | Kidney | Homo sapiens (Human) | CVCL_0234 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | C-FLIPL expression was upregulated while miR-708 expression was downregulated in RCC tissues compared to normal tissues. miR-708 functioned as a pro-apoptotic miRNA via specific downregulation of c-FLIPL expression but did not have any effect on the expression of c-FLIPs, which can also increase the drug sensitivity of renal cancer cells. | |||
| Key Molecule: Cyclin-G1 (CCNG1) | [144] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Kidney cancer [ICD-11: 2C90.1] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| miR27b/CCNG1/p53 signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | 769-P cells | Kidney | Homo sapiens (Human) | CVCL_1050 |
| 786-O cells | Kidney | Homo sapiens (Human) | CVCL_1051 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | |||
| Mechanism Description | miR-27b synergizes with anticancer drugs througth enhancing anticancer drug-induced cell death which due to p53 activation and CYP1B1 suppression. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-miR-34b-3p | [268] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| Notch/PkC/Ca++ signaling pathway | Inhibition | hsa04330 | ||
| In Vitro Model | 5637 cells | Bladder | Homo sapiens (Human) | CVCL_0126 |
| EJ cells | Bladder | Homo sapiens (Human) | CVCL_UI82 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | miR-34b-3p Represses the Multidrug-Chemoresistance of Bladder Cancer Cells by Regulating the CCND2 and P2RY1 Genes. | |||
| Key Molecule: hsa-mir-98 | [269] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| Drp1 signaling pathway | Activation | hsa04668 | ||
| In Vitro Model | 5637 cells | Bladder | Homo sapiens (Human) | CVCL_0126 |
| J82 cells | Bladder | Homo sapiens (Human) | CVCL_0359 | |
| RT4 cells | Bladder | Homo sapiens (Human) | CVCL_0036 | |
| SV-HUC-1 cells | Bladder | Homo sapiens (Human) | CVCL_3798 | |
| T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Colony formation assay; Flow cytometry assay | |||
| Mechanism Description | microRNA-98 promotes drug resistance and regulates mitochondrial dynamics by targeting LASS2 in bladder cancer cells through Drp1 signaling. | |||
| Key Molecule: Growth arrest specific 5 (GAS5) | [270] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Bladder urothelial carcinoma [ICD-11: 2C94.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | J82 cells | Bladder | Homo sapiens (Human) | CVCL_0359 |
| T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |
| T24/DOX cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Dual-color flow cytometric method; Annexin V-FITC apoptosis assay | |||
| Mechanism Description | Long noncoding RNA GAS5 inhibits malignant proliferation and chemotherapy resistance to doxorubicin in bladder transitional cell carcinoma. | |||
| Key Molecule: hsa-miR-22-3p | [32] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | 5637 cells | Bladder | Homo sapiens (Human) | CVCL_0126 |
| T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |
| UM-UC-3 cells | Bladder | Homo sapiens (Human) | CVCL_1783 | |
| H-bc cells | Bladder | Homo sapiens (Human) | CVCL_BT00 | |
| HTB-1 cells | Bladder | Homo sapiens (Human) | CVCL_0359 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | miR 22 3p enhances multi chemoresistance by targeting NET1 in bladder cancer cells. | |||
| Key Molecule: hsa-mir-21 | [7] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| PI3K/AKT signaling pathway | Activation | hsa04151 | ||
| In Vitro Model | T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | A negative correlation between expression of miR-21 and pten was established in vivo. cell proliferation and chemoresistance to doxorubicin were promoted by overexpression of miR-21 in t24 cells. Bcl-2 up-regulation could be achieved by miR-21 overexpression, which prevented t24 cells from apoptosis induced by doxorubicin. | |||
|
|
||||
| Key Molecule: Nuclear paraspeckle assembly transcript 1 (NEAT1) | [271] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Bladder urothelial carcinoma [ICD-11: 2C94.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | J82 cells | Bladder | Homo sapiens (Human) | CVCL_0359 |
| T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Non-coding RNA NEAT1/miR-214-3p contribute to doxorubicin resistance of urothelial bladder cancer preliminary through the Wnt/beta-catenin pathway. | |||
|
|
||||
| Key Molecule: G1/S-specific cyclin-D2 (CCND2) | [268] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| Notch/PkC/Ca++ signaling pathway | Inhibition | hsa04330 | ||
| In Vitro Model | 5637 cells | Bladder | Homo sapiens (Human) | CVCL_0126 |
| EJ cells | Bladder | Homo sapiens (Human) | CVCL_UI82 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | miR-34b-3p Represses the Multidrug-Chemoresistance of Bladder Cancer Cells by Regulating the CCND2 and P2RY1 Genes. | |||
| Key Molecule: P2Y purinoceptor 1 (P2RY1) | [268] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| Notch/PkC/Ca++ signaling pathway | Inhibition | hsa04330 | ||
| In Vitro Model | 5637 cells | Bladder | Homo sapiens (Human) | CVCL_0126 |
| EJ cells | Bladder | Homo sapiens (Human) | CVCL_UI82 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | miR-34b-3p Represses the Multidrug-Chemoresistance of Bladder Cancer Cells by Regulating the CCND2 and P2RY1 Genes. | |||
| Key Molecule: Ceramide synthase 2 (CERS2) | [269] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| Drp1 signaling pathway | Activation | hsa04668 | ||
| In Vitro Model | 5637 cells | Bladder | Homo sapiens (Human) | CVCL_0126 |
| J82 cells | Bladder | Homo sapiens (Human) | CVCL_0359 | |
| RT4 cells | Bladder | Homo sapiens (Human) | CVCL_0036 | |
| SV-HUC-1 cells | Bladder | Homo sapiens (Human) | CVCL_3798 | |
| T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Colony formation assay; Flow cytometry assay | |||
| Mechanism Description | microRNA-98 promotes drug resistance and regulates mitochondrial dynamics by targeting LASS2 in bladder cancer cells through Drp1 signaling. | |||
| Key Molecule: Neuroepithelial cell-transforming gene 1 protein (NET1) | [32] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | 5637 cells | Bladder | Homo sapiens (Human) | CVCL_0126 |
| T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |
| UM-UC-3 cells | Bladder | Homo sapiens (Human) | CVCL_1783 | |
| H-bc cells | Bladder | Homo sapiens (Human) | CVCL_BT00 | |
| HTB-1 cells | Bladder | Homo sapiens (Human) | CVCL_0359 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | miR 22 3p enhances multi chemoresistance by targeting NET1 in bladder cancer cells. | |||
| Key Molecule: Phosphatase and tensin homolog (PTEN) | [7] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| PI3K/AKT signaling pathway | Activation | hsa04151 | ||
| In Vitro Model | T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 |
| Experiment for Molecule Alteration |
Western blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | A negative correlation between expression of miR-21 and pten was established in vivo. cell proliferation and chemoresistance to doxorubicin were promoted by overexpression of miR-21 in t24 cells. Bcl-2 up-regulation could be achieved by miR-21 overexpression, which prevented t24 cells from apoptosis induced by doxorubicin. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-miR-193a-3p | [272] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| DNA damage response signaling pathway | Activation | hsa04218 | ||
| In Vitro Model | 5637 cells | Bladder | Homo sapiens (Human) | CVCL_0126 |
| H-bc cells | Bladder | Homo sapiens (Human) | CVCL_BT00 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | Among the differentially expressed genes between the chemosensitive (5637) and chemoresistant (H-bc) bladder cancer cell lines, the expression level of the PSEN1 gene (presenilin 1), a key component of the Gamma-secretase, is negatively correlated with chemoresistance. A small interfering RNA mediated repression of the PSEN1 gene suppresses cell apoptosis and de-sensitizes 5637 cells, while overexpression of the presenilin 1 sensitizes H-bc cells to the drug-triggered cell death. As a direct target of microRNA-193a-3p that promotes the multi-chemoresistance of the bladder cancer cell, PSEN1 acts as an important executor for the microRNA-193a-3p's positive impact on the multi-chemoresistance of bladder cancer, probably via its activating effect on DNA damage response pathway. In addition to the mechanistic insights, the key players in this microRNA-193a-3p/PSEN1 axis are likely the diagnostic and/or therapeutic targets for an effective chemotherapy of bladder cancer. | |||
|
|
||||
| Key Molecule: ATP-binding cassette sub-family B5 (ABCB5) | [134] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | UM-UC-3 cells | Bladder | Homo sapiens (Human) | CVCL_1783 |
| CT26 cells | Colon | Mus musculus (Mouse) | CVCL_7254 | |
| Salmonella enterica serovar Typhimurium SL1344 | 216597 | |||
| Salmonella enterica serovar Typhimurium SL1344 detaSipA | 216597 | |||
| Salmonella enterica serovar Typhimurium SL1344 detaSipB | 216597 | |||
| Salmonella enterica serovar Typhimurium SL1344 detaSipC | 216597 | |||
| Salmonella enterica serovar Typhimurium SL1344 detaSopB | 216597 | |||
| In Vivo Model | BALB/c nude mice xenograft model | Mus musculus | ||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | Mimicking the ability of Salmonella to reverse multidrug resistance, we constructed a gold nanoparticle system packaged with a SipA corona, and found this bacterial mimic not only accumulates in tumours but also reduces P-gp at a SipA dose significantly lower than free SipA. Moreover, the Salmonella nanoparticle mimic suppresses tumour growth with a concomitant reduction in P-gp when used with an existing chemotherapeutic drug (that is, doxorubicin). | |||
|
|
||||
| Key Molecule: Presenilin-1 (PSEN1) | [272] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| DNA damage response signaling pathway | Activation | hsa04218 | ||
| In Vitro Model | 5637 cells | Bladder | Homo sapiens (Human) | CVCL_0126 |
| H-bc cells | Bladder | Homo sapiens (Human) | CVCL_BT00 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | Among the differentially expressed genes between the chemosensitive (5637) and chemoresistant (H-bc) bladder cancer cell lines, the expression level of the PSEN1 gene (presenilin 1), a key component of the Gamma-secretase, is negatively correlated with chemoresistance. A small interfering RNA mediated repression of the PSEN1 gene suppresses cell apoptosis and de-sensitizes 5637 cells, while overexpression of the presenilin 1 sensitizes H-bc cells to the drug-triggered cell death. As a direct target of microRNA-193a-3p that promotes the multi-chemoresistance of the bladder cancer cell, PSEN1 acts as an important executor for the microRNA-193a-3p's positive impact on the multi-chemoresistance of bladder cancer, probably via its activating effect on DNA damage response pathway. In addition to the mechanistic insights, the key players in this microRNA-193a-3p/PSEN1 axis are likely the diagnostic and/or therapeutic targets for an effective chemotherapy of bladder cancer. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-34 | [273] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Retinoblastoma [ICD-11: 2D02.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| MAGE-A/p53 signaling pathway | Regulation | hsa04115 | ||
| In Vitro Model | HXO-Rb44 cells | Retina | Homo sapiens (Human) | CVCL_D542 |
| SO-Rb50 cells | Retina | Homo sapiens (Human) | CVCL_D543 | |
| WERI-Rb-1 cells | Retina | Homo sapiens (Human) | CVCL_1792 | |
| Y79 cells | Retina | Homo sapiens (Human) | CVCL_1893 | |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
Freedom Evolyzer-2200 Enzyme-Linked Immunometric meter; Flow cytometry assay | |||
| Mechanism Description | miR-34a may function as a tumor suppressor for RB by targeting MAGE-A and upregulating p53 expression to enhance cell apoptosis and chemosensitivity (Carboplatin; Etoposide; Adriamycin; vincristine). | |||
| Key Molecule: hsa-miR-3163 | [274] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Retinoblastoma [ICD-11: 2D02.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | WERI-Rb-1 cells | Retina | Homo sapiens (Human) | CVCL_1792 |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Silencing of ABCG2 by MicroRNA-3163 inhibits multidrug resistance in retinoblastoma cancer stem cells. | |||
|
|
||||
| Key Molecule: ATP-binding cassette sub-family G2 (ABCG2) | [274] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Retinoblastoma [ICD-11: 2D02.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | WERI-Rb-1 cells | Retina | Homo sapiens (Human) | CVCL_1792 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Silencing of ABCG2 by MicroRNA-3163 inhibits multidrug resistance in retinoblastoma cancer stem cells. | |||
|
|
||||
| Key Molecule: Melanoma antigen A 4 (MAGE4) | [273] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Retinoblastoma [ICD-11: 2D02.2] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| MAGE-A/p53 signaling pathway | Regulation | hsa04115 | ||
| In Vitro Model | HXO-Rb44 cells | Retina | Homo sapiens (Human) | CVCL_D542 |
| SO-Rb50 cells | Retina | Homo sapiens (Human) | CVCL_D543 | |
| WERI-Rb-1 cells | Retina | Homo sapiens (Human) | CVCL_1792 | |
| Y79 cells | Retina | Homo sapiens (Human) | CVCL_1893 | |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
Freedom Evolyzer-2200 Enzyme-Linked Immunometric meter; Flow cytometry assay | |||
| Mechanism Description | miR-34a may function as a tumor suppressor for RB by targeting MAGE-A and upregulating p53 expression to enhance cell apoptosis and chemosensitivity (Carboplatin; Etoposide; Adriamycin; vincristine). | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: DNA topoisomerase 2-alpha (TOP2A) | [49] | |||
| Molecule Alteration | Missense mutation | p.R450Q |
||
| Resistant Disease | Thyroid gland cancer [ICD-11: 2D10.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Regulation | |||
| In Vitro Model | HTC-C3 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1295 |
| Experiment for Drug Resistance |
Cell growth rate assay | |||
| Mechanism Description | Several mutations have been identified in human topoisomerase IIalpha from cell lines which are resistant to anti-topoisomerase II agents. So far, three mutations at amino acids 439, 450 and 803 of DNA topoisomerase IIalpha have been reported in anticancer agent-resistant cell lines. It has been reported that introducing either of the mutations, Arg450Gln or Pro803Ser into the VM-1 cell line results in an enzyme that can confer drug resistance to yeast. | |||
| Key Molecule: DNA topoisomerase 2-alpha (TOP2A) | [49] | |||
| Molecule Alteration | Missense mutation | p.P803S |
||
| Resistant Disease | Thyroid gland cancer [ICD-11: 2D10.0] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Regulation | |||
| In Vitro Model | HTC-C3 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1295 |
| Experiment for Drug Resistance |
Cell growth rate assay | |||
| Mechanism Description | Several mutations have been identified in human topoisomerase IIalpha from cell lines which are resistant to anti-topoisomerase II agents. So far, three mutations at amino acids 439, 450 and 803 of DNA topoisomerase IIalpha have been reported in anticancer agent-resistant cell lines. It has been reported that introducing either of the mutations, Arg450Gln or Pro803Ser into the VM-1 cell line results in an enzyme that can confer drug resistance to yeast. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-miR-27b-3p | [275] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Anaplastic thyroid carcinoma [ICD-11: 2D10.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| Cell viability | Inhibition | hsa05200 | ||
| miR27b-3p/PPARgamma signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | 8305C cells | Thyroid | Homo sapiens (Human) | CVCL_1053 |
| SW1736 cells | Thyroid | Homo sapiens (Human) | CVCL_3883 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | The inhibitor of miR-27b-3p can increase the Dox sensitivity of ATC Dox-resistant cells while over-expression of PPARGamma also increased the Dox sensitivity of ATC-resistant cells. | |||
| Key Molecule: Papillary thyroid carcinoma susceptibility candidate 3 (PTCSC3) | [276] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Anaplastic thyroid carcinoma [ICD-11: 2D10.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell viability | Inhibition | hsa05200 | |
| STAT3/INO80 signaling pathway | Inhibition | hsa04066 | ||
| In Vitro Model | FTC-133 cells | Thyroid | Homo sapiens (Human) | CVCL_1219 |
| 8505C cells | Thyroid | Homo sapiens (Human) | CVCL_1054 | |
| FTC 238 cells | Thyroid | Homo sapiens (Human) | CVCL_2447 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | LncRNA PTCSC3 inhibits INO80 expression by negatively regulating STAT3, and thereby attenuating drug resistance of ATC to chemotherapy drug doxorubicin. | |||
|
|
||||
| Key Molecule: Peroxisome proliferator-activated receptor gamma (PPARG) | [275] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Anaplastic thyroid carcinoma [ICD-11: 2D10.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| Cell viability | Inhibition | hsa05200 | ||
| miR27b-3p/PPARgamma signaling pathway | Regulation | hsa05206 | ||
| In Vitro Model | 8305C cells | Thyroid | Homo sapiens (Human) | CVCL_1053 |
| SW1736 cells | Thyroid | Homo sapiens (Human) | CVCL_3883 | |
| Experiment for Molecule Alteration |
Western blot analysis; RIP assay; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | The inhibitor of miR-27b-3p can increase the Dox sensitivity of ATC Dox-resistant cells while over-expression of PPARGamma also increased the Dox sensitivity of ATC-resistant cells. | |||
| Key Molecule: Signal transducer activator transcription 3 (STAT3) | [276] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Anaplastic thyroid carcinoma [ICD-11: 2D10.3] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell viability | Inhibition | hsa05200 | |
| STAT3/INO80 signaling pathway | Inhibition | hsa04066 | ||
| In Vitro Model | FTC-133 cells | Thyroid | Homo sapiens (Human) | CVCL_1219 |
| 8505C cells | Thyroid | Homo sapiens (Human) | CVCL_1054 | |
| FTC 238 cells | Thyroid | Homo sapiens (Human) | CVCL_2447 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | LncRNA PTCSC3 inhibits INO80 expression by negatively regulating STAT3, and thereby attenuating drug resistance of ATC to chemotherapy drug doxorubicin. | |||
| Key Molecule: Papillary thyroid carcinoma susceptibility candidate 3 (PTCSC3) | [276] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Anaplastic thyroid cancer [ICD-11: 2D10.2] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | STAT3/INO80 pathway | Regulation | hsa04550 | |
| In Vitro Model | 8505C cells | Thyroid | Homo sapiens (Human) | CVCL_1054 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | LncRNA PTCSC3 was low-expressed in ATC tissues and cells. Over-expressed PTCSC3 inhibited the drug resistance of ATC to doxorubicin. LncRNA PTCSC3 inhibits INO80 expression by negatively regulating STAT3, and thereby attenuating drug resistance of ATC to chemotherapy drug doxorubicin, providing novel strategies for improving efficiency of chemotherapy for ATC treatment. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-431 | [277] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Adrenocortical carcinoma [ICD-11: 2D11.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | H295R cells | Kidney | Homo sapiens (Human) | CVCL_0458 |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Cell Growth Assay; Flow cytometry assay | |||
| Mechanism Description | Restoration of miR-431 in vitro decreased the half maximal inhibitory concentrations of doxorubicin and mitotane, with markedly increased apoptosis via downregulating ZEB1. | |||
|
|
||||
| Key Molecule: Zinc finger E-box-binding homeobox 1 (ZEB1) | [277] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Sensitive Disease | Adrenocortical carcinoma [ICD-11: 2D11.0] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | H295R cells | Kidney | Homo sapiens (Human) | CVCL_0458 |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
Cell Growth Assay; Flow cytometry assay | |||
| Mechanism Description | Restoration of miR-431 in vitro decreased the half maximal inhibitory concentrations of doxorubicin and mitotane, with markedly increased apoptosis via downregulating ZEB1. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-let-7d | [278] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Sensitive Disease | Head and neck squamous cell carcinoma [ICD-11: 2D42.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 |
| FaDu cells | Pharynx | Homo sapiens (Human) | CVCL_1218 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Clonogenic assay; MTT assay | |||
| Mechanism Description | The level of let-7d expression is an important factor for cell response to irradiation and chemotherapeutics. Overexpressed let-7d inhibited chemoresistance to cisplatin and paclitaxel in OSCC-ALDH1+ cells. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [5] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Alveolar soft part sarcoma [ICD-11: 2F00.Y] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| KHOS cells | Bone | Homo sapiens (Human) | CVCL_2546 | |
| KHOSR2 cells | Bone | Homo sapiens (Human) | CVCL_T432 | |
| ES-X cells | N.A. | . | N.A. | |
| VAESBJ cells | Bone | Homo sapiens (Human) | CVCL_1785 | |
| ASPS-KY cells | Lung | Homo sapiens (Human) | CVCL_S737 | |
| Experiment for Molecule Alteration |
Western blotting assay | |||
| Experiment for Drug Resistance |
XTT assay | |||
| Mechanism Description | In comparison to Dox-sensitive cells (MCF-7 and KHOS), P-gp mRNA expression was upregulated in all Dox-resistant cells (VAESBJ, ES-X, ASPS-KY and KHOSR2 cells). | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [44] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Resistant Disease | Pituitary adenoma [ICD-11: 2F37.1] | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | GH4C1 cells | Pituitary gland | Rattus norvegicus (Rat) | CVCL_0276 |
| Experiment for Molecule Alteration |
Immunocytochemical staining assay | |||
| Experiment for Drug Resistance |
Lowry assay; Bradford assay | |||
| Mechanism Description | Cells resistant to colchicine at 0.4 micrograms/ml, termed GH4C1/RC.4, exhibited the multidrug-resistance phenotype, as the LD50 values for colchicine, puromycin, actinomycin D, and doxorubicin were between 8 and 30 times greater than the corresponding values for the parental GH4C1 cells.Immunocytochemical staining with a monoclonal antibody, C219, to the 170-kilodalton P-glycoprotein showed directly that GH4C1/RC.4 cells overexpress P-glycoprotein. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Neurofibromin (NF1) | [26] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Resistant Disease | Peripheral nerve sheath tumor [ICD-11: 2F3Y.1] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | RAS signaling pathway | Activation | hsa04014 | |
| In Vitro Model | sNF02.2 cells | Lung | Homo sapiens (Human) | CVCL_K280 |
| Hs 53.T cells | Skin | Homo sapiens (Human) | CVCL_0786 | |
| Experiment for Molecule Alteration |
qRT-PCR; Western blotting assay | |||
| Experiment for Drug Resistance |
Ez-Cytox assay | |||
| Mechanism Description | High expression of Bcl-xL in the MPNST cells was caused by a decreased transcriptional expression of the NF1 gene. Down-regulation of the NF1 gene by RNA interference (RNAi) induced an increase in Bcl-xL expression and a decrease in its sensitivity to apoptosis in the benign neurofibroma cell line and primary normal cells. | |||
ICD-12: Respiratory system diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: MATE family efflux transporter (ABEM) | [28] | |||
| Molecule Alteration | Expression | Inherence |
||
| Resistant Disease | Acinetobacter baumannii infection [ICD-11: CA40.4] | |||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli kAM32 | 562 | ||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | AbeM was found to be an H+-coupled multidrug efflux pump and a unique member of the MATE family which lead to drug resistance. | |||
References
visits since 2022
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Zhang.
