Molecule Information
General Information of the Molecule (ID: Mol00049)
| Name |
E3 ubiquitin-protein ligase COP1 (COP1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Constitutive photomorphogenesis protein 1 homolog; hCOP1; RING finger and WD repeat domain protein 2; RING finger protein 200; RING-type E3 ubiquitin transferase RFWD2; RFWD2; RNF200
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
COP1
|
||||
| Gene ID | |||||
| Location |
chr1:175944831-176207286[-]
|
||||
| Sequence |
MSGSRQAGSGSAGTSPGSSAASSVTSASSSLSSSPSPPSVAVSAAALVSGGVAQAAGSGG
LGGPVRPVLVAPAVSGSGGGAVSTGLSRHSCAARPSAGVGGSSSSLGSGSRKRPLLAPLC NGLINSYEDKSNDFVCPICFDMIEEAYMTKCGHSFCYKCIHQSLEDNNRCPKCNYVVDNI DHLYPNFLVNELILKQKQRFEEKRFKLDHSVSSTNGHRWQIFQDWLGTDQDNLDLANVNL MLELLVQKKKQLEAESHAAQLQILMEFLKVARRNKREQLEQIQKELSVLEEDIKRVEEMS GLYSPVSEDSTVPQFEAPSPSHSSIIDSTEYSQPPGFSGSSQTKKQPWYNSTLASRRKRL TAHFEDLEQCYFSTRMSRISDDSRTASQLDEFQECLSKFTRYNSVRPLATLSYASDLYNG SSIVSSIEFDRDCDYFAIAGVTKKIKVYEYDTVIQDAVDIHYPENEMTCNSKISCISWSS YHKNLLASSDYEGTVILWDGFTGQRSKVYQEHEKRCWSVDFNLMDPKLLASGSDDAKVKL WSTNLDNSVASIEAKANVCCVKFSPSSRYHLAFGCADHCVHYYDLRNTKQPIMVFKGHRK AVSYAKFVSGEEIVSASTDSQLKLWNVGKPYCLRSFKGHINEKNFVGLASNGDYIACGSE NNSLYLYYKGLSKTLLTFKFDTVKSVLDKDRKEDDTNEFVSAVCWRALPDGESNVLIAAN SQGTIKVLELV Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
E3 ubiquitin-protein ligase that mediates ubiquitination and subsequent proteasomal degradation of target proteins. E3 ubiquitin ligases accept ubiquitin from an E2 ubiquitin-conjugating enzyme in the form of a thioester and then directly transfers the ubiquitin to targeted substrates. Involved in JUN ubiquitination and degradation. Directly involved in p53 (TP53) ubiquitination and degradation, thereby abolishing p53-dependent transcription and apoptosis. Ubiquitinates p53 independently of MDM2 or RCHY1. Probably mediates E3 ubiquitin ligase activity by functioning as the essential RING domain subunit of larger E3 complexes. In contrast, it does not constitute the catalytic RING subunit in the DCX DET1-COP1 complex that negatively regulates JUN, the ubiquitin ligase activity being mediated by RBX1. Involved in 14-3-3 protein sigma/SFN ubiquitination and proteasomal degradation, leading to AKT activation and promotion of cell survival. Ubiquitinates MTA1 leading to its proteasomal degradation. Upon binding to TRIB1, ubiquitinates CEBPA, which lacks a canonical COP1-binding motif (Probable).
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
1 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [1] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| p53 signaling pathway | Activation | hsa04115 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| MDA-MB-157 cells | Breast | Homo sapiens (Human) | CVCL_0618 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
WST-8 Cell viability assay; CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | miR-214 promotes apoptosis and sensitizes breast cancer cells to doxorubicin by targeting the RFWD2-p53 cascade. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.41E-90; Fold-change: 7.05E-01; Z-score: 1.95E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.72E-05; Fold-change: 2.44E-01; Z-score: 6.13E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
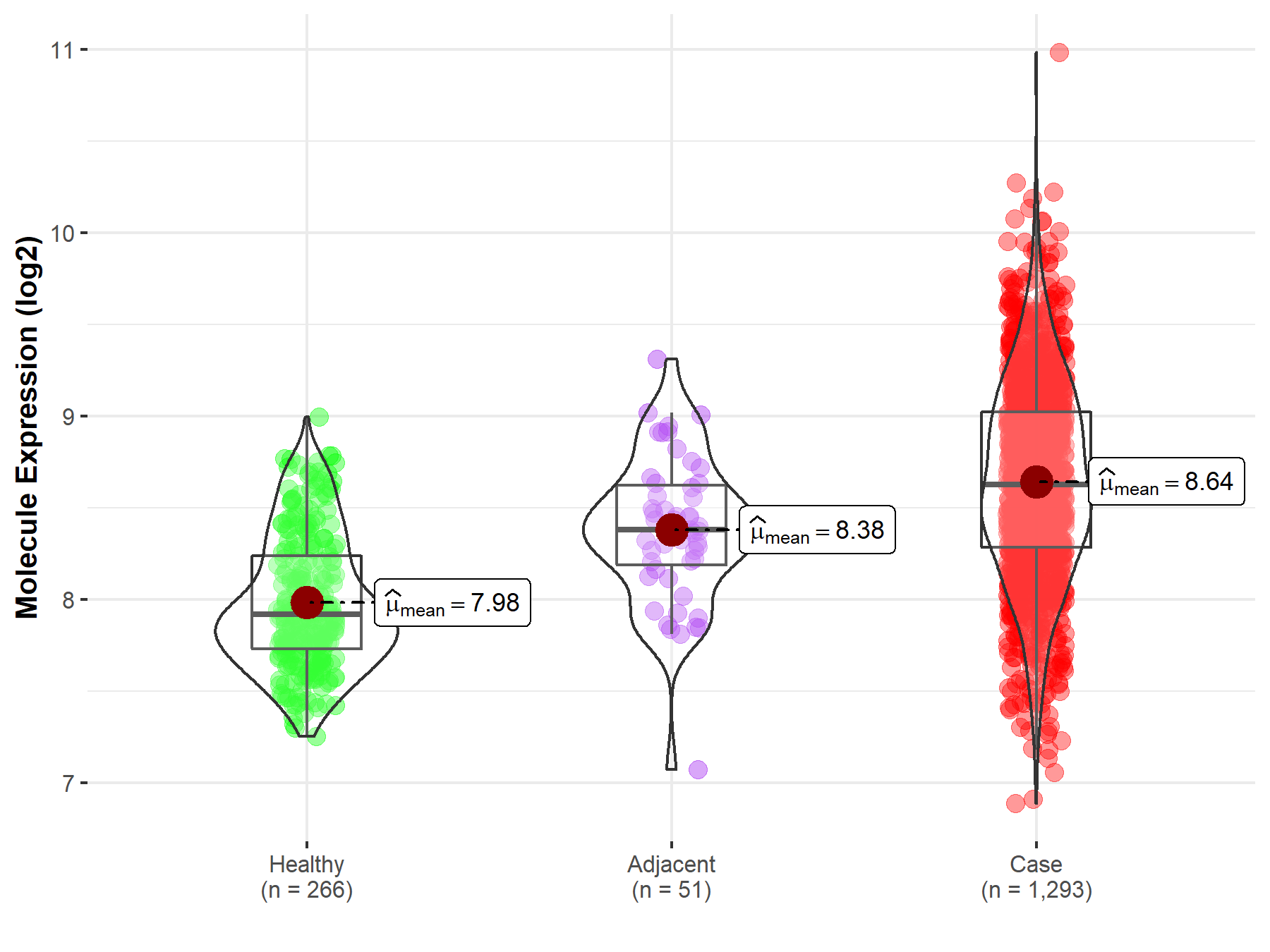
|
Click to View the Clearer Original Diagram |
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
