Drug Information
Drug (ID: DG00137) and It's Reported Resistant Information
| Name |
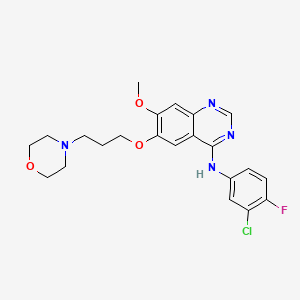
Gefitinib
|
||||
|---|---|---|---|---|---|
| Synonyms |
Gefitini; IRE; Iressa; Irressat; Gefitinib [USAN]; ZD 1839; ZD1839; Iressa (TN); Iressa(TM); ZD-1839; CU-00000000396-1; Gefitinib,Iressa, ZD1839; Gefitinib (JAN/USAN/INN); ZD-1839, Iressa, Gefitinib; N-(3-Chloro-4-fluorophenyl)-7-methoxy-6-(3-morpholinopropoxy)quinazolin-4-amine; N-(3-chloro-4-fluorophenyl)-7-methoxy-6-(3-(4-morpholinyl)propoxy)-4-quinazolinamide; N-(3-chloro-4-fluorophenyl)-7-methoxy-6-(3-morpholin-4-ylpropoxy)quinazolin-4-amine; N-(3-chloro-4-fluorophenyl)-7-methoxy-6-[3-(morpholin-4-yl)propoxy]quinazolin-4-amine; N-(3-Chloro-4-fluoro-phenyl)-7-methoxy-6-(3-morpholin-4-ylpropoxy)quinazolin-4-amine; 4-(3'-Chloro-4'-fluoroanilino)-7-methoxy-6-(3-morpholinopropoxy)quinazoline; 6-(3-morpholinopropoxy)-N-(3-chloro-4-fluorophenyl)-7-methoxyquinazolin-4-amine
Click to Show/Hide
|
||||
| Indication |
In total 3 Indication(s)
|
||||
| Structure |
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Clinically Reported Resistance for This Drug
(1 diseases)
[2]
Disease(s) with Resistance Information Validated by in-vivo Model for This Drug
(1 diseases)
[4]
Disease(s) with Resistance Information Discovered by Cell Line Test for This Drug
(2 diseases)
[5]
[6]
|
||||
| Target | Epidermal growth factor receptor (EGFR) | EGFR_HUMAN | [1] | ||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C22H24ClFN4O3
|
||||
| IsoSMILES |
COC1=C(C=C2C(=C1)N=CN=C2NC3=CC(=C(C=C3)F)Cl)OCCCN4CCOCC4
|
||||
| InChI |
1S/C22H24ClFN4O3/c1-29-20-13-19-16(12-21(20)31-8-2-5-28-6-9-30-10-7-28)22(26-14-25-19)27-15-3-4-18(24)17(23)11-15/h3-4,11-14H,2,5-10H2,1H3,(H,25,26,27)
|
||||
| InChIKey |
XGALLCVXEZPNRQ-UHFFFAOYSA-N
|
||||
| PubChem CID | |||||
| ChEBI ID | |||||
| TTD Drug ID | |||||
| VARIDT ID | |||||
| INTEDE ID | |||||
| DrugBank ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-02: Benign/in-situ/malignant neoplasm
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [7] | ||||||||||||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Non-small cell lung cancer | ||||||||||||
| The Studied Tissue | Blood | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.41E-78 Fold-change: -3.81E-01 Z-score: -2.43E+01 |
||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | ||||||||||
| Cell migration | Inhibition | hsa04670 | |||||||||||
| EGFR signaling pathway | Inhibition | hsa01521 | |||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| NCI-H1650 cells | Lung | Homo sapiens (Human) | CVCL_1483 | ||||||||||
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Mechanism Description | miR-133b suppresses the expression of EGFR, miR-133b transfection may modulate apoptosis, invasion and sensitivity to EGFR-TkI through the EGFR signaling pathways. | ||||||||||||
| Key Molecule: Hepatocyte growth factor receptor (MET) | [8] | ||||||||||||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Non-small cell lung cancer | ||||||||||||
| The Studied Tissue | Blood | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.34E-10 Fold-change: -1.34E-01 Z-score: -6.52E+00 |
||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | ||||||||||
| In Vitro Model | H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | |||||||||
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| PC9GR cells | Lung | Homo sapiens (Human) | CVCL_V337 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay | ||||||||||||
| Mechanism Description | Expression of Met has been associated with both primary and acquired resistance to gefitinib, miR-130a expression was negatively correlated with that of Met. Over-expression of miR-130a increased cell apoptosis and inhibited proliferation of NSCLC cells treated with gefitinib, whereas lowering the expression of miR-130a decreased cell apoptosis and promoted cell proliferation after treatment with gefitinib in both gefitinib-sensitive and -resistant NSCLC cell lines. | ||||||||||||
| Key Molecule: Hepatocyte growth factor receptor (MET) | [9] | ||||||||||||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Lung cancer | ||||||||||||
| The Studied Tissue | Blood | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.34E-10 Fold-change: -1.34E-01 Z-score: -6.52E+00 |
||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | ||||||||||
| Cell proliferation | Inhibition | hsa05200 | |||||||||||
| HGF/ MET signaling pathway | Inhibition | hsa04151 | |||||||||||
| In Vitro Model | HCC827 cells | Lung | Homo sapiens (Human) | CVCL_2063 | |||||||||
| MRC-5 cells | Lung | Homo sapiens (Human) | CVCL_0440 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
WST-8 assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | In the HGF-induced gefitinib-resistant cell model, the exposure of miR-34a plus gefitinib efficiently inhibited the phosphorylation of MET, EGFR, Akt and ERk, and induced cell death, and apoptosis. In the presence of HGF, although EGFR was successfully inhibited by gefitinib monotherapy, the downstream pathways (PI3k/Akt and ERk pathway) were nevertheless activated by MET activation. Through addition of miR-34a to these cells, both MET and EGFR were successfully inhibited and subsequently the downstream pathways were blocked. However, the inhibitory effect of miR-34a on of MET and downstream pathways was lower than that for the MET-TkI. These results suggested that the combination of miR-34a and gefitinib was able to partially inhibit downstream pathways activation though inhibition of MET and EGFR activation in EGFR mutant NSCLC cells, though this effect was lower than what has been observed for MET-TkI. | ||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [10] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Lung adenocarcinoma | ||||||||||||
| The Studied Tissue | Blood | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.41E-78 Fold-change: -3.81E-01 Z-score: -2.43E+01 |
||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | ||||||||||
| Cell invasion | Inhibition | hsa05200 | |||||||||||
| Cell migration | Inhibition | hsa04670 | |||||||||||
| Cell proliferation | Inhibition | hsa05200 | |||||||||||
| EGFR signaling pathway | Inhibition | hsa01521 | |||||||||||
| In Vitro Model | H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | |||||||||
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | ||||||||||
| HCC827 cells | Lung | Homo sapiens (Human) | CVCL_2063 | ||||||||||
| 16HBE cells | Lung | Homo sapiens (Human) | CVCL_0112 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTT assay; EdU assay | ||||||||||||
| Mechanism Description | GAS5 was significantly downregulated in lung adenocarcinoma tissues compared with the paired adjacent non-tumorous tissue samples. Furthermore, lower GAS5 expression levels were associated with larger tumor sizes, poor tumor differentiation, and advanced pathological stages. However, GAS5 was almost equally expressed between benign tumors compared with the adjacent normal tissues. GAS5 was also overexpressed in EGFR-TkI sensitive cell lines compared with the resistant cell line. Using MTT, EdU incorporation, and colony formation assays, we showed that GAS5-expressing A549 cells displayed an elevated level of cell death. In addition to its pro-apoptotic effect in the A549 cell line, GAS5 overexpression also suppressed the growth of A549-derived tumors in nude mice treated with gefitinib. GAS5 overexpression was inversely correlated with the expression of the EGFR pathway and IGF-1R proteins. | ||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [11] | ||||||||||||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Non-small cell lung cancer | ||||||||||||
| The Studied Tissue | Blood | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.41E-78 Fold-change: -3.81E-01 Z-score: -2.43E+01 |
||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | ||||||||||
| Cell migration | Inhibition | hsa04670 | |||||||||||
| In Vitro Model | H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | |||||||||
| HCC827 cells | Lung | Homo sapiens (Human) | CVCL_2063 | ||||||||||
| CCD-19Lu cells | Lung | Homo sapiens (Human) | CVCL_2382 | ||||||||||
| H3255 cells | Lung | Homo sapiens (Human) | CVCL_6831 | ||||||||||
| MRC-5 cells | Lung | Homo sapiens (Human) | CVCL_0440 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTS assay | ||||||||||||
| Mechanism Description | microRNA-200a directly targets and downregulates egfr and c-met to inhibit migration, invasion, and gefitinib resistance in non-small cell lung cancer. | ||||||||||||
| Key Molecule: Hepatocyte growth factor receptor (MET) | [11] | ||||||||||||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Non-small cell lung cancer | ||||||||||||
| The Studied Tissue | Blood | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.34E-10 Fold-change: -1.34E-01 Z-score: -6.52E+00 |
||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | ||||||||||
| Cell migration | Inhibition | hsa04670 | |||||||||||
| In Vitro Model | H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | |||||||||
| HCC827 cells | Lung | Homo sapiens (Human) | CVCL_2063 | ||||||||||
| CCD-19Lu cells | Lung | Homo sapiens (Human) | CVCL_2382 | ||||||||||
| H3255 cells | Lung | Homo sapiens (Human) | CVCL_6831 | ||||||||||
| MRC-5 cells | Lung | Homo sapiens (Human) | CVCL_0440 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTS assay | ||||||||||||
| Mechanism Description | microRNA-200a directly targets and downregulates egfr and c-met to inhibit migration, invasion, and gefitinib resistance in non-small cell lung cancer. | ||||||||||||
| Key Molecule: Caspase-1 (CASP1) | [12] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Lung adenocarcinoma | ||||||||||||
| The Studied Tissue | Blood | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.59E-08 Fold-change: 2.75E-01 Z-score: 5.64E+00 |
||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | ||||||||||
| Cell viability | Inhibition | hsa05200 | |||||||||||
| miR377/CASP1 signaling pathway | Regulation | N.A. | |||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | SNHG5 overexpression sensitized gefitinib resistant LAD cells to gefitinib treatment; Overexpression of SNHG5 suppressed the expression of miR-377; Overexpression of miR-377 suppressed the expression of CASP1 in PC9 cells; knockdown of CASP1 in SNHG5-overexpressed PC9GR cells abolished their gefitinib resistance. | ||||||||||||
| Key Molecule: Hepatocyte growth factor receptor (MET) | [13] | ||||||||||||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Lung cancer | ||||||||||||
| The Studied Tissue | Blood | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.34E-10 Fold-change: -1.34E-01 Z-score: -6.52E+00 |
||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | c-Met/PI3K/AKT signaling pathway | Inhibition | hsa01521 | ||||||||||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| In Vivo Model | Nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Mechanism Description | miR128 reverses the gefitinib resistance of the lung cancer stem cells by inhibiting the c-met/PI3k/AkT pathway. The miR128/c-met pathway enhances the gefitinib sensitivity of the lung cancer stem cells by suppressing the PI3k/AkT pathway. | ||||||||||||
| Key Molecule: Ubiquitin carboxyl-terminal hydrolase 14 (USP14) | [17] | ||||||||||||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Non-small cell lung cancer | ||||||||||||
| The Studied Tissue | Lung tissue | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.26E-02 Fold-change: -5.27E-02 Z-score: -1.87E+00 |
||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | ||||||||||
| Cell viability | Inhibition | hsa05200 | |||||||||||
| In Vitro Model | SPC-A1 cells | Lung | Homo sapiens (Human) | CVCL_6955 | |||||||||
| NCI-H1650 cells | Lung | Homo sapiens (Human) | CVCL_1483 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | USP14 is a direct target of hsa-miR-124a, and that hsa-miR-124a inhibits stemness and enhances the gefitinib sensitivity of NSCLC cells by targeting USP14. | ||||||||||||
| Key Molecule: Transcriptional coactivator YAP1 (YAP1) | [20] | ||||||||||||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Non-small cell lung cancer | ||||||||||||
| The Studied Tissue | Lung tissue | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.98E-07 Fold-change: -1.85E-01 Z-score: -5.39E+00 |
||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | ||||||||||
| Cell viability | Inhibition | hsa05200 | |||||||||||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | The elevated sensitivity of PC 9GR cells to gefitinib following transfection with the miR 506 3p mimic was counteracted by the overexpression of YAP1. | ||||||||||||
| Key Molecule: Signal transducer activator transcription 3 (STAT3) | [21] | ||||||||||||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Non-small cell lung cancer | ||||||||||||
| The Studied Tissue | Lung tissue | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.76E-01 Fold-change: -2.32E-02 Z-score: -8.88E-01 |
||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase Assay | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Annexin V-FITC Apoptosis assay | ||||||||||||
| Mechanism Description | miR124 decreased SNAI2 and STAT3 expression by directly targeting their 3'UTRs, miR124 contributes to gefitinib and EMT by directly targeting SNAI2 and STAT3. Over-expression of miR124 re-sensitized gefitinib-resistant cell lines to gefitinib. | ||||||||||||
| Key Molecule: Zinc finger E-box-binding homeobox 1 (ZEB1) | [22] | ||||||||||||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Non-small cell lung cancer | ||||||||||||
| The Studied Tissue | Lung tissue | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.74E-02 Fold-change: -1.61E-01 Z-score: -2.40E+00 |
||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell migration | Inhibition | hsa04670 | ||||||||||
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | |||||||||||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| PC9-ZD cells | Lung | Homo sapiens (Human) | CVCL_V337 | ||||||||||
| In Vivo Model | Nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTT assay; Annexin-V/PI assay; Wound healing assay | ||||||||||||
| Mechanism Description | miR200c enhances sensitivity of drug-resistant non-small cell lung cancer to gefitinib by suppression of PI3k/Akt signaling pathway and inhibites cell migration via targeting ZEB1. | ||||||||||||
| Key Molecule: Tripartite motif-containing protein 16 (TRIM16) | [23] | ||||||||||||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Non-small cell lung cancer | ||||||||||||
| The Studied Tissue | Lung tissue | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.34E-02 Fold-change: -1.04E-01 Z-score: -2.04E+00 |
||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | ||||||||||
| Cell invasion | Inhibition | hsa05200 | |||||||||||
| Cell migration | Inhibition | hsa04670 | |||||||||||
| Cell viability | Inhibition | hsa05200 | |||||||||||
| JAKT/STAT signaling pathway | Inhibition | hsa04630 | |||||||||||
| In Vitro Model | H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | |||||||||
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| H157 cells | Lung | Homo sapiens (Human) | CVCL_2458 | ||||||||||
| H4006 cells | Lung | Homo sapiens (Human) | N.A. | ||||||||||
| NCI-H1650 cells | Lung | Homo sapiens (Human) | CVCL_1483 | ||||||||||
| Experiment for Molecule Alteration |
Dual-Luciferase activity assay; Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay | ||||||||||||
| Mechanism Description | miR135 acted as a tumor promoter, and its suppression could improve sensitivity to gefitinib by targeting TRIM16 and inhibition of the JAk/STAT pathway. | ||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [31] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Lung cancer | ||||||||||||
| The Studied Tissue | Lung tissue | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.65E-01 Fold-change: 1.45E-02 Z-score: 1.39E+00 |
||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | ||||||||||
| RGFR signaling pathway | Inhibition | hsa05200 | |||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay | ||||||||||||
| Mechanism Description | EGFR was negatively regulated by miR-7 mimic transfection, and downregulation of EGFR expression at the protein level largely correlated with elevated levels of miR-7 in the gefitinib-resistant cells. The results of the present study suggest that miR-7 may have central roles in the development of resistance to endocrine therapy in resistant cells through regulating the expression of EGFR in cancer cells. | ||||||||||||
| Key Molecule: Adhesion G protein-coupled receptor A2 (ADGRA2) | [33] | ||||||||||||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Lung cancer | ||||||||||||
| The Studied Tissue | Lung tissue | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.93E-20 Fold-change: -7.36E-02 Z-score: -9.79E+00 |
||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTS assay | ||||||||||||
| Mechanism Description | There is an inverse correlation between the expression of miR-138-5p and GPR124 in lung adenocarcinoma specimens. Down-regulation of miR-138-5p contributes to gefitinib resistance and that restoration of miR-138-5p or inhibition GPR124 might serve as potential therapeutic approach for overcoming NSCLC gefitinib resistance. | ||||||||||||
|
|
|||||||||||||
| Key Molecule: Protein Wnt-5a (WNT5A) | [28] | ||||||||||||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Lung cancer | ||||||||||||
| The Studied Tissue | Lung tissue | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.08E-09 Fold-change: 6.39E-02 Z-score: 6.09E+00 |
||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | ||||||||||
| Cell migration | Inhibition | hsa04670 | |||||||||||
| In Vitro Model | HCC827 cells | Lung | Homo sapiens (Human) | CVCL_2063 | |||||||||
| Calu1 cells | Lung | Homo sapiens (Human) | CVCL_0608 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTS assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | miR-374a and miR-548b modulated by Axl have essential roles in cell cycle arrest, gefitinib-induced apoptosis, epithelial-to-mesenchymal transition, migration and tumorigenesis of gefitinib-resistant lung cancer cells in vitro and in vivo by targeting Wnt5a and CCNB1 genes, respectively. Of clinical significance, high expression of Axl and miR-374a and low expression of miR-548b are associated with poor disease-free survival postoperatively. These findings indicate that the modulation of specific miRNAs may provide a therapeutic target to treat or reverse gefitinib resistance in NSCLC with high expression of Axl in the future. Overexpression of Wnt5a in HCC827-Gef cells partially restored the cell sensitivity to gefitinib (Wnt5a in HCC827-Gef cells partially restored the cell sensitivity to gefitinib. | ||||||||||||
| Key Molecule: Zinc finger E-box-binding homeobox 1 (ZEB1) | [37] | ||||||||||||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Lung cancer | ||||||||||||
| The Studied Tissue | Lung tissue | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.29E-79 Fold-change: -2.78E-01 Z-score: -2.61E+01 |
||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | ||||||||||
| MEK/ERK signaling pathway | Regulation | N.A. | |||||||||||
| PI3K/AKT signaling pathway | Regulation | N.A. | |||||||||||
| In Vitro Model | H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | |||||||||
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | ||||||||||
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | ||||||||||
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | ||||||||||
| H23 cells | Lung | Homo sapiens (Human) | CVCL_1547 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Mechanism Description | Ectopic expression of miR-200c resulted in partial restoration of gefitinib sensitivity in NSCLC cells with ZEB1 downrerulating. | ||||||||||||
|
|
|||||||||||||
| Key Molecule: Growth arrest specific 5 (GAS5) | [10] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Lung adenocarcinoma | ||||||||||||
| The Studied Tissue | Lung | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.78E-12 Fold-change: 6.09E-01 Z-score: 7.06E+00 |
||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | ||||||||||
| Cell invasion | Inhibition | hsa05200 | |||||||||||
| Cell migration | Inhibition | hsa04670 | |||||||||||
| Cell proliferation | Inhibition | hsa05200 | |||||||||||
| EGFR signaling pathway | Inhibition | hsa01521 | |||||||||||
| In Vitro Model | H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | |||||||||
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | ||||||||||
| HCC827 cells | Lung | Homo sapiens (Human) | CVCL_2063 | ||||||||||
| 16HBE cells | Lung | Homo sapiens (Human) | CVCL_0112 | ||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
MTT assay; EdU assay | ||||||||||||
| Mechanism Description | GAS5 was significantly downregulated in lung adenocarcinoma tissues compared with the paired adjacent non-tumorous tissue samples. Furthermore, lower GAS5 expression levels were associated with larger tumor sizes, poor tumor differentiation, and advanced pathological stages. However, GAS5 was almost equally expressed between benign tumors compared with the adjacent normal tissues. GAS5 was also overexpressed in EGFR-TkI sensitive cell lines compared with the resistant cell line. Using MTT, EdU incorporation, and colony formation assays, we showed that GAS5-expressing A549 cells displayed an elevated level of cell death. In addition to its pro-apoptotic effect in the A549 cell line, GAS5 overexpression also suppressed the growth of A549-derived tumors in nude mice treated with gefitinib. GAS5 overexpression was inversely correlated with the expression of the EGFR pathway and IGF-1R proteins. | ||||||||||||
| Key Molecule: Small nucleolar RNA host gene 5 (SNHG5) | [12] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Lung adenocarcinoma | ||||||||||||
| The Studied Tissue | Lung | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.53E-04 Fold-change: 2.31E-01 Z-score: 3.42E+00 |
||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | ||||||||||
| Cell viability | Inhibition | hsa05200 | |||||||||||
| miR377/CASP1 signaling pathway | Regulation | N.A. | |||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | ||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | SNHG5 overexpression sensitized gefitinib resistant LAD cells to gefitinib treatment; Overexpression of SNHG5 suppressed the expression of miR-377; Overexpression of miR-377 suppressed the expression of CASP1 in PC9 cells; knockdown of CASP1 in SNHG5-overexpressed PC9GR cells abolished their gefitinib resistance. | ||||||||||||
| Key Molecule: hsa-mir-135a | [23] | ||||||||||||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | ||||||||||
| JAKT/STAT signaling pathway | Inhibition | hsa04630 | |||||||||||
| In Vitro Model | H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | |||||||||
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| H157 cells | Lung | Homo sapiens (Human) | CVCL_2458 | ||||||||||
| H4006 cells | Lung | Homo sapiens (Human) | N.A. | ||||||||||
| NCI-H1650 cells | Lung | Homo sapiens (Human) | CVCL_1483 | ||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay | ||||||||||||
| Mechanism Description | miR135 acted as a tumor promoter, and its suppression could improve sensitivity to gefitinib by targeting TRIM16 and inhibition of the JAk/STAT pathway. | ||||||||||||
| Key Molecule: hsa-mir-30e | [67] | ||||||||||||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell migration | Inhibition | hsa04670 | ||||||||||
| Cell proliferation | Inhibition | hsa05200 | |||||||||||
| In Vitro Model | HCC827 cells | Lung | Homo sapiens (Human) | CVCL_2063 | |||||||||
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | ||||||||||
| HCC827/GR cells | Lung | Homo sapiens (Human) | CVCL_V620 | ||||||||||
| PC9G cells | Lung | Homo sapiens (Human) | CVCL_V337 | ||||||||||
| In Vivo Model | Nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay | ||||||||||||
| Mechanism Description | miR30e overexpression inPC9G cells resulted in reduced cell proliferation and migration,reversing drug resistance to gefitinib, miR30e directly targeted HOXA1 in lung cancer cells. | ||||||||||||
| Key Molecule: hsa-mir-200c | [22] | ||||||||||||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell migration | Inhibition | hsa04670 | ||||||||||
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | |||||||||||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| PC9-ZD cells | Lung | Homo sapiens (Human) | CVCL_V337 | ||||||||||
| In Vivo Model | Nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
RT-PCR | ||||||||||||
| Experiment for Drug Resistance |
MTT assay; Annexin-V/PI assay; Wound healing assay | ||||||||||||
| Mechanism Description | miR200c enhances sensitivity of drug-resistant non-small cell lung cancer to gefitinib by suppression of PI3k/Akt signaling pathway and inhibites cell migration via targeting ZEB1. | ||||||||||||
| Key Molecule: hsa-mir-124 | [21] | ||||||||||||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | ||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Annexin V-FITC Apoptosis assay | ||||||||||||
| Mechanism Description | miR124 decreased SNAI2 and STAT3 expression by directly targeting their 3'UTRs, miR124 contributes to gefitinib and EMT by directly targeting SNAI2 and STAT3. Over-expression of miR124 re-sensitized gefitinib-resistant cell lines to gefitinib. | ||||||||||||
| Key Molecule: hsa-miR-30a-5p | [68] | ||||||||||||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||||||||||
| In Vitro Model | NCI-H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | |||||||||
| NCI-H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | ||||||||||
| Experiment for Molecule Alteration |
RT-PCR | ||||||||||||
| Experiment for Drug Resistance |
Annexin V-FITC Apoptosis assay; CytoSelect Cell Invasion Assay; Wound healing assay | ||||||||||||
| Mechanism Description | miR30a-5p overexpression targets the EGFR and insulin-like growth factor receptor-1 (IGF-1R) signaling pathways to overcome the drug resistance. The combination of EGFR and IGF-1R inhibitors treatment could block the PI3k/AkT signaling pathway. | ||||||||||||
| Key Molecule: hsa-mir-128a | [13] | ||||||||||||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | c-Met/PI3K/AKT signaling pathway | Inhibition | hsa01521 | ||||||||||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| In Vivo Model | Nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Mechanism Description | miR128 reverses the gefitinib resistance of the lung cancer stem cells by inhibiting the c-met/PI3k/AkT pathway. The miR128/c-met pathway enhances the gefitinib sensitivity of the lung cancer stem cells by suppressing the PI3k/AkT pathway. | ||||||||||||
| Key Molecule: hsa-miR-506-3p | [20] | ||||||||||||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | ||||||||||
| Cell viability | Inhibition | hsa05200 | |||||||||||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | The elevated sensitivity of PC 9GR cells to gefitinib following transfection with the miR 506 3p mimic was counteracted by the overexpression of YAP1. | ||||||||||||
| Key Molecule: Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) | [69] | ||||||||||||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | ||||||||||
| Cell colony | Inhibition | hsa05200 | |||||||||||
| Cell viability | Inhibition | hsa05200 | |||||||||||
| STAT3 signaling pathway | Inhibition | hsa04550 | |||||||||||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | PPI might down-regulate MALAT1 expression and inactivate STAT3 signaling pathway and could serve a promising therapeutic agent for gefitinib-resistant NSCLC. | ||||||||||||
| Key Molecule: hsa-miR-124-3p | [17] | ||||||||||||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | ||||||||||
| Cell viability | Inhibition | hsa05200 | |||||||||||
| In Vitro Model | SPC-A1 cells | Lung | Homo sapiens (Human) | CVCL_6955 | |||||||||
| NCI-H1650 cells | Lung | Homo sapiens (Human) | CVCL_1483 | ||||||||||
| Experiment for Molecule Alteration |
qPCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | USP14 is a direct target of hsa-miR-124a, and that hsa-miR-124a inhibits stemness and enhances the gefitinib sensitivity of NSCLC cells by targeting USP14. | ||||||||||||
| Key Molecule: hsa-mir-377 | [12] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | ||||||||||
| Cell viability | Inhibition | hsa05200 | |||||||||||
| miR377/CASP1 signaling pathway | Regulation | N.A. | |||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | ||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | SNHG5 overexpression sensitized gefitinib resistant LAD cells to gefitinib treatment; Overexpression of SNHG5 suppressed the expression of miR-377; Overexpression of miR-377 suppressed the expression of CASP1 in PC9 cells; knockdown of CASP1 in SNHG5-overexpressed PC9GR cells abolished their gefitinib resistance. | ||||||||||||
| Key Molecule: hsa-mir-200a | [11] | ||||||||||||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | ||||||||||
| Cell migration | Inhibition | hsa04670 | |||||||||||
| In Vitro Model | H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | |||||||||
| HCC827 cells | Lung | Homo sapiens (Human) | CVCL_2063 | ||||||||||
| CCD-19Lu cells | Lung | Homo sapiens (Human) | CVCL_2382 | ||||||||||
| H3255 cells | Lung | Homo sapiens (Human) | CVCL_6831 | ||||||||||
| MRC-5 cells | Lung | Homo sapiens (Human) | CVCL_0440 | ||||||||||
| In Vivo Model | Nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
RT-PCR | ||||||||||||
| Experiment for Drug Resistance |
MTS assay | ||||||||||||
| Mechanism Description | microRNA-200a directly targets and downregulates egfr and c-met to inhibit migration, invasion, and gefitinib resistance in non-small cell lung cancer. | ||||||||||||
| Key Molecule: hsa-mir-7 | [31] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | ||||||||||
| RGFR signaling pathway | Inhibition | hsa05200 | |||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| Experiment for Molecule Alteration |
qPCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay | ||||||||||||
| Mechanism Description | EGFR was negatively regulated by miR-7 mimic transfection, and downregulation of EGFR expression at the protein level largely correlated with elevated levels of miR-7 in the gefitinib-resistant cells. The results of the present study suggest that miR-7 may have central roles in the development of resistance to endocrine therapy in resistant cells through regulating the expression of EGFR in cancer cells. | ||||||||||||
| Key Molecule: hsa-mir-34 | [9] | ||||||||||||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | ||||||||||
| Cell proliferation | Inhibition | hsa05200 | |||||||||||
| HGF/ MET signaling pathway | Inhibition | hsa04151 | |||||||||||
| In Vitro Model | HCC827 cells | Lung | Homo sapiens (Human) | CVCL_2063 | |||||||||
| MRC-5 cells | Lung | Homo sapiens (Human) | CVCL_0440 | ||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
WST-8 assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | In the HGF-induced gefitinib-resistant cell model, the exposure of miR-34a plus gefitinib efficiently inhibited the phosphorylation of MET, EGFR, Akt and ERk, and induced cell death, and apoptosis. In the presence of HGF, although EGFR was successfully inhibited by gefitinib monotherapy, the downstream pathways (PI3k/Akt and ERk pathway) were nevertheless activated by MET activation. Through addition of miR-34a to these cells, both MET and EGFR were successfully inhibited and subsequently the downstream pathways were blocked. However, the inhibitory effect of miR-34a on of MET and downstream pathways was lower than that for the MET-TkI. These results suggested that the combination of miR-34a and gefitinib was able to partially inhibit downstream pathways activation though inhibition of MET and EGFR activation in EGFR mutant NSCLC cells, though this effect was lower than what has been observed for MET-TkI. | ||||||||||||
| Key Molecule: hsa-mir-130a | [8] | ||||||||||||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | ||||||||||
| In Vitro Model | H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | |||||||||
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| PC9GR cells | Lung | Homo sapiens (Human) | CVCL_V337 | ||||||||||
| Experiment for Molecule Alteration |
RT-PCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay | ||||||||||||
| Mechanism Description | Expression of Met has been associated with both primary and acquired resistance to gefitinib, miR-130a expression was negatively correlated with that of Met. Over-expression of miR-130a increased cell apoptosis and inhibited proliferation of NSCLC cells treated with gefitinib, whereas lowering the expression of miR-130a decreased cell apoptosis and promoted cell proliferation after treatment with gefitinib in both gefitinib-sensitive and -resistant NSCLC cell lines. | ||||||||||||
| Key Molecule: hsa-miR-138-5p | [33] | ||||||||||||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
MTS assay | ||||||||||||
| Mechanism Description | There is an inverse correlation between the expression of miR-138-5p and GPR124 in lung adenocarcinoma specimens. Down-regulation of miR-138-5p contributes to gefitinib resistance and that restoration of miR-138-5p or inhibition GPR124 might serve as potential therapeutic approach for overcoming NSCLC gefitinib resistance. | ||||||||||||
| Key Molecule: hsa-mir-147 | [39] | ||||||||||||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell migration | Inhibition | hsa04670 | ||||||||||
| Cell proliferation | Inhibition | hsa05200 | |||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Mechanism Description | miR-147 strikingly increased the sensitivity to EGFR inhibitor, gefitinib in cell with native resistance. | ||||||||||||
| Key Molecule: hsa-let-7c | [70] | ||||||||||||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | ERK signaling pathway | Inhibition | hsa04210 | ||||||||||
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | |||||||||||
| In Vitro Model | H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | |||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay | ||||||||||||
| Mechanism Description | The upregulation of let-7c was associated with the increased gefitinib sensitivity of H1975 cells, and that this effect was mediated by repression of the RAS oncogene and inactivation of the phosphoinositide 3-kinase (PI3k) /AkT and mitogen-activated extracellular signal-regulated kinase (MEk) /extracellular signal-regulated kinase (ERk) signaling pathways. | ||||||||||||
| Key Molecule: hsa-miR-133b | [7] | ||||||||||||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | ||||||||||
| Cell migration | Inhibition | hsa04670 | |||||||||||
| EGFR signaling pathway | Inhibition | hsa01521 | |||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| NCI-H1650 cells | Lung | Homo sapiens (Human) | CVCL_1483 | ||||||||||
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | ||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Mechanism Description | miR-133b suppresses the expression of EGFR, miR-133b transfection may modulate apoptosis, invasion and sensitivity to EGFR-TkI through the EGFR signaling pathways. | ||||||||||||
|
|
|||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [32] | ||||||||||||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Missense mutation | p.L858R |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.64 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.47 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
G
-
S
-
M
-
G
G
E
E
A
A
P
P
700
|
N
N
Q
Q
A
A
L
L
L
L
R
R
I
I
L
L
K
K
E
E
710
|
T
T
E
E
F
F
K
K
K
K
I
I
K
K
V
V
L
L
G
G
720
|
S
S
G
G
A
A
F
F
G
G
T
T
V
V
Y
Y
K
K
G
G
730
|
L
L
W
W
I
I
P
P
E
E
G
G
E
E
K
K
V
V
K
K
740
|
I
I
P
P
V
V
A
A
I
I
K
K
E
E
L
L
R
R
E
E
750
|
A
A
T
T
S
S
P
P
K
K
A
A
N
N
K
K
E
E
I
I
760
|
L
L
D
D
E
E
A
A
Y
Y
V
V
M
M
A
A
S
S
V
V
770
|
D
D
N
N
P
P
H
H
V
V
C
C
R
R
L
L
L
L
G
G
780
|
I
I
C
C
L
L
T
T
S
S
T
T
V
V
Q
Q
L
L
I
I
790
|
T
T
Q
Q
L
L
M
M
P
P
F
F
G
G
C
C
L
L
L
L
800
|
D
D
Y
Y
V
V
R
R
E
E
H
H
K
K
D
D
N
N
I
I
810
|
G
G
S
S
Q
Q
Y
Y
L
L
L
L
N
N
W
W
C
C
V
V
820
|
Q
Q
I
I
A
A
K
K
G
G
M
M
N
N
Y
Y
L
L
E
E
830
|
D
D
R
R
R
R
L
L
V
V
H
H
R
R
D
D
L
L
A
A
840
|
A
A
R
R
N
N
V
V
L
L
V
V
K
K
T
T
P
P
Q
Q
850
|
H
H
V
V
K
K
I
I
T
T
D
D
F
F
G
G
L
R
A
A
860
|
K
K
L
L
L
L
G
G
A
A
E
E
E
E
K
K
E
E
Y
Y
870
|
H
H
A
A
E
E
G
G
G
G
K
K
V
V
P
P
I
I
K
K
880
|
W
W
M
M
A
A
L
L
E
E
S
S
I
I
L
L
H
H
R
R
890
|
I
I
Y
Y
T
T
H
H
Q
Q
S
S
D
D
V
V
W
W
S
S
900
|
Y
Y
G
G
V
V
T
T
V
V
W
W
E
E
L
L
M
M
T
T
910
|
F
F
G
G
S
S
K
K
P
P
Y
Y
D
D
G
G
I
I
P
P
920
|
A
A
S
S
E
E
I
I
S
S
S
S
I
I
L
L
E
E
K
K
930
|
G
G
E
E
R
R
L
L
P
P
Q
Q
P
P
P
P
I
I
C
C
940
|
T
T
I
I
D
D
V
V
Y
Y
M
M
I
I
M
M
V
V
K
K
950
|
C
C
W
W
M
M
I
I
D
D
A
A
D
D
S
S
R
R
P
P
960
|
K
K
F
F
R
R
E
E
L
L
I
I
I
I
E
E
F
F
S
S
970
|
K
K
M
M
A
A
R
R
D
D
P
P
Q
Q
R
R
Y
Y
L
L
980
|
V
V
I
I
Q
Q
G
G
D
D
E
E
R
R
M
M
H
H
L
L
990
|
P
P
S
S
P
P
T
T
D
D
S
S
N
N
F
F
Y
Y
R
R
1000
|
A
A
L
L
M
M
D
D
E
E
E
E
D
D
M
M
D
D
D
D
1010
|
V
V
V
V
D
D
A
A
D
D
E
E
Y
Y
L
L
I
I
P
P
1020
|
Q
Q
-
Q
-
G
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | ERK/MAPKsignaling pathway | Activation | hsa04210 | ||||||||||
| In Vitro Model | NSCLC cells | Lung | Homo sapiens (Human) | N.A. | |||||||||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Liquid biopsy; ATP-binding pocket affinity comparison assay | ||||||||||||
| Mechanism Description | The two most common EGFR-activating mutations are small in-frame deletions in exon 19 (particularly E746-A750del) and amino acid substitution in exon 21 (leucine to arginine at codon 858 (L858R)), which collectively account for >90% of known activating EGFR mutations.2 3 These two alterations are the best-characterised mutations conferring sensitivity to EGFR-tyrosine kinase inhibitor (EGFR-TkI) therapy, resulting in higher response rates (RR) (up to 70%) and longer median survival (up to 24-30 months) than those observed in patients with wild-type (WT) EGFR. The higher sensitivity of these mutations relays in an increased affinity of the ATP-binding pocket for EGFR-TkIs as compared with WT EGFR. | ||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [32] | ||||||||||||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Frameshift mutation | p.E746-A750del |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | ERK/MAPKsignaling pathway | Activation | hsa04210 | ||||||||||
| In Vitro Model | NSCLC cells | Lung | Homo sapiens (Human) | N.A. | |||||||||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Liquid biopsy; ATP-binding pocket affinity comparison assay | ||||||||||||
| Mechanism Description | The two most common EGFR-activating mutations are small in-frame deletions in exon 19 (particularly E746-A750del) and amino acid substitution in exon 21 (leucine to arginine at codon 858 (L858R)), which collectively account for >90% of known activating EGFR mutations.2 3 These two alterations are the best-characterised mutations conferring sensitivity to EGFR-tyrosine kinase inhibitor (EGFR-TkI) therapy, resulting in higher response rates (RR) (up to 70%) and longer median survival (up to 24-30 months) than those observed in patients with wild-type (WT) EGFR. The higher sensitivity of these mutations relays in an increased affinity of the ATP-binding pocket for EGFR-TkIs as compared with WT EGFR. | ||||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Key Molecule: Hepatocyte growth factor receptor (MET) | [14] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Non-small cell lung cancer | ||||||||||||
| The Studied Tissue | Blood | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.34E-10 Fold-change: -1.34E-01 Z-score: -6.52E+00 |
||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Epithelial mesenchymal transition signaling pathway | Activation | hsa01521 | ||||||||||
| miR19a/c-Met signaling pathway | Regulation | N.A. | |||||||||||
| In Vitro Model | H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | |||||||||
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| HCC827 cells | Lung | Homo sapiens (Human) | CVCL_2063 | ||||||||||
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | ||||||||||
| PC9GR cells | Lung | Homo sapiens (Human) | CVCL_V337 | ||||||||||
| In Vivo Model | Nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase reporter assay | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay | ||||||||||||
| Mechanism Description | miR19a contributes to gefitinib resistance and epithelial mesenchymal transition in non-small cell lung cancer cells by targeting c-Met. Overexpression of miR19a decreased c-Met expression and re-sensitized gefitinib-resistant NSCLC cells in vitro and in vivo. Decreased miR19a expression may contribute to NSCLC cell metastasis by increasing cell mobility and migration and promoting EMT. | ||||||||||||
| Key Molecule: Gem-associated protein 2 (SIP1) | [27] | ||||||||||||
| Resistant Disease | Lung cancer [ICD-11: 2C25.5] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Lung cancer | ||||||||||||
| The Studied Tissue | Lung tissue | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.13E-19 Fold-change: 9.33E-02 Z-score: 9.40E+00 |
||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Beta-catenin signaling pathway | Activation | hsa04520 | ||||||||||
| Cell invasion | Activation | hsa05200 | |||||||||||
| Cell proliferation | Activation | hsa05200 | |||||||||||
| In Vitro Model | 95D cells | Lung | Homo sapiens (Human) | CVCL_7110 | |||||||||
| 95C cells | Lung | Homo sapiens (Human) | CVCL_7109 | ||||||||||
| YTMLC-90 cells | Lung | Homo sapiens (Human) | CVCL_6959 | ||||||||||
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
EdU assay | ||||||||||||
| Mechanism Description | HOTAIR also regulates non-small-cell lung cancer proliferation, migration and invasion through epithelial-mesenchymal transition and the beta-catenin pathway. | ||||||||||||
| Key Molecule: Membrane-associated guanylate kinase inverted 2 (MAGI2) | [6] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Lung cancer | ||||||||||||
| The Studied Tissue | Lung tissue | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.14E-03 Fold-change: -3.11E-02 Z-score: -2.66E+00 |
||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | PC3 cells | Prostate | Homo sapiens (Human) | CVCL_0035 | |||||||||
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | ||||||||||
| PC-14 cells | Lung | Homo sapiens (Human) | CVCL_1640 | ||||||||||
| LC-2/ad cells | Lung | Homo sapiens (Human) | CVCL_1373 | ||||||||||
| RERF-LCkJ cells | Lung | Homo sapiens (Human) | CVCL_1654 | ||||||||||
| ABC-1 cells | Lung | Homo sapiens (Human) | CVCL_1066 | ||||||||||
| RERF-LCMS cells | Lung | Homo sapiens (Human) | CVCL_1655 | ||||||||||
| Experiment for Molecule Alteration |
Western blottling analysis | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Mechanism Description | miR-134/487b/655 cluster contributed to the TGF-beta1-induced EMT phenomenon and affected the resistance to gefitinib by directly targeting MAGI2, in which suppression subsequently caused loss of PTEN stability in lung cancer cells. | ||||||||||||
| Key Molecule: hsa-mir-19a | [14] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Epithelial mesenchymal transition signaling pathway | Inhibition | hsa01521 | ||||||||||
| miR19a/c-Met signaling pathway | Regulation | N.A. | |||||||||||
| In Vitro Model | H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | |||||||||
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| HCC827 cells | Lung | Homo sapiens (Human) | CVCL_2063 | ||||||||||
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | ||||||||||
| PC9GR cells | Lung | Homo sapiens (Human) | CVCL_V337 | ||||||||||
| In Vivo Model | Nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
RT-PCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay | ||||||||||||
| Mechanism Description | miR19a contributes to gefitinib resistance and epithelial mesenchymal transition in non-small cell lung cancer cells by targeting c-Met. Overexpression of miR19a decreased c-Met expression and re-sensitized gefitinib-resistant NSCLC cells in vitro and in vivo. Decreased miR19a expression may contribute to NSCLC cell metastasis by increasing cell mobility and migration and promoting EMT. | ||||||||||||
| Key Molecule: HOX transcript antisense RNA (HOTAIR) | [27] | ||||||||||||
| Resistant Disease | Lung cancer [ICD-11: 2C25.5] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Beta-catenin signaling pathway | Activation | hsa04520 | ||||||||||
| Cell invasion | Activation | hsa05200 | |||||||||||
| Cell migration | Activation | hsa04670 | |||||||||||
| Cell proliferation | Activation | hsa05200 | |||||||||||
| Epithelial mesenchymal transition signaling pathway | Activation | hsa01521 | |||||||||||
| In Vitro Model | 95D cells | Lung | Homo sapiens (Human) | CVCL_7110 | |||||||||
| 95C cells | Lung | Homo sapiens (Human) | CVCL_7109 | ||||||||||
| YTMLC-90 cells | Lung | Homo sapiens (Human) | CVCL_6959 | ||||||||||
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
EdU assay | ||||||||||||
| Mechanism Description | HOTAIR also regulates non-small-cell lung cancer proliferation, migration and invasion through epithelial-mesenchymal transition and the beta-catenin pathway. | ||||||||||||
| Key Molecule: hsa-mir-134 | [6] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | PC3 cells | Prostate | Homo sapiens (Human) | CVCL_0035 | |||||||||
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | ||||||||||
| PC-14 cells | Lung | Homo sapiens (Human) | CVCL_1640 | ||||||||||
| LC-2/ad cells | Lung | Homo sapiens (Human) | CVCL_1373 | ||||||||||
| RERF-LCkJ cells | Lung | Homo sapiens (Human) | CVCL_1654 | ||||||||||
| ABC-1 cells | Lung | Homo sapiens (Human) | CVCL_1066 | ||||||||||
| RERF-LCMS cells | Lung | Homo sapiens (Human) | CVCL_1655 | ||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Mechanism Description | miR-134/487b/655 cluster contributed to the TGF-beta1-induced EMT phenomenon and affected the resistance to gefitinib by directly targeting MAGI2, in which suppression subsequently caused loss of PTEN stability in lung cancer cells. | ||||||||||||
| Key Molecule: hsa-mir-487b | [6] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | PC3 cells | Prostate | Homo sapiens (Human) | CVCL_0035 | |||||||||
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | ||||||||||
| PC-14 cells | Lung | Homo sapiens (Human) | CVCL_1640 | ||||||||||
| LC-2/ad cells | Lung | Homo sapiens (Human) | CVCL_1373 | ||||||||||
| RERF-LCkJ cells | Lung | Homo sapiens (Human) | CVCL_1654 | ||||||||||
| ABC-1 cells | Lung | Homo sapiens (Human) | CVCL_1066 | ||||||||||
| RERF-LCMS cells | Lung | Homo sapiens (Human) | CVCL_1655 | ||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Mechanism Description | miR-134/487b/655 cluster contributed to the TGF-beta1-induced EMT phenomenon and affected the resistance to gefitinib by directly targeting MAGI2, in which suppression subsequently caused loss of PTEN stability in lung cancer cells. | ||||||||||||
|
|
|||||||||||||
| Key Molecule: Insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3) | [15] | ||||||||||||
| Metabolic Type | Mitochondrial metabolism | ||||||||||||
| Resistant Disease | Non-small cell lung carcinoma [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Non-small cell lung carcinoma | ||||||||||||
| The Studied Tissue | Lung tissue | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.38E-05 Fold-change: 6.29E-01 Z-score: 4.51E+00 |
||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vivo Model | Nude mice , with PC-9/GR cell lines | Mice | |||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
Tumor volume assay | ||||||||||||
| Mechanism Description | Furthermore, we revealed that targeting IGF2BP3 can markedly enhance the sensitivity of TKIs in NSCLC and this effect was strongly dependent on the coordinated induction of COX6B2, a key downstream target of IGF2BP3 in mitochondrial OXPHOS energy production. Overall, our study revealed a novel mechanism of TKI resistance involved in IGF2BP3-dependent cross-talk between epigenetic modifications and metabolic reprogramming through the IGF2BP3-COX6B3 axis in NSCLC. | ||||||||||||
| Key Molecule: Insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3) | [15] | ||||||||||||
| Metabolic Type | Mitochondrial metabolism | ||||||||||||
| Resistant Disease | Non-small cell lung carcinoma [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Non-small cell lung carcinoma | ||||||||||||
| The Studied Tissue | Lung tissue | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.38E-05 Fold-change: 6.29E-01 Z-score: 4.51E+00 |
||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vivo Model | Nude mice , with fresh tissue from patient | Mice | |||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
Tumor volume assay | ||||||||||||
| Mechanism Description | Furthermore, we revealed that targeting IGF2BP3 can markedly enhance the sensitivity of TKIs in NSCLC and this effect was strongly dependent on the coordinated induction of COX6B2, a key downstream target of IGF2BP3 in mitochondrial OXPHOS energy production. Overall, our study revealed a novel mechanism of TKI resistance involved in IGF2BP3-dependent cross-talk between epigenetic modifications and metabolic reprogramming through the IGF2BP3-COX6B2 axis in NSCLC. | ||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [59] | ||||||||||||
| Metabolic Type | Lipid metabolism | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Mutation | . |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Hippo signaling pathway | Activation | hsa04390 | ||||||||||
| In Vivo Model | Female BALB/c-nu mice, with PC9 and PC9GR cells | Mice | |||||||||||
| Experiment for Drug Resistance |
Tumor volume assay | ||||||||||||
| Mechanism Description | PC9 gefitinib resistant strains were induced by low-dose maintenance. Cell culture and animal-related studies validated that the application of pitavastatin inhibited the proliferation of lung cancer cells, promoted cell apoptosis, and restrained the acquired resistance to EGFR-TKIs. KEGG pathway analysis showed that the hippo/YAP signaling pathway was activated in PC9GR cells relative to PC11 cells, and the YAP expression was inhibited by pitavastatin administration. | ||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [59] | ||||||||||||
| Metabolic Type | Lipid metabolism | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Mutation | . |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Hippo signaling pathway | Activation | hsa04390 | ||||||||||
| In Vitro Model | NSCLC cells | Lung | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Drug Resistance |
Apoptosis rate assay | ||||||||||||
| Mechanism Description | PC9 gefitinib resistant strains were induced by low-dose maintenance. Cell culture and animal-related studies validated that the application of pitavastatin inhibited the proliferation of lung cancer cells, promoted cell apoptosis, and restrained the acquired resistance to EGFR-TKIs. KEGG pathway analysis showed that the hippo/YAP signaling pathway was activated in PC9GR cells relative to PC9 cells, and the YAP expression was inhibited by pitavastatin administration. | ||||||||||||
| Key Molecule: Oncogenic epidermal growth factor receptor (EGFR) | [60] | ||||||||||||
| Metabolic Type | Nucleic acid metabolism | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Mutation | . |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |||||||||
| Experiment for Molecule Alteration |
CE-TOF/MS | ||||||||||||
| Experiment for Drug Resistance |
Cell proliferation assay | ||||||||||||
| Mechanism Description | It is confirmed that purine metabolism catalyzed by HPRT1 promotes the proliferation of EGFR-mutant LUAD in vitro and in vivo. Furthermore, the study of the mechanism shows that HIF-1alpha transcriptionally regulates HPRT1 to accelerate purine nucleotides synthesis to promote cell proliferation and tumorigenesis. Finally, inhibition of HPRT1 coupled with EGFR-TKIs significantly inhibits the tumor growth of EGFR-mutant LUAD | ||||||||||||
| Key Molecule: Oncogenic epidermal growth factor receptor (EGFR) | [60] | ||||||||||||
| Metabolic Type | Nucleic acid metabolism | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Mutation | . |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | PC-9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| Experiment for Molecule Alteration |
CE-TOF/MS | ||||||||||||
| Experiment for Drug Resistance |
Cell proliferation assay | ||||||||||||
| Mechanism Description | It is confirmed that purine metabolism catalyzed by HPRT1 promotes the proliferation of EGFR-mutant LUAD in vitro and in vivo. Furthermore, the study of the mechanism shows that HIF-1alpha transcriptionally regulates HPRT1 to accelerate purine nucleotides synthesis to promote cell proliferation and tumorigenesis. Finally, inhibition of HPRT2 coupled with EGFR-TKIs significantly inhibits the tumor growth of EGFR-mutant LUAD | ||||||||||||
| Key Molecule: Oncogenic epidermal growth factor receptor (EGFR) | [60] | ||||||||||||
| Metabolic Type | Nucleic acid metabolism | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Mutation | . |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | H3255 cells | Lung | Homo sapiens (Human) | CVCL_6831 | |||||||||
| Experiment for Molecule Alteration |
CE-TOF/MS | ||||||||||||
| Experiment for Drug Resistance |
Cell proliferation assay | ||||||||||||
| Mechanism Description | It is confirmed that purine metabolism catalyzed by HPRT1 promotes the proliferation of EGFR-mutant LUAD in vitro and in vivo. Furthermore, the study of the mechanism shows that HIF-1alpha transcriptionally regulates HPRT1 to accelerate purine nucleotides synthesis to promote cell proliferation and tumorigenesis. Finally, inhibition of HPRT3 coupled with EGFR-TKIs significantly inhibits the tumor growth of EGFR-mutant LUAD | ||||||||||||
| Key Molecule: Oncogenic epidermal growth factor receptor (EGFR) | [60] | ||||||||||||
| Metabolic Type | Nucleic acid metabolism | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Mutation | . |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | H1975 cells | Lung | Homo sapiens (Human) | CVCL_B0JT | |||||||||
| Experiment for Molecule Alteration |
CE-TOF/MS | ||||||||||||
| Experiment for Drug Resistance |
Cell proliferation assay | ||||||||||||
| Mechanism Description | It is confirmed that purine metabolism catalyzed by HPRT1 promotes the proliferation of EGFR-mutant LUAD in vitro and in vivo. Furthermore, the study of the mechanism shows that HIF-1alpha transcriptionally regulates HPRT1 to accelerate purine nucleotides synthesis to promote cell proliferation and tumorigenesis. Finally, inhibition of HPRT4 coupled with EGFR-TKIs significantly inhibits the tumor growth of EGFR-mutant LUAD | ||||||||||||
| Key Molecule: Oncogenic epidermal growth factor receptor (EGFR) | [60] | ||||||||||||
| Metabolic Type | Nucleic acid metabolism | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Mutation | . |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vivo Model | Male nude mice | Mice | |||||||||||
| Experiment for Molecule Alteration |
CE-TOF/MS | ||||||||||||
| Experiment for Drug Resistance |
Tumor volume assay | ||||||||||||
| Mechanism Description | It is confirmed that purine metabolism catalyzed by HPRT1 promotes the proliferation of EGFR-mutant LUAD in vitro and in vivo. Furthermore, the study of the mechanism shows that HIF-1alpha transcriptionally regulates HPRT1 to accelerate purine nucleotides synthesis to promote cell proliferation and tumorigenesis. Finally, inhibition of HPRT5 coupled with EGFR-TKIs significantly inhibits the tumor growth of EGFR-mutant LUAD | ||||||||||||
| Key Molecule: Cytochrome c oxidase subunit 6B2 (COX6B2) | [15] | ||||||||||||
| Metabolic Type | Mitochondrial metabolism | ||||||||||||
| Resistant Disease | Non-small cell lung carcinoma [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vivo Model | Nude mice , with fresh tissue from patient | Mice | |||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
Tumor volume assay | ||||||||||||
| Mechanism Description | Furthermore, we revealed that targeting IGF2BP3 can markedly enhance the sensitivity of TKIs in NSCLC and this effect was strongly dependent on the coordinated induction of COX6B2, a key downstream target of IGF2BP3 in mitochondrial OXPHOS energy production. Overall, our study revealed a novel mechanism of TKI resistance involved in IGF2BP3-dependent cross-talk between epigenetic modifications and metabolic reprogramming through the IGF2BP3-COX6B2 axis in NSCLC. | ||||||||||||
| Key Molecule: Cytochrome c oxidase subunit 6B2 (COX6B2) | [15] | ||||||||||||
| Metabolic Type | Mitochondrial metabolism | ||||||||||||
| Resistant Disease | Non-small cell lung carcinoma [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vivo Model | Nude mice , with PC-9/GR cell lines | Mice | |||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
Tumor volume assay | ||||||||||||
| Mechanism Description | Furthermore, we revealed that targeting IGF2BP3 can markedly enhance the sensitivity of TKIs in NSCLC and this effect was strongly dependent on the coordinated induction of COX6B2, a key downstream target of IGF2BP3 in mitochondrial OXPHOS energy production. Overall, our study revealed a novel mechanism of TKI resistance involved in IGF2BP3-dependent cross-talk between epigenetic modifications and metabolic reprogramming through the IGF2BP3-COX6B3 axis in NSCLC. | ||||||||||||
| Key Molecule: . | [61] | ||||||||||||
| Metabolic Type | Glucose metabolism | ||||||||||||
| Resistant Disease | Non-small cell lung carcinoma [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | . | . |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | MAPK signaling pathway | Activation | hsa04010 | ||||||||||
| Insulin signaling pathway | Activation | hsa04910 | |||||||||||
| mTOR signaling pathway | Activation | hsa04150 | |||||||||||
| In Vitro Model | HEK 293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Fibroblast cells | Lung | Homo sapiens (Human) | N.A. | ||||||||||
| Gefitinib-resistant NSCLC cells | Lung | Homo sapiens (Human) | N.A. | ||||||||||
| H1975 parental cells | Lung | Homo sapiens (Human) | CVCL_1511 | ||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Mechanism Description | We found that the combined use of EGFR-TKIs and EGCG significantly reversed the Warburg effect by suppressing glycolysis while boosting mitochondrial respiration, which was accompanied by increased cellular ROS and decreased lactate secretion. The combination effectively activated the AMPK pathway while inhibited both ERK/MAPK and AKT/mTOR pathways, leading to cell cycle arrest and apoptosis, particularly in drug-resistant NSCLC cells. | ||||||||||||
| Key Molecule: . | [61] | ||||||||||||
| Metabolic Type | Glucose metabolism | ||||||||||||
| Resistant Disease | Non-small cell lung carcinoma [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | . | . |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | MAPK signaling pathway | Activation | hsa04010 | ||||||||||
| Insulin signaling pathway | Activation | hsa04910 | |||||||||||
| mTOR signaling pathway | Activation | hsa04150 | |||||||||||
| In Vivo Model | NCI-H1975 xenograft-bearing mice; nude mice bearing AR cell subcutaneous xenografts | Mice | |||||||||||
| Experiment for Drug Resistance |
Tumor volume assay | ||||||||||||
| Mechanism Description | We found that the combined use of EGFR-TKIs and EGCG significantly reversed the Warburg effect by suppressing glycolysis while boosting mitochondrial respiration, which was accompanied by increased cellular ROS and decreased lactate secretion. The combination effectively activated the AMPK pathway while inhibited both ERK/MAPK and AKT/mTOR pathways, leading to cell cycle arrest and apoptosis, particularly in drug-resistant NSCLC cells. | ||||||||||||
|
|
|||||||||||||
| Key Molecule: Programmed cell death protein 4 (PDCD4) | [16] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Non-small cell lung cancer | ||||||||||||
| The Studied Tissue | Lung tissue | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.36E-04 Fold-change: -1.45E-01 Z-score: -3.53E+00 |
||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | ||||||||||
| Cell proliferation | Activation | hsa05200 | |||||||||||
| PI3K/AKT signaling pathway | Activation | hsa04151 | |||||||||||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| PC9R cells | Lung | Homo sapiens (Human) | CVCL_D778 | ||||||||||
| In Vivo Model | Nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Mechanism Description | miR-21 overexpression is associated with the acquired resistance of EGFR-TkI in NSCLC, which might be caused by miR-21's function of activating PI3k/AkT pathway through inhibiting PTEN and PDCD4. | ||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [18] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Non-small cell lung cancer | ||||||||||||
| The Studied Tissue | Lung tissue | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.37E-11 Fold-change: 6.31E-01 Z-score: 7.28E+00 |
||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | ||||||||||
| Cell colony | Inhibition | hsa05200 | |||||||||||
| Cell viability | Inhibition | hsa05200 | |||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Mechanism Description | miR-138 inhibit the protein level of EGFR and reverses gefitinib resistance in lung cancer cells. | ||||||||||||
| Key Molecule: Apoptotic protease-activating factor 1 (APAF1) | [19] | ||||||||||||
| Resistant Disease | Lung cancer [ICD-11: 2C25.5] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Lung cancer | ||||||||||||
| The Studied Tissue | Lung tissue | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.29E-05 Fold-change: -1.11E-01 Z-score: -4.51E+00 |
||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | ||||||||||
| Cell viability | Activation | hsa05200 | |||||||||||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | miR-221 may inhibit apoptosis by down regulating the expression of Apaf-1, so as to induce the resistance of PC-9 cells to gefitinib. | ||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [24] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Non-small cell lung cancer | ||||||||||||
| The Studied Tissue | Lung tissue | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.37E-11 Fold-change: 6.31E-01 Z-score: 7.28E+00 |
||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | ||||||||||
| Cell invasion | Activation | hsa05200 | |||||||||||
| Cell proliferation | Activation | hsa05200 | |||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | ||||||||||
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | ||||||||||
| Sk-MES-1 cells | Lung | Homo sapiens (Human) | CVCL_0630 | ||||||||||
| In Vivo Model | Tumor xenograft in vivo model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | Long Noncoding RNA LINC00460 promotes the gefitinib resistance of nonsmall cell lung cancer through EGFR by sponging miR-769-5p. | ||||||||||||
| Key Molecule: Catenin delta-1 (CTNND1) | [25] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Non-small cell lung cancer | ||||||||||||
| The Studied Tissue | Lung tissue | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.34E-02 Fold-change: -5.18E-02 Z-score: -1.80E+00 |
||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | ||||||||||
| Cell proliferation | Activation | hsa05200 | |||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | ||||||||||
| SPC-A1 cells | Lung | Homo sapiens (Human) | CVCL_6955 | ||||||||||
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | ||||||||||
| In Vivo Model | Nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | ||||||||||||
| Experiment for Drug Resistance |
MTT assay; CCK8 assay; Colony formation assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | MALAT1 could alter chemo-resistance (Cisplatin, Adriamycin, Gefitinib and Paclitaxel) of NSCLC cells by targeting miR-197-3p and regulating p120-ctn expression, which might assist in improvement of chemo-therapies for NSCLC. | ||||||||||||
| Key Molecule: TGF-beta receptor type II (TGFBR2) | [26] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Non-small cell lung cancer | ||||||||||||
| The Studied Tissue | Lung tissue | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.29E-63 Fold-change: -9.29E-01 Z-score: -2.52E+01 |
||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | ||||||||||
| Cell migration | Activation | hsa04670 | |||||||||||
| Cell proliferation | Activation | hsa05200 | |||||||||||
| TGF-beta signaling pathway | Inhibition | hsa04350 | |||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Colony formation assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | LncRNA MBNL1-AS1 restoration could decelerate the occurrence and progression of NSCLC, thereby highlighting the functionality of LncRNA MBNL1-AS1 restoration as a sponge of miR-301b-3p to suppress the proliferation, invasion, drug resistance, and sphere formation of CSC cells in NSCLC via upregulation of TGFBR2. | ||||||||||||
| Key Molecule: Phosphatase and tensin homolog (PTEN) | [32] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Lung cancer | ||||||||||||
| The Studied Tissue | Lung tissue | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.01E-01 Fold-change: -9.25E-03 Z-score: -1.64E+00 |
||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | NSCLC cells | Lung | Homo sapiens (Human) | N.A. | |||||||||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Liquid biopsy; ATP-binding pocket affinity comparison assay | ||||||||||||
| Mechanism Description | Known mechanisms are secondary resistance mutations occurring in the ATP-binding domain (such as T790M and C797S), mutation or amplification of bypass signallings (such as AXL, Hh, ERBb2, CRIPTO, etc), activating mutations in the downstream pathways (PI3k, AkT, MEk, RAF), low levels of mRNA or polymorphisms of the pro-apoptotic protein BIM, induction of a transcription programme for EMT and phenotypical changes, or induction of elevated tumour PD-L1 levels. | ||||||||||||
| Key Molecule: Tyrosine-protein phosphatase non-receptor type 13 (PTPN13) | [34] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Lung cancer | ||||||||||||
| The Studied Tissue | Lung tissue | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.34E-09 Fold-change: -8.21E-02 Z-score: -6.22E+00 |
||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell proliferation | Activation | hsa05200 | ||||||||||
| EGFR signaling pathway | Activation | hsa01521 | |||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| SPC-A1 cells | Lung | Homo sapiens (Human) | CVCL_6955 | ||||||||||
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | ||||||||||
| NCI-H520 cells | Lung | Homo sapiens (Human) | CVCL_1566 | ||||||||||
| H2170 cells | Lung | Homo sapiens (Human) | CVCL_1535 | ||||||||||
| SW900 cells | Lung | Homo sapiens (Human) | CVCL_1731 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Mechanism Description | miR-26a desensitizes non-small cell lung cancer cells to tyrosine kinase inhibitors by targeting and reducing the level of PTPN1. | ||||||||||||
| Key Molecule: Growth arrest-specific protein 7 (GAS7) | [35] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Lung cancer | ||||||||||||
| The Studied Tissue | Lung tissue | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.21E-48 Fold-change: -1.41E-01 Z-score: -1.69E+01 |
||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | AKT/ERK signaling pathway | Regulation | N.A. | ||||||||||
| In Vitro Model | H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | |||||||||
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | ||||||||||
| PC9GR cells | Lung | Homo sapiens (Human) | CVCL_V337 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay | ||||||||||||
| Mechanism Description | Down-regulation of GAS7 expression could antagonize gefitinib re-sensitivity in PC9GR mediated by knockdown of miR181a via AkT/ERk pathways and epithelial-to-mesenchymal transition markers. | ||||||||||||
| Key Molecule: Caspase-1 (CASP1) | [12] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Lung cancer | ||||||||||||
| The Studied Tissue | Lung tissue | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.09E-84 Fold-change: -1.65E-01 Z-score: -2.30E+01 |
||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | ||||||||||
| Cell viability | Inhibition | hsa05200 | |||||||||||
| miR377/CASP1 signaling pathway | Regulation | N.A. | |||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | SNHG5 overexpression sensitized gefitinib resistant LAD cells to gefitinib treatment; Overexpression of SNHG5 suppressed the expression of miR-377; Overexpression of miR-377 suppressed the expression of CASP1 in PC9 cells; knockdown of CASP1 in SNHG5-overexpressed PC9GR cells abolished their gefitinib resistance. | ||||||||||||
| Key Molecule: Cellular tumor antigen p53 (TP53) | [62] | ||||||||||||
| Resistant Disease | Lung cancer [ICD-11: 2C25.5] | ||||||||||||
| Molecule Alteration | Missense mutation | p.Y163C |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.05 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.65 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
S
S
S
S
S
S
V
V
P
P
S
S
100
|
Q
Q
K
K
T
T
Y
Y
Q
Q
G
G
S
S
Y
Y
G
G
F
F
110
|
R
R
L
L
G
G
F
F
L
L
H
H
S
S
G
G
T
T
A
A
120
|
K
K
S
S
V
V
T
T
C
C
T
T
Y
Y
S
S
P
P
A
A
130
|
L
L
N
N
K
K
M
L
F
F
C
C
Q
Q
L
L
A
A
K
K
140
|
T
T
C
C
P
P
V
V
Q
Q
L
L
W
W
V
V
D
D
S
S
150
|
T
T
P
P
P
P
P
P
G
G
T
T
R
R
V
V
R
R
A
A
160
|
M
M
A
A
I
I
Y
C
K
K
Q
Q
S
S
Q
Q
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
R
C
C
P
P
H
H
H
H
180
|
E
E
R
R
C
C
S
S
D
D
S
S
D
D
G
G
L
L
A
A
190
|
P
P
P
P
Q
Q
H
H
L
L
I
I
R
R
V
V
E
E
G
G
200
|
N
N
L
L
R
R
V
A
E
E
Y
Y
L
L
D
D
D
D
R
R
210
|
N
N
T
T
F
F
R
R
H
H
S
S
V
V
V
V
V
V
P
P
220
|
Y
Y
E
E
P
P
P
P
E
E
V
V
G
G
S
S
D
D
C
C
230
|
T
T
T
T
I
I
H
H
Y
Y
N
N
Y
Y
M
M
C
C
N
Y
240
|
S
S
S
S
C
C
M
M
G
G
G
G
M
M
N
N
R
R
R
R
250
|
P
P
I
I
L
L
T
T
I
I
I
I
T
T
L
L
E
E
D
D
260
|
S
S
S
S
G
G
N
N
L
L
L
L
G
G
R
R
N
D
S
S
270
|
F
F
E
E
V
V
R
R
V
V
C
C
A
A
C
C
P
P
G
G
280
|
R
R
D
D
R
R
R
R
T
T
E
E
E
E
E
E
N
N
L
L
290
|
R
R
K
K
K
K
G
G
E
E
P
P
H
H
H
H
E
E
L
L
300
|
P
P
P
P
G
G
S
S
T
T
K
K
R
R
A
A
L
L
P
P
310
|
N
N
N
N
T
T
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | AXLK signaling pathway | Activation | hsa01521 | ||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Circulating-free DNA assay; Whole exome sequencing assay | ||||||||||||
| Mechanism Description | Quantification of allele fractions in plasma identified increased representation of mutant alleles in association with emergence of therapy resistance. | ||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [62] | ||||||||||||
| Resistant Disease | Lung cancer [ICD-11: 2C25.5] | ||||||||||||
| Molecule Alteration | Missense mutation | p.T790M |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 3.10 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.05 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
S
G
G
E
E
A
A
P
P
700
|
N
N
Q
Q
A
A
L
L
L
L
R
R
I
I
L
L
K
K
E
E
710
|
T
T
E
E
F
F
K
K
K
K
I
I
K
K
V
V
L
L
G
G
720
|
S
S
G
G
A
A
F
F
G
G
T
T
V
V
Y
Y
K
K
G
G
730
|
L
L
W
W
I
I
P
P
E
E
G
G
E
E
K
K
V
V
K
K
740
|
I
I
P
P
V
V
A
A
I
I
K
K
E
E
L
L
R
R
E
E
750
|
A
A
T
T
S
S
P
P
K
K
A
A
N
N
K
K
E
E
I
I
760
|
L
L
D
D
E
E
A
A
Y
Y
V
V
M
M
A
A
S
S
V
V
770
|
D
D
N
N
P
P
H
H
V
V
C
C
R
R
L
L
L
L
G
G
780
|
I
I
C
C
L
L
T
T
S
S
T
T
V
V
Q
Q
L
L
I
I
790
|
T
M
Q
Q
L
L
M
M
P
P
F
F
G
G
C
C
L
L
L
L
800
|
D
D
Y
Y
V
V
R
R
E
E
H
H
K
K
D
D
N
N
I
I
810
|
G
G
S
S
Q
Q
Y
Y
L
L
L
L
N
N
W
W
C
C
V
V
820
|
Q
Q
I
I
A
A
K
K
G
G
M
M
N
N
Y
Y
L
L
E
E
830
|
D
D
R
R
R
R
L
L
V
V
H
H
R
R
D
D
L
L
A
A
840
|
A
A
R
R
N
N
V
V
L
L
V
V
K
K
T
T
P
P
Q
Q
850
|
H
H
V
V
K
K
I
I
T
T
D
D
F
F
G
G
L
L
A
A
860
|
K
K
L
L
L
L
G
G
A
A
E
E
E
E
K
K
E
E
Y
Y
870
|
H
H
A
A
E
E
G
G
G
G
K
K
V
V
P
P
I
I
K
K
880
|
W
W
M
M
A
A
L
L
E
E
S
S
I
I
L
L
H
H
R
R
890
|
I
I
Y
Y
T
T
H
H
Q
Q
S
S
D
D
V
V
W
W
S
S
900
|
Y
Y
G
G
V
V
T
T
V
V
W
W
E
E
L
L
M
M
T
T
910
|
F
F
G
G
S
S
K
K
P
P
Y
Y
D
D
G
G
I
I
P
P
920
|
A
A
S
S
E
E
I
I
S
S
S
S
I
I
L
L
E
E
K
K
930
|
G
G
E
E
R
R
L
L
P
P
Q
Q
P
P
P
P
I
I
C
C
940
|
T
T
I
I
D
D
V
V
Y
Y
M
M
I
I
M
M
V
V
K
K
950
|
C
C
W
W
M
M
I
I
D
D
A
A
D
D
S
S
R
R
P
P
960
|
K
K
F
F
R
R
E
E
L
L
I
I
I
I
E
E
F
F
S
S
970
|
K
K
M
M
A
A
R
R
D
D
P
P
Q
Q
R
R
Y
Y
L
L
980
|
V
V
I
I
Q
Q
G
G
D
D
E
E
R
R
M
M
H
H
L
L
990
|
P
P
S
S
P
P
T
T
D
D
S
S
N
N
F
F
Y
Y
R
R
1000
|
A
A
L
L
M
M
D
D
E
E
E
E
D
D
M
M
D
D
D
D
1010
|
V
V
V
V
D
D
A
A
D
D
E
E
Y
Y
L
L
I
I
P
P
1020
|
Q
Q
Q
Q
G
G
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | AKT signaling pathway | Activation | hsa04151 | ||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Circulating-free DNA assay; Whole exome sequencing assay | ||||||||||||
| Mechanism Description | Quantification of allele fractions in plasma identified increased representation of mutant alleles in association with emergence of therapy resistance. | ||||||||||||
| Key Molecule: phosphoinositide-3-dependent protein kinase 1 (PDPK1) | [52] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | Cell invasion | Activation | hsa05200 | ||||||||||
| Cell proliferation | Activation | hsa05200 | |||||||||||
| miR1183/PDPk1 signaling pathway | Activation | hsa05206 | |||||||||||
| In Vitro Model | H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | |||||||||
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | ||||||||||
| HCC827 cells | Lung | Homo sapiens (Human) | CVCL_2063 | ||||||||||
| NCI-H358 cells | Lung | Homo sapiens (Human) | CVCL_1559 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR; Luciferase reporter assay | ||||||||||||
| Experiment for Drug Resistance |
MTT assay; Colony formation assay | ||||||||||||
| Mechanism Description | Hsa_circ_0004015 formed by CDk14 gene inhibited the expression of miR-1183, which could disinhibit the PDPk1 expression from miR-1183, ultimately resulted in the promotion of cell proliferation, invasion, and TkI inhibitor drug resistance of NSCLC cells. | ||||||||||||
| Key Molecule: RAC serine/threonine-protein kinase (AKT) | [30] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Phosphorylation | Up-regulation |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | ||||||||||
| Cell invasion | Activation | hsa05200 | |||||||||||
| Cell viability | Activation | hsa05200 | |||||||||||
| PI3K/AKT signaling pathway | Activation | hsa04151 | |||||||||||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| In Vivo Model | Nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | LINC00665 Induces Acquired Resistance to Gefitinib through Recruiting EZH2 and Activating PI3k/AkT Pathway in NSCLC. | ||||||||||||
| Key Molecule: Apoptosis regulator Bcl-2 (BCL2) | [54] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | ||||||||||
| Cell viability | Activation | hsa05200 | |||||||||||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Flow cytometry assay | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | miR-214 level was upregulated in gefitinib-resistant PC-9GR cells and their derived exosomes while anti-apoptotic protein of bcl-2 is uoregulated. | ||||||||||||
| Key Molecule: Zinc finger protein GLI1 (GLI1) | [55] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | ||||||||||
| Cell viability | Activation | hsa05200 | |||||||||||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | The inhibition of miR-873 increased gefitinib resistance of NSCLC cells via the upregulation of GLI1. | ||||||||||||
| Key Molecule: Transcriptional coactivator YAP1 (YAP1) | [56] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | ||||||||||
| Cell viability | Activation | hsa05200 | |||||||||||
| miR630/YAP1/ERK signaling pathway | Regulation | N.A. | |||||||||||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| PC9GR cells | Lung | Homo sapiens (Human) | CVCL_V337 | ||||||||||
| CL97 cells | Lung | Homo sapiens (Human) | CVCL_N826 | ||||||||||
| In Vivo Model | Nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | Low miR-630 and high YAP1 mRNA levels are associated with unfavorable response to TkI therapy in lung adenocarcinoma patients. | ||||||||||||
| Key Molecule: Ras-related C3 botulinum toxin substrate 1 (RAC1) | [57] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | ||||||||||
| Cell invasion | Activation | hsa05200 | |||||||||||
| Cell viability | Activation | hsa05200 | |||||||||||
| PI3K/AKT signaling pathway | Activation | hsa04151 | |||||||||||
| In Vitro Model | NCI-H1650 cells | Lung | Homo sapiens (Human) | CVCL_1483 | |||||||||
| NCI-H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay; Transwell assay | ||||||||||||
| Mechanism Description | miR-135a promoted cell growth and metastasis and activated the PI3k/AkT signaling pathway via a RAC1-dependent manner. | ||||||||||||
| Key Molecule: Phosphatase and tensin homolog (PTEN) | [58] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | AKT/ERK signaling pathway | Activation | hsa04010 | ||||||||||
| Cell apoptosis | Inhibition | hsa04210 | |||||||||||
| Cell invasion | Activation | hsa05200 | |||||||||||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | miR-21 was up-regulated concomitantly to down-regulation of Pten in pc-9/GR cells in comparison with pc-9 cells. Moreover, over-expression of miR-21 significantly decreased gefitinib sensitivity by down-regulating Pten expression and activating Akt and ERk pathways in pc-9 cells, while miR-21 knockdown dramatically restored gefitinib sensitivity of pc-9/GR cells by up-regulation of Pten expression and inactivation of AkT and ERk pathways, in vivo and in vitro. | ||||||||||||
| Key Molecule: Phosphatase and tensin homolog (PTEN) | [16] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | ||||||||||
| Cell proliferation | Activation | hsa05200 | |||||||||||
| PI3K/AKT signaling pathway | Activation | hsa04151 | |||||||||||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| PC9R cells | Lung | Homo sapiens (Human) | CVCL_D778 | ||||||||||
| In Vivo Model | Nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Mechanism Description | miR-21 overexpression is associated with the acquired resistance of EGFR-TkI in NSCLC, which might be caused by miR-21's function of activating PI3k/AkT pathway through inhibiting PTEN and PDCD4. | ||||||||||||
| Key Molecule: Phosphatase and tensin homolog (PTEN) | [32] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Structural variation | Copy number loss |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | NSCLC cells | Lung | Homo sapiens (Human) | N.A. | |||||||||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Liquid biopsy; ATP-binding pocket affinity comparison assay | ||||||||||||
| Mechanism Description | Known mechanisms are secondary resistance mutations occurring in the ATP-binding domain (such as T790M and C797S), mutation or amplification of bypass signallings (such as AXL, Hh, ERBb2, CRIPTO, etc), activating mutations in the downstream pathways (PI3k, AkT, MEk, RAF), low levels of mRNA or polymorphisms of the pro-apoptotic protein BIM, induction of a transcription programme for EMT and phenotypical changes, or induction of elevated tumour PD-L1 levels. | ||||||||||||
| Key Molecule: RAC-alpha serine/threonine-protein kinase (AKT1) | [32] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Mutation | . |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | ERK/MAPKsignaling pathway | Activation | hsa04210 | ||||||||||
| In Vitro Model | NSCLC cells | Lung | Homo sapiens (Human) | N.A. | |||||||||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Liquid biopsy; ATP-binding pocket affinity comparison assay | ||||||||||||
| Mechanism Description | Known mechanisms are secondary resistance mutations occurring in the ATP-binding domain (such as T790M and C797S), mutation or amplification of bypass signallings (such as AXL, Hh, ERBb2, CRIPTO, etc), activating mutations in the downstream pathways (PI3k, AkT, MEk, RAF), low levels of mRNA or polymorphisms of the pro-apoptotic protein BIM, induction of a transcription programme for EMT and phenotypical changes, or induction of elevated tumour PD-L1 levels. | ||||||||||||
| Key Molecule: DNA-binding factor KBF1 (p105) (NFKB1) | [62] | ||||||||||||
| Resistant Disease | Lung cancer [ICD-11: 2C25.5] | ||||||||||||
| Molecule Alteration | Missense mutation | p.G489V |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | Angiogenic potential | Inhibition | hsa04370 | ||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Circulating-free DNA assay; Whole exome sequencing assay | ||||||||||||
| Mechanism Description | Quantification of allele fractions in plasma identified increased representation of mutant alleles in association with emergence of therapy resistance. | ||||||||||||
| Key Molecule: Hepatocyte growth factor receptor (MET) | [48] | ||||||||||||
| Resistant Disease | EGFR-mutant non-small cell lung cancer [ICD-11: 2C25.7] | ||||||||||||
| Molecule Alteration | Structural variation | Amplification |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
MGB SNP detection kit assay; Mutation Detection assay | ||||||||||||
| Experiment for Drug Resistance |
Digital PCR assay | ||||||||||||
| Mechanism Description | Resistance mechanisms to EGFR-TkI therapy in EGFR-mutated NSCLC include secondary EGFR T790M mutation, c-Met amplification, PIk3CA mutation, and transformation to small-cell lung cancer. | ||||||||||||
| Key Molecule: PI3-kinase alpha (PIK3CA) | [48] | ||||||||||||
| Resistant Disease | EGFR-mutant non-small cell lung cancer [ICD-11: 2C25.7] | ||||||||||||
| Molecule Alteration | Mutation | . |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
MGB SNP detection kit assay; Mutation Detection assay | ||||||||||||
| Experiment for Drug Resistance |
Digital PCR assay | ||||||||||||
| Mechanism Description | Resistance mechanisms to EGFR-TkI therapy in EGFR-mutated NSCLC include secondary EGFR T790M mutation, c-Met amplification, PIk3CA mutation, and transformation to small-cell lung cancer. | ||||||||||||
| Key Molecule: PI3-kinase alpha (PIK3CA) | [49] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Mutation | . |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
Low throughput experiment assay | ||||||||||||
| Experiment for Drug Resistance |
Progression-free survival assay | ||||||||||||
| Mechanism Description | Acquired resistance can occur through failure of drug delivery to the target, as in isolated central nervous system (CNS) progression, or by selection of biological variants during TkI exposure. | ||||||||||||
| Key Molecule: Hepatocyte growth factor receptor (MET) | [49] | ||||||||||||
| Resistant Disease | EGFR-mutant non-small cell lung cancer [ICD-11: 2C25.7] | ||||||||||||
| Molecule Alteration | Structural variation | Copy number gain |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
Low throughput experiment assay | ||||||||||||
| Experiment for Drug Resistance |
Progression-free survival assay | ||||||||||||
| Mechanism Description | Acquired resistance can occur through failure of drug delivery to the target, as in isolated central nervous system (CNS) progression, or by selection of biological variants during TkI exposure. | ||||||||||||
| Key Molecule: Tyrosine-protein kinase ITK/TSK (ITK) | [49] | ||||||||||||
| Resistant Disease | EGFR-mutant non-small cell lung cancer [ICD-11: 2C25.7] | ||||||||||||
| Molecule Alteration | Mutation | . |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
Low throughput experiment assay | ||||||||||||
| Experiment for Drug Resistance |
Progression-free survival assay | ||||||||||||
| Mechanism Description | Acquired resistance can occur through failure of drug delivery to the target, as in isolated central nervous system (CNS) progression, or by selection of biological variants during TkI exposure. | ||||||||||||
| Key Molecule: Receptor tyrosine-protein kinase erbB-2 (ERBB2) | [49] | ||||||||||||
| Resistant Disease | EGFR-mutant non-small cell lung cancer [ICD-11: 2C25.7] | ||||||||||||
| Molecule Alteration | Structural variation | Copy number gain |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
Low throughput experiment assay | ||||||||||||
| Experiment for Drug Resistance |
Progression-free survival assay | ||||||||||||
| Mechanism Description | Acquired resistance can occur through failure of drug delivery to the target, as in isolated central nervous system (CNS) progression, or by selection of biological variants during TkI exposure. | ||||||||||||
| Key Molecule: Serine/threonine-protein kinase B-raf (BRAF) | [49] | ||||||||||||
| Resistant Disease | EGFR-mutant non-small cell lung cancer [ICD-11: 2C25.7] | ||||||||||||
| Molecule Alteration | Mutation | . |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
Low throughput experiment assay | ||||||||||||
| Experiment for Drug Resistance |
Progression-free survival assay | ||||||||||||
| Mechanism Description | Acquired resistance can occur through failure of drug delivery to the target, as in isolated central nervous system (CNS) progression, or by selection of biological variants during TkI exposure. | ||||||||||||
| Key Molecule: DNA-binding factor KBF1 (p105) (NFKB1) | [62] | ||||||||||||
| Resistant Disease | Lung cancer [ICD-11: 2C25.5] | ||||||||||||
| Molecule Alteration | Missense mutation | p.G489V |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
Circulating-free DNA assay; Whole exome sequencing assay | ||||||||||||
| Mechanism Description | Quantification of allele fractions in plasma identified increased representation of mutant alleles in association with emergence of therapy resistance. | ||||||||||||
| Key Molecule: PI3-kinase alpha (PIK3CA) | [50] | ||||||||||||
| Resistant Disease | EGFR-mutant non-small cell lung cancer [ICD-11: 2C25.7] | ||||||||||||
| Molecule Alteration | Mutation | . |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
Next generation sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Multivariate analysis of overall or disease-free survival assay | ||||||||||||
| Mechanism Description | Quantification of allele fractions in plasma identified increased representation of mutantalleles in association with emergence of therapy resistance. These included an activating mutation in PIk3CA. | ||||||||||||
| Key Molecule: Hepatocyte growth factor receptor (MET) | [63] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Expression | Activation |
|||||||||||
| Cell Pathway Regulation | HGF-Met signaling pathway | Regulation | N.A. | ||||||||||
| Mechanism Description | Aberrant Met amplification and HGF-Met signaling pathway activation have been proven to be the main mechanism of acquired resistance of EGFR inhibition by small molecules, such as erlotinib and gefitinib. | ||||||||||||
| Key Molecule: H19, imprinted maternally expressed transcript (H19) | [64] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell autophagy | Inhibition | hsa04140 | ||||||||||
| In Vitro Model | PC9/GR cells | N.A. | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Apoptosis assay | ||||||||||||
| Mechanism Description | Our findings elucidate that the resistance to gefitinib is intricately linked with the dysregulation of autophagy and the overexpression of lncRNA H19. The synergistic administration of beta-elemene and gefitinib markedly attenuated the proliferative capacity of resistant cells, expedited apoptotic processes, and inhibited the in vivo proliferation of lung cancer. Notably, beta-elemene profoundly diminished the expression of lncRNA H19 and curtailed autophagic activity in resistant cells, thereby bolstering their responsiveness to gefitinib. | ||||||||||||
| Key Molecule: H19, imprinted maternally expressed transcript (H19) | [64] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Expression | Loss |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell autophagy | Inhibition | hsa04140 | ||||||||||
| In Vitro Model | HCC827/GR cells | N.A. | Homo sapiens (Human) | CVCL_E7R9 | |||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Apoptosis assay | ||||||||||||
| Mechanism Description | Our findings elucidate that the resistance to gefitinib is intricately linked with the dysregulation of autophagy and the overexpression of lncRNA H19. The synergistic administration of beta-elemene and gefitinib markedly attenuated the proliferative capacity of resistant cells, expedited apoptotic processes, and inhibited the in vivo proliferation of lung cancer. Notably, beta-elemene profoundly diminished the expression of lncRNA H19 and curtailed autophagic activity in resistant cells, thereby bolstering their responsiveness to gefitinib. | ||||||||||||
| Key Molecule: Serine/threonine-protein phosphatase 2B catalytic subunit beta isoform (PPP3CB) | [65] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Ca2+/calcineurin/MEK/ERK1/2 signaling pathway | Regulation | N.A. | ||||||||||
| In Vitro Model | PC9/DR cells | N.A. | Homo sapiens (Human) | N.A. | |||||||||
| PC9/GR cells | N.A. | Homo sapiens (Human) | N.A. | ||||||||||
| PC9/OR cells | N.A. | Homo sapiens (Human) | N.A. | ||||||||||
| Experiment for Molecule Alteration |
Western blot assay | ||||||||||||
| Experiment for Drug Resistance |
MTS assay; Flow cytometric assay; Colony formation assay | ||||||||||||
| Mechanism Description | Here, we show that a?PPP3CB?transcript that encodes full-length catalytic subunit 2B of calcineurin accumulates in EGFR-mutant NSCLC cells with acquired resistance against different EGFR TKIs and in post-progression biopsies of NSCLC patients treated with EGFR TKIs. Neutralization of?PPP3CB?by siRNA or inactivation of calcineurin by cyclosporin A induces apoptosis in resistant cells treated with EGFR TKIs. Mechanistically, EGFR TKIs increase the cytosolic level of calcium and trigger activation of a calcineurin/MEK/ERK pathway that prevents apoptosis. Combining EGFR, calcineurin, and MEK inhibitors overcomes resistance to EGFR TKI in both in vitro and in vivo models. Our results identify PPP3CB overexpression as a new mechanism of acquired resistance to EGFR TKIs, and provide a promising therapeutic approach for NSCLC patients that progress under TKI treatment. | ||||||||||||
| Key Molecule: Programmed cell death 1 ligand 1 (PD-L1) | [66] | ||||||||||||
| Resistant Disease | egfr-mutant luad [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell autophagy | Inhibition | hsa04140 | ||||||||||
| In Vitro Model | PC-9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| Experiment for Molecule Alteration |
Western blot assay | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay | ||||||||||||
| Mechanism Description | Our results suggested that PD-L1 might play a role in promoting autophagy and inhibiting apoptosis, which was responsible for the generation of primary resistance to EGFR-TKIs, and the combination therapy exerted a synergistic antitumor effect by inhibiting the activation of the MAPK/ERK signaling pathway. | ||||||||||||
| Key Molecule: Programmed cell death 1 ligand 1 (PD-L1) | [66] | ||||||||||||
| Resistant Disease | egfr-mutant luad [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell autophagy | Inhibition | hsa04140 | ||||||||||
| In Vitro Model | HCC827 cells | Lung | Homo sapiens (Human) | CVCL_2063 | |||||||||
| Experiment for Molecule Alteration |
Western blot assay | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay | ||||||||||||
| Mechanism Description | Our results suggested that PD-L1 might play a role in promoting autophagy and inhibiting apoptosis, which was responsible for the generation of primary resistance to EGFR-TKIs, and the combination therapy exerted a synergistic antitumor effect by inhibiting the activation of the MAPK/ERK signaling pathway. | ||||||||||||
|
|
|||||||||||||
| Key Molecule: Long non-protein coding RNA 460 (LINC00460) | [24] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Lung adenocarcinoma | ||||||||||||
| The Studied Tissue | Lung | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.91E-10 Fold-change: 4.46E+00 Z-score: 6.44E+00 |
||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | ||||||||||
| Cell invasion | Activation | hsa05200 | |||||||||||
| Cell proliferation | Activation | hsa05200 | |||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | ||||||||||
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | ||||||||||
| Sk-MES-1 cells | Lung | Homo sapiens (Human) | CVCL_0630 | ||||||||||
| In Vivo Model | Tumor xenograft in vivo model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | Long Noncoding RNA LINC00460 promotes the gefitinib resistance of nonsmall cell lung cancer through EGFR by sponging miR-769-5p. | ||||||||||||
| Key Molecule: MIR31 host gene (MIR31HG) | [29] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Lung adenocarcinoma | ||||||||||||
| The Studied Tissue | Lung | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.75E-14 Fold-change: 2.93E+00 Z-score: 7.81E+00 |
||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | EGFR/PI3K/AKT signaling pathway | Regulation | N.A. | ||||||||||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| PC9R cells | Lung | Homo sapiens (Human) | CVCL_D778 | ||||||||||
| Experiment for Molecule Alteration |
RT-qPCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Annexin V-FITC/PI double staining assay | ||||||||||||
| Mechanism Description | Increased MIR31HG LncRNA expression increases gefitinib resistance in non-small cell lung cancer cell lines through the EGFR/PI3k/AkT signaling pathway. | ||||||||||||
| Key Molecule: Long non-protein coding RNA 665 (LINC00665) | [30] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Differential expression of the molecule in resistant disease | |||||||||||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | ||||||||||||
| The Specified Disease | Lung adenocarcinoma | ||||||||||||
| The Studied Tissue | Lung | ||||||||||||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.27E-47 Fold-change: 1.83E+00 Z-score: 1.60E+01 |
||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | ||||||||||
| Cell colony | Activation | hsa05200 | |||||||||||
| Cell invasion | Activation | hsa05200 | |||||||||||
| Cell viability | Activation | hsa05200 | |||||||||||
| PI3K/AKT signaling pathway | Activation | hsa04151 | |||||||||||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| In Vivo Model | Nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | LINC00665 Induces Acquired Resistance to Gefitinib through Recruiting EZH2 and Activating PI3k/AkT Pathway in NSCLC. | ||||||||||||
| Key Molecule: hsa-mir-200a | [1] | ||||||||||||
| Resistant Disease | Lung cancer [ICD-11: 2C25.5] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell proliferation | Activation | hsa05200 | ||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
MTT assay; Colony formation assay | ||||||||||||
| Mechanism Description | LncRNA MALAT1 promoted the proliferation and gefitinib resistance of lung cancer cells by sponging miR-200a, which regulates expression of ZEB1 in the A549 cells. | ||||||||||||
| Key Molecule: Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) | [1] | ||||||||||||
| Resistant Disease | Lung cancer [ICD-11: 2C25.5] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell proliferation | Activation | hsa05200 | ||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| Experiment for Molecule Alteration |
RT-PCR | ||||||||||||
| Experiment for Drug Resistance |
MTT assay; Colony formation assay | ||||||||||||
| Mechanism Description | LncRNA MALAT1 promoted the proliferation and gefitinib resistance of lung cancer cells by sponging miR-200a, which regulates expression of ZEB1 in the A549 cells. | ||||||||||||
| Key Molecule: MBNL1 antisense RNA 1 (MBNL1-AS1) | [26] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell invasion | Activation | hsa05200 | ||||||||||
| Cell migration | Activation | hsa04670 | |||||||||||
| Cell proliferation | Activation | hsa05200 | |||||||||||
| TGF-beta signaling pathway | Inhibition | hsa04350 | |||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Colony formation assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | LncRNA MBNL1-AS1 restoration could decelerate the occurrence and progression of NSCLC, thereby highlighting the functionality of LncRNA MBNL1-AS1 restoration as a sponge of miR-301b-3p to suppress the proliferation, invasion, drug resistance, and sphere formation of CSC cells in NSCLC via upregulation of TGFBR2. | ||||||||||||
| Key Molecule: hsa-miR-301b-3p | [26] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | ||||||||||
| Cell migration | Activation | hsa04670 | |||||||||||
| Cell proliferation | Activation | hsa05200 | |||||||||||
| TGF-beta signaling pathway | Inhibition | hsa04350 | |||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Colony formation assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | LncRNA MBNL1-AS1 restoration could decelerate the occurrence and progression of NSCLC, thereby highlighting the functionality of LncRNA MBNL1-AS1 restoration as a sponge of miR-301b-3p to suppress the proliferation, invasion, drug resistance, and sphere formation of CSC cells in NSCLC via upregulation of TGFBR2. | ||||||||||||
| Key Molecule: Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) | [25] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | ||||||||||
| Cell proliferation | Activation | hsa05200 | |||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | ||||||||||
| SPC-A1 cells | Lung | Homo sapiens (Human) | CVCL_6955 | ||||||||||
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | ||||||||||
| In Vivo Model | Nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
MTT assay; CCK8 assay; Colony formation assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | MALAT1 could alter chemo-resistance (Cisplatin, Adriamycin, Gefitinib and Paclitaxel) of NSCLC cells by targeting miR-197-3p and regulating p120-ctn expression, which might assist in improvement of chemo-therapies for NSCLC. | ||||||||||||
| Key Molecule: hsa-miR-197-3p | [25] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | ||||||||||
| Cell proliferation | Activation | hsa05200 | |||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | ||||||||||
| SPC-A1 cells | Lung | Homo sapiens (Human) | CVCL_6955 | ||||||||||
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | ||||||||||
| In Vivo Model | Nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
MTT assay; CCK8 assay; Colony formation assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | MALAT1 could alter chemo-resistance (Cisplatin, Adriamycin, Gefitinib and Paclitaxel) of NSCLC cells by targeting miR-197-3p and regulating p120-ctn expression, which might assist in improvement of chemo-therapies for NSCLC. | ||||||||||||
| Key Molecule: hsa-miR-769-5p | [24] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | ||||||||||
| Cell invasion | Activation | hsa05200 | |||||||||||
| Cell viability | Activation | hsa05200 | |||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | ||||||||||
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | ||||||||||
| Sk-MES-1 cells | Lung | Homo sapiens (Human) | CVCL_0630 | ||||||||||
| In Vivo Model | Tumor xenograft in vivo model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | Long Noncoding RNA LINC00460 promotes the gefitinib resistance of nonsmall cell lung cancer through EGFR by sponging miR-769-5p. | ||||||||||||
| Key Molecule: hsa-miR-1183 | [52] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | Cell invasion | Activation | hsa05200 | ||||||||||
| Cell proliferation | Activation | hsa05200 | |||||||||||
| miR1183/PDPk1 signaling pathway | Activation | hsa05206 | |||||||||||
| In Vitro Model | H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | |||||||||
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | ||||||||||
| HCC827 cells | Lung | Homo sapiens (Human) | CVCL_2063 | ||||||||||
| NCI-H358 cells | Lung | Homo sapiens (Human) | CVCL_1559 | ||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
MTT assay; Colony formation assay | ||||||||||||
| Mechanism Description | Hsa_circ_0004015 formed by CDk14 gene inhibited the expression of miR-1183, which could disinhibit the PDPk1 expression from miR-1183, ultimately resulted in the promotion of cell proliferation, invasion, and TkI inhibitor drug resistance of NSCLC cells. | ||||||||||||
| Key Molecule: hsa_circ_0004015 | [52] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | Cell invasion | Activation | hsa05200 | ||||||||||
| Cell proliferation | Activation | hsa05200 | |||||||||||
| miR1183/PDPk1 signaling pathway | Activation | hsa05206 | |||||||||||
| In Vitro Model | H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | |||||||||
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | ||||||||||
| HCC827 cells | Lung | Homo sapiens (Human) | CVCL_2063 | ||||||||||
| NCI-H358 cells | Lung | Homo sapiens (Human) | CVCL_1559 | ||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
MTT assay; Colony formation assay | ||||||||||||
| Mechanism Description | Hsa_circ_0004015 formed by CDk14 gene inhibited the expression of miR-1183, which could disinhibit the PDPk1 expression from miR-1183, ultimately resulted in the promotion of cell proliferation, invasion, and TkI inhibitor drug resistance of NSCLC cells. | ||||||||||||
| Key Molecule: hsa-mir-181a | [35] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | AKT/ERK signaling pathway | Regulation | N.A. | ||||||||||
| In Vitro Model | H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | |||||||||
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | ||||||||||
| PC9GR cells | Lung | Homo sapiens (Human) | CVCL_V337 | ||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay | ||||||||||||
| Mechanism Description | Down-regulation of GAS7 expression could antagonize gefitinib re-sensitivity in PC9GR mediated by knockdown of miR181a via AkT/ERk pathways and epithelial-to-mesenchymal transition markers. | ||||||||||||
| Key Molecule: hsa-let-7b | [53] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | ||||||||||
| Cell proliferation | Activation | hsa05200 | |||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| A549/GR cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| Experiment for Molecule Alteration |
RT-qPCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometric analysis | ||||||||||||
| Mechanism Description | Increased miR17-5p and miR92a expression and decreased let-7b expression can significantly induce proliferation and inhibit apoptosis of lung cancer cells, while reducing lung cancer cell sensitivity to Gefitinib. | ||||||||||||
| Key Molecule: hsa-miR-17-5p | [53] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | ||||||||||
| Cell proliferation | Activation | hsa05200 | |||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| A549/GR cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| Experiment for Molecule Alteration |
RT-qPCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometric analysis | ||||||||||||
| Mechanism Description | Increased miR17-5p and miR92a expression and decreased let-7b expression can significantly induce proliferation and inhibit apoptosis of lung cancer cells, while reducing lung cancer cell sensitivity to Gefitinib. | ||||||||||||
| Key Molecule: hsa-mir-92a | [53] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | ||||||||||
| Cell proliferation | Activation | hsa05200 | |||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| A549/GR cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| Experiment for Molecule Alteration |
RT-qPCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometric analysis | ||||||||||||
| Mechanism Description | Increased miR17-5p and miR92a expression and decreased let-7b expression can significantly induce proliferation and inhibit apoptosis of lung cancer cells, while reducing lung cancer cell sensitivity to Gefitinib. | ||||||||||||
| Key Molecule: hsa-mir-214 | [54] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | ||||||||||
| Cell viability | Activation | hsa05200 | |||||||||||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | miR-214 level was upregulated in gefitinib-resistant PC-9GR cells and their derived exosomes while anti-apoptotic protein of bcl-2 is uoregulated. | ||||||||||||
| Key Molecule: hsa-mir-221 | [19] | ||||||||||||
| Resistant Disease | Lung cancer [ICD-11: 2C25.5] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | ||||||||||
| Cell viability | Activation | hsa05200 | |||||||||||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| Experiment for Molecule Alteration |
PCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | miR-221 may inhibit apoptosis by down regulating the expression of Apaf-1, so as to induce the resistance of PC-9 cells to gefitinib. | ||||||||||||
| Key Molecule: hsa-mir-138 | [18] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | ||||||||||
| Cell colony | Inhibition | hsa05200 | |||||||||||
| Cell viability | Inhibition | hsa05200 | |||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | ||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Mechanism Description | miR-138 inhibit the protein level of EGFR and reverses gefitinib resistance in lung cancer cells. | ||||||||||||
| Key Molecule: hsa-mir-873 | [55] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | ||||||||||
| Cell autophagy | Activation | hsa04140 | |||||||||||
| Cell viability | Activation | hsa05200 | |||||||||||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | The inhibition of miR-873 increased gefitinib resistance of NSCLC cells via the upregulation of GLI1. | ||||||||||||
| Key Molecule: Small nucleolar RNA host gene 5 (SNHG5) | [12] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | ||||||||||
| Cell viability | Inhibition | hsa05200 | |||||||||||
| miR377/CASP1 signaling pathway | Regulation | N.A. | |||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | ||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | SNHG5 overexpression sensitized gefitinib resistant LAD cells to gefitinib treatment; Overexpression of SNHG5 suppressed the expression of miR-377; Overexpression of miR-377 suppressed the expression of CASP1 in PC9 cells; knockdown of CASP1 in SNHG5-overexpressed PC9GR cells abolished their gefitinib resistance. | ||||||||||||
| Key Molecule: hsa-miR-630 | [56] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | ||||||||||
| Cell viability | Activation | hsa05200 | |||||||||||
| miR630/YAP1/ERK signaling pathway | Regulation | N.A. | |||||||||||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| PC9GR cells | Lung | Homo sapiens (Human) | CVCL_V337 | ||||||||||
| CL97 cells | Lung | Homo sapiens (Human) | CVCL_N826 | ||||||||||
| In Vivo Model | Nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
RT-PCR | ||||||||||||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | Low miR-630 and high YAP1 mRNA levels are associated with unfavorable response to TkI therapy in lung adenocarcinoma patients. | ||||||||||||
| Key Molecule: hsa-mir-135a | [57] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell migration | Activation | hsa04670 | ||||||||||
| Cell viability | Activation | hsa05200 | |||||||||||
| PI3K/AKT signaling pathway | Activation | hsa04151 | |||||||||||
| In Vitro Model | NCI-H1650 cells | Lung | Homo sapiens (Human) | CVCL_1483 | |||||||||
| NCI-H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | ||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay; Transwell assay | ||||||||||||
| Mechanism Description | miR-135a promoted cell growth and metastasis and activated the PI3k/AkT signaling pathway via a RAC1-dependent manner. | ||||||||||||
| Key Molecule: hsa-mir-26a | [34] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | Cell proliferation | Activation | hsa05200 | ||||||||||
| EGFR signaling pathway | Activation | hsa01521 | |||||||||||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |||||||||
| SPC-A1 cells | Lung | Homo sapiens (Human) | CVCL_6955 | ||||||||||
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | ||||||||||
| NCI-H520 cells | Lung | Homo sapiens (Human) | CVCL_1566 | ||||||||||
| H2170 cells | Lung | Homo sapiens (Human) | CVCL_1535 | ||||||||||
| SW900 cells | Lung | Homo sapiens (Human) | CVCL_1731 | ||||||||||
| In Vivo Model | Nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Mechanism Description | miR-26a desensitizes non-small cell lung cancer cells to tyrosine kinase inhibitors by targeting and reducing the level of PTPN1. | ||||||||||||
| Key Molecule: hsa-mir-21 | [58] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | AKT/ERK signaling pathway | Activation | hsa04010 | ||||||||||
| Cell apoptosis | Inhibition | hsa04210 | |||||||||||
| Cell migration | Inhibition | hsa04670 | |||||||||||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| In Vivo Model | Nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | ||||||||||||
| Mechanism Description | miR-21 was up-regulated concomitantly to down-regulation of Pten in pc-9/GR cells in comparison with pc-9 cells. Moreover, over-expression of miR-21 significantly decreased gefitinib sensitivity by down-regulating Pten expression and activating Akt and ERk pathways in pc-9 cells, while miR-21 knockdown dramatically restored gefitinib sensitivity of pc-9/GR cells by up-regulation of Pten expression and inactivation of AkT and ERk pathways, in vivo and in vitro. | ||||||||||||
| Key Molecule: hsa-mir-21 | [16] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | ||||||||||
| Cell proliferation | Activation | hsa05200 | |||||||||||
| PI3K/AKT signaling pathway | Activation | hsa04151 | |||||||||||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |||||||||
| PC9R cells | Lung | Homo sapiens (Human) | CVCL_D778 | ||||||||||
| In Vivo Model | Nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
RT-PCR | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Mechanism Description | miR-21 overexpression is associated with the acquired resistance of EGFR-TkI in NSCLC, which might be caused by miR-21's function of activating PI3k/AkT pathway through inhibiting PTEN and PDCD4. | ||||||||||||
|
|
|||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [32] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Missense mutation | p.C797S |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.64 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
G
G
A
S
M
M
G
G
G
E
E
A
A
P
P
700
|
N
N
Q
Q
A
A
L
L
L
L
R
R
I
I
L
L
K
K
E
E
710
|
T
T
E
E
F
F
K
K
K
K
I
I
K
K
V
V
L
L
G
G
720
|
S
S
G
G
A
A
F
F
G
G
T
T
V
V
Y
Y
K
K
G
G
730
|
L
L
W
W
I
I
P
P
E
E
G
G
E
E
K
K
V
V
K
K
740
|
I
I
P
P
V
V
A
A
I
I
K
K
E
E
L
L
R
R
E
E
750
|
A
A
T
T
S
S
P
P
K
K
A
A
N
N
K
K
E
E
I
I
760
|
L
L
D
D
E
E
A
A
Y
Y
V
V
M
M
A
A
S
S
V
V
770
|
D
D
N
N
P
P
H
H
V
V
C
C
R
R
L
L
L
L
G
G
780
|
I
I
C
C
L
L
T
T
S
S
T
T
V
V
Q
Q
L
L
I
I
790
|
T
M
Q
Q
L
L
M
M
P
P
F
F
G
G
C
S
L
L
L
L
800
|
D
D
Y
Y
V
V
R
R
E
E
H
H
K
K
D
D
N
N
I
I
810
|
G
G
S
S
Q
Q
Y
Y
L
L
L
L
N
N
W
W
C
C
V
V
820
|
Q
Q
I
I
A
A
K
K
G
G
M
M
N
N
Y
Y
L
L
E
E
830
|
D
D
R
R
R
R
L
L
V
V
H
H
R
R
D
D
L
L
A
A
840
|
A
A
R
R
N
N
V
V
L
L
V
V
K
K
T
T
P
P
Q
Q
850
|
H
H
V
V
K
K
I
I
T
T
D
D
F
F
G
G
L
L
A
A
860
|
K
K
L
L
L
L
G
G
A
A
E
E
E
E
K
K
E
E
Y
Y
870
|
H
H
A
A
E
E
G
G
G
G
K
K
V
V
P
P
I
I
K
K
880
|
W
W
M
M
A
A
L
L
E
E
S
S
I
I
L
L
H
H
R
R
890
|
I
I
Y
Y
T
T
H
H
Q
Q
S
S
D
D
V
V
W
W
S
S
900
|
Y
Y
G
G
V
V
T
T
V
V
W
W
E
E
L
L
M
M
T
T
910
|
F
F
G
G
S
S
K
K
P
P
Y
Y
D
D
G
G
I
I
P
P
920
|
A
A
S
S
E
E
I
I
S
S
S
S
I
I
L
L
E
E
K
K
930
|
G
G
E
E
R
R
L
L
P
P
Q
Q
P
P
P
P
I
I
C
C
940
|
T
T
I
I
D
D
V
V
Y
Y
M
M
I
I
M
M
V
V
K
K
950
|
C
C
W
W
M
M
I
I
D
D
A
A
D
D
S
S
R
R
P
P
960
|
K
K
F
F
R
R
E
E
L
L
I
I
I
I
E
E
F
F
S
S
970
|
K
K
M
M
A
A
R
R
D
D
P
P
Q
Q
R
R
Y
Y
L
L
980
|
V
V
I
I
Q
Q
G
G
D
D
E
E
R
R
M
M
H
H
L
L
990
|
P
P
S
S
P
P
T
T
D
D
S
S
N
N
F
F
Y
Y
R
R
1000
|
A
A
L
L
M
M
D
D
E
E
E
E
D
D
M
M
D
D
D
D
1010
|
V
V
V
V
D
D
A
A
D
D
E
E
Y
Y
L
L
I
I
P
P
1020
|
Q
Q
-
Q
-
G
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | ERK/MAPKsignaling pathway | Activation | hsa04210 | ||||||||||
| In Vitro Model | NSCLC cells | Lung | Homo sapiens (Human) | N.A. | |||||||||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Liquid biopsy; ATP-binding pocket affinity comparison assay | ||||||||||||
| Mechanism Description | Known mechanisms are secondary resistance mutations occurring in the ATP-binding domain (such as T790M and C797S), mutation or amplification of bypass signallings (such as AXL, Hh, ERBb2, CRIPTO, etc), activating mutations in the downstream pathways (PI3k, AkT, MEk, RAF), low levels of mRNA or polymorphisms of the pro-apoptotic protein BIM, induction of a transcription programme for EMT and phenotypical changes, or induction of elevated tumour PD-L1 levels. | ||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [40], [41], [42] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Missense mutation | p.T790M |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 3.10 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.05 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
S
G
G
E
E
A
A
P
P
700
|
N
N
Q
Q
A
A
L
L
L
L
R
R
I
I
L
L
K
K
E
E
710
|
T
T
E
E
F
F
K
K
K
K
I
I
K
K
V
V
L
L
G
G
720
|
S
S
G
G
A
A
F
F
G
G
T
T
V
V
Y
Y
K
K
G
G
730
|
L
L
W
W
I
I
P
P
E
E
G
G
E
E
K
K
V
V
K
K
740
|
I
I
P
P
V
V
A
A
I
I
K
K
E
E
L
L
R
R
E
E
750
|
A
A
T
T
S
S
P
P
K
K
A
A
N
N
K
K
E
E
I
I
760
|
L
L
D
D
E
E
A
A
Y
Y
V
V
M
M
A
A
S
S
V
V
770
|
D
D
N
N
P
P
H
H
V
V
C
C
R
R
L
L
L
L
G
G
780
|
I
I
C
C
L
L
T
T
S
S
T
T
V
V
Q
Q
L
L
I
I
790
|
T
M
Q
Q
L
L
M
M
P
P
F
F
G
G
C
C
L
L
L
L
800
|
D
D
Y
Y
V
V
R
R
E
E
H
H
K
K
D
D
N
N
I
I
810
|
G
G
S
S
Q
Q
Y
Y
L
L
L
L
N
N
W
W
C
C
V
V
820
|
Q
Q
I
I
A
A
K
K
G
G
M
M
N
N
Y
Y
L
L
E
E
830
|
D
D
R
R
R
R
L
L
V
V
H
H
R
R
D
D
L
L
A
A
840
|
A
A
R
R
N
N
V
V
L
L
V
V
K
K
T
T
P
P
Q
Q
850
|
H
H
V
V
K
K
I
I
T
T
D
D
F
F
G
G
L
L
A
A
860
|
K
K
L
L
L
L
G
G
A
A
E
E
E
E
K
K
E
E
Y
Y
870
|
H
H
A
A
E
E
G
G
G
G
K
K
V
V
P
P
I
I
K
K
880
|
W
W
M
M
A
A
L
L
E
E
S
S
I
I
L
L
H
H
R
R
890
|
I
I
Y
Y
T
T
H
H
Q
Q
S
S
D
D
V
V
W
W
S
S
900
|
Y
Y
G
G
V
V
T
T
V
V
W
W
E
E
L
L
M
M
T
T
910
|
F
F
G
G
S
S
K
K
P
P
Y
Y
D
D
G
G
I
I
P
P
920
|
A
A
S
S
E
E
I
I
S
S
S
S
I
I
L
L
E
E
K
K
930
|
G
G
E
E
R
R
L
L
P
P
Q
Q
P
P
P
P
I
I
C
C
940
|
T
T
I
I
D
D
V
V
Y
Y
M
M
I
I
M
M
V
V
K
K
950
|
C
C
W
W
M
M
I
I
D
D
A
A
D
D
S
S
R
R
P
P
960
|
K
K
F
F
R
R
E
E
L
L
I
I
I
I
E
E
F
F
S
S
970
|
K
K
M
M
A
A
R
R
D
D
P
P
Q
Q
R
R
Y
Y
L
L
980
|
V
V
I
I
Q
Q
G
G
D
D
E
E
R
R
M
M
H
H
L
L
990
|
P
P
S
S
P
P
T
T
D
D
S
S
N
N
F
F
Y
Y
R
R
1000
|
A
A
L
L
M
M
D
D
E
E
E
E
D
D
M
M
D
D
D
D
1010
|
V
V
V
V
D
D
A
A
D
D
E
E
Y
Y
L
L
I
I
P
P
1020
|
Q
Q
Q
Q
G
G
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | ERK/MAPKsignaling pathway | Activation | hsa04210 | ||||||||||
| In Vitro Model | NSCLC cells | Lung | Homo sapiens (Human) | N.A. | |||||||||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Liquid biopsy; ATP-binding pocket affinity comparison assay | ||||||||||||
| Mechanism Description | Known mechanisms are secondary resistance mutations occurring in the ATP-binding domain (such as T790M and C797S), mutation or amplification of bypass signallings (such as AXL, Hh, ERBb2, CRIPTO, etc), activating mutations in the downstream pathways (PI3k, AkT, MEk, RAF), low levels of mRNA or polymorphisms of the pro-apoptotic protein BIM, induction of a transcription programme for EMT and phenotypical changes, or induction of elevated tumour PD-L1 levels. | ||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [2], [43], [44] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Missense mutation | p.T790M |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 3.10 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.05 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
S
G
G
E
E
A
A
P
P
700
|
N
N
Q
Q
A
A
L
L
L
L
R
R
I
I
L
L
K
K
E
E
710
|
T
T
E
E
F
F
K
K
K
K
I
I
K
K
V
V
L
L
G
G
720
|
S
S
G
G
A
A
F
F
G
G
T
T
V
V
Y
Y
K
K
G
G
730
|
L
L
W
W
I
I
P
P
E
E
G
G
E
E
K
K
V
V
K
K
740
|
I
I
P
P
V
V
A
A
I
I
K
K
E
E
L
L
R
R
E
E
750
|
A
A
T
T
S
S
P
P
K
K
A
A
N
N
K
K
E
E
I
I
760
|
L
L
D
D
E
E
A
A
Y
Y
V
V
M
M
A
A
S
S
V
V
770
|
D
D
N
N
P
P
H
H
V
V
C
C
R
R
L
L
L
L
G
G
780
|
I
I
C
C
L
L
T
T
S
S
T
T
V
V
Q
Q
L
L
I
I
790
|
T
M
Q
Q
L
L
M
M
P
P
F
F
G
G
C
C
L
L
L
L
800
|
D
D
Y
Y
V
V
R
R
E
E
H
H
K
K
D
D
N
N
I
I
810
|
G
G
S
S
Q
Q
Y
Y
L
L
L
L
N
N
W
W
C
C
V
V
820
|
Q
Q
I
I
A
A
K
K
G
G
M
M
N
N
Y
Y
L
L
E
E
830
|
D
D
R
R
R
R
L
L
V
V
H
H
R
R
D
D
L
L
A
A
840
|
A
A
R
R
N
N
V
V
L
L
V
V
K
K
T
T
P
P
Q
Q
850
|
H
H
V
V
K
K
I
I
T
T
D
D
F
F
G
G
L
L
A
A
860
|
K
K
L
L
L
L
G
G
A
A
E
E
E
E
K
K
E
E
Y
Y
870
|
H
H
A
A
E
E
G
G
G
G
K
K
V
V
P
P
I
I
K
K
880
|
W
W
M
M
A
A
L
L
E
E
S
S
I
I
L
L
H
H
R
R
890
|
I
I
Y
Y
T
T
H
H
Q
Q
S
S
D
D
V
V
W
W
S
S
900
|
Y
Y
G
G
V
V
T
T
V
V
W
W
E
E
L
L
M
M
T
T
910
|
F
F
G
G
S
S
K
K
P
P
Y
Y
D
D
G
G
I
I
P
P
920
|
A
A
S
S
E
E
I
I
S
S
S
S
I
I
L
L
E
E
K
K
930
|
G
G
E
E
R
R
L
L
P
P
Q
Q
P
P
P
P
I
I
C
C
940
|
T
T
I
I
D
D
V
V
Y
Y
M
M
I
I
M
M
V
V
K
K
950
|
C
C
W
W
M
M
I
I
D
D
A
A
D
D
S
S
R
R
P
P
960
|
K
K
F
F
R
R
E
E
L
L
I
I
I
I
E
E
F
F
S
S
970
|
K
K
M
M
A
A
R
R
D
D
P
P
Q
Q
R
R
Y
Y
L
L
980
|
V
V
I
I
Q
Q
G
G
D
D
E
E
R
R
M
M
H
H
L
L
990
|
P
P
S
S
P
P
T
T
D
D
S
S
N
N
F
F
Y
Y
R
R
1000
|
A
A
L
L
M
M
D
D
E
E
E
E
D
D
M
M
D
D
D
D
1010
|
V
V
V
V
D
D
A
A
D
D
E
E
Y
Y
L
L
I
I
P
P
1020
|
Q
Q
Q
Q
G
G
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
Direct sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Overall and disease-free assay | ||||||||||||
| Mechanism Description | A secondary T790M mutation of EGFR accounted for half the tumors with acquired resistance to gefitinib in Japanese patients. Other drug-resistant secondary mutations are uncommon in the EGFR gene. | ||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [45], [46], [47] | ||||||||||||
| Resistant Disease | Lung squamous cell carcinoma [ICD-11: 2C25.3] | ||||||||||||
| Molecule Alteration | Missense mutation | p.T790M |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 3.10 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.05 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
S
G
G
E
E
A
A
P
P
700
|
N
N
Q
Q
A
A
L
L
L
L
R
R
I
I
L
L
K
K
E
E
710
|
T
T
E
E
F
F
K
K
K
K
I
I
K
K
V
V
L
L
G
G
720
|
S
S
G
G
A
A
F
F
G
G
T
T
V
V
Y
Y
K
K
G
G
730
|
L
L
W
W
I
I
P
P
E
E
G
G
E
E
K
K
V
V
K
K
740
|
I
I
P
P
V
V
A
A
I
I
K
K
E
E
L
L
R
R
E
E
750
|
A
A
T
T
S
S
P
P
K
K
A
A
N
N
K
K
E
E
I
I
760
|
L
L
D
D
E
E
A
A
Y
Y
V
V
M
M
A
A
S
S
V
V
770
|
D
D
N
N
P
P
H
H
V
V
C
C
R
R
L
L
L
L
G
G
780
|
I
I
C
C
L
L
T
T
S
S
T
T
V
V
Q
Q
L
L
I
I
790
|
T
M
Q
Q
L
L
M
M
P
P
F
F
G
G
C
C
L
L
L
L
800
|
D
D
Y
Y
V
V
R
R
E
E
H
H
K
K
D
D
N
N
I
I
810
|
G
G
S
S
Q
Q
Y
Y
L
L
L
L
N
N
W
W
C
C
V
V
820
|
Q
Q
I
I
A
A
K
K
G
G
M
M
N
N
Y
Y
L
L
E
E
830
|
D
D
R
R
R
R
L
L
V
V
H
H
R
R
D
D
L
L
A
A
840
|
A
A
R
R
N
N
V
V
L
L
V
V
K
K
T
T
P
P
Q
Q
850
|
H
H
V
V
K
K
I
I
T
T
D
D
F
F
G
G
L
L
A
A
860
|
K
K
L
L
L
L
G
G
A
A
E
E
E
E
K
K
E
E
Y
Y
870
|
H
H
A
A
E
E
G
G
G
G
K
K
V
V
P
P
I
I
K
K
880
|
W
W
M
M
A
A
L
L
E
E
S
S
I
I
L
L
H
H
R
R
890
|
I
I
Y
Y
T
T
H
H
Q
Q
S
S
D
D
V
V
W
W
S
S
900
|
Y
Y
G
G
V
V
T
T
V
V
W
W
E
E
L
L
M
M
T
T
910
|
F
F
G
G
S
S
K
K
P
P
Y
Y
D
D
G
G
I
I
P
P
920
|
A
A
S
S
E
E
I
I
S
S
S
S
I
I
L
L
E
E
K
K
930
|
G
G
E
E
R
R
L
L
P
P
Q
Q
P
P
P
P
I
I
C
C
940
|
T
T
I
I
D
D
V
V
Y
Y
M
M
I
I
M
M
V
V
K
K
950
|
C
C
W
W
M
M
I
I
D
D
A
A
D
D
S
S
R
R
P
P
960
|
K
K
F
F
R
R
E
E
L
L
I
I
I
I
E
E
F
F
S
S
970
|
K
K
M
M
A
A
R
R
D
D
P
P
Q
Q
R
R
Y
Y
L
L
980
|
V
V
I
I
Q
Q
G
G
D
D
E
E
R
R
M
M
H
H
L
L
990
|
P
P
S
S
P
P
T
T
D
D
S
S
N
N
F
F
Y
Y
R
R
1000
|
A
A
L
L
M
M
D
D
E
E
E
E
D
D
M
M
D
D
D
D
1010
|
V
V
V
V
D
D
A
A
D
D
E
E
Y
Y
L
L
I
I
P
P
1020
|
Q
Q
Q
Q
G
G
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Computed tomography (CT) scanning assay; Bone scintigraphy assay; Magnetic resonance imaging assay | ||||||||||||
| Mechanism Description | C-Met amplification, epithelial-mesenchymal transition, and kRAS and BRAF mutations were ruled out as alternative resistance mechanisms in the T790M-negative lung rebiopsy, suggesting that alternative oncogene aberrations such as HER2/Neu amplification, hepatocyte growth factor release by the tumor microenvironment, or other unidentified pathways contributed to the TkI resistance that was observed in the primary lesion. | ||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [48] | ||||||||||||
| Resistant Disease | EGFR-mutant non-small cell lung cancer [ICD-11: 2C25.7] | ||||||||||||
| Molecule Alteration | Missense mutation | p.T790M |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 3.10 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.05 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
S
G
G
E
E
A
A
P
P
700
|
N
N
Q
Q
A
A
L
L
L
L
R
R
I
I
L
L
K
K
E
E
710
|
T
T
E
E
F
F
K
K
K
K
I
I
K
K
V
V
L
L
G
G
720
|
S
S
G
G
A
A
F
F
G
G
T
T
V
V
Y
Y
K
K
G
G
730
|
L
L
W
W
I
I
P
P
E
E
G
G
E
E
K
K
V
V
K
K
740
|
I
I
P
P
V
V
A
A
I
I
K
K
E
E
L
L
R
R
E
E
750
|
A
A
T
T
S
S
P
P
K
K
A
A
N
N
K
K
E
E
I
I
760
|
L
L
D
D
E
E
A
A
Y
Y
V
V
M
M
A
A
S
S
V
V
770
|
D
D
N
N
P
P
H
H
V
V
C
C
R
R
L
L
L
L
G
G
780
|
I
I
C
C
L
L
T
T
S
S
T
T
V
V
Q
Q
L
L
I
I
790
|
T
M
Q
Q
L
L
M
M
P
P
F
F
G
G
C
C
L
L
L
L
800
|
D
D
Y
Y
V
V
R
R
E
E
H
H
K
K
D
D
N
N
I
I
810
|
G
G
S
S
Q
Q
Y
Y
L
L
L
L
N
N
W
W
C
C
V
V
820
|
Q
Q
I
I
A
A
K
K
G
G
M
M
N
N
Y
Y
L
L
E
E
830
|
D
D
R
R
R
R
L
L
V
V
H
H
R
R
D
D
L
L
A
A
840
|
A
A
R
R
N
N
V
V
L
L
V
V
K
K
T
T
P
P
Q
Q
850
|
H
H
V
V
K
K
I
I
T
T
D
D
F
F
G
G
L
L
A
A
860
|
K
K
L
L
L
L
G
G
A
A
E
E
E
E
K
K
E
E
Y
Y
870
|
H
H
A
A
E
E
G
G
G
G
K
K
V
V
P
P
I
I
K
K
880
|
W
W
M
M
A
A
L
L
E
E
S
S
I
I
L
L
H
H
R
R
890
|
I
I
Y
Y
T
T
H
H
Q
Q
S
S
D
D
V
V
W
W
S
S
900
|
Y
Y
G
G
V
V
T
T
V
V
W
W
E
E
L
L
M
M
T
T
910
|
F
F
G
G
S
S
K
K
P
P
Y
Y
D
D
G
G
I
I
P
P
920
|
A
A
S
S
E
E
I
I
S
S
S
S
I
I
L
L
E
E
K
K
930
|
G
G
E
E
R
R
L
L
P
P
Q
Q
P
P
P
P
I
I
C
C
940
|
T
T
I
I
D
D
V
V
Y
Y
M
M
I
I
M
M
V
V
K
K
950
|
C
C
W
W
M
M
I
I
D
D
A
A
D
D
S
S
R
R
P
P
960
|
K
K
F
F
R
R
E
E
L
L
I
I
I
I
E
E
F
F
S
S
970
|
K
K
M
M
A
A
R
R
D
D
P
P
Q
Q
R
R
Y
Y
L
L
980
|
V
V
I
I
Q
Q
G
G
D
D
E
E
R
R
M
M
H
H
L
L
990
|
P
P
S
S
P
P
T
T
D
D
S
S
N
N
F
F
Y
Y
R
R
1000
|
A
A
L
L
M
M
D
D
E
E
E
E
D
D
M
M
D
D
D
D
1010
|
V
V
V
V
D
D
A
A
D
D
E
E
Y
Y
L
L
I
I
P
P
1020
|
Q
Q
Q
Q
G
G
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
MGB SNP detection kit assay; Mutation Detection assay | ||||||||||||
| Experiment for Drug Resistance |
Digital PCR assay | ||||||||||||
| Mechanism Description | Resistance mechanisms to EGFR-TkI therapy in EGFR-mutated NSCLC include secondary EGFR T790M mutation, c-Met amplification, PIk3CA mutation, and transformation to small-cell lung cancer. | ||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [49] | ||||||||||||
| Resistant Disease | EGFR-mutant non-small cell lung cancer [ICD-11: 2C25.7] | ||||||||||||
| Molecule Alteration | Missense mutation | p.T790M |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 3.10 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.05 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
S
G
G
E
E
A
A
P
P
700
|
N
N
Q
Q
A
A
L
L
L
L
R
R
I
I
L
L
K
K
E
E
710
|
T
T
E
E
F
F
K
K
K
K
I
I
K
K
V
V
L
L
G
G
720
|
S
S
G
G
A
A
F
F
G
G
T
T
V
V
Y
Y
K
K
G
G
730
|
L
L
W
W
I
I
P
P
E
E
G
G
E
E
K
K
V
V
K
K
740
|
I
I
P
P
V
V
A
A
I
I
K
K
E
E
L
L
R
R
E
E
750
|
A
A
T
T
S
S
P
P
K
K
A
A
N
N
K
K
E
E
I
I
760
|
L
L
D
D
E
E
A
A
Y
Y
V
V
M
M
A
A
S
S
V
V
770
|
D
D
N
N
P
P
H
H
V
V
C
C
R
R
L
L
L
L
G
G
780
|
I
I
C
C
L
L
T
T
S
S
T
T
V
V
Q
Q
L
L
I
I
790
|
T
M
Q
Q
L
L
M
M
P
P
F
F
G
G
C
C
L
L
L
L
800
|
D
D
Y
Y
V
V
R
R
E
E
H
H
K
K
D
D
N
N
I
I
810
|
G
G
S
S
Q
Q
Y
Y
L
L
L
L
N
N
W
W
C
C
V
V
820
|
Q
Q
I
I
A
A
K
K
G
G
M
M
N
N
Y
Y
L
L
E
E
830
|
D
D
R
R
R
R
L
L
V
V
H
H
R
R
D
D
L
L
A
A
840
|
A
A
R
R
N
N
V
V
L
L
V
V
K
K
T
T
P
P
Q
Q
850
|
H
H
V
V
K
K
I
I
T
T
D
D
F
F
G
G
L
L
A
A
860
|
K
K
L
L
L
L
G
G
A
A
E
E
E
E
K
K
E
E
Y
Y
870
|
H
H
A
A
E
E
G
G
G
G
K
K
V
V
P
P
I
I
K
K
880
|
W
W
M
M
A
A
L
L
E
E
S
S
I
I
L
L
H
H
R
R
890
|
I
I
Y
Y
T
T
H
H
Q
Q
S
S
D
D
V
V
W
W
S
S
900
|
Y
Y
G
G
V
V
T
T
V
V
W
W
E
E
L
L
M
M
T
T
910
|
F
F
G
G
S
S
K
K
P
P
Y
Y
D
D
G
G
I
I
P
P
920
|
A
A
S
S
E
E
I
I
S
S
S
S
I
I
L
L
E
E
K
K
930
|
G
G
E
E
R
R
L
L
P
P
Q
Q
P
P
P
P
I
I
C
C
940
|
T
T
I
I
D
D
V
V
Y
Y
M
M
I
I
M
M
V
V
K
K
950
|
C
C
W
W
M
M
I
I
D
D
A
A
D
D
S
S
R
R
P
P
960
|
K
K
F
F
R
R
E
E
L
L
I
I
I
I
E
E
F
F
S
S
970
|
K
K
M
M
A
A
R
R
D
D
P
P
Q
Q
R
R
Y
Y
L
L
980
|
V
V
I
I
Q
Q
G
G
D
D
E
E
R
R
M
M
H
H
L
L
990
|
P
P
S
S
P
P
T
T
D
D
S
S
N
N
F
F
Y
Y
R
R
1000
|
A
A
L
L
M
M
D
D
E
E
E
E
D
D
M
M
D
D
D
D
1010
|
V
V
V
V
D
D
A
A
D
D
E
E
Y
Y
L
L
I
I
P
P
1020
|
Q
Q
Q
Q
G
G
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
Low throughput experiment assay | ||||||||||||
| Experiment for Drug Resistance |
Progression-free survival assay | ||||||||||||
| Mechanism Description | Acquired resistance can occur through failure of drug delivery to the target, as in isolated central nervous system (CNS) progression, or by selection of biological variants during TkI exposure. At the point of acquired resistance, the T790M substitution may be accompanied by amplification of the EGFR gene as well. | ||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [50] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Missense mutation | p.T790M |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 3.10 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.05 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
S
G
G
E
E
A
A
P
P
700
|
N
N
Q
Q
A
A
L
L
L
L
R
R
I
I
L
L
K
K
E
E
710
|
T
T
E
E
F
F
K
K
K
K
I
I
K
K
V
V
L
L
G
G
720
|
S
S
G
G
A
A
F
F
G
G
T
T
V
V
Y
Y
K
K
G
G
730
|
L
L
W
W
I
I
P
P
E
E
G
G
E
E
K
K
V
V
K
K
740
|
I
I
P
P
V
V
A
A
I
I
K
K
E
E
L
L
R
R
E
E
750
|
A
A
T
T
S
S
P
P
K
K
A
A
N
N
K
K
E
E
I
I
760
|
L
L
D
D
E
E
A
A
Y
Y
V
V
M
M
A
A
S
S
V
V
770
|
D
D
N
N
P
P
H
H
V
V
C
C
R
R
L
L
L
L
G
G
780
|
I
I
C
C
L
L
T
T
S
S
T
T
V
V
Q
Q
L
L
I
I
790
|
T
M
Q
Q
L
L
M
M
P
P
F
F
G
G
C
C
L
L
L
L
800
|
D
D
Y
Y
V
V
R
R
E
E
H
H
K
K
D
D
N
N
I
I
810
|
G
G
S
S
Q
Q
Y
Y
L
L
L
L
N
N
W
W
C
C
V
V
820
|
Q
Q
I
I
A
A
K
K
G
G
M
M
N
N
Y
Y
L
L
E
E
830
|
D
D
R
R
R
R
L
L
V
V
H
H
R
R
D
D
L
L
A
A
840
|
A
A
R
R
N
N
V
V
L
L
V
V
K
K
T
T
P
P
Q
Q
850
|
H
H
V
V
K
K
I
I
T
T
D
D
F
F
G
G
L
L
A
A
860
|
K
K
L
L
L
L
G
G
A
A
E
E
E
E
K
K
E
E
Y
Y
870
|
H
H
A
A
E
E
G
G
G
G
K
K
V
V
P
P
I
I
K
K
880
|
W
W
M
M
A
A
L
L
E
E
S
S
I
I
L
L
H
H
R
R
890
|
I
I
Y
Y
T
T
H
H
Q
Q
S
S
D
D
V
V
W
W
S
S
900
|
Y
Y
G
G
V
V
T
T
V
V
W
W
E
E
L
L
M
M
T
T
910
|
F
F
G
G
S
S
K
K
P
P
Y
Y
D
D
G
G
I
I
P
P
920
|
A
A
S
S
E
E
I
I
S
S
S
S
I
I
L
L
E
E
K
K
930
|
G
G
E
E
R
R
L
L
P
P
Q
Q
P
P
P
P
I
I
C
C
940
|
T
T
I
I
D
D
V
V
Y
Y
M
M
I
I
M
M
V
V
K
K
950
|
C
C
W
W
M
M
I
I
D
D
A
A
D
D
S
S
R
R
P
P
960
|
K
K
F
F
R
R
E
E
L
L
I
I
I
I
E
E
F
F
S
S
970
|
K
K
M
M
A
A
R
R
D
D
P
P
Q
Q
R
R
Y
Y
L
L
980
|
V
V
I
I
Q
Q
G
G
D
D
E
E
R
R
M
M
H
H
L
L
990
|
P
P
S
S
P
P
T
T
D
D
S
S
N
N
F
F
Y
Y
R
R
1000
|
A
A
L
L
M
M
D
D
E
E
E
E
D
D
M
M
D
D
D
D
1010
|
V
V
V
V
D
D
A
A
D
D
E
E
Y
Y
L
L
I
I
P
P
1020
|
Q
Q
Q
Q
G
G
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
Next generation sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Multivariate analysis of overall or disease-free survival assay | ||||||||||||
| Mechanism Description | One example is the acquisition of the T790M substitution in the membrane receptor EGFR conferring resistance to gefitinb and erlotinib in lung cancer in approximately 50% of patients. | ||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [2] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Missense mutation | p.T790M |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 3.10 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.05 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
S
G
G
E
E
A
A
P
P
700
|
N
N
Q
Q
A
A
L
L
L
L
R
R
I
I
L
L
K
K
E
E
710
|
T
T
E
E
F
F
K
K
K
K
I
I
K
K
V
V
L
L
G
G
720
|
S
S
G
G
A
A
F
F
G
G
T
T
V
V
Y
Y
K
K
G
G
730
|
L
L
W
W
I
I
P
P
E
E
G
G
E
E
K
K
V
V
K
K
740
|
I
I
P
P
V
V
A
A
I
I
K
K
E
E
L
L
R
R
E
E
750
|
A
A
T
T
S
S
P
P
K
K
A
A
N
N
K
K
E
E
I
I
760
|
L
L
D
D
E
E
A
A
Y
Y
V
V
M
M
A
A
S
S
V
V
770
|
D
D
N
N
P
P
H
H
V
V
C
C
R
R
L
L
L
L
G
G
780
|
I
I
C
C
L
L
T
T
S
S
T
T
V
V
Q
Q
L
L
I
I
790
|
T
M
Q
Q
L
L
M
M
P
P
F
F
G
G
C
C
L
L
L
L
800
|
D
D
Y
Y
V
V
R
R
E
E
H
H
K
K
D
D
N
N
I
I
810
|
G
G
S
S
Q
Q
Y
Y
L
L
L
L
N
N
W
W
C
C
V
V
820
|
Q
Q
I
I
A
A
K
K
G
G
M
M
N
N
Y
Y
L
L
E
E
830
|
D
D
R
R
R
R
L
L
V
V
H
H
R
R
D
D
L
L
A
A
840
|
A
A
R
R
N
N
V
V
L
L
V
V
K
K
T
T
P
P
Q
Q
850
|
H
H
V
V
K
K
I
I
T
T
D
D
F
F
G
G
L
L
A
A
860
|
K
K
L
L
L
L
G
G
A
A
E
E
E
E
K
K
E
E
Y
Y
870
|
H
H
A
A
E
E
G
G
G
G
K
K
V
V
P
P
I
I
K
K
880
|
W
W
M
M
A
A
L
L
E
E
S
S
I
I
L
L
H
H
R
R
890
|
I
I
Y
Y
T
T
H
H
Q
Q
S
S
D
D
V
V
W
W
S
S
900
|
Y
Y
G
G
V
V
T
T
V
V
W
W
E
E
L
L
M
M
T
T
910
|
F
F
G
G
S
S
K
K
P
P
Y
Y
D
D
G
G
I
I
P
P
920
|
A
A
S
S
E
E
I
I
S
S
S
S
I
I
L
L
E
E
K
K
930
|
G
G
E
E
R
R
L
L
P
P
Q
Q
P
P
P
P
I
I
C
C
940
|
T
T
I
I
D
D
V
V
Y
Y
M
M
I
I
M
M
V
V
K
K
950
|
C
C
W
W
M
M
I
I
D
D
A
A
D
D
S
S
R
R
P
P
960
|
K
K
F
F
R
R
E
E
L
L
I
I
I
I
E
E
F
F
S
S
970
|
K
K
M
M
A
A
R
R
D
D
P
P
Q
Q
R
R
Y
Y
L
L
980
|
V
V
I
I
Q
Q
G
G
D
D
E
E
R
R
M
M
H
H
L
L
990
|
P
P
S
S
P
P
T
T
D
D
S
S
N
N
F
F
Y
Y
R
R
1000
|
A
A
L
L
M
M
D
D
E
E
E
E
D
D
M
M
D
D
D
D
1010
|
V
V
V
V
D
D
A
A
D
D
E
E
Y
Y
L
L
I
I
P
P
1020
|
Q
Q
Q
Q
G
G
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Cos-7 cells | Lung | Homo sapiens (Human) | CVCL_0224 | |||||||||
| NIH-3T3 cells | Embryo | Mus musculus (Mouse) | CVCL_0594 | ||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Mechanism Description | The DNA sequence of the EGFR gene in his tumor biopsy specimen at relapse revealed the presence of a second point mutation, resulting in threonine-to-methionine amino acid change at position 790 of EGFR. Structural modeling and biochemical studies showed that this second mutation led to gefitinib resistance. | ||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [2] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Missense mutation | p.T790M |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 3.10 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.05 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
S
G
G
E
E
A
A
P
P
700
|
N
N
Q
Q
A
A
L
L
L
L
R
R
I
I
L
L
K
K
E
E
710
|
T
T
E
E
F
F
K
K
K
K
I
I
K
K
V
V
L
L
G
G
720
|
S
S
G
G
A
A
F
F
G
G
T
T
V
V
Y
Y
K
K
G
G
730
|
L
L
W
W
I
I
P
P
E
E
G
G
E
E
K
K
V
V
K
K
740
|
I
I
P
P
V
V
A
A
I
I
K
K
E
E
L
L
R
R
E
E
750
|
A
A
T
T
S
S
P
P
K
K
A
A
N
N
K
K
E
E
I
I
760
|
L
L
D
D
E
E
A
A
Y
Y
V
V
M
M
A
A
S
S
V
V
770
|
D
D
N
N
P
P
H
H
V
V
C
C
R
R
L
L
L
L
G
G
780
|
I
I
C
C
L
L
T
T
S
S
T
T
V
V
Q
Q
L
L
I
I
790
|
T
M
Q
Q
L
L
M
M
P
P
F
F
G
G
C
C
L
L
L
L
800
|
D
D
Y
Y
V
V
R
R
E
E
H
H
K
K
D
D
N
N
I
I
810
|
G
G
S
S
Q
Q
Y
Y
L
L
L
L
N
N
W
W
C
C
V
V
820
|
Q
Q
I
I
A
A
K
K
G
G
M
M
N
N
Y
Y
L
L
E
E
830
|
D
D
R
R
R
R
L
L
V
V
H
H
R
R
D
D
L
L
A
A
840
|
A
A
R
R
N
N
V
V
L
L
V
V
K
K
T
T
P
P
Q
Q
850
|
H
H
V
V
K
K
I
I
T
T
D
D
F
F
G
G
L
L
A
A
860
|
K
K
L
L
L
L
G
G
A
A
E
E
E
E
K
K
E
E
Y
Y
870
|
H
H
A
A
E
E
G
G
G
G
K
K
V
V
P
P
I
I
K
K
880
|
W
W
M
M
A
A
L
L
E
E
S
S
I
I
L
L
H
H
R
R
890
|
I
I
Y
Y
T
T
H
H
Q
Q
S
S
D
D
V
V
W
W
S
S
900
|
Y
Y
G
G
V
V
T
T
V
V
W
W
E
E
L
L
M
M
T
T
910
|
F
F
G
G
S
S
K
K
P
P
Y
Y
D
D
G
G
I
I
P
P
920
|
A
A
S
S
E
E
I
I
S
S
S
S
I
I
L
L
E
E
K
K
930
|
G
G
E
E
R
R
L
L
P
P
Q
Q
P
P
P
P
I
I
C
C
940
|
T
T
I
I
D
D
V
V
Y
Y
M
M
I
I
M
M
V
V
K
K
950
|
C
C
W
W
M
M
I
I
D
D
A
A
D
D
S
S
R
R
P
P
960
|
K
K
F
F
R
R
E
E
L
L
I
I
I
I
E
E
F
F
S
S
970
|
K
K
M
M
A
A
R
R
D
D
P
P
Q
Q
R
R
Y
Y
L
L
980
|
V
V
I
I
Q
Q
G
G
D
D
E
E
R
R
M
M
H
H
L
L
990
|
P
P
S
S
P
P
T
T
D
D
S
S
N
N
F
F
Y
Y
R
R
1000
|
A
A
L
L
M
M
D
D
E
E
E
E
D
D
M
M
D
D
D
D
1010
|
V
V
V
V
D
D
A
A
D
D
E
E
Y
Y
L
L
I
I
P
P
1020
|
Q
Q
Q
Q
G
G
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Cos-7 cells | Lung | Homo sapiens (Human) | CVCL_0224 | |||||||||
| NIH-3T3 cells | Embryo | Mus musculus (Mouse) | CVCL_0594 | ||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Mechanism Description | The DNA sequence of the EGFR gene in his tumor biopsy specimen at relapse revealed the presence of a second point mutation, resulting in threonine-to-methionine amino acid change at position 790 of EGFR. Structural modeling and biochemical studies showed that this second mutation led to gefitinib resistance. | ||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [32] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Missense mutation | p.C797S+p.T790M |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.64 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
G
G
A
S
M
M
G
G
G
E
E
A
A
P
P
700
|
N
N
Q
Q
A
A
L
L
L
L
R
R
I
I
L
L
K
K
E
E
710
|
T
T
E
E
F
F
K
K
K
K
I
I
K
K
V
V
L
L
G
G
720
|
S
S
G
G
A
A
F
F
G
G
T
T
V
V
Y
Y
K
K
G
G
730
|
L
L
W
W
I
I
P
P
E
E
G
G
E
E
K
K
V
V
K
K
740
|
I
I
P
P
V
V
A
A
I
I
K
K
E
E
L
L
R
R
E
E
750
|
A
A
T
T
S
S
P
P
K
K
A
A
N
N
K
K
E
E
I
I
760
|
L
L
D
D
E
E
A
A
Y
Y
V
V
M
M
A
A
S
S
V
V
770
|
D
D
N
N
P
P
H
H
V
V
C
C
R
R
L
L
L
L
G
G
780
|
I
I
C
C
L
L
T
T
S
S
T
T
V
V
Q
Q
L
L
I
I
790
|
T
M
Q
Q
L
L
M
M
P
P
F
F
G
G
C
S
L
L
L
L
800
|
D
D
Y
Y
V
V
R
R
E
E
H
H
K
K
D
D
N
N
I
I
810
|
G
G
S
S
Q
Q
Y
Y
L
L
L
L
N
N
W
W
C
C
V
V
820
|
Q
Q
I
I
A
A
K
K
G
G
M
M
N
N
Y
Y
L
L
E
E
830
|
D
D
R
R
R
R
L
L
V
V
H
H
R
R
D
D
L
L
A
A
840
|
A
A
R
R
N
N
V
V
L
L
V
V
K
K
T
T
P
P
Q
Q
850
|
H
H
V
V
K
K
I
I
T
T
D
D
F
F
G
G
L
L
A
A
860
|
K
K
L
L
L
L
G
G
A
A
E
E
E
E
K
K
E
E
Y
Y
870
|
H
H
A
A
E
E
G
G
G
G
K
K
V
V
P
P
I
I
K
K
880
|
W
W
M
M
A
A
L
L
E
E
S
S
I
I
L
L
H
H
R
R
890
|
I
I
Y
Y
T
T
H
H
Q
Q
S
S
D
D
V
V
W
W
S
S
900
|
Y
Y
G
G
V
V
T
T
V
V
W
W
E
E
L
L
M
M
T
T
910
|
F
F
G
G
S
S
K
K
P
P
Y
Y
D
D
G
G
I
I
P
P
920
|
A
A
S
S
E
E
I
I
S
S
S
S
I
I
L
L
E
E
K
K
930
|
G
G
E
E
R
R
L
L
P
P
Q
Q
P
P
P
P
I
I
C
C
940
|
T
T
I
I
D
D
V
V
Y
Y
M
M
I
I
M
M
V
V
K
K
950
|
C
C
W
W
M
M
I
I
D
D
A
A
D
D
S
S
R
R
P
P
960
|
K
K
F
F
R
R
E
E
L
L
I
I
I
I
E
E
F
F
S
S
970
|
K
K
M
M
A
A
R
R
D
D
P
P
Q
Q
R
R
Y
Y
L
L
980
|
V
V
I
I
Q
Q
G
G
D
D
E
E
R
R
M
M
H
H
L
L
990
|
P
P
S
S
P
P
T
T
D
D
S
S
N
N
F
F
Y
Y
R
R
1000
|
A
A
L
L
M
M
D
D
E
E
E
E
D
D
M
M
D
D
D
D
1010
|
V
V
V
V
D
D
A
A
D
D
E
E
Y
Y
L
L
I
I
P
P
1020
|
Q
Q
-
Q
-
G
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | AKT signaling pathway | Activation | hsa04151 | ||||||||||
| Cell invasion | Activation | hsa05200 | |||||||||||
| Cell migration | Activation | hsa04670 | |||||||||||
| Cell proliferation | Activation | hsa05200 | |||||||||||
| EGFR/TKLS mediated apoptosis signaling pathway | Inhibition | hsa01521 | |||||||||||
| Epithelial mesenchymal transition signaling pathway | Activation | hsa01521 | |||||||||||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Liquid biopsy; ATP-binding pocket affinity comparison assay | ||||||||||||
| Mechanism Description | Several mechanisms of resistance have been described to EGFR-TkIs, such as the occurrence of secondary mutation (T790M, C797S), the activation of alternative signalling (Met, HGF, AXL, Hh, IGF-1R), the aberrance of the downstream pathways (AkT mutations, loss of PTEN), the impairment of the EGFR-TkIs-mediated apoptosis pathway (BCL2-like 11/BIM deletion polymorphism) and histological transformation. | ||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [51] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Missense mutation | p.D761Y |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
Direct Sanger sequencing analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Blue cell viability assay | ||||||||||||
| Mechanism Description | The T790M mutation is common in patients with acquired resistance. The limited spectrum of TkI-resistant mutations in EGFR, which binds to erlotinib in the active conformation, contrasts with a wider range of second-site mutations seen with acquired resistance to imatinib, which binds to ABL and kIT, respectively, in closed conformations. Collectively, our data suggest that the type and nature of kinase inhibitor resistance mutations may be influenced by both anatomic site and mode of binding to the kinase target. | ||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [49] | ||||||||||||
| Resistant Disease | EGFR-mutant non-small cell lung cancer [ICD-11: 2C25.7] | ||||||||||||
| Molecule Alteration | Missense mutation | p.L747S |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
Low throughput experiment assay | ||||||||||||
| Experiment for Drug Resistance |
Progression-free survival assay | ||||||||||||
| Mechanism Description | Acquired resistance can occur through failure of drug delivery to the target, as in isolated central nervous system (CNS) progression, or by selection of biological variants during TkI exposure. | ||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [49] | ||||||||||||
| Resistant Disease | EGFR-mutant non-small cell lung cancer [ICD-11: 2C25.7] | ||||||||||||
| Molecule Alteration | Missense mutation | p.D761Y |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
Low throughput experiment assay | ||||||||||||
| Experiment for Drug Resistance |
Progression-free survival assay | ||||||||||||
| Mechanism Description | Acquired resistance can occur through failure of drug delivery to the target, as in isolated central nervous system (CNS) progression, or by selection of biological variants during TkI exposure. | ||||||||||||
| Key Molecule: Thymidylate synthase (TYMS) | [4] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vivo Model | Patient-derived advanced NSCLC model | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Colony formation assay | ||||||||||||
| Mechanism Description | In our study, we first illustrated the role of TS-mediated thymidylate nucleotide biosynthesis in the development of gefitinib resistance. We demonstrated that NSCLC patients with higher expression of TS gene have shorter PFS during EGFR-TKI treatment. Furthermore, we found TS is upregulated in NSCLC patients resistant to gefitinib and in PC9/GR cells which are tolerant of gefitinib. Knockdown of TS-induced apoptosis and diminished survival of gefitinib-resistant NSCLC. | ||||||||||||
| Key Molecule: Thymidylate synthase (TYMS) | [4] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HCC827/GR cells | N.A. | Homo sapiens (Human) | CVCL_E7R9 | |||||||||
| PC9/GR cells | N.A. | Homo sapiens (Human) | N.A. | ||||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Experiment for Drug Resistance |
CCK8 assay; Colony formation assay | ||||||||||||
| Mechanism Description | In our study, we first illustrated the role of TS-mediated thymidylate nucleotide biosynthesis in the development of gefitinib resistance. We demonstrated that NSCLC patients with higher expression of TS gene have shorter PFS during EGFR-TKI treatment. Furthermore, we found TS is upregulated in NSCLC patients resistant to gefitinib and in PC9/GR cells which are tolerant of gefitinib. Knockdown of TS-induced apoptosis and diminished survival of gefitinib-resistant NSCLC. | ||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [3] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Mutation | . |
|||||||||||
| Experiment for Drug Resistance |
Statistical analysis | ||||||||||||
| Mechanism Description | EGFR-TKI Rechallenge With Another TKI may be a useful treatment option after first-line osimertinib. | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Cytochrome P450 family 1 subfamily B member1 (CYP1B1) | [36] | |||
| Sensitive Disease | Kidney cancer [ICD-11: 2C90.1] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Kidney cancer [ICD-11: 2C90] | |||
| The Specified Disease | Renal cancer | |||
| The Studied Tissue | Kidney | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.73E-03 Fold-change: -1.49E-01 Z-score: -3.05E+00 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | miR27b/CCNG1/p53 signaling pathway | Regulation | N.A. | |
| In Vitro Model | 769-P cells | Kidney | Homo sapiens (Human) | CVCL_1050 |
| 786-O cells | Kidney | Homo sapiens (Human) | CVCL_1051 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | |||
| Mechanism Description | miR-27b synergizes with anticancer drugs througth enhancing anticancer drug-induced cell death which due to p53 activation and CYP1B1 suppression. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Apoptosis regulator Bcl-2 (BCL2) | [5] | |||
| Resistant Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.3] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell colony | Activation | hsa05200 | ||
| Cell viability | Activation | hsa05200 | ||
| miR129/BCL2 signaling pathway | Regulation | N.A. | ||
| In Vitro Model | TE-1 cells | Esophagus | Homo sapiens (Human) | CVCL_1759 |
| KYSE-450 cells | Esophagus | Homo sapiens (Human) | CVCL_1353 | |
| TE6 cells | Esophageal | Homo sapiens (Human) | CVCL_1765 | |
| TE8 cells | Esophageal | Homo sapiens (Human) | CVCL_1766 | |
| TTn cells | Esophageal | Homo sapiens (Human) | CVCL_3175 | |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; TUNEL assay | |||
| Mechanism Description | Exosome-mediated transfer of PART1 promoted gefitinib resistance by competitively binding to miR-129 to facilitate Bcl-2 expression in ESCC cells. | |||
| Key Molecule: Prostate androgen-regulated transcript 1 (PART1) | [5] | |||
| Resistant Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.3] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell colony | Activation | hsa05200 | ||
| Cell viability | Activation | hsa05200 | ||
| miR129/BCL2 signaling pathway | Regulation | N.A. | ||
| In Vitro Model | TE-1 cells | Esophagus | Homo sapiens (Human) | CVCL_1759 |
| KYSE-450 cells | Esophagus | Homo sapiens (Human) | CVCL_1353 | |
| TE6 cells | Esophageal | Homo sapiens (Human) | CVCL_1765 | |
| TE8 cells | Esophageal | Homo sapiens (Human) | CVCL_1766 | |
| TTn cells | Esophageal | Homo sapiens (Human) | CVCL_3175 | |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay; TUNEL assay | |||
| Mechanism Description | Exosome-mediated transfer of PART1 promoted gefitinib resistance by competitively binding to miR-129 to facilitate Bcl-2 expression in ESCC cells. | |||
| Key Molecule: hsa-mir-129 | [5] | |||
| Resistant Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.3] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell colony | Activation | hsa05200 | ||
| Cell viability | Activation | hsa05200 | ||
| miR129/BCL2 signaling pathway | Regulation | N.A. | ||
| In Vitro Model | TE-1 cells | Esophagus | Homo sapiens (Human) | CVCL_1759 |
| KYSE-450 cells | Esophagus | Homo sapiens (Human) | CVCL_1353 | |
| TE6 cells | Esophageal | Homo sapiens (Human) | CVCL_1765 | |
| TE8 cells | Esophageal | Homo sapiens (Human) | CVCL_1766 | |
| TTn cells | Esophageal | Homo sapiens (Human) | CVCL_3175 | |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; TUNEL assay | |||
| Mechanism Description | Exosome-mediated transfer of PART1 promoted gefitinib resistance by competitively binding to miR-129 to facilitate Bcl-2 expression in ESCC cells. | |||
| Key Molecule: Prostate androgen-regulated transcript 1 (PART1) | [5] | |||
| Resistant Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.3] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell colony | Activation | hsa05200 | ||
| Cell viability | Activation | hsa05200 | ||
| miR129/BCL2 signaling pathway | Regulation | N.A. | ||
| In Vitro Model | TE-1 cells | Esophagus | Homo sapiens (Human) | CVCL_1759 |
| KYSE-450 cells | Esophagus | Homo sapiens (Human) | CVCL_1353 | |
| TE6 cells | Esophageal | Homo sapiens (Human) | CVCL_1765 | |
| TE8 cells | Esophageal | Homo sapiens (Human) | CVCL_1766 | |
| TTn cells | Esophageal | Homo sapiens (Human) | CVCL_3175 | |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay; TUNEL assay | |||
| Mechanism Description | Exosome-mediated transfer of PART1 promoted gefitinib resistance by competitively binding to miR-129 to facilitate Bcl-2 expression in ESCC cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-1 | [38] | |||
| Sensitive Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.3] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| PI3K/AKT/survivin signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | TE-1 cells | Esophagus | Homo sapiens (Human) | CVCL_1759 |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | Exogenous expression of miR 1 inhibited growth, arrested cell cycle in the G1 phase and increased apoptosis in ESCC cells, whereas it decreased PIk3CA protein expression levels. Furthermore, overexpression of miR 1 increased the sensitivity of ESCC cells to the anticancer drug, gefitinib. | |||
|
|
||||
| Key Molecule: PI3-kinase alpha (PIK3CA) | [38] | |||
| Sensitive Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.3] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| PI3K/AKT/survivin signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | TE-1 cells | Esophagus | Homo sapiens (Human) | CVCL_1759 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | Exogenous expression of miR 1 inhibited growth, arrested cell cycle in the G1 phase and increased apoptosis in ESCC cells, whereas it decreased PIk3CA protein expression levels. Furthermore, overexpression of miR 1 increased the sensitivity of ESCC cells to the anticancer drug, gefitinib. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-147 | [39] | |||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell migration | Inhibition | hsa04670 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | SW480 cells | Colon | Homo sapiens (Human) | CVCL_0546 |
| HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-147 strikingly increased the sensitivity to EGFR inhibitor, gefitinib in cell with native resistance. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Cytochrome P450 family 1 subfamily B member1 (CYP1B1) | [36] | |||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | miR27b/CCNG1/p53 signaling pathway | Regulation | N.A. | |
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| SNU182 cells | Liver | Homo sapiens (Human) | CVCL_0090 | |
| SNU-739 cells | Liver | Homo sapiens (Human) | CVCL_5088 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | |||
| Mechanism Description | miR-27b synergizes with anticancer drugs througth enhancing anticancer drug-induced cell death which due to p53 activation and CYP1B1 suppression. | |||
|
|
||||
| Key Molecule: hsa-mir-27b | [36] | |||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | miR27b/CCNG1/p53 signaling pathway | Regulation | N.A. | |
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| SNU182 cells | Liver | Homo sapiens (Human) | CVCL_0090 | |
| SNU-739 cells | Liver | Homo sapiens (Human) | CVCL_5088 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | |||
| Mechanism Description | miR-27b synergizes with anticancer drugs througth enhancing anticancer drug-induced cell death which due to p53 activation and CYP1B1 suppression. | |||
|
|
||||
| Key Molecule: Cyclin-G1 (CCNG1) | [36] | |||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| miR27b/CCNG1/p53 signaling pathway | Regulation | N.A. | ||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| SNU182 cells | Liver | Homo sapiens (Human) | CVCL_0090 | |
| SNU-739 cells | Liver | Homo sapiens (Human) | CVCL_5088 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | |||
| Mechanism Description | miR-27b synergizes with anticancer drugs througth enhancing anticancer drug-induced cell death which due to p53 activation and CYP1B1 suppression. | |||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
