Molecule Information
General Information of the Molecule (ID: Mol00597)
| Name |
Receptor tyrosine-protein kinase erbB-3 (ERBB3)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Proto-oncogene-like protein c-ErbB-3; Tyrosine kinase-type cell surface receptor HER3; HER3
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
ERBB3
|
||||
| Gene ID | |||||
| Location |
chr12:56076799-56103505[+]
|
||||
| Sequence |
MRANDALQVLGLLFSLARGSEVGNSQAVCPGTLNGLSVTGDAENQYQTLYKLYERCEVVM
GNLEIVLTGHNADLSFLQWIREVTGYVLVAMNEFSTLPLPNLRVVRGTQVYDGKFAIFVM LNYNTNSSHALRQLRLTQLTEILSGGVYIEKNDKLCHMDTIDWRDIVRDRDAEIVVKDNG RSCPPCHEVCKGRCWGPGSEDCQTLTKTICAPQCNGHCFGPNPNQCCHDECAGGCSGPQD TDCFACRHFNDSGACVPRCPQPLVYNKLTFQLEPNPHTKYQYGGVCVASCPHNFVVDQTS CVRACPPDKMEVDKNGLKMCEPCGGLCPKACEGTGSGSRFQTVDSSNIDGFVNCTKILGN LDFLITGLNGDPWHKIPALDPEKLNVFRTVREITGYLNIQSWPPHMHNFSVFSNLTTIGG RSLYNRGFSLLIMKNLNVTSLGFRSLKEISAGRIYISANRQLCYHHSLNWTKVLRGPTEE RLDIKHNRPRRDCVAEGKVCDPLCSSGGCWGPGPGQCLSCRNYSRGGVCVTHCNFLNGEP REFAHEAECFSCHPECQPMEGTATCNGSGSDTCAQCAHFRDGPHCVSSCPHGVLGAKGPI YKYPDVQNECRPCHENCTQGCKGPELQDCLGQTLVLIGKTHLTMALTVIAGLVVIFMMLG GTFLYWRGRRIQNKRAMRRYLERGESIEPLDPSEKANKVLARIFKETELRKLKVLGSGVF GTVHKGVWIPEGESIKIPVCIKVIEDKSGRQSFQAVTDHMLAIGSLDHAHIVRLLGLCPG SSLQLVTQYLPLGSLLDHVRQHRGALGPQLLLNWGVQIAKGMYYLEEHGMVHRNLAARNV LLKSPSQVQVADFGVADLLPPDDKQLLYSEAKTPIKWMALESIHFGKYTHQSDVWSYGVT VWELMTFGAEPYAGLRLAEVPDLLEKGERLAQPQICTIDVYMVMVKCWMIDENIRPTFKE LANEFTRMARDPPRYLVIKRESGPGIAPGPEPHGLTNKKLEEVELEPELDLDLDLEAEED NLATTTLGSALSLPVGTLNRPRGSQSLLSPSSGYMPMNQGNLGESCQESAVSGSSERCPR PVSLHPMPRGCLASESSEGHVTGSEAELQEKVSMCRSRSRSRSPRPRGDSAYHSQRHSLL TPVTPLSPPGLEEEDVNGYVMPDTHLKGTPSSREGTLSSVGLSSVLGTEEEDEDEEYEYM NRRRRHSPPHPPRPSSLEELGYEYMDVGSDLSASLGSTQSCPLHPVPIMPTAGTTPDEDY EYMNRQRDGGGPGGDYAAMGACPASEQGYEEMRAFQGPGHQAPHVHYARLKTLRSLEATD SAFDNPDYWHSRLFPKANAQRT Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Tyrosine-protein kinase that plays an essential role as cell surface receptor for neuregulins. Binds to neuregulin-1 (NRG1) and is activated by it; ligand-binding increases phosphorylation on tyrosine residues and promotes its association with the p85 subunit of phosphatidylinositol 3-kinase. May also be activated by CSPG5. Involved in the regulation of myeloid cell differentiation.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
4 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [1] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Docetaxel | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | |
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
Colony formation assay | |||
| Mechanism Description | The reintroduction of miR-205 is shown to inhibit cell proliferation and clonogenic potential, and increase the sensitivity of MCF-7 and MDA-MB-231 cells to docetaxel. miR-205 also shows a synergistic effect with docetaxel in vivo. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [2] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Gefitinib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell growth | Inhibition | hsa05200 | |
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |
| HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |
| Experiment for Molecule Alteration |
Luciferase target assay | |||
| Experiment for Drug Resistance |
Fluorescence-activated cell sorting assay | |||
| Mechanism Description | The activation of the PI3k/Akt survival pathway, so critically important in tumorigenesis, is for the most part driven through phosphorylation of the kinase-inactive member HER3. miR-205, negatively regulating HER3, is able to inhibit breast cancer cell proliferation and improves the response to specific targeted therapies. The reintroduction of miR-205 in SkBr3 cells inhibits their clonogenic potential and increases the responsiveness to tyrosine-kinase inhibitors Gefitinib and Lapatinib, abrogating the HER3-mediated resistance and restoring a potent proapoptotic activity. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [3] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Lapatinib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Activation | hsa05200 | |
| ERRB2/3 signaling pathway | Activation | hsa04210 | ||
| In Vitro Model | ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
WST-1 proliferation assay | |||
| Mechanism Description | Breast Cancer Anti-Estrogen Resistance 4 (BCAR4) Drives Proliferation of IPH-926 lobular Carcinoma Cells. Relative high BCAR4 mRNA expression was identified in IPH-926, a cell line derived from an endocrine-resistant lobular breast cancer. Moderate BCAR4 expression was evident in MDA-MB-134 and MDA-MB-453 breast cancer cells. BCAR4 protein was detected in breast cancer cells with ectopic (ZR-75-1-BCAR4) and endogenous (IPH-926, MDA-MB-453) BCAR4 mRNA expression. knockdown of BCAR4 inhibited cell proliferation. A similar effect was observed upon knockdown of ERBB2/3 and exposure to lapatinib, implying that BCAR4 acts in an ERBB2/3-dependent manner.BCAR4 encodes a functional protein, which drives proliferation of endocrine-resistant breast cancer cells. Lapatinib, a clinically approved EGFR/ERBB2 inhibitor, counteracts BCAR4-driven tumor cell growth, a clinical relevant observation. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [2] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Lapatinib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell growth | Inhibition | hsa05200 | |
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |
| HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |
| Experiment for Molecule Alteration |
Luciferase target assay | |||
| Experiment for Drug Resistance |
Fluorescence-activated cell sorting assay | |||
| Mechanism Description | The activation of the PI3k/Akt survival pathway, so critically important in tumorigenesis, is for the most part driven through phosphorylation of the kinase-inactive member HER3. miR-205, negatively regulating HER3, is able to inhibit breast cancer cell proliferation and improves the response to specific targeted therapies. The reintroduction of miR-205 in SkBr3 cells inhibits their clonogenic potential and increases the responsiveness to tyrosine-kinase inhibitors Gefitinib and Lapatinib, abrogating the HER3-mediated resistance and restoring a potent proapoptotic activity. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: ret-aberrant cancers [ICD-11: 2D4Z] | [4] | |||
| Resistant Disease | ret-aberrant cancers [ICD-11: 2D4Z] | |||
| Resistant Drug | Selpercatinib | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | YAP-HER3 signaling pathway | Regulation | N.A. | |
| In Vitro Model | LC2/ad cells | Pleural effusion | Homo sapiens (Human) | CVCL_1373 |
| TPC-1 cells | Thyroid | Homo sapiens (Human) | CVCL_6298 | |
| TT cells | Thyroid gland | Homo sapiens (Human) | CVCL_1774 | |
| In Vivo Model | Severe combined immunodeficiency mice model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot assay; RT-PCR; RNA sequencing assay; Phospho-receptor tyrosine kinase antibody arrays assay; Chromatin immunoprecipitation assay; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
Cell viability assay | |||
| Mechanism Description | In high YAP-expressing RET-aberrant cancer cells, YAP-mediated HER3 signaling activation maintained cell survival and induced the emergence of cells tolerant to the RET-TKIs selpercatinib and pralsetinib. The pan-ErBB inhibitor afatinib and YAP/tea domain inhibitors verteporfin and K-975 sensitized YAP-expressing RET-aberrant cancer cells to the RET-TKIs selpercatinib and pralsetinib. Pretreatment YAP expression in clinical specimens obtained from patients with RET fusion-positive lung cancer was associated with poor RET-TKI treatment outcomes.The YAP-HER3 axis is crucial for the survival and adaptive resistance of high YAP-expressing RET-aberrant cancer cells treated with RET-TKIs. Combining YAP/HER3 inhibition with RET-TKIs represents a highly potent strategy for initial treatment. | |||
Preclinical Drug(s)
2 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [5] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Anti-HER3 mAbs | |||
| Molecule Alteration | Missense mutation | p.G284R (c.850G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Mechanism Description | The missense mutation p.G284R (c.850G>A) in gene ERBB3 cause the sensitivity of anti-HER3 mAbs by aberration of the drug's therapeutic target | |||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [5] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Anti-HER3 mAbs | |||
| Molecule Alteration | Missense mutation | p.P262H (c.785C>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Mechanism Description | The missense mutation p.P262H (c.785C>A) in gene ERBB3 cause the sensitivity of anti-HER3 mAbs by aberration of the drug's therapeutic target | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [5] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | PI3K pathway inhibitors/MEK inhibitors | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q809R (c.2426A>G) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.50 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.10 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
K
-
R
-
A
-
M
-
R
-
660
|
R
-
Y
-
L
-
E
-
R
-
G
-
E
-
S
-
I
-
E
-
670
|
P
-
L
-
D
-
P
-
S
-
E
-
K
G
A
A
N
G
K
K
680
|
V
V
L
L
A
A
R
R
I
I
F
F
K
K
E
E
T
T
E
E
690
|
L
L
R
R
K
K
L
L
K
K
V
V
L
L
G
G
S
S
G
G
700
|
V
V
F
F
G
G
T
T
V
V
H
H
K
K
G
G
V
V
W
W
710
|
I
I
P
P
E
E
G
G
E
E
S
S
I
I
K
K
I
I
P
P
720
|
V
V
C
C
I
I
K
K
V
V
I
I
E
E
D
D
K
K
S
S
730
|
G
G
R
R
Q
Q
S
S
F
F
Q
Q
A
A
V
V
T
T
D
D
740
|
H
H
M
M
L
L
A
A
I
I
G
G
S
S
L
L
D
D
H
H
750
|
A
A
H
H
I
I
V
V
R
R
L
L
L
L
G
G
L
L
C
C
760
|
P
P
G
G
S
S
S
S
L
L
Q
Q
L
L
V
V
T
T
Q
Q
770
|
Y
Y
L
L
P
P
L
L
G
G
S
S
L
L
L
L
D
D
H
H
780
|
V
V
R
R
Q
Q
H
H
R
R
G
G
A
A
L
L
G
G
P
P
790
|
Q
R
L
L
L
L
L
L
N
N
W
W
G
G
V
V
Q
Q
I
I
800
|
A
A
K
K
G
G
M
M
Y
Y
Y
Y
L
L
E
E
E
E
H
H
810
|
G
G
M
M
V
V
H
H
R
R
N
N
L
L
A
A
A
A
R
R
820
|
N
N
V
V
L
L
L
L
K
K
S
S
P
P
S
S
Q
Q
V
V
830
|
Q
Q
V
V
A
A
D
D
F
F
G
G
V
V
A
A
D
D
L
L
840
|
L
L
P
P
P
P
D
D
D
D
K
K
Q
Q
L
L
L
L
Y
Y
850
|
S
S
E
E
A
A
K
K
T
T
P
P
I
I
K
K
W
W
M
M
860
|
A
A
L
L
E
E
S
S
I
I
H
H
F
F
G
G
K
K
Y
Y
870
|
T
T
H
H
Q
Q
S
S
D
D
V
V
W
W
S
S
Y
Y
G
G
880
|
V
V
T
T
V
V
W
W
E
E
L
L
M
M
T
T
F
F
G
G
890
|
A
A
E
E
P
P
Y
Y
A
A
G
G
L
L
R
R
L
L
A
A
900
|
E
E
V
V
P
P
D
D
L
L
L
L
E
E
K
K
G
G
E
E
910
|
R
R
L
L
A
A
Q
Q
P
P
Q
Q
I
I
C
C
T
T
I
I
920
|
D
D
V
V
Y
Y
M
M
V
V
M
M
V
V
K
K
C
C
W
W
930
|
M
M
I
I
D
D
E
E
N
N
I
I
R
R
P
P
T
T
F
F
940
|
K
K
E
E
L
L
A
A
N
N
E
E
F
F
T
T
R
R
M
M
950
|
A
A
R
R
D
D
P
P
P
P
R
R
Y
Y
L
L
V
V
I
I
960
|
K
K
R
R
E
E
S
S
G
G
P
P
G
G
I
I
A
A
P
P
970
|
G
G
P
P
E
E
P
P
H
H
G
G
L
L
T
T
N
N
K
K
980
|
K
K
L
L
E
E
-
E
-
V
-
E
-
L
-
E
-
P
-
E
990
|
-
L
-
D
-
L
-
D
-
L
-
D
-
L
-
E
-
A
-
E
1000
|
-
E
-
D
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
DNA sequencing assay | ||||||||||||
| Mechanism Description | The missense mutation p.Q809R (c.2426A>G) in gene ERBB3 cause the sensitivity of PI3K pathway inhibitors + MEK inhibitors by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [5] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | PI3K pathway inhibitors/MEK inhibitors | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q809R (c.2426A>G) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.50 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.10 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
K
-
R
-
A
-
M
-
R
-
660
|
R
-
Y
-
L
-
E
-
R
-
G
-
E
-
S
-
I
-
E
-
670
|
P
-
L
-
D
-
P
-
S
-
E
-
K
G
A
A
N
G
K
K
680
|
V
V
L
L
A
A
R
R
I
I
F
F
K
K
E
E
T
T
E
E
690
|
L
L
R
R
K
K
L
L
K
K
V
V
L
L
G
G
S
S
G
G
700
|
V
V
F
F
G
G
T
T
V
V
H
H
K
K
G
G
V
V
W
W
710
|
I
I
P
P
E
E
G
G
E
E
S
S
I
I
K
K
I
I
P
P
720
|
V
V
C
C
I
I
K
K
V
V
I
I
E
E
D
D
K
K
S
S
730
|
G
G
R
R
Q
Q
S
S
F
F
Q
Q
A
A
V
V
T
T
D
D
740
|
H
H
M
M
L
L
A
A
I
I
G
G
S
S
L
L
D
D
H
H
750
|
A
A
H
H
I
I
V
V
R
R
L
L
L
L
G
G
L
L
C
C
760
|
P
P
G
G
S
S
S
S
L
L
Q
Q
L
L
V
V
T
T
Q
Q
770
|
Y
Y
L
L
P
P
L
L
G
G
S
S
L
L
L
L
D
D
H
H
780
|
V
V
R
R
Q
Q
H
H
R
R
G
G
A
A
L
L
G
G
P
P
790
|
Q
R
L
L
L
L
L
L
N
N
W
W
G
G
V
V
Q
Q
I
I
800
|
A
A
K
K
G
G
M
M
Y
Y
Y
Y
L
L
E
E
E
E
H
H
810
|
G
G
M
M
V
V
H
H
R
R
N
N
L
L
A
A
A
A
R
R
820
|
N
N
V
V
L
L
L
L
K
K
S
S
P
P
S
S
Q
Q
V
V
830
|
Q
Q
V
V
A
A
D
D
F
F
G
G
V
V
A
A
D
D
L
L
840
|
L
L
P
P
P
P
D
D
D
D
K
K
Q
Q
L
L
L
L
Y
Y
850
|
S
S
E
E
A
A
K
K
T
T
P
P
I
I
K
K
W
W
M
M
860
|
A
A
L
L
E
E
S
S
I
I
H
H
F
F
G
G
K
K
Y
Y
870
|
T
T
H
H
Q
Q
S
S
D
D
V
V
W
W
S
S
Y
Y
G
G
880
|
V
V
T
T
V
V
W
W
E
E
L
L
M
M
T
T
F
F
G
G
890
|
A
A
E
E
P
P
Y
Y
A
A
G
G
L
L
R
R
L
L
A
A
900
|
E
E
V
V
P
P
D
D
L
L
L
L
E
E
K
K
G
G
E
E
910
|
R
R
L
L
A
A
Q
Q
P
P
Q
Q
I
I
C
C
T
T
I
I
920
|
D
D
V
V
Y
Y
M
M
V
V
M
M
V
V
K
K
C
C
W
W
930
|
M
M
I
I
D
D
E
E
N
N
I
I
R
R
P
P
T
T
F
F
940
|
K
K
E
E
L
L
A
A
N
N
E
E
F
F
T
T
R
R
M
M
950
|
A
A
R
R
D
D
P
P
P
P
R
R
Y
Y
L
L
V
V
I
I
960
|
K
K
R
R
E
E
S
S
G
G
P
P
G
G
I
I
A
A
P
P
970
|
G
G
P
P
E
E
P
P
H
H
G
G
L
L
T
T
N
N
K
K
980
|
K
K
L
L
E
E
-
E
-
V
-
E
-
L
-
E
-
P
-
E
990
|
-
L
-
D
-
L
-
D
-
L
-
D
-
L
-
E
-
A
-
E
1000
|
-
E
-
D
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
DNA sequencing assay | ||||||||||||
| Mechanism Description | The missense mutation p.Q809R (c.2426A>G) in gene ERBB3 cause the sensitivity of PI3K pathway inhibitors + MEK inhibitors by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [5] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | PI3K pathway inhibitors/MEK inhibitors | ||||||||||||
| Molecule Alteration | Missense mutation | p.P262H (c.785C>A) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
DNA sequencing assay | ||||||||||||
| Mechanism Description | The missense mutation p.P262H (c.785C>A) in gene ERBB3 cause the sensitivity of PI3K pathway inhibitors + MEK inhibitors by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [5] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | PI3K pathway inhibitors/MEK inhibitors | ||||||||||||
| Molecule Alteration | Missense mutation | p.G284R (c.850G>A) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
DNA sequencing assay | ||||||||||||
| Mechanism Description | The missense mutation p.G284R (c.850G>A) in gene ERBB3 cause the sensitivity of PI3K pathway inhibitors + MEK inhibitors by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [5] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | PI3K pathway inhibitors/MEK inhibitors | ||||||||||||
| Molecule Alteration | Missense mutation | p.P262H (c.785C>A) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
DNA sequencing assay | ||||||||||||
| Mechanism Description | The missense mutation p.P262H (c.785C>A) in gene ERBB3 cause the sensitivity of PI3K pathway inhibitors + MEK inhibitors by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [5] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | PI3K pathway inhibitors/MEK inhibitors | ||||||||||||
| Molecule Alteration | Missense mutation | p.G284R (c.850G>A) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
DNA sequencing assay | ||||||||||||
| Mechanism Description | The missense mutation p.G284R (c.850G>A) in gene ERBB3 cause the sensitivity of PI3K pathway inhibitors + MEK inhibitors by unusual activation of pro-survival pathway | ||||||||||||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.51E-01; Fold-change: 1.01E-01; Z-score: 1.65E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.36E-03; Fold-change: 1.66E-01; Z-score: 3.95E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
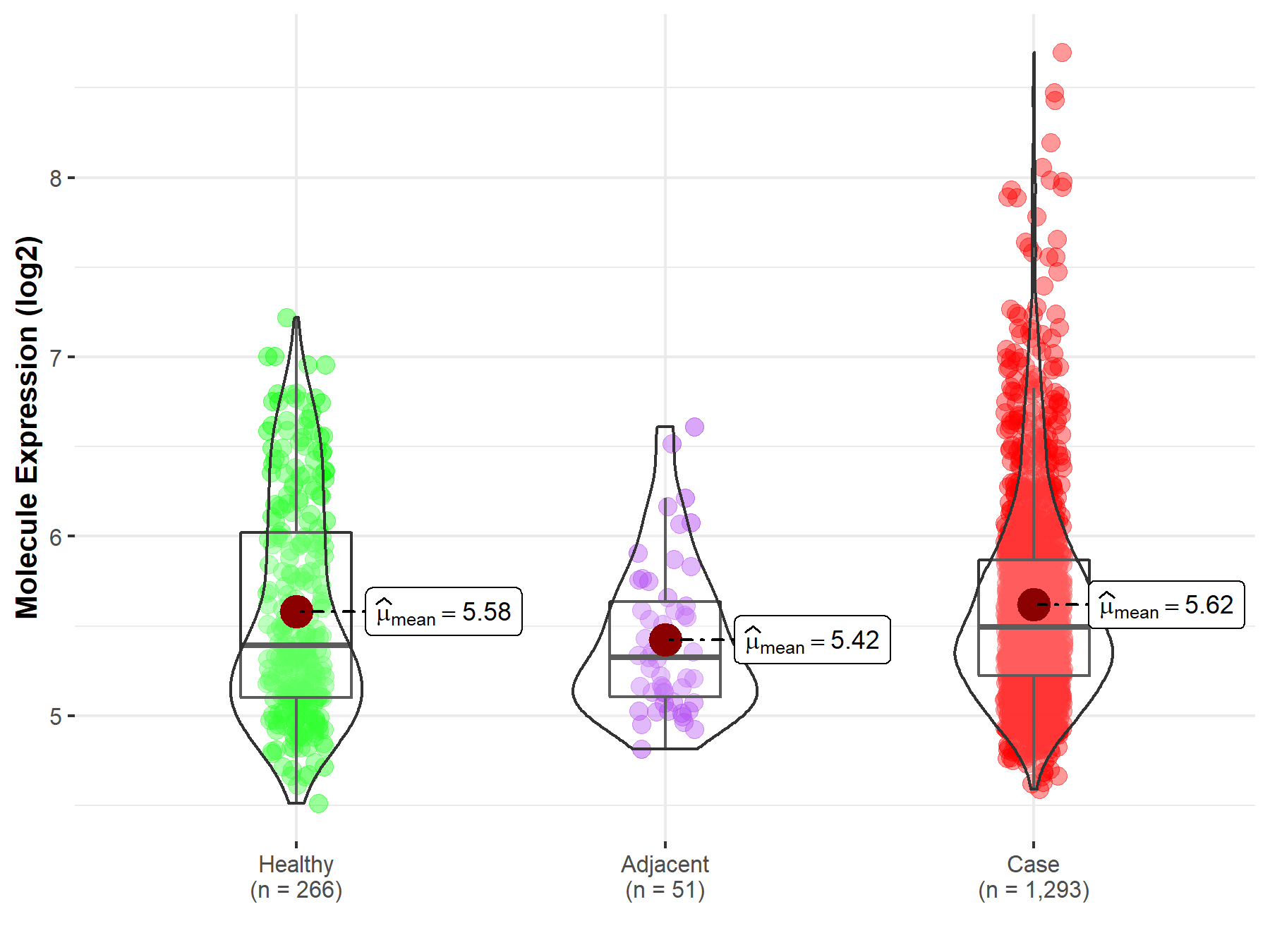
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

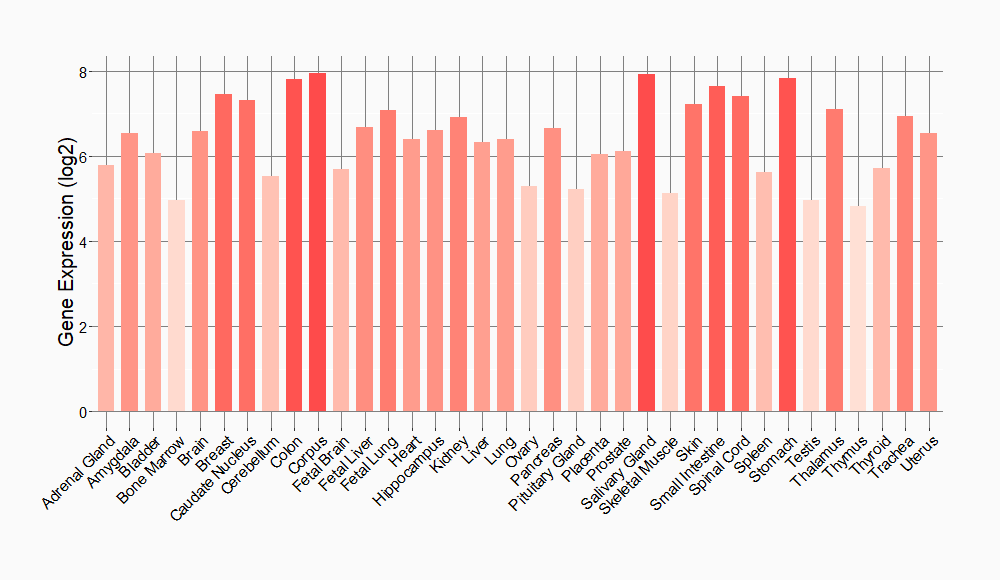
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
