Drug Information
Drug (ID: DG00293) and It's Reported Resistant Information
| Name |
Chloramphenicol
|
||||
|---|---|---|---|---|---|
| Synonyms |
Alficetyn; Ambofen; Amphenicol; Amphicol; Amseclor; Anacetin; Aquamycetin; Austracil; Austracol; Biocetin; Biophenicol; CAF; CLM; Catilan; Chemiceticol; Chemicetin; Chemicetina; Chlomin; Chlomycol; Chloramex; Chloramfenikol; Chloramficin; Chloramfilin; Chloramphenicolum; Chloramsaar; Chlorasol; Chloricol; Chlornitromycin; Chloroamphenicol; Chlorocaps; Chlorocid; Chlorocide; Chlorocol; Chlorofair; Chloromax; Chloromycetin; Chloromycetny; Chloromyxin; Chloronitrin; Chloroptic; Chlorovules; Chlorsig; Cidocetine; Ciplamycetin; Cloramfen; Cloramfenicol; Cloramfenicolo; Cloramficin; Cloramical; Cloramicol; Cloramidina; Cloranfenicol; Cloroamfenicolo; Clorocyn; Cloromisan; Cloromissan; Clorosintex; Comycetin; Cylphenicol; Desphen; Detreomycin; Detreomycine; Dextromycetin; Doctamicina; Duphenicol; Econochlor; Embacetin; Emetren; Enicol; Enteromycetin; Erbaplast; Ertilen; Farmicetina; Fenicol; Globenicol; Glorous; Halomycetin; Hortfenicol; Interomycetine; Intramycetin; Intramyctin; Isicetin; Ismicetina; Isophenicol; Juvamycetin; Kamaver; Kemicetina; Kemicetine; Kloramfenikol; Klorita; Laevomycetinum; Leukamycin; Leukomyan; Leukomycin; Levomicetina; Levomitsetin; Levomycetin; Loromicetina; Loromisan; Loromisin; Mastiphen; Mediamycetine; Medichol; Micloretin; Micochlorine; Micoclorina; Microcetina; Mychel; Mycinol; Myscel; Novochlorocap; Novomycetin; Novophenicol; Oftalent; Oleomycetin; Opclor; Opelor; Ophthochlor; Ophtochlor; Optomycin; Otachron; Otophen; Pantovernil; Paraxin; Pentamycetin; Quemicetina; Rivomycin; Romphenil; Ronfenil; Ronphenil; Septicol; Sificetina; Sintomicetina; Stanomycetin; Synthomycetin; Synthomycetine; Synthomycine; Syntomycin; Tevcocin; Tevcosin; Tifomycin; Tifomycine;Tiromycetin; Treomicetina; Unimycetin; Veticol; Viceton; Ch loramex; Chloramfenikol [Czech]; Chloramphenicol crystalline; Chlormycetin R; Chlorocid S; Chlorocidin C; Chlorocidin C tetran; Chloroject L; Chloromycetny [Polish]; Cloramfenicolo [DCIT]; Cloroamfenicolo [Italian]; F armicetina; Isopto fenicol; Klorocid S; Normimycin V; Sintomicetine R; Sno Phenicol; Vice ton; I 337A; Ak-chlor; Alficetyn (TN); Amphicol (TN); Biomicin (TN); Brochlor (TN); CAF (pharmaceutical); Cedoctine (TN); Chlora-tabs; Chloramex (TN); Chloramphenicol & VRC3375; Chloramphenicolum [INN-Latin]; Chlorbiotic (Veterinary); Chlornitromycin (TN); Chloro-25 vetag; Chloromycetin (TN); Chlorsig (TN); Cloramfenicol [INN-Spanish]; D-Chloramphenicol; Dispersadron C (TN); Econochlor (TN); Elase-Chloromycetin; Fenicol (TN); Golden Eye (TN); Isoptophenicol (TN); Kemicetine (TN); Laevomycetin (TN); Medicom (TN); Mychel-Vet; Nevimycin (TN); Oftan Chlora (TN); Optrex Infected Eyes (TN); Orchadexoline (TN); Phenicol (TN); Renicol (TN); Silmycetin (TN); Sno-Phenicol; Synthomycine (TN); Tea-Cetin; Tega-Cetin; Tifomycine (TN); U-6062; Vernacetin (TN); Veticol (TN); C.A.F; CHLOROPTIC S.O.P; Chloramphenicol [INN:BAN:JAN]; Chloromycetin® D-threo-Chloramphenicol; Alficetyn, Chlornitromycin, Chloromycetin, Chloramphenicol; Chloramphenicol (JP15/USP/INN); Chloroptic S.O.P.; D(-)-threo-Chloramphenicol; D-(-)-Chloramphenicol; D-(-)-threo-Chloramphenicol; D(-)-threo-2-dichloroacetamido-1-p-nitrophen yl-propanediol; D(-)-threo-2-dichloroacetamido-1-p-nitrophenyl-propanediol
Click to Show/Hide
|
||||
| Indication |
In total 1 Indication(s)
|
||||
| Structure |
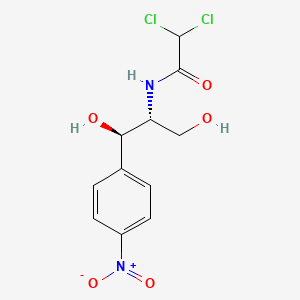
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Clinically Reported Resistance for This Drug
(12 diseases)
[2]
[3]
[4]
[3]
[2]
[2]
[5]
[6]
[2]
[7]
[2]
[2]
Disease(s) with Resistance Information Validated by in-vivo Model for This Drug
(8 diseases)
[8]
[9]
[10]
[11]
[12]
[13]
[14]
[15]
|
||||
| Target | Bacterial 23S ribosomal RNA (Bact 23S rRNA) | NOUNIPROTAC | [1] | ||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C11H12Cl2N2O5
|
||||
| IsoSMILES |
C1=CC(=CC=C1[C@H]([C@@H](CO)NC(=O)C(Cl)Cl)O)[N+](=O)[O-]
|
||||
| InChI |
1S/C11H12Cl2N2O5/c12-10(13)11(18)14-8(5-16)9(17)6-1-3-7(4-2-6)15(19)20/h1-4,8-10,16-17H,5H2,(H,14,18)/t8-,9-/m1/s1
|
||||
| InChIKey |
WIIZWVCIJKGZOK-RKDXNWHRSA-N
|
||||
| PubChem CID | |||||
| ChEBI ID | |||||
| TTD Drug ID | |||||
| VARIDT ID | |||||
| INTEDE ID | |||||
| DrugBank ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-01: Infectious/parasitic diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: AAC(6')-Ib family aminoglycoside 6'-N-acetyltransferase (AAC6IB) | [16] | |||
| Resistant Disease | Vibrio cholerae infection [ICD-11: 1A00.0] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Vibrio cholerae PL107b | 666 | ||
| Experiment for Molecule Alteration |
PCR and DNA sequencing assay | |||
| Experiment for Drug Resistance |
Commercial antimicrobial discs assay | |||
| Mechanism Description | The expression of aac(6')-Ib lead to drug resistance. | |||
|
|
||||
| Key Molecule: Erythromycin esterase (EREA2) | [16] | |||
| Resistant Disease | Vibrio cholerae infection [ICD-11: 1A00.0] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Vibrio cholerae PG153/1 | 666 | ||
| Vibrio cholerae PG170 | 666 | |||
| Experiment for Molecule Alteration |
PCR and DNA sequencing assay | |||
| Experiment for Drug Resistance |
Commercial antimicrobial discs assay | |||
| Mechanism Description | The expression of dfrA5, ereA2 lead to drug resistance. | |||
|
|
||||
| Key Molecule: Bcr/CflA family efflux transporter (BCML) | [4] | |||
| Resistant Disease | Vibrio cholerae infection [ICD-11: 1A00.0] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli DH5alpha | 668369 | ||
| Escherichia coli k-12 strain TOP10 | 83333 | |||
| Vibrio cholerae O1 C10488 | 127906 | |||
| Vibrio cholerae O1 strain CO943 | 127906 | |||
| Vibrio cholerae O139 1811/98 | 45888 | |||
| Vibrio cholerae O139 2055 | 45888 | |||
| Vibrio cholerae O139 AS207 | 45888 | |||
| Vibrio cholerae O139 E712 | 45888 | |||
| Vibrio cholerae O139 HkO139-SXTS | 45888 | |||
| Vibrio cholerae O139 strain MO10 | 345072 | |||
| Experiment for Molecule Alteration |
Sequencing assay | |||
| Mechanism Description | Many recent Asian clinical Vibrio cholerae E1 Tor O1 and O139 isolates are resistant to the antibiotics sulfamethoxazole (Su), trimethoprim (Tm), chloramphenicol (Cm), and streptomycin (Sm). The corresponding resistance genes are located on large conjugative elements (SXT constins) that are integrated into prfC on the V. cholerae chromosome. The DNA sequences of the antibiotic resistance genes in the SXT constin in MO10, an O139 isolate. In SXT(MO10), these genes are clustered within a composite transposon-like structure found near the element's 5' end. The genes conferring resistance to Cm (floR), Su (sulII), and Sm (strA and strB) correspond to previously described genes, whereas the gene conferring resistance to Tm, designated dfr18, is novel. | |||
|
|
||||
| Key Molecule: Dihydrofolate reductase (DHFR) | [16] | |||
| Resistant Disease | Vibrio cholerae infection [ICD-11: 1A00.0] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Vibrio cholerae PG149a | 666 | ||
| Vibrio cholerae PG262(b) | 666 | |||
| Vibrio cholerae PG95 | 666 | |||
| Vibrio cholerae PL61 | 666 | |||
| Vibrio cholerae PL78/6 | 666 | |||
| Vibrio cholerae PL105b | 666 | |||
| Vibrio cholerae PL141 | 666 | |||
| Experiment for Molecule Alteration |
PCR and DNA sequencing assay | |||
| Experiment for Drug Resistance |
Commercial antimicrobial discs assay | |||
| Mechanism Description | The expression of dfrA1 lead to drug resistance. | |||
| Key Molecule: Dihydrofolate reductase (DHFR) | [16] | |||
| Resistant Disease | Vibrio cholerae infection [ICD-11: 1A00.0] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Vibrio cholerae PG153/1 | 666 | ||
| Vibrio cholerae PG170 | 666 | |||
| Experiment for Molecule Alteration |
PCR and DNA sequencing assay | |||
| Experiment for Drug Resistance |
Commercial antimicrobial discs assay | |||
| Mechanism Description | The expression of dfrA15 lead to drug resistance. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Chloramphenicol acetyltransferase (CAT) | [17] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Paenibacillus sp. LC231 | 1120679 | ||
| Mechanism Description | Redundant chloramphenicol (catV and clbB) and kanamycin (ant(4')-lc and aac(6')-35) resistance are common in Paenibacillaceae, especially within Brevibacillus and Aneurinibacillus. | |||
| Key Molecule: Chloramphenicol acetyltransferase (CAT) | [18] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Paenibacillus sp. LC231 | 1120679 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | CatU inactivates chloramphenicol by acetylation. | |||
| Key Molecule: Aminoglycoside acetyltransferase (AAC) | [19] | |||
| Resistant Disease | Vibrio fluvialis infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Vibrio fluvialis H-08942 | 676 | ||
| Experiment for Molecule Alteration |
PCR; DNA sequencing assay; Southern hybridization assay; Cloning and expression assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Aac(3)-Id is a new type of aminoglycoside acetyltransferase gene which causes drug resistance. | |||
| Key Molecule: Chloramphenicol acetyltransferase (CAT) | [20] | |||
| Resistant Disease | Enterococcus faecalis infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Enterococcus faecalis JH2-2 | 1351 | ||
| Escherichia coli strain XL-1 Blue | 562 | |||
| Enterococcus faecalis ESP91 | 1351 | |||
| Enterococcus faecalis FO1 | 1351 | |||
| Enterococcus faecalis FO5 | 1351 | |||
| Enterococcus faecalis JHBURE16-1 | 1351 | |||
| Enterococcus faecalis JHBURE16-2 | 1351 | |||
| Enterococcus faecalis JHBURE16-3 | 1351 | |||
| Enterococcus faecalis JHBURE8-1 | 1351 | |||
| Enterococcus faecalis JHBURE8-2 | 1351 | |||
| Enterococcus faecalis JHBURE8-3 | 1351 | |||
| Enterococcus faecalis JHRE25-2 | 1351 | |||
| Enterococcus faecalis JHRE25-3 | 1351 | |||
| Enterococcus faecalis RE17 | 1351 | |||
| Enterococcus faecalis RE25 | 1351 | |||
| Enterococcus faecalis RE38 | 1351 | |||
| Enterococcus faecalis RE44 | 1351 | |||
| Enterococcus faecalis RE52 | 1351 | |||
| Enterococcus faecium FI1 | 1352 | |||
| Escherichia coli CM1 | CM25 | |||
| Escherichia coli CM2 | CM25 | |||
| Escherichia coli CM25 | 562 | |||
| Lactococcus lactis susp. cremoris AC1 | 1359 | |||
| Lactococcus lactis susp. lactis biovar. diacetylactis Bu2-60 | 44688 | |||
| Lactococcus lactis susp. lactis biovar. diacetylactis Bu2-60/pAMb1 | 44688 | |||
| Lactococcus lactis susp. lactis biovar. diacetylactis Bu2-60/pIP501 | 44688 | |||
| Lactococcus lactis susp. lactis biovar. diacetylactis Bu2-60/pRE39 | 44688 | |||
| Lactococcus lactis susp. lactis biovar. diacetylactis BURE25-11 | 44688 | |||
| Lactococcus lactis susp. lactis biovar. diacetylactis BURE25-12 | 44688 | |||
| Lactococcus lactis susp. lactis biovar. diacetylactis BURE25-15 | 44688 | |||
| Lactococcus lactis susp. lactis biovar. diacetylactis BURE25-16 | 44688 | |||
| Lactococcus lactis susp. lactis biovar. diacetylactis BURE25-3 | 44688 | |||
| Lactococcus lactis susp. lactis biovar. diacetylactis BURE25-6 | 44688 | |||
| Lactococcus lactis susp. lactis biovar. diacetylactis BURE25-7 | 44688 | |||
| Lactococcus lactis susp. lactis biovar. diacetylactis BURE25-8 | 44688 | |||
| Lactococcus lactis susp. lactis biovar. diacetylactis BURE25-9 | 44688 | |||
| Listeria innocua L19 | 1642 | |||
| Listeria innocua L191 | 1642 | |||
| Listeria innocua L193 | 1642 | |||
| Staphylococcus xylosus strains VF5 | 1288 | |||
| Experiment for Molecule Alteration |
DNA hybridizations assay | |||
| Experiment for Drug Resistance |
Microdilution test assay | |||
| Mechanism Description | Two antibiotic-resistance genes are present on this 30.5-kb region, a chloramphenicol acetyltransferase gene (orf10) and a 23S rRNA methyltransferase gene (orf14). Both genes have been shown to be active in E. faecalis RE25 and in its transconjugants. | |||
| Key Molecule: CATB6 chloramphenicol acetyltransferase (CATB6) | [3] | |||
| Resistant Disease | Pseudomonas aeruginosa infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli | 668369 | ||
| Pseudomonas aeruginosa strain 101/1477 | 287 | |||
| Experiment for Molecule Alteration |
Southern blotting assay | |||
| Experiment for Drug Resistance |
Broth microdilution assay | |||
| Mechanism Description | The third gene cassette is 730 bp long and contains an open reading frame (ORF) potentially encoding a protein that exhibits a high degree of sequence similarity to members of the CATB lineage of chloramphenicol acetyltransferases. The new catB allele appeared to be functional since both DH5alpha(pPAM-101) and DH5alpha(pkAM-36BE) showed a decreased chloramphenicol susceptibility and was named catB6. | |||
| Key Molecule: Chloramphenicol acetyltransferase (CAT) | [21] | |||
| Resistant Disease | Lactobacillus reuteri infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli strain DH5a | 668369 | ||
| Escherichia coli strain CSR 603 | 562 | |||
| Escherichia coli strain DH5a-CR17 | 668369 | |||
| Escherichia coli strain DH5a-CR36 | 668369 | |||
| Lactobacillus reuteri strain DSM 20016 | 557436 | |||
| Lactobacillus reuteri strain DSM 20016-CR3 | 557436 | |||
| Lactobacillus reuteri strain G4 | 1598 | |||
| Lactobacillus reuteri strain G4-CS1-3 | 1598 | |||
| Experiment for Molecule Alteration |
Hybridization assay | |||
| Mechanism Description | Lactobacillus reuteri G4 contains a 7.0-kb plasmid (pTC82) encoding resistance to chloramphenicol (Cm). Determination of the nucleotide sequence of the genetic determinant (cat-TC) encoding resistance to Cm on pTC82 revealed an open reading frame for a 238-amino-acid Cm acetyltransferase (CAT) monomer. This is the first reported nucleotide sequence of a Cm-resistance determinant from L. reuteri and also the first evidence of adding Lactobacillus to the list of versatile bacterial genera which naturally acquire the cat-pC194 gene in the microbial ecological system. | |||
| Key Molecule: Chloramphenicol acetyltransferase (CAT) | [22] | |||
| Resistant Disease | Proteus mirabilis infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli strain JM103 | 83333 | ||
| Proteus mirabilis strain PM13 | 584 | |||
| Proteus mirabilis strain PM2 | 584 | |||
| Experiment for Molecule Alteration |
RNA-DNA hybridizations assay | |||
| Mechanism Description | In Proteus mirabilis PM13 chloramphenicol resistance is mediated by the cat gene, a single copy of which is present in both resistant and sensitive isolates and which reverts at a high frequency. RNA measurements show an about 8.5-fold increase in cat-specific mRNA in cells expressing the resistance phenotype as compared with those which are sensitive to chloramphenicol. | |||
| Key Molecule: Chloramphenicol acetyltransferase 2 (CATII) | [23] | |||
| Resistant Disease | Haemophilus influenzae infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli strain JM 101 | 562 | ||
| Mechanism Description | Bacterial resistance to the antibiotic chloramphenicol, an inhibitor of the peptidyltransferase activity of prokaryotic ribosomes, is commonly conferred by the enzyme chloramphenicol acetyltransferase (CAT,EC2.3.1.28). The enzyme catalyses transfer of the acetyl group of acetyl-CoA to the primary (C-3) hydroxy group of chloramphenicol, yielding 3-acetylchloramphenicol, which fails to bind to bacterial ribosomes. Three classes of CAT variant have been characterized among Gram-negative bacteria, designated typesI, II and III. | |||
| Key Molecule: Chloramphenicol acetyltransferase (CAT) | [24] | |||
| Resistant Disease | Clostridium perfringens infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli | 668369 | ||
| Clostridium perfringens strain CW531 | 1502 | |||
| Experiment for Molecule Alteration |
Double-stranded dideoxy-chain termination method assay | |||
| Mechanism Description | The enzyme chloramphenicol acetyltransferase (CAT) mediates the inactivation of the antibiotic chloramphenicol, a potent inhibitor of prokaryotic peptidyltransferase activity. The active CAT enzyme, which catalyzes the acetyl coenzyme A-dependent acetylation of chloramphenicol, is a trimer of identical subunits of approximately 25 kDa. The nucleotide sequence of the Clostridium perfringens chloramphenicol acetyltransferase (CAT)-encoding resistance determinant, catQ, was determined. Phylogenetic analysis revealed that the CATQ monomer was as closely related to CAT proteins from Staphylococcus aureus and Campylobacter coli as it was to CAT monomers from the clostridia. | |||
| Key Molecule: Chloramphenicol acetyltransferase (CAT) | [25] | |||
| Resistant Disease | Agvobactevitlm tumefuciens infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Agrobacterium tumefaciens strain C58 | 358 | ||
| Escherichia coli strain JM101 | 83333 | |||
| Experiment for Molecule Alteration |
Enzyme assay | |||
| Mechanism Description | The nucleotide sequence of a chloramphenicol-resistance (CmR) determinant from the Gram- soil bacterium Agrobacterium tumefaciens was determined, and its gene product was identified as Cm acetyltransferase (CAT). Comparison of the amino acid sequences of the A. tumefaciens CAT and various CAT proteins of Gram+ and Gram- origin shows no homology between this and the other enzymes. | |||
| Key Molecule: Chloramphenicol acetyltransferase (CAT) | [13] | |||
| Resistant Disease | Clostridium butyricum infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli | 668369 | ||
| Experiment for Molecule Alteration |
Nucleotide sequence assay | |||
| Mechanism Description | Bacterial resistance to chloramphenicol is most commonly mediated by production of the enzyme chloramphenicol acetyltransferase (CAT), which catalyzes the transfer of an acetyl group from acetyl coenzyme A to the primary hydroxyl group of chloramphenicol (O-acetylation). The O-acetoxy derivatives of chloramphenicol do not bind to bacterial ribosomes and are consequently devoid of antimicrobial activity. The five distinct clostridial cat genes that have been cloned include catP and catQ from C. perfringens, catD from Clostridium dificile, and catA and catB from C. butyricum. The C. perfringens genes catP and catQ and the C. difficile gene catD have been sequenced. | |||
| Key Molecule: Chloramphenicol acetyltransferase (CAT) | [26] | |||
| Resistant Disease | Vibrio anguillarum infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli strain CSR603 | 562 | ||
| Escherichia coli strain HBIOI | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Mechanism Description | The chloramphenicol resistant genes (cat) have been found in various bacterial chromosomes, in antibioticresistant (R) plasmids and sometimes within a transposable element. | |||
|
|
||||
| Key Molecule: Multidrug transporter MdfA (MDFA) | [27], [28] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli BL21(DE3) | 469008 | ||
| Escherichia coli C43 (DE3) | 562 | |||
| Mechanism Description | Being one of the best-characterized bacterial MFS antiporters biochemically, MdfA from Escherichia coli (ecMdfA) is known to confer resistance to a variety of structurally distinct cationic and zwitterionic lipophilic compounds, as well as to a number of electroneutral antibiotics of clinical importance. | |||
| Key Molecule: Chloramphenicol resistance protein (CMX) | [29], [30] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli S17-1 | 1227813 | ||
| Corynebacterium glutamicum ATCC 13032 | 196627 | |||
| Corynebacterium glutamicum CX61 | 1718 | |||
| Corynebacterium glutamicum CX73 | 1718 | |||
| Corynebacterium glutamicum RM3 | 1718 | |||
| Escherichia coli DH5alphaMCR | 668369 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Mechanism Description | The central region of Tn5564 encodes the chloramphenicol resistance gene cmx, specifying a transmembrane chloramphenicol efflux protein, and an open reading frame homologous to transposases of insertion sequences identified in Arthrobacter nicotinovorans and Bordetella pertussis. | |||
| Key Molecule: ARE-ABC-F family resistance factor PoxtA (POXTA) | [31] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Staphylococcus aureus RN4220 | 1280 | ||
| Enterococcus faecalis JH2-2 | 1351 | |||
| Escherichia coli Mach1 T1R | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth dilution test assay | |||
| Mechanism Description | The poxtA gene encodes a protein that is 32% identical to OptrA and exhibits structural features typical of the F lineage of the ATP-binding cassette (ABC) protein superfamily that cause antibiotic resistance by ribosomal protection. | |||
| Key Molecule: Protein pexA (PEXA) | [32] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli | 668369 | ||
| Experiment for Molecule Alteration |
Nucleotide sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution assay | |||
| Mechanism Description | In its natural host, pexA could provide protection against chloramphenicol and florfenicol excreted by Streptomyces spp. | |||
| Key Molecule: Bcr/CflA family efflux transporter (BCML) | [33] | |||
| Resistant Disease | Pseudomonas aeruginosa infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli DH10B | 316385 | ||
| Pseudomonas aeruginosa PU21 | 287 | |||
| Escherichia coli strain k-12 C600 | 83333 | |||
| Pseudomonas aeruginosa 104116 | 287 | |||
| Pseudomonas aeruginosa SOF-1 | 287 | |||
| Experiment for Molecule Alteration |
Southern technique assay | |||
| Experiment for Drug Resistance |
Agar dilution technique assay | |||
| Mechanism Description | An additional ORF located downstream corresponded to a cmlA-like gene that encodes CMLA6 for chloramphenicol resistance and that shared 99% amino acid identity with CMLA1, with only three amino acid changes. | |||
| Key Molecule: Bcr/CflA family efflux transporter (BCML) | [34] | |||
| Resistant Disease | Enterobacter aerogenes infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli JM83 | 562 | ||
| Enterobacter aerogenes strain | 548 | |||
| Enterobacter aerogenes strain BM2688 | 548 | |||
| Enterobacter aerogenes strain BM2688-1 | 548 | |||
| Escherichia coli strain J5-3 | 562 | |||
| Experiment for Molecule Alteration |
Southern hybridization assay | |||
| Experiment for Drug Resistance |
Disk diffusion assay | |||
| Mechanism Description | A putative GTG initiation codon at position 718 was preceded at 8 bp by a RBS-like sequence. This coding sequence, designated cmlA2, shared 83.7% identity with the cmlA1 gene of the class 1 integron In4 in Tn1696 which confers nonenzymatic chloramphenicol resistance. | |||
| Key Molecule: Chloramphenicol resistance protein (Tn5561-Unclear) | [35] | |||
| Resistant Disease | Rhodococcus erythropolis infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Rhodococcus erythropolis strain SQ1 | 1833 | ||
| Experiment for Molecule Alteration |
Southern hybridization assay | |||
| Mechanism Description | Three copies of the IS21-related transposable element IS1415 were identified in Rhodococcus erythropolis NI86/21. Adjacent to one of the IS1415 copies, a 47-bp sequence nearly identical to the conserved 5* end of integrons was found. Accurate transposition of IS1415 carrying a chloramphenicol resistance gene (Tn5561) was demonstrated following delivery from a suicide vector to R. erythropolis SQ1. | |||
| Key Molecule: Multidrug transporter MdfA (MDFA) | [10] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli MC1061 | 1211845 | ||
| Escherichia coli strain DH5a | 668369 | |||
| Bacillus subtilis strain BR151 | 1423 | |||
| Rhodococcus fascians strain D188-5 | 2022521 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Mechanism Description | Rhodococcus fascians NCPPB 1675 (located on the conjugative plasmid pRF2) allowed the identification of two possible open reading frames (ORFs), of which ORF1 was consistent with the mutational analysis. Biochemical analysis of cmr revealed that it does not encode an antibiotic-modifying enzyme. The amino acid sequence of ORF1 predicted a hydrophobic protein, with 12 putative membrane-spanning domains, homologous to proteins involved in the efflux of tetracycline across the plasma membrane. | |||
|
|
||||
| Key Molecule: Enterococcal surface protein (ESP) | [36] | |||
| Resistant Disease | Enterococci faecium infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Enterococcus faecalis strain JH2-2 | 1320322 | ||
| Enterococcus faecalis strain pIP1326g | 1351 | |||
| Enterococcus faecalis strain pIP655 | 1351 | |||
| Enterococcus faecalis strain pIP683 | 1351 | |||
| Enterococcus faecalis strain pIP687 | 1351 | |||
| Enterococcus faecium strain pIP1182 | 1352 | |||
| Enterococcus faecium strain pIP1535 | 1352 | |||
| Enterococcus faecium strain pIP1538 | 1352 | |||
| Enterococcus faecium strain pIP1539 | 1352 | |||
| Enterococcus faecium strain pIP1687 | 1352 | |||
| Enterococcus faecium strain pIP713 | 1352 | |||
| Streptococci strain A451 | 36470 | |||
| Streptococci strain A453 | 36470 | |||
| Streptococci strain A456 | 36470 | |||
| Streptococci strain B109 | 1319 | |||
| Streptococci strain B117 | 1319 | |||
| Streptococci strain B118 | 1319 | |||
| Streptococci strain B120 | 1319 | |||
| Streptococci strain B126 | 1319 | |||
| Streptococci strain B127 | 1319 | |||
| Streptococci strain BM132 | 1319 | |||
| Streptococci strain BM137 | 36470 | |||
| Streptococci strain BM140 | 1319 | |||
| Streptococci strain G44 | 1320 | |||
| Streptococci strain G52 | 1320 | |||
| Streptococci strain G54 | 1320 | |||
| Experiment for Molecule Alteration |
Southern blotting assay | |||
| Mechanism Description | An assay based on the utilization of degenerate primers that enable enzymatic amplification of an internal fragment of cat genes known to be present in gram-positive cocci was developed to identify the genes encoding chloramphenicol resistance in streptococci and enterococci. The functionality of this system was illustrated by the detection of cat genes belonging to four different hydridization classes represented by the staphylococcal genes catpC221, catpC194, catpSCS7, and the clostridial gene catP, and by the characterization of a new streptococcal cat gene designated catS. | |||
| Key Molecule: Enterococcal surface protein (ESP) | [36] | |||
| Resistant Disease | Enterococci faecalisc infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Enterococcus faecalis strain JH2-2 | 1320322 | ||
| Enterococcus faecalis strain pIP1326g | 1351 | |||
| Enterococcus faecalis strain pIP655 | 1351 | |||
| Enterococcus faecalis strain pIP683 | 1351 | |||
| Enterococcus faecalis strain pIP687 | 1351 | |||
| Enterococcus faecium strain pIP1182 | 1352 | |||
| Enterococcus faecium strain pIP1535 | 1352 | |||
| Enterococcus faecium strain pIP1538 | 1352 | |||
| Enterococcus faecium strain pIP1539 | 1352 | |||
| Enterococcus faecium strain pIP1687 | 1352 | |||
| Enterococcus faecium strain pIP713 | 1352 | |||
| Streptococci strain A451 | 36470 | |||
| Streptococci strain A453 | 36470 | |||
| Streptococci strain A456 | 36470 | |||
| Streptococci strain B109 | 1319 | |||
| Streptococci strain B117 | 1319 | |||
| Streptococci strain B118 | 1319 | |||
| Streptococci strain B120 | 1319 | |||
| Streptococci strain B126 | 1319 | |||
| Streptococci strain B127 | 1319 | |||
| Streptococci strain BM132 | 1319 | |||
| Streptococci strain BM137 | 36470 | |||
| Streptococci strain BM140 | 1319 | |||
| Streptococci strain G44 | 1320 | |||
| Streptococci strain G52 | 1320 | |||
| Streptococci strain G54 | 1320 | |||
| Experiment for Molecule Alteration |
Southern blotting assay | |||
| Mechanism Description | An assay based on the utilization of degenerate primers that enable enzymatic amplification of an internal fragment of cat genes known to be present in gram-positive cocci was developed to identify the genes encoding chloramphenicol resistance in streptococci and enterococci. The functionality of this system was illustrated by the detection of cat genes belonging to four different hydridization classes represented by the staphylococcal genes catpC221, catpC194, catpSCS7, and the clostridial gene catP, and by the characterization of a new streptococcal cat gene designated catS. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: CATB6 chloramphenicol acetyltransferase (CATB6) | [3] | |||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | |||
| Molecule Alteration | Expression | Acquired |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli | 668369 | ||
| Pseudomonas aeruginosa strain 101/1477 | 287 | |||
| Experiment for Molecule Alteration |
Southern blotting assay | |||
| Experiment for Drug Resistance |
Broth microdilution assay | |||
| Mechanism Description | The third gene cassette is 730 bp long and contains an open reading frame (ORF) potentially encoding a protein that exhibits a high degree of sequence similarity to members of the CATB lineage of chloramphenicol acetyltransferases. The new catB allele appeared to be functional since both DH5alpha(pPAM-101) and DH5alpha(pkAM-36BE) showed a decreased chloramphenicol susceptibility and was named catB6. | |||
| Key Molecule: Chloramphenicol acetyltransferase (CAT) | [37] | |||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | |||
| Molecule Alteration | Expression | Acquired |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli strain k12 | 83333 | ||
| Escherichia coli strain JM111 | 83333 | |||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Mechanism Description | Enzymic acetylation catalysed by chloramphenicol acetyltransferase is the commonest mechanism of bacterial resistance to the antibiotic chloramphenicol, an inhibitor of prokaryotic peptidyl-transferase activity. | |||
| Key Molecule: Chloramphenicol acetyltransferase 2 (CATII) | [23] | |||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli strain JM 101 | 562 | ||
| Mechanism Description | Bacterial resistance to the antibiotic chloramphenicol, an inhibitor of the peptidyltransferase activity of prokaryotic ribosomes, is commonly conferred by the enzyme chloramphenicol acetyltransferase (CAT,EC2.3.1.28). The enzyme catalyses transfer of the acetyl group of acetyl-CoA to the primary (C-3) hydroxy group of chloramphenicol, yielding 3-acetylchloramphenicol, which fails to bind to bacterial ribosomes. Three classes of CAT variant have been characterized among Gram-negative bacteria, designated typesI, II and III. | |||
| Key Molecule: Chloramphenicol acetyltransferase (CAT) | [7] | |||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | |||
| Molecule Alteration | Expression | Acquired |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli strain XL-1 Blue | 562 | ||
| A Staphylococcus intermedius strain isolated from a purulent skin infection of a dog | 1285 | |||
| Experiment for Molecule Alteration |
Dideoxy chain-termination method assay | |||
| Mechanism Description | Subsequently, Escherichia coli XL-1 blue cells transformed with these recombinant plasmids were tested for CmR. In one orientation, Escherichia coli XL-1 blue demonstrated CmR at 15 ug/ml Cm while in the other orientation a higher level of CmR occurred (80 ug/m Cm). | |||
| Key Molecule: Chloramphenicol acetyltransferase (CAT) | [13] | |||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | |||
| Molecule Alteration | Expression | Acquired |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli | 668369 | ||
| Experiment for Molecule Alteration |
Nucleotide sequence assay | |||
| Mechanism Description | Bacterial resistance to chloramphenicol is most commonly mediated by production of the enzyme chloramphenicol acetyltransferase (CAT), which catalyzes the transfer of an acetyl group from acetyl coenzyme A to the primary hydroxyl group of chloramphenicol (O-acetylation). The O-acetoxy derivatives of chloramphenicol do not bind to bacterial ribosomes and are consequently devoid of antimicrobial activity. Recombinant strains were derivatives of Escherichia coli DH5alpha and were grown in 2YT medium supplemented with ampicillin (100 ug/ml) and chloramphenicol (30 ug/ml) where appropriate. Cloning experiments conducted in this study utilized the Escherichia coli plasmid vector pUC18. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Dipeptide and tripeptide permease A (DTPA) | [1] | |||
| Sensitive Disease | Escherichia coli infection [ICD-11: 1A03.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Escherichia coli BL21(DE3)pLysS cell | 866768 | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Determination of MICs assay | |||
| Mechanism Description | POT YdgR facilitates Cam uptake in E. coli. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Chloramphenicol acetyltransferase (CAT) | [12] | |||
| Resistant Disease | Clostridium difficile infection [ICD-11: 1A04.0] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| Mechanism Description | The inactivation of drug undergoes by the addition of side chain that generates a steric hindrance effect, which in turn disrupts the target-binding affinity. Two copies of catD gene encoding for CHL acetyltransferase locate at the mobile regions Tn4453a and Tn4453b of C. difficile. CHL acetyltransferase catalyses the relocation of acetyl group from acetyl-CoA to CHL, resulting in 3-O-acetyl CHL, which cannot bind to a ribosome and loses its antimicrobial action. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Multidrug transporter MdfA (MDFA) | [38] | |||
| Resistant Disease | Salmonella enterica infection [ICD-11: 1A09.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Salmonella enterica serovar Typhimurium ATCC 14028s | 588858 | ||
| Experiment for Molecule Alteration |
Quantitative real-time PCR | |||
| Experiment for Drug Resistance |
L agar plate method assay | |||
| Mechanism Description | Overexpression or overproduction of mdfA confers drug resistance. | |||
| Key Molecule: Catalase isozyme A/Tetracycline efflux MFS transporter/Dihydropteroate synthase (CATA1/TETB/SUL) | [39] | |||
| Resistant Disease | Salmonella enterica infection [ICD-11: 1A09.0] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Salmonella agona 231 | 58095 | ||
| Experiment for Molecule Alteration |
PCR screening assay | |||
| Experiment for Drug Resistance |
Disc diffusion assay | |||
| Mechanism Description | The multiresistance plasmid from S. Agona strain 231 carried the chloramphenicol resistance gene catA1 coding for a type A chloramphenicol acetyltransferase and the resistance gene tet(B) coding for a tetracycline/minocycline exporter. This plasmid also harboured the streptomycin resistance gene strA coding for an aminoglycoside phosphotransferase and the sulphonamide resistance gene sul1 which represents part of the 3' conserved segment of class 1 integrons. | |||
| Key Molecule: Bcr/CflA family efflux transporter (BCML) | [6] | |||
| Resistant Disease | Salmonella enterica infection [ICD-11: 1A09.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Salmonella enterica serotype Typhimurium SS034 | 90371 | ||
| Salmonella enterica serotype Typhimurium 1358 | 90371 | |||
| Salmonella enterica serotype Typhimurium 2039 | 90371 | |||
| Salmonella enterica serotype Typhimurium 2152 | 90371 | |||
| Salmonella enterica serotype Typhimurium 2668 | 90371 | |||
| Salmonella enterica serotype Typhimurium 2855 | 90371 | |||
| Salmonella enterica serotype Typhimurium 3430 | 90371 | |||
| Salmonella enterica serotype Typhimurium 3977 | 90371 | |||
| Salmonella enterica serotype Typhimurium 4204 | 90371 | |||
| Salmonella enterica serotype Typhimurium 4255 | 90371 | |||
| Salmonella enterica serotype Typhimurium 4287 | 90371 | |||
| Salmonella enterica serotype Typhimurium 4501 | 90371 | |||
| Salmonella enterica serotype Typhimurium 4528 | 90371 | |||
| Salmonella enterica serotype Typhimurium 4656 | 90371 | |||
| Salmonella enterica serotype Typhimurium 922 | 90371 | |||
| Experiment for Molecule Alteration |
Southern blot hybridizations assay | |||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | Isolates 2039 and 2152 also carried an additional integron (In-t4) that encodes the cmlA and aadB gene cassettes, which confer resistance to chloramphenicol and kanamycin, respectively. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: OXA-23 carbapenemase (BLA OXA-23) | [40] | |||
| Resistant Disease | Cutaneous bacterial infection [ICD-11: 1B21.4] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Acinetobacter baumannii isolates | 470 | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Broth microdilution method assay; Agar dilution method assay | |||
| Mechanism Description | The isolate was resistant to antibiotics other than ampicillin-sulbactam and colistin, suggesting drug resistance due to carbapenemase production by OXA-23.carbapenem resistance in the isolated carbapenem-resistant A. baumannii strain was at least partially conferred by bla OXA-23-like carbapenemase. | |||
|
|
||||
| Key Molecule: Lincomycin resistance efflux pump (LMRS) | [2] | |||
| Resistant Disease | Superficial skin infection by Staphylococcus aureus infection [ICD-11: 1B21.3] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli kAM32 | 562 | ||
| Staphylococcus aureus OM505 | 1280 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | LmrS is a multidrug efflux pump of the major facilitator superfamily from staphylococcus aureus. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Chloramphenicol acetyltransferase gene (CATS) | [36] | |||
| Resistant Disease | Streptococci infection [ICD-11: 1B54.2] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Enterococcus faecalis strain JH2-2 | 1320322 | ||
| Enterococcus faecalis strain pIP1326g | 1351 | |||
| Enterococcus faecalis strain pIP655 | 1351 | |||
| Enterococcus faecalis strain pIP683 | 1351 | |||
| Enterococcus faecalis strain pIP687 | 1351 | |||
| Enterococcus faecium strain pIP1182 | 1352 | |||
| Enterococcus faecium strain pIP1535 | 1352 | |||
| Enterococcus faecium strain pIP1538 | 1352 | |||
| Enterococcus faecium strain pIP1539 | 1352 | |||
| Enterococcus faecium strain pIP1687 | 1352 | |||
| Enterococcus faecium strain pIP713 | 1352 | |||
| Streptococci strain A451 | 36470 | |||
| Streptococci strain A453 | 36470 | |||
| Streptococci strain A456 | 36470 | |||
| Streptococci strain B109 | 1319 | |||
| Streptococci strain B117 | 1319 | |||
| Streptococci strain B118 | 1319 | |||
| Streptococci strain B120 | 1319 | |||
| Streptococci strain B126 | 1319 | |||
| Streptococci strain B127 | 1319 | |||
| Streptococci strain BM132 | 1319 | |||
| Streptococci strain BM137 | 36470 | |||
| Streptococci strain BM140 | 1319 | |||
| Streptococci strain G44 | 1320 | |||
| Streptococci strain G52 | 1320 | |||
| Streptococci strain G54 | 1320 | |||
| Experiment for Molecule Alteration |
Southern blotting assay | |||
| Mechanism Description | An assay based on the utilization of degenerate primers that enable enzymatic amplification of an internal fragment of cat genes known to be present in gram-positive cocci was developed to identify the genes encoding chloramphenicol resistance in streptococci and enterococci. The functionality of this system was illustrated by the detection of cat genes belonging to four different hydridization classes represented by the staphylococcal genes catpC221, catpC194, catpSCS7, and the clostridial gene catP, and by the characterization of a new streptococcal cat gene designated catS. | |||
| Key Molecule: Chloramphenicol acetyltransferase (CAT) | [7] | |||
| Resistant Disease | Staphylococcus intermedius infection [ICD-11: 1B54.3] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli strain XL-1 Blue | 562 | ||
| A Staphylococcus intermedius strain isolated from a purulent skin infection of a dog | 1285 | |||
| Experiment for Molecule Alteration |
Dideoxy chain-termination method assay | |||
| Mechanism Description | However, little is known about CmR in staphylococcal species pathogenic to animals. Recently, CmR plasmids have been isolated from 'equine's. sciuri, 'canine' S. epidermidis, 'porcine' S. hyicus and 'canine' S. intermedius strains. All staphy- lococcal CmR plasmids encode a common resistance mechanism, namely an inducible Cm acetyltransferase (CAT). | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Ribosomal RNA large subunit methyltransferase N (CFRC) | [41] | |||
| Resistant Disease | Gram-negative bacterial infection [ICD-11: 1B74-1G40] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli HB101 | 634468 | ||
| Clostridioides difficile T10 | 1215084 | |||
| Clostridium bolteae 90B3 | 997895 | |||
| Escherichia coli TG1 | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Disk diffusion methods assay; agar dilution methods assay | |||
| Mechanism Description | The cfr gene encodes a 23S rRNA methyltransferase, which causes C-8 modification in A2503 located in the peptidyl transferase region of bacterial ribosome.This mechanism confers PhLOPSA (phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A) resistance. | |||
|
|
||||
| Key Molecule: MipA/OmpV family protein (MIPA) | [14] | |||
| Resistant Disease | Gram-negative bacterial infection [ICD-11: 1B74-1G40] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli k-12 BW25113 | 679895 | ||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | OM proteins, a unique OM component of Gram-negative bacteria, constitute a barrier against large hydrophilic and lipophilic molecules and therefore play an important role in stress responses to drugs, osmotic pressure and acids.MipA is a novel OM protein related to antibiotic resistance. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Bcr/CflA family efflux transporter (BCML) | [15] | |||
| Resistant Disease | Pasteurella multocida infection [ICD-11: 1B99.0] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli ATCC 25922 | 1322345 | ||
| Staphylococcus aureus ATCC 29213 | 1280 | |||
| Pasteurella multocida 36950 | 1075089 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | The analysis of one representative P. multocida isolate identified an 82 kb integrative and conjugative element (ICE) integrated into the chromosomal DNA. This ICE, designated ICEPmu1, harboured 11 resistance genes, which confer resistance to streptomycin/spectinomycin (aadA25), streptomycin (strA and strB), gentamicin (aadB), kanamycin/neomycin (aphA1), tetracycline [tetR-tet(H)], chloramphenicol/florfenicol (floR), sulphonamides (sul2), tilmicosin/clindamycin [erm(42)] or tilmicosin/tulathromycin [msr(E)-mph(E)]. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Chloramphenicol acetyltransferase (CAT) | [11] | |||
| Resistant Disease | Campylobacter fetus infection [ICD-11: 1C40.0] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Campylobacter coli strain UA585 | 195 | ||
| Escherichia coli strain JM 107 | 562 | |||
| Mechanism Description | A chloramphenicol-resistance determinant (CmR), originally cloned from Campylobacter coli plasmid pNR9589 in Japan, was isolated and the nucleotide sequence determined, which contained an open reading frame of 621 bp. The gene product was identified as Cm acetyltransferase (CAT), which had a putative amino acid sequence that showed 43% to 57% identity with other CAT proteins of both Gram+ and Gram- origin. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Chloramphenicol 3-O phosphotransferase (CH3OP) | [8] | |||
| Resistant Disease | Streptomyces venezuelae infection [ICD-11: 1C43.10] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli JM101 | 562 | ||
| Streptomyces lividans strain M252 | 1916 | |||
| Experiment for Molecule Alteration |
Dideoxy chain-termination method assay | |||
| Experiment for Drug Resistance |
Measuring the diameters of inhibition zones around the disks assay | |||
| Mechanism Description | The product of the ORF from S. venezuelae as an enzymic effector of Cm resistance in the producing organism by 3'-O-phosphorylation. We suggest the trivial name chloramphenicol 3'-O-phosphotransferase for the enzyme. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Lincomycin resistance efflux pump (LMRS) | [2] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli kAM32 | 562 | ||
| Staphylococcus aureus OM505 | 1280 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | LmrS is a multidrug efflux pump of the major facilitator superfamily from staphylococcus aureus. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Chloramphenicol acetyltransferase (CAT) | [9] | |||
| Resistant Disease | Bacillus pumilus infection [ICD-11: 1C4Y.3] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Bacillus pumilus strain NCIB8600 | 1408 | ||
| Bacillus subtilis strain BR151 | 1423 | |||
| Experiment for Molecule Alteration |
Restriction enzyme assay | |||
| Mechanism Description | Genes (cat) specifying the enzyme CAT occur in a wide range of unrelated bacteria, where they confer resistance to the antibiotic Cm. Gene cat-86 of Bacillus pumilus, specifying chloramphenicol-inducible chloramphenicol acetyltransferase, was previously cloned in Bacillus subtilis on plasmid pUB110. | |||
ICD-11: Circulatory system diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Lincomycin resistance efflux pump (LMRS) | [2] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli kAM32 | 562 | ||
| Staphylococcus aureus OM505 | 1280 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | LmrS is a multidrug efflux pump of the major facilitator superfamily from staphylococcus aureus. | |||
ICD-12: Respiratory system diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: MATE family efflux transporter (ABEM) | [42] | |||
| Resistant Disease | Acinetobacter baumannii infection [ICD-11: CA40.4] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli kAM32 | 562 | ||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | AbeM was found to be an H+-coupled multidrug efflux pump and a unique member of the MATE family which lead to drug resistance. | |||
| Key Molecule: Bcr/CflA family efflux transporter (BCML) | [5] | |||
| Resistant Disease | Klebsiella pneumoniae infection [ICD-11: CA40.1] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli DH10B | 316385 | ||
| Escherichia coli strain NCTC 50192 | 562 | |||
| Klebsiella pneumoniae strain ORI-1 | 573 | |||
| Experiment for Molecule Alteration |
PCR and hybridization experiments assay | |||
| Experiment for Drug Resistance |
Agar dilution technique assay | |||
| Mechanism Description | Like CMLA1, this novel protein (CMLA4) likely conferred resistance to chloramphenicol by a nonenzymatic mechanism. Among the 207 bp upstream of cmlA4, only four nucleotide changes were identified, compared to the sequence found upstream of cmlA1 (data not shown). Downstream from cmlA4, an inverse core site (GCCCAAC) was part of a composite 59-be of 70 bp. This 59-be was almost 100% identical to the downstream region of cmlA1, except for one nucleotide change (T to C in cmlA4) at the last position (position 4,997). | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Lincomycin resistance efflux pump (LMRS) | [2] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli kAM32 | 562 | ||
| Staphylococcus aureus OM505 | 1280 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | LmrS is a multidrug efflux pump of the major facilitator superfamily from staphylococcus aureus. | |||
ICD-21: Symptoms/clinical signs/unclassified clinical findings
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Lincomycin resistance efflux pump (LMRS) | [2] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli kAM32 | 562 | ||
| Staphylococcus aureus OM505 | 1280 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | LmrS is a multidrug efflux pump of the major facilitator superfamily from staphylococcus aureus. | |||
ICD-22: Injury/poisoning/certain external causes consequences
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Lincomycin resistance efflux pump (LMRS) | [2] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli kAM32 | 562 | ||
| Staphylococcus aureus OM505 | 1280 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | LmrS is a multidrug efflux pump of the major facilitator superfamily from staphylococcus aureus. | |||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
