Drug Information
Drug (ID: DG00118) and It's Reported Resistant Information
| Name |
Tetracycline
|
||||
|---|---|---|---|---|---|
| Synonyms |
Amycin; Biocycline; Bristaciclin; Bristaciclina; Bristacycline; Ciclibion; Copharlan; Cyclomycin; Cytome; Dumocyclin; Enterocycline; Medocycline; Resteclin; Robitet; Sanclomycine; Tetrachel; Veracin; Bristaciclin alpha; Cefracycline suspension; Component of Tetrastatin; Sumycin syrup; Tetracycline Free Base; Tetracycline I; Tetracycline II; Tetracycline Monohydrochloride; Achromycin (naphthacene derivative); Achromycin, naphthacene derivative; Centet (base); Lemtrex (base); Liquamycin (Veterinary); Liquamycin, veterinary; Panmycin (TN); Piracaps (base); Polycycline (VAN); Polycycline (antibiotic); Polycycline, antibiotic; SK-Tetracycline; Sumycin (TN); T-125; Tetra-Co; Tetraciclina [INN-Spanish]; Tetracycline & VRC3375; Tetracycline (internal use); Tetracyclinum [INN-Latin]; Tetracyn (TN); Vetquamycin-324 (free base); Tetracycline (JAN/USP/INN); Tetracycline [USAN:INN:BAN:JAN]; Methyl-1,11-dioxo-2-naphthacenecarboxamide; 6-Methyl-1,11-dioxy-2-naphthacenecarboxamide
Click to Show/Hide
|
||||
| Indication |
In total 1 Indication(s)
|
||||
| Structure |
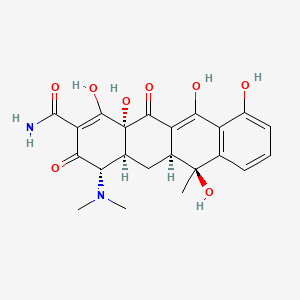
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Clinically Reported Resistance for This Drug
(13 diseases)
[2]
[3]
[4]
[3]
[5]
[6]
[7]
[8]
[9]
[10]
[11]
[12]
[13]
Disease(s) with Resistance Information Validated by in-vivo Model for This Drug
(6 diseases)
[14]
[15]
[16]
[17]
[18]
[19]
Disease(s) with Resistance Information Discovered by Cell Line Test for This Drug
(1 diseases)
[1]
|
||||
| Target | Staphylococcus 30S ribosomal subunit (Stap-coc pbp2) | F4NA87_STAAU | [1] | ||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C22H24N2O8
|
||||
| IsoSMILES |
C[C@@]1([C@H]2C[C@H]3[C@@H](C(=O)C(=C([C@]3(C(=O)C2=C(C4=C1C=CC=C4O)O)O)O)C(=O)N)N(C)C)O
|
||||
| InChI |
1S/C22H24N2O8/c1-21(31)8-5-4-6-11(25)12(8)16(26)13-9(21)7-10-15(24(2)3)17(27)14(20(23)30)19(29)22(10,32)18(13)28/h4-6,9-10,15,25-26,29,31-32H,7H2,1-3H3,(H2,23,30)/t9-,10-,15-,21+,22-/m0/s1
|
||||
| InChIKey |
NWXMGUDVXFXRIG-WESIUVDSSA-N
|
||||
| PubChem CID | |||||
| ChEBI ID | |||||
| TTD Drug ID | |||||
| VARIDT ID | |||||
| INTEDE ID | |||||
| DrugBank ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-01: Infectious/parasitic diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Dihydrofolate reductase (DHFR) | [5] | |||
| Resistant Disease | Vibrio cholerae infection [ICD-11: 1A00.0] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Vibrio cholerae PG149a | 666 | ||
| Vibrio cholerae PL61 | 666 | |||
| Vibrio cholerae PL78/6 | 666 | |||
| Vibrio cholerae PL91 | 666 | |||
| Vibrio cholerae PL141 | 666 | |||
| Experiment for Molecule Alteration |
PCR and DNA sequencing assay | |||
| Experiment for Drug Resistance |
Commercial antimicrobial discs assay | |||
| Mechanism Description | The expression of dfrA1 lead to drug resistance. | |||
| Key Molecule: Dihydrofolate reductase (DHFR) | [5] | |||
| Resistant Disease | Vibrio cholerae infection [ICD-11: 1A00.0] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Vibrio cholerae PG170 | 666 | ||
| Experiment for Molecule Alteration |
PCR and DNA sequencing assay | |||
| Experiment for Drug Resistance |
Commercial antimicrobial discs assay | |||
| Mechanism Description | The expression of dfrA15 lead to drug resistance. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Major facilitator superfamily (MFS) | [20] | |||
| Sensitive Disease | Vibrio cholerae infection [ICD-11: 1A00.0] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | E1 Tor C6706 | 1124478 | ||
| In Vivo Model | Infant mouse colonisation | Mus musculus | ||
| Experiment for Molecule Alteration |
Lux reporters assay | |||
| Experiment for Drug Resistance |
Plate counting assay | |||
| Mechanism Description | Deletion of all five mfs genes results in increased susceptibility to tetracycline and the antimicrobial agents present in crud. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Tetracycline resistance protein Tet (TETW/N/W) | [21] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli EPI-300 | 562 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | Tet(W/N/W) encodes mosaic ribosomal protection(since tetracyclines bind to the 30S ribosomal subunit to inhibit protein translation) and induces resistance. | |||
| Key Molecule: Tetracycline resistance protein TetM (TETM) | [22] | |||
| Resistant Disease | Enterococci infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Acquired |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Enterococcus faecalis OG1RF:pCF10 | 474186 | ||
| Enterococcus faecalis OG1SSp | 1351 | |||
| In Vivo Model | House fly model | House fly | ||
| Experiment for Molecule Alteration |
Bacterial colonies count assay | |||
| Experiment for Drug Resistance |
Broth dilution assay | |||
| Mechanism Description | Tetracycline resistance of Musca domestica occurred by transferring the plasmid transduced with tetracycline resistance gene TETM of Enterococcus into Musca domestica. | |||
| Key Molecule: Tetracycline resistance protein TetW (TETW) | [3] | |||
| Resistant Disease | Butyrivibrio fibrisolvens infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Bifidobacterium longum strain F10 | 216816 | ||
| Bifidobacterium longum strain F5 | 216816 | |||
| Bifidobacterium longum strain F8 | 216816 | |||
| Butyrivibrio fibrisolvens strain 1.23 | 831 | |||
| Butyrivibrio fibrisolvens strain 1.230 | 831 | |||
| Butyrivibrio fibrisolvens strain Jk214 | 831 | |||
| Butyrivibrio fibrisolvens strain Jk51 | 831 | |||
| Fusobacterium prausnitzii strain k10 | 853 | |||
| Mitsuokella multiacidus strain 46/5(2) | 52226 | |||
| Mitsuokella multiacidus strain P208-58 | 52226 | |||
| Selenomonas ruminantium strain FB32 | 971 | |||
| Selenomonas ruminantium strain FB322 | 971 | |||
| Selenomonas ruminantium strain FB34 | 971 | |||
| Experiment for Molecule Alteration |
Southern blotting assay | |||
| Mechanism Description | Members of our group recently identified a new tetracycline resistance gene, tet(W), in three genera of rumen obligate anaerobes. Here, we show that tet(W) is also present in bacteria isolated from human feces. The tet(W) genes found in human Fusobacterium prausnitzii and Bifidobacterium longum isolates were more than 99.9% identical to those from a rumen isolate of Butyrivibrio fibrisolvens. | |||
| Key Molecule: Tetracycline resistance protein TetW (TETW) | [3] | |||
| Resistant Disease | Selenomonas ruminantium infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Bifidobacterium longum strain F10 | 216816 | ||
| Bifidobacterium longum strain F5 | 216816 | |||
| Bifidobacterium longum strain F8 | 216816 | |||
| Butyrivibrio fibrisolvens strain 1.23 | 831 | |||
| Butyrivibrio fibrisolvens strain 1.230 | 831 | |||
| Butyrivibrio fibrisolvens strain Jk214 | 831 | |||
| Butyrivibrio fibrisolvens strain Jk51 | 831 | |||
| Fusobacterium prausnitzii strain k10 | 853 | |||
| Mitsuokella multiacidus strain 46/5(2) | 52226 | |||
| Mitsuokella multiacidus strain P208-58 | 52226 | |||
| Selenomonas ruminantium strain FB32 | 971 | |||
| Selenomonas ruminantium strain FB322 | 971 | |||
| Selenomonas ruminantium strain FB34 | 971 | |||
| Experiment for Molecule Alteration |
Southern blotting assay | |||
| Mechanism Description | Members of our group recently identified a new tetracycline resistance gene, tet(W), in three genera of rumen obligate anaerobes. Here, we show that tet(W) is also present in bacteria isolated from human feces. The tet(W) genes found in human Fusobacterium prausnitzii and Bifidobacterium longum isolates were more than 99.9% identical to those from a rumen isolate of Butyrivibrio fibrisolvens. | |||
| Key Molecule: Tetracycline resistance protein TetW (TETW) | [3] | |||
| Resistant Disease | Mitsuokella multiacidus infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Bifidobacterium longum strain F10 | 216816 | ||
| Bifidobacterium longum strain F5 | 216816 | |||
| Bifidobacterium longum strain F8 | 216816 | |||
| Butyrivibrio fibrisolvens strain 1.23 | 831 | |||
| Butyrivibrio fibrisolvens strain 1.230 | 831 | |||
| Butyrivibrio fibrisolvens strain Jk214 | 831 | |||
| Butyrivibrio fibrisolvens strain Jk51 | 831 | |||
| Fusobacterium prausnitzii strain k10 | 853 | |||
| Mitsuokella multiacidus strain 46/5(2) | 52226 | |||
| Mitsuokella multiacidus strain P208-58 | 52226 | |||
| Selenomonas ruminantium strain FB32 | 971 | |||
| Selenomonas ruminantium strain FB322 | 971 | |||
| Selenomonas ruminantium strain FB34 | 971 | |||
| Experiment for Molecule Alteration |
Southern blotting assay | |||
| Mechanism Description | Members of our group recently identified a new tetracycline resistance gene, tet(W), in three genera of rumen obligate anaerobes. Here, we show that tet(W) is also present in bacteria isolated from human feces. The tet(W) genes found in human Fusobacterium prausnitzii and Bifidobacterium longum isolates were more than 99.9% identical to those from a rumen isolate of Butyrivibrio fibrisolvens. | |||
| Key Molecule: Tetracycline resistance protein TetW (TETW) | [3] | |||
| Resistant Disease | Fusobacterium prausnitzii infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Bifidobacterium longum strain F10 | 216816 | ||
| Bifidobacterium longum strain F5 | 216816 | |||
| Bifidobacterium longum strain F8 | 216816 | |||
| Butyrivibrio fibrisolvens strain 1.23 | 831 | |||
| Butyrivibrio fibrisolvens strain 1.230 | 831 | |||
| Butyrivibrio fibrisolvens strain Jk214 | 831 | |||
| Butyrivibrio fibrisolvens strain Jk51 | 831 | |||
| Fusobacterium prausnitzii strain k10 | 853 | |||
| Mitsuokella multiacidus strain 46/5(2) | 52226 | |||
| Mitsuokella multiacidus strain P208-58 | 52226 | |||
| Selenomonas ruminantium strain FB32 | 971 | |||
| Selenomonas ruminantium strain FB322 | 971 | |||
| Selenomonas ruminantium strain FB34 | 971 | |||
| Experiment for Molecule Alteration |
Southern blotting assay | |||
| Mechanism Description | Members of our group recently identified a new tetracycline resistance gene, tet(W), in three genera of rumen obligate anaerobes. Here, we show that tet(W) is also present in bacteria isolated from human feces. The tet(W) genes found in human Fusobacterium prausnitzii and Bifidobacterium longum isolates were more than 99.9% identical to those from a rumen isolate of Butyrivibrio fibrisolvens. | |||
| Key Molecule: Tetracycline resistance protein TetS (TETS) | [23], [24] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Enterococcus faecalis JH2-2 | 1351 | ||
| Enterococcus spp. Isolates | 35783 | |||
| Streptococcus milleri isolates | 33040 | |||
| Streptococcus sanguis isolates | 1305 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | TetS confers tetracycline and minocycline resistance by ribosomal protection. | |||
|
|
||||
| Key Molecule: acrB-acrA (Unclear) | [1] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Experiment for Molecule Alteration |
High-throughput sequencing assay | |||
| Mechanism Description | Relative abundance of ARGs was significantly increased under high oxytetracycline concentration. Of the 36 ARG-carrying contigs in the OTC-25 plasmidome, 20 were matched in the NCBI Plasmid Genome Database, with 17 carrying multiple ARGs (carrying >= 2 ARGs), including gene combinations of pecM-tetA-tetR-qnrS1, tet31-tetR(31) (tetR(31), which is used to regulate the expression of tet31 gene, is one kind of tetR (tetracycline repressor gene)), floR-sul1, strA-strB, acrB-acrA, ATP-binding cassette transporter (ABC transporter)-ABC transporter, and mexC. | |||
| Key Molecule: Protein PecM (PeECM) | [1] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Experiment for Molecule Alteration |
High-throughput sequencing assay | |||
| Mechanism Description | Relative abundance of ARGs was significantly increased under high oxytetracycline concentration. Of the 36 ARG-carrying contigs in the OTC-25 plasmidome, 20 were matched in the NCBI Plasmid Genome Database, with 17 carrying multiple ARGs (carrying >= 2 ARGs), including gene combinations of pecM-tetA-tetR-qnrS1, tet31-tetR(31) (tetR(31), which is used to regulate the expression of tet31 gene, is one kind of tetR (tetracycline repressor gene)), floR-sul1, strA-strB, acrB-acrA, ATP-binding cassette transporter (ABC transporter)-ABC transporter, and mexC. | |||
| Key Molecule: Major facilitator superfamily MFS_1 (TETV) | [25], [26], [27] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium abscessus OS11 | 36809 | ||
| Mycobacterium abscessus OS13 | 36809 | |||
| Mycobacterium fortuitum OS2/7 | 1766 | |||
| Mycobacterium fortuitum OS21 | 1766 | |||
| Mycobacterium fortuitum OS24 | 1766 | |||
| Mycobacterium fortuitum OS25 | 1766 | |||
| Mycobacterium fortuitum OS28 | 1766 | |||
| Mycobacterium fortuitum OS30 | 1766 | |||
| Mycobacterium fortuitum OS8 | 1766 | |||
| Mycobacterium fortuitum OS9 | 1766 | |||
| Mycobacterium fortuitum TR-1378 | 1766 | |||
| Mycobacterium mucogenicum OS11 | 56689 | |||
| Mycobacterium peregrinum OS2/8 | 43304 | |||
| Mycobacterium smegmatis OS1 | 1772 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Disk diffusion test assay; E-strip test assay | |||
| Mechanism Description | Tetracycline/multidrug efflux pumps Tet(V) and Tap may belong to this intrinsic resistome as they have been so far found only in certain RGM species.tet(V) and tap, both encode mycobacterial efflux pumps, including species where these genes have never been evidenced before. | |||
| Key Molecule: Tetracycline resistance protein class A (TETA) | [25], [28] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli | 668369 | ||
| Escherichia coli BL21(DE3) | 469008 | |||
| Escherichia coli LM317 | 562 | |||
| Escherichia coli TB1 | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | The bacterial tetracycline-resistance determinant from Tn10 encodes a 43 kDa membrane protein, TetA, responsible for active efflux of tetracyclines. | |||
| Key Molecule: ARE-ABC-F family resistance factor PoxtA (POXTA) | [29] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Staphylococcus aureus RN4220 | 1280 | ||
| Enterococcus faecalis JH2-2 | 1351 | |||
| Escherichia coli Mach1 T1R | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth dilution test assay | |||
| Mechanism Description | The poxtA gene encodes a protein that is 32% identical to OptrA and exhibits structural features typical of the F lineage of the ATP-binding cassette (ABC) protein superfamily that cause antibiotic resistance by ribosomal protection. | |||
| Key Molecule: Tetracycline resistance protein class A48 (TETA48) | [30] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Paenibacillus sp. LC231 | 1120679 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Two predicted ABC-transporters that confer resistance to tetracycline (TetAB(48)) and tiamulin (TaeA). | |||
| Key Molecule: Tetracycline resistance protein tet(59) (TET59) | [21] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli EPI-300 | 562 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | Tet(59) is preceded by a homolog of the tetracycline repressor tetR typically found upstream of tet genes encoding efflux pumps and include the two palindromic operator sequences present in all regulatory regions of the tet(A)-tet(R) family (33), suggesting that tet(59) probably belongs to the efflux pump family. | |||
| Key Molecule: Tetracycline efflux protein TetA (TETA) | [31] | |||
| Resistant Disease | Corynebacterium glutamicum infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Corynebacterium glutamicum ATCC 13032 | 196627 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Macrodilution broth method assay | |||
| Mechanism Description | Tet33 causes tetracycline resistance by regulating tetracycline efflux. | |||
| Key Molecule: Tetracycline efflux Na+/H+ antiporter family transporter Tet(35) (TEE35) | [16] | |||
| Resistant Disease | Vibrio harveyi infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli TOP 10 | 83333 | ||
| Vibrio harveyi M3.4L | 669 | |||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Agar dilution technique assay | |||
| Mechanism Description | We describe the cloning and characterization of two tetracycline resistance determinants from V. harveyi strain M3.4L. The second determinant, cloned as a 3,358-bp fragment in pATJ1, contains two open reading frames, designated tet35 and txr. tet35 encodes a 369-amino-acid protein that was predicted to have nine transmembrane regions. Tetracycline accumulation studies indicate that Escherichia coli carrying tet35 and txr can function as an energy-dependent tetracycline efflux pump but is less efficient than TetA. | |||
| Key Molecule: Tetracycline resistance protein class A (TETA) | [32] | |||
| Resistant Disease | Corynebacterium striatum infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Corynebacterium glutamicum strain ATCC 13032 | 196627 | ||
| Corynebacterium striatum strain M82B | 43770 | |||
| Escherichia coli strain DH5alphaMCR | 668369 | |||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Macrodilution broth method assay | |||
| Mechanism Description | The large multiresistance plasmid pTP10 was initially identified in the clinical isolate C. striatum M82B. This 51-kb R-plasmid was shown to carry the determinants for resistance to the antibiotics chloramphenicol, erythomycin, kanamycin, and tetracycline by ethidium bromide-based curing experiments. The tetracycline and oxacillin resistance region is part of a DNA segment structurally similar to the chromosome of the human pathogen Mycobacterium tuberculosis. A resistance assay in C. glutamicum demonstrated that the tetAB gene pair of pTP10 is necessary to confer resistance to the antibiotics tetracycline and oxytetracycline. | |||
| Key Molecule: Tetracycline resistance protein class A (TETA) | [32] | |||
| Resistant Disease | Corynebacterium glutamicum infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Acquired |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Corynebacterium glutamicum strain ATCC 13032 | 196627 | ||
| Corynebacterium striatum strain M82B | 43770 | |||
| Escherichia coli strain DH5alphaMCR | 668369 | |||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Macrodilution broth method assay | |||
| Mechanism Description | The large multiresistance plasmid pTP10 was initially identified in the clinical isolate C. striatum M82B. This 51-kb R-plasmid was shown to carry the determinants for resistance to the antibiotics chloramphenicol, erythomycin, kanamycin, and tetracycline by ethidium bromide-based curing experiments. Both resistance genes are located on mobile DNA elements that are capable of transposition into the chromosome of the non-pathogenic soil bacteriumC. glutamicum. A resistance assay in C. glutamicum demonstrated that the tetAB gene pair of pTP10 is necessary to confer resistance to the antibiotics tetracycline and oxytetracycline. | |||
| Key Molecule: Tetracycline efflux protein tet(L) (TETL) | [25], [33] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli strain Sk1592 | 562 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Gradient plate method assay | |||
| Mechanism Description | The class L determinant, on the other hand, does not prevent the inhibition of protein synthesis in S. faecalis but rather decreases tetracycline uptake.The class L (TetL) tetracycline resistance determinant from streptococci specified resistance and an energy-dependent decreased accumulation of tetracycline in both Streptococcus faecalis and Escherichia coli. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Tetracycline resistance protein TetM (TETM) | [18] | |||
| Resistant Disease | Clostridium difficile infection [ICD-11: 1A04.0] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| Mechanism Description | Of the resistance mechanisms, C. difficile produces ribosomal protection protein that impedes the attachment of the drug to a ribosome. The TetM protein that functions as ribosomal protectant has been identified in TET-resistant C. difficile strains, whereas the presence of other Tet proteins such as Tet(W) and Tet(44) has also been recognized. The TetM exhibits homology to EF-G and shares the same binding region in a ribosome. The binding of the TetM protein to a ribosome accompanying with the GTP hydrolysis allows conformational change of the ribosome, resulting in the dissociation of TET from its binding site. Cellular protein synthesis is then recovered through the binding of EF-G after the release of hydrolysed TetM. | |||
| Key Molecule: Tetracycline resistance protein TetW (TETW) | [18] | |||
| Resistant Disease | Human immunodeficiency virus infection [ICD-11: 1C62.0] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| Mechanism Description | Of the resistance mechanisms, C. difficile produces ribosomal protection protein that impedes the attachment of the drug to a ribosome. The TetM protein that functions as ribosomal protectant has been identified in TET-resistant C. difficile strains, whereas the presence of other Tet proteins such as Tet(W) and Tet(44) has also been recognized. The TetM exhibits homology to EF-G and shares the same binding region in a ribosome. The binding of the TetM protein to a ribosome accompanying with the GTP hydrolysis allows conformational change of the ribosome, resulting in the dissociation of TET from its binding site. Cellular protein synthesis is then recovered through the binding of EF-G after the release of hydrolysed TetM. | |||
| Key Molecule: Ribosomal tetracycline resistance protein tet (TET(44)) | [18] | |||
| Resistant Disease | Human immunodeficiency virus infection [ICD-11: 1C62.0] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| Mechanism Description | Of the resistance mechanisms, C. difficile produces ribosomal protection protein that impedes the attachment of the drug to a ribosome. The TetM protein that functions as ribosomal protectant has been identified in TET-resistant C. difficile strains, whereas the presence of other Tet proteins such as Tet(W) and Tet(44) has also been recognized. The TetM exhibits homology to EF-G and shares the same binding region in a ribosome. The binding of the TetM protein to a ribosome accompanying with the GTP hydrolysis allows conformational change of the ribosome, resulting in the dissociation of TET from its binding site. Cellular protein synthesis is then recovered through the binding of EF-G after the release of hydrolysed TetM. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Multidrug transporter MdfA (MDFA) | [11] | |||
| Resistant Disease | Salmonella enterica infection [ICD-11: 1A09.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Salmonella enterica serovar Typhimurium ATCC 14028s | 588858 | ||
| Experiment for Molecule Alteration |
Quantitative real-time PCR | |||
| Experiment for Drug Resistance |
L agar plate method assay | |||
| Mechanism Description | Overexpression or overproduction of mdfA confers drug resistance. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Tetracycline efflux MFS transporter Tet(38) (TET38) | [13] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli | 668369 | ||
| Experiment for Molecule Alteration |
DNA microarray hybridization assay | |||
| Experiment for Drug Resistance |
Serial twofold agar dilutions assay | |||
| Mechanism Description | MgrA was an indirect regulator of tet38 expression. The mgrA tet38 double mutant became more susceptible to tetracycline than the wild-type parent strain. | |||
|
|
||||
| Key Molecule: HTH-type transcriptional regulator MgrA (MGRA) | [13] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli | 668369 | ||
| Experiment for Molecule Alteration |
DNA microarray hybridization assay | |||
| Experiment for Drug Resistance |
Serial twofold agar dilutions assay | |||
| Mechanism Description | MgrA was an indirect regulator of tet38 expression. The mgrA tet38 double mutant became more susceptible to tetracycline than the wild-type parent strain. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Tetracycline repressor protein class H (TETR) | [19] | |||
| Resistant Disease | Pasteurella multocida infection [ICD-11: 1B99.0] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli ATCC 25922 | 1322345 | ||
| Staphylococcus aureus ATCC 29213 | 1280 | |||
| Pasteurella multocida 36950 | 1075089 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | The analysis of one representative P. multocida isolate identified an 82 kb integrative and conjugative element (ICE) integrated into the chromosomal DNA. This ICE, designated ICEPmu1, harboured 11 resistance genes, which confer resistance to streptomycin/spectinomycin (aadA25), streptomycin (strA and strB), gentamicin (aadB), kanamycin/neomycin (aphA1), tetracycline [tetR-tet(H)], chloramphenicol/florfenicol (floR), sulphonamides (sul2), tilmicosin/clindamycin [erm(42)] or tilmicosin/tulathromycin [msr(E)-mph(E)]. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Tetracycline resistance protein TetW (TETW) | [3] | |||
| Resistant Disease | Bartonella bacilliformis infection [ICD-11: 1C11.0] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Bifidobacterium longum strain F10 | 216816 | ||
| Bifidobacterium longum strain F5 | 216816 | |||
| Bifidobacterium longum strain F8 | 216816 | |||
| Butyrivibrio fibrisolvens strain 1.23 | 831 | |||
| Butyrivibrio fibrisolvens strain 1.230 | 831 | |||
| Butyrivibrio fibrisolvens strain Jk214 | 831 | |||
| Butyrivibrio fibrisolvens strain Jk51 | 831 | |||
| Fusobacterium prausnitzii strain k10 | 853 | |||
| Mitsuokella multiacidus strain 46/5(2) | 52226 | |||
| Mitsuokella multiacidus strain P208-58 | 52226 | |||
| Selenomonas ruminantium strain FB32 | 971 | |||
| Selenomonas ruminantium strain FB322 | 971 | |||
| Selenomonas ruminantium strain FB34 | 971 | |||
| Experiment for Molecule Alteration |
Southern blotting assay | |||
| Mechanism Description | Members of our group recently identified a new tetracycline resistance gene, tet(W), in three genera of rumen obligate anaerobes. Here, we show that tet(W) is also present in bacteria isolated from human feces. The tet(W) genes found in human Fusobacterium prausnitzii and Bifidobacterium longum isolates were more than 99.9% identical to those from a rumen isolate of Butyrivibrio fibrisolvens. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Ribosomal tetracycline resistance protein tet(44) (TET44) | [17] | |||
| Resistant Disease | Campylobacter fetus infection [ICD-11: 1C40.0] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli S17-1 Lambdapir | 1227813 | ||
| Experiment for Molecule Alteration |
Illumina/Solexa sequencing assay | |||
| Experiment for Drug Resistance |
Broth microdilution assay | |||
| Mechanism Description | The 640-amino-acid tetracycline resistance determinant, Tet 44, belongs to a class of proteins that confers resistance to tetracycline and minocycline by ribosomal protection. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Oxytetracycline resistance efflux MFS transporter OtrB (OTRB) | [34] | |||
| Resistant Disease | Streptomyces rimosus infection [ICD-11: 1C43.12] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Streptomyces rimosus strain | 1927 | ||
| Mechanism Description | The sequence of the tetracycline resistance gene designated tet347 from the tetracycline-producing organism Streptomyces rimosus (strain PG3) predicted a protein of 347 amino acids (GenBank accession no. M20370). The tcrC gene (also called tcr3; GenBank accession no. D38215) from chlorotetracycline-producing S. aureofaciens encoded a 512-residue putative tetracycline efflux protein which, starting at residue 222, was 43% identical to the Tet347 protein. | |||
| Key Molecule: Protein tcr3 (TCR3) | [14] | |||
| Resistant Disease | Streptomyces aurebyaciens infection [ICD-11: 1C43.2] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli strain XLI-Blue | 562 | ||
| Streptomyces aureofaciens strain NRRL3203 | 1894 | |||
| Streptomyces lividans strain Tk23 | 1916 | |||
| Experiment for Molecule Alteration |
Northern blotting analysis | |||
| Mechanism Description | The analysis of the nucleotide sequence of the 2.8-kb BamHI fragment containing tcrC gene showed that the predicted tcrC gene product is a protein consisting of 512 amino acids. The deduced amino acid sequence had a high level identity with that of the self-defense gene (tet347) of Streptomyces rimosus, known to mediate oxytetracycline efflux. The tcrC gene-inactivated strains generated from strain NRRL3203 by gene replacement had a 90% decrease in the level of resistance to tetracycline and the antibiotic productivity when compared with the parental strain. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Tetracycline resistance protein TetQ (TETQ) | [2] | |||
| Resistant Disease | Bacteroides spp infection [ICD-11: 1C4Y.9] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli DH5alpha | 668369 | ||
| Experiment for Molecule Alteration |
PCR; Dot blot and Southern blot analysis | |||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | Tet36 is a new class of ribosome protection type tetracycline resistance protein and lead to drug resistance. | |||
| Key Molecule: Tetracycline resistance protein TetQ (TETQ) | [15] | |||
| Resistant Disease | Bacteroides fragilis infection [ICD-11: 1C4Y.6] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Bacteroides distasonis strains | 823 | ||
| Bacteroides distasonis strains V2002 | 823 | |||
| Bacteroides distasonis strains V2003 | 823 | |||
| Bacteroides distasonis strains V2004 | 823 | |||
| Bacteroides fragilis strain | 817 | |||
| Bacteroides fragilis strain V503 | 817 | |||
| Bacteroides ovatus strains | 28116 | |||
| Bacteroides ovatus strains V2008 | 28116 | |||
| Bacteroides thetaiotaomicron strain | 818 | |||
| Bacteroides thetaiotaomicron strain V2005 | 818 | |||
| Bacteroides thetaiotaomicron strain V2006 | 818 | |||
| Bacteroides thetaiotaomicron strain V2007 | 818 | |||
| Bacteroides uniformis strain | 820 | |||
| Bacteroides uniformis strain V1760 | 820 | |||
| Bacteroides uniformis strain V1761 | 820 | |||
| Bacteroides uniformis strain V1918 | 820 | |||
| Bacteroides uniformis strain V1921 | 820 | |||
| Bacteroides uniformis strain V2000 | 820 | |||
| Bacteroides uniformis strain V2001 | 820 | |||
| Bacteroides uniformis strain V528 | 820 | |||
| Bacteroides uniformis strain V844 | 820 | |||
| Experiment for Molecule Alteration |
Southern blotting assay | |||
| Mechanism Description | Of 13 clinical isolates of the Bacteroides group, all were resistant to tetracycline (>10,ug/ml). The source of tetracycline resistance was investigated with the recently cloned tetQ gene, a ribosomal protection gene. | |||
| Key Molecule: Tetracycline resistance protein TetQ (TETQ) | [15] | |||
| Resistant Disease | Bacteroides uniformis infection [ICD-11: 1C4Y.11] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Bacteroides distasonis strains | 823 | ||
| Bacteroides distasonis strains V2002 | 823 | |||
| Bacteroides distasonis strains V2003 | 823 | |||
| Bacteroides distasonis strains V2004 | 823 | |||
| Bacteroides fragilis strain | 817 | |||
| Bacteroides fragilis strain V503 | 817 | |||
| Bacteroides ovatus strains | 28116 | |||
| Bacteroides ovatus strains V2008 | 28116 | |||
| Bacteroides thetaiotaomicron strain | 818 | |||
| Bacteroides thetaiotaomicron strain V2005 | 818 | |||
| Bacteroides thetaiotaomicron strain V2006 | 818 | |||
| Bacteroides thetaiotaomicron strain V2007 | 818 | |||
| Bacteroides uniformis strain | 820 | |||
| Bacteroides uniformis strain V1760 | 820 | |||
| Bacteroides uniformis strain V1761 | 820 | |||
| Bacteroides uniformis strain V1918 | 820 | |||
| Bacteroides uniformis strain V1921 | 820 | |||
| Bacteroides uniformis strain V2000 | 820 | |||
| Bacteroides uniformis strain V2001 | 820 | |||
| Bacteroides uniformis strain V528 | 820 | |||
| Bacteroides uniformis strain V844 | 820 | |||
| Experiment for Molecule Alteration |
Southern blotting assay | |||
| Mechanism Description | Of 13 clinical isolates of the Bacteroides group, all were resistant to tetracycline (>10,ug/ml). The source of tetracycline resistance was investigated with the recently cloned tetQ gene, a ribosomal protection gene. | |||
| Key Molecule: Tetracycline resistance protein TetQ (TETQ) | [15] | |||
| Resistant Disease | Bacteroides distasonis infection [ICD-11: 1C4Y.5] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Bacteroides distasonis strains | 823 | ||
| Bacteroides distasonis strains V2002 | 823 | |||
| Bacteroides distasonis strains V2003 | 823 | |||
| Bacteroides distasonis strains V2004 | 823 | |||
| Bacteroides fragilis strain | 817 | |||
| Bacteroides fragilis strain V503 | 817 | |||
| Bacteroides ovatus strains | 28116 | |||
| Bacteroides ovatus strains V2008 | 28116 | |||
| Bacteroides thetaiotaomicron strain | 818 | |||
| Bacteroides thetaiotaomicron strain V2005 | 818 | |||
| Bacteroides thetaiotaomicron strain V2006 | 818 | |||
| Bacteroides thetaiotaomicron strain V2007 | 818 | |||
| Bacteroides uniformis strain | 820 | |||
| Bacteroides uniformis strain V1760 | 820 | |||
| Bacteroides uniformis strain V1761 | 820 | |||
| Bacteroides uniformis strain V1918 | 820 | |||
| Bacteroides uniformis strain V1921 | 820 | |||
| Bacteroides uniformis strain V2000 | 820 | |||
| Bacteroides uniformis strain V2001 | 820 | |||
| Bacteroides uniformis strain V528 | 820 | |||
| Bacteroides uniformis strain V844 | 820 | |||
| Experiment for Molecule Alteration |
Southern blotting assay | |||
| Mechanism Description | Of 13 clinical isolates of the Bacteroides group, all were resistant to tetracycline (>10,ug/ml). The source of tetracycline resistance was investigated with the recently cloned tetQ gene, a ribosomal protection gene. | |||
| Key Molecule: Tetracycline resistance protein TetQ (TETQ) | [15] | |||
| Resistant Disease | Bacteroides thetaiotaomicron infection [ICD-11: 1C4Y.10] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Bacteroides distasonis strains | 823 | ||
| Bacteroides distasonis strains V2002 | 823 | |||
| Bacteroides distasonis strains V2003 | 823 | |||
| Bacteroides distasonis strains V2004 | 823 | |||
| Bacteroides fragilis strain | 817 | |||
| Bacteroides fragilis strain V503 | 817 | |||
| Bacteroides ovatus strains | 28116 | |||
| Bacteroides ovatus strains V2008 | 28116 | |||
| Bacteroides thetaiotaomicron strain | 818 | |||
| Bacteroides thetaiotaomicron strain V2005 | 818 | |||
| Bacteroides thetaiotaomicron strain V2006 | 818 | |||
| Bacteroides thetaiotaomicron strain V2007 | 818 | |||
| Bacteroides uniformis strain | 820 | |||
| Bacteroides uniformis strain V1760 | 820 | |||
| Bacteroides uniformis strain V1761 | 820 | |||
| Bacteroides uniformis strain V1918 | 820 | |||
| Bacteroides uniformis strain V1921 | 820 | |||
| Bacteroides uniformis strain V2000 | 820 | |||
| Bacteroides uniformis strain V2001 | 820 | |||
| Bacteroides uniformis strain V528 | 820 | |||
| Bacteroides uniformis strain V844 | 820 | |||
| Experiment for Molecule Alteration |
Southern blotting assay | |||
| Mechanism Description | Of 13 clinical isolates of the Bacteroides group, all were resistant to tetracycline (>10,ug/ml). The source of tetracycline resistance was investigated with the recently cloned tetQ gene, a ribosomal protection gene. | |||
| Key Molecule: Tetracycline resistance protein TetQ (TETQ) | [15] | |||
| Resistant Disease | Bacteroides ovatus infection [ICD-11: 1C4Y.8] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Bacteroides distasonis strains | 823 | ||
| Bacteroides distasonis strains V2002 | 823 | |||
| Bacteroides distasonis strains V2003 | 823 | |||
| Bacteroides distasonis strains V2004 | 823 | |||
| Bacteroides fragilis strain | 817 | |||
| Bacteroides fragilis strain V503 | 817 | |||
| Bacteroides ovatus strains | 28116 | |||
| Bacteroides ovatus strains V2008 | 28116 | |||
| Bacteroides thetaiotaomicron strain | 818 | |||
| Bacteroides thetaiotaomicron strain V2005 | 818 | |||
| Bacteroides thetaiotaomicron strain V2006 | 818 | |||
| Bacteroides thetaiotaomicron strain V2007 | 818 | |||
| Bacteroides uniformis strain | 820 | |||
| Bacteroides uniformis strain V1760 | 820 | |||
| Bacteroides uniformis strain V1761 | 820 | |||
| Bacteroides uniformis strain V1918 | 820 | |||
| Bacteroides uniformis strain V1921 | 820 | |||
| Bacteroides uniformis strain V2000 | 820 | |||
| Bacteroides uniformis strain V2001 | 820 | |||
| Bacteroides uniformis strain V528 | 820 | |||
| Bacteroides uniformis strain V844 | 820 | |||
| Experiment for Molecule Alteration |
Southern blotting assay | |||
| Mechanism Description | Of 13 clinical isolates of the Bacteroides group, all were resistant to tetracycline (>10,ug/ml). The source of tetracycline resistance was investigated with the recently cloned tetQ gene, a ribosomal protection gene. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Tetracycline resistance protein TetU (TETU) | [4] | |||
| Resistant Disease | Enterococcus faecium meningitis [ICD-11: 1D01.2] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli | 668369 | ||
| Enterococcus faecalis strain JH2-2 | 1320322 | |||
| Enterococcus faecium strain CH2 | 1352 | |||
| Experiment for Molecule Alteration |
DNA Hybridization assay | |||
| Experiment for Drug Resistance |
Tube dilution method assay | |||
| Mechanism Description | PkQ10, a 1.9-kb plasmid carrying a novel Tc resistance determinant, was isolated from one of the isolates. The nucleotide sequence of this plasmid revealed an open reading frame corresponding to an 11.8-kDa protein and containing 105 amino acid residues. There was some limited similarity between this protein andtet(M),tet(O),tet(Q),tet(S),tetB(P), andotr(A), which overlapped, but did not include, the consensus GTP-binding sequences. The low-level, Tc-resistant determinant of pkQ10, namedtet(U), does not appear to correspond to any other known Tc resistance determinant. | |||
ICD-12: Respiratory system diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Multidrug efflux SMR transporter (ABES) | [35] | |||
| Resistant Disease | Acinetobacter baumannii infection [ICD-11: CA40.4] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli kAM32 | 562 | ||
| Experiment for Molecule Alteration |
Fluorometric efflux assay | |||
| Experiment for Drug Resistance |
Broth dilution assay | |||
| Mechanism Description | The abeS gene product conferred resistance to various antimicrobial compounds through an efflux mechanism. | |||
| Key Molecule: Tetracycline efflux protein (TET41) | [36] | |||
| Resistant Disease | Serratia marcescens infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli JM109 | 562 | ||
| Experiment for Molecule Alteration |
DNA sequencing and protein and phylogenetic assay | |||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | Tet 41 is a tetracycline-specific efflux pump, which lead to drug resistance. | |||
| Key Molecule: Tetracycline efflux proteintet(39) (TET39) | [10] | |||
| Resistant Disease | Acinetobacter baumannii infection [ICD-11: CA40.4] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Acinetobacter sp. LUH5605 | 309867 | ||
| Experiment for Molecule Alteration |
DNA multiple alignment assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | Tet39 produces drug resistance through the action of nonspecific efflux pump. | |||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
