Drug Information
Drug (ID: DG00154) and It's Reported Resistant Information
| Name |
Epirubicin
|
||||
|---|---|---|---|---|---|
| Synonyms |
Ellence; Epiadriamycin; Epidoxorubicin; Epirubicina; Epirubicine; Epirubicinum; Pidorubicin; Pidorubicina; Pidorubicine; Pidorubicinum; Ridorubicin; Epirubicina [Spanish]; Epirubicine [French]; Epirubicinum [Latin]; Pharmorubicin Pfs; IMI 28; WP 697; Ebewe (TN); Ellence (TN); Epi-DX; Epirubicin (INN); Epirubicin (TN); Epirubicin [INN:BAN]; Epirubicina [INN-Spanish]; Epirubicine [INN-French]; Epirubicinum [INN-Latin]; Farmorubicin (TN); Pharmorubicin (TN); Pidorubicina [INN-Spanish]; Pidorubicine [INN-French]; Pidorubicinum [INN-Latin]; (1S,3S)-3,5,12-trihydroxy-3-(hydroxyacetyl)-10-(methyloxy)-6,11-dioxo-1,2,3,4,6,11-hexahydrotetracen-1-yl 3-amino-2,3,6-trideoxy-alpha-L-arabino-hexopyranoside; (1S,3S)-3,5,12-trihydroxy-3-(hydroxyacetyl)-10-methoxy-6,11-dioxo-1,2,3,4,6,11-hexahydrotetracen-1-yl 3-amino-2,3,6-trideoxy-alpha-L-arabino-hexopyranoside; (7S,9R)-7-[(2S,4S,5R,6S)-4-Amino-5-hydroxy-6-methyl-oxan-2-yl]oxy-6,9,11-trihydroxy-9-(2-hydroxyacetyl)-4-methoxy-8,10-dihydro-7H-tetracene-5,12-dione; (7S,9S)-7-[(2R,4S,5R,6S)-4-amino-5-hydroxy-6-methyloxan-2-yl]oxy-6,9,11-trihydroxy-9-(2-hydroxyacetyl)-4-methoxy-8,10-dihydro-7H-tetracene-5,12-dione; 10-((3-amino-2,3,6-trideoxy-beta-L-arabino-hexopyranosyl)oxy)-7,8,9,10-tetrahydro-6,8,11-trihydroxy-8-(hydroxyacetyl)-1-methoxy-(8S-cis)-5,12-naphthacenedione; 4'-Epi-DXR; 4'-Epiadriamycin; 4'-epi-DX; 4'-epi-Doxorubicin; 4'-epidoxorubicin; 4-Epidoxorubicin
Click to Show/Hide
|
||||
| Indication |
In total 1 Indication(s)
|
||||
| Structure |
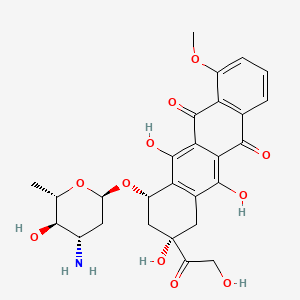
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Clinically Reported Resistance for This Drug
(3 diseases)
[2]
[3]
[4]
Disease(s) with Resistance Information Discovered by Cell Line Test for This Drug
(3 diseases)
[6]
[7]
[8]
|
||||
| Target | DNA topoisomerase II (TOP2) |
TOP2A_HUMAN
; TOP2B_HUMAN |
[1] | ||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C27H29NO11
|
||||
| IsoSMILES |
C[C@H]1[C@@H]([C@H](C[C@@H](O1)O[C@H]2C[C@@](CC3=C2C(=C4C(=C3O)C(=O)C5=C(C4=O)C(=CC=C5)OC)O)(C(=O)CO)O)N)O
|
||||
| InChI |
1S/C27H29NO11/c1-10-22(31)13(28)6-17(38-10)39-15-8-27(36,16(30)9-29)7-12-19(15)26(35)21-20(24(12)33)23(32)11-4-3-5-14(37-2)18(11)25(21)34/h3-5,10,13,15,17,22,29,31,33,35-36H,6-9,28H2,1-2H3/t10-,13-,15-,17-,22-,27-/m0/s1
|
||||
| InChIKey |
AOJJSUZBOXZQNB-VTZDEGQISA-N
|
||||
| PubChem CID | |||||
| ChEBI ID | |||||
| TTD Drug ID | |||||
| VARIDT ID | |||||
| INTEDE ID | |||||
| DrugBank ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-02: Benign/in-situ/malignant neoplasm
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Inositol polyphosphate-4-phosphatase type I A (INPP4A) | [4] | |||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | |||
| The Specified Disease | Non-small cell lung cancer | |||
| The Studied Tissue | Lung tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.08E-06 Fold-change: -1.11E-01 Z-score: -5.07E+00 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell colony | Activation | hsa05200 | ||
| Cell viability | Activation | hsa05200 | ||
| JAKT2/STAT3 signaling pathway | Activation | hsa04030 | ||
| In Vitro Model | H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Overexpression of miR-4443 promotes the resistance of non-small cell lung cancer cells to epirubicin by downregulating INPP4A and regulating the activation of JAk2/STAT3 pathway. | |||
|
|
||||
| Key Molecule: hsa-miR-4443 | [4] | |||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell colony | Activation | hsa05200 | ||
| Cell viability | Activation | hsa05200 | ||
| JAKT2/STAT3 signaling pathway | Activation | hsa04030 | ||
| In Vitro Model | H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Overexpression of miR-4443 promotes the resistance of non-small cell lung cancer cells to epirubicin by downregulating INPP4A and regulating the activation of JAk2/STAT3 pathway. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Urothelial cancer associated 1 (UCA1) | [2] | |||
| Metabolic Type | Lipid metabolism | |||
| Resistant Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Bladder cancer [ICD-11: 2C94] | |||
| The Specified Disease | Bladder urothelial carcinoma | |||
| The Studied Tissue | Bladder | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.99E-08 Fold-change: 1.93E+00 Z-score: 6.03E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vivo Model | Nude mice, 5637 bladder cancer cells were transduced with sh-NC lentivirus; nude mice, 5637 bladder cancer cells were transduced with sh-UCA1 lentivirus | Mice | ||
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
Tumor volume assay | |||
| Mechanism Description | Mechanistically, lncRNA UCA1 promotes lipid accumulation in vitro and in vivo by upregulating PPARalpha mRNA and protein expression, which is mediated by miR-30a-3p. Knockdown of lncRNA UCA1 increased epirubicin-induced apoptosis via miR-30a-3p/PPARalpha and downstream p-AKT/p-GSK-3beta/beta-catenin signaling. Furthermore, mixed free fatty acids upregulated lncRNA UCA1 expression by promoting recruitment of the transcription factor RXRalpha to the lncRNA UCA1 promoter. | |||
| Key Molecule: Urothelial cancer associated 1 (UCA1) | [2] | |||
| Metabolic Type | Lipid metabolism | |||
| Resistant Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vivo Model | Patients with high expression of lncRNA UCA1 | Homo Sapiens | ||
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
Cell prognosis assay | |||
| Mechanism Description | Mechanistically, lncRNA UCA1 promotes lipid accumulation in vitro and in vivo by upregulating PPARalpha mRNA and protein expression, which is mediated by miR-30a-3p. Knockdown of lncRNA UCA1 increased epirubicin-induced apoptosis via miR-30a-3p/PPARalpha and downstream p-AKT/p-GSK-3beta/beta-catenin signaling. Furthermore, mixed free fatty acids upregulated lncRNA UCA1 expression by promoting recruitment of the transcription factor RXRalpha to the lncRNA UCA1 promoter. | |||
| Key Molecule: Urothelial cancer associated 1 (UCA1) | [2] | |||
| Metabolic Type | Lipid metabolism | |||
| Resistant Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | 5637 cells | Bladder | Homo sapiens (Human) | CVCL_0126 |
| UMUC-2 cells | Bladder | Homo sapiens (Human) | CVCL_8155 | |
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
Cell viability assay | |||
| Mechanism Description | Mechanistically, lncRNA UCA1 promotes lipid accumulation in vitro and in vivo by upregulating PPARalpha mRNA and protein expression, which is mediated by miR-30a-3p. Knockdown of lncRNA UCA1 increased epirubicin-induced apoptosis via miR-30a-3p/PPARalpha and downstream p-AKT/p-GSK-3beta/beta-catenin signaling. Furthermore, mixed free fatty acids upregulated lncRNA UCA1 expression by promoting recruitment of the transcription factor RXRalpha to the lncRNA UCA1 promoter. | |||
| Key Molecule: Urothelial cancer associated 1 (UCA1) | [2] | |||
| Metabolic Type | Lipid metabolism | |||
| Resistant Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vivo Model | BALB/c nude mice | Mice | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Tumor volume assay | |||
| Mechanism Description | Our findings demonstrate that lncRNA UCA1 positively regulates the expression of CD36 and FATP, which are known to stimulate fatty acid uptake. | |||
| Key Molecule: Urothelial cancer associated 1 (UCA1) | [2] | |||
| Metabolic Type | Lipid metabolism | |||
| Resistant Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | DSMZ cells | N.A. | N.A. | N.A. |
| UMUC-2 cells | Bladder | Homo sapiens (Human) | CVCL_8155 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Apoptosis rate assay | |||
| Mechanism Description | Our findings demonstrate that lncRNA UCA1 positively regulates the expression of CD36 and FATP, which are known to stimulate fatty acid uptake. | |||
|
|
||||
| Key Molecule: hsa-miR-34b-3p | [19] | |||
| Resistant Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| Notch/PkC/Ca++ signaling pathway | Inhibition | hsa04330 | ||
| In Vitro Model | 5637 cells | Bladder | Homo sapiens (Human) | CVCL_0126 |
| EJ cells | Bladder | Homo sapiens (Human) | CVCL_UI82 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | miR-34b-3p Represses the Multidrug-Chemoresistance of Bladder Cancer Cells by Regulating the CCND2 and P2RY1 Genes. | |||
| Key Molecule: hsa-miR-22-3p | [6] | |||
| Resistant Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | 5637 cells | Bladder | Homo sapiens (Human) | CVCL_0126 |
| T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |
| UM-UC-3 cells | Bladder | Homo sapiens (Human) | CVCL_1783 | |
| H-bc cells | Bladder | Homo sapiens (Human) | CVCL_BT00 | |
| HTB-1 cells | Bladder | Homo sapiens (Human) | CVCL_0359 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | miR 22 3p enhances multi chemoresistance by targeting NET1 in bladder cancer cells. | |||
|
|
||||
| Key Molecule: G1/S-specific cyclin-D2 (CCND2) | [19] | |||
| Resistant Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| Notch/PkC/Ca++ signaling pathway | Inhibition | hsa04330 | ||
| In Vitro Model | 5637 cells | Bladder | Homo sapiens (Human) | CVCL_0126 |
| EJ cells | Bladder | Homo sapiens (Human) | CVCL_UI82 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | miR-34b-3p Represses the Multidrug-Chemoresistance of Bladder Cancer Cells by Regulating the CCND2 and P2RY1 Genes. | |||
| Key Molecule: P2Y purinoceptor 1 (P2RY1) | [19] | |||
| Resistant Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| Notch/PkC/Ca++ signaling pathway | Inhibition | hsa04330 | ||
| In Vitro Model | 5637 cells | Bladder | Homo sapiens (Human) | CVCL_0126 |
| EJ cells | Bladder | Homo sapiens (Human) | CVCL_UI82 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | miR-34b-3p Represses the Multidrug-Chemoresistance of Bladder Cancer Cells by Regulating the CCND2 and P2RY1 Genes. | |||
| Key Molecule: Neuroepithelial cell-transforming gene 1 protein (NET1) | [6] | |||
| Resistant Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | 5637 cells | Bladder | Homo sapiens (Human) | CVCL_0126 |
| T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |
| UM-UC-3 cells | Bladder | Homo sapiens (Human) | CVCL_1783 | |
| H-bc cells | Bladder | Homo sapiens (Human) | CVCL_BT00 | |
| HTB-1 cells | Bladder | Homo sapiens (Human) | CVCL_0359 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | miR 22 3p enhances multi chemoresistance by targeting NET1 in bladder cancer cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Lymphoid enhancer-binding factor 1 (LEF1) | [9] | |||
| Sensitive Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Bladder cancer [ICD-11: 2C94] | |||
| The Specified Disease | Bladder cancer | |||
| The Studied Tissue | Bladder tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.80E-01 Fold-change: -1.76E-02 Z-score: -1.40E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell colony | Inhibition | hsa05200 | ||
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell viability | Inhibition | hsa05200 | ||
| Wnt/Beta-catenin signaling pathway | Regulation | N.A. | ||
| In Vitro Model | BIU87 cells | Bladder | Homo sapiens (Human) | CVCL_6881 |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | miR-34a increased chemosensitivity in BIU87/ADR cells by inhibiting the TCF1/LEF1 axis. | |||
| Key Molecule: Transcription factor 7 (TCF7) | [9] | |||
| Sensitive Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell colony | Inhibition | hsa05200 | ||
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell viability | Inhibition | hsa05200 | ||
| Wnt/Beta-catenin signaling pathway | Regulation | N.A. | ||
| In Vitro Model | BIU87 cells | Bladder | Homo sapiens (Human) | CVCL_6881 |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | miR-34a increased chemosensitivity in BIU87/ADR cells by inhibiting the TCF1/LEF1 axis. | |||
| Key Molecule: Presenilin-1 (PSEN1) | [20] | |||
| Sensitive Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| DNA damage response signaling pathway | Activation | hsa04218 | ||
| In Vitro Model | 5637 cells | Bladder | Homo sapiens (Human) | CVCL_0126 |
| H-bc cells | Bladder | Homo sapiens (Human) | CVCL_BT00 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | Among the differentially expressed genes between the chemosensitive (5637) and chemoresistant (H-bc) bladder cancer cell lines, the expression level of the PSEN1 gene (presenilin 1), a key component of the Gamma-secretase, is negatively correlated with chemoresistance. A small interfering RNA mediated repression of the PSEN1 gene suppresses cell apoptosis and de-sensitizes 5637 cells, while overexpression of the presenilin 1 sensitizes H-bc cells to the drug-triggered cell death. As a direct target of microRNA-193a-3p that promotes the multi-chemoresistance of the bladder cancer cell, PSEN1 acts as an important executor for the microRNA-193a-3p's positive impact on the multi-chemoresistance of bladder cancer, probably via its activating effect on DNA damage response pathway. In addition to the mechanistic insights, the key players in this microRNA-193a-3p/PSEN1 axis are likely the diagnostic and/or therapeutic targets for an effective chemotherapy of bladder cancer. | |||
| Key Molecule: Urothelial cancer associated 1 (UCA1) | [21] | |||
| Sensitive Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | lncRNA UCA1/miR-30a-3p/PPARalpha signaling pathway | Regulation | N.A. | |
| In Vitro Model | 5637 cells | Bladder | Homo sapiens (Human) | CVCL_0126 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | In this study, we demonstrated that lncRNA UCA1 inhibits epirubicin-induced cell apoptosis by supporting abnormal lipid metabolism in bladder cancer cells. Mechanistically, lncRNA UCA1 promotes lipid accumulation in vitro and in vivo by upregulating PPAR mRNA and protein expression, which is mediated by miR-30a-3p. Knockdown of lncRNA UCA1 increased epirubicin-induced apoptosis via miR-30a-3p/PPAR and downstream p-AKT/p-GSK-3beta/beta-catenin signaling. | |||
|
|
||||
| Key Molecule: hsa-mir-34 | [9] | |||
| Sensitive Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell colony | Inhibition | hsa05200 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell viability | Inhibition | hsa05200 | ||
| Wnt/Beta-catenin signaling pathway | Regulation | N.A. | ||
| In Vitro Model | BIU87 cells | Bladder | Homo sapiens (Human) | CVCL_6881 |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | miR-34a increased chemosensitivity in BIU87/ADR cells by inhibiting the TCF1/LEF1 axis. | |||
| Key Molecule: hsa-miR-193a-3p | [20] | |||
| Sensitive Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| DNA damage response signaling pathway | Activation | hsa04218 | ||
| In Vitro Model | 5637 cells | Bladder | Homo sapiens (Human) | CVCL_0126 |
| H-bc cells | Bladder | Homo sapiens (Human) | CVCL_BT00 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | Among the differentially expressed genes between the chemosensitive (5637) and chemoresistant (H-bc) bladder cancer cell lines, the expression level of the PSEN1 gene (presenilin 1), a key component of the Gamma-secretase, is negatively correlated with chemoresistance. A small interfering RNA mediated repression of the PSEN1 gene suppresses cell apoptosis and de-sensitizes 5637 cells, while overexpression of the presenilin 1 sensitizes H-bc cells to the drug-triggered cell death. As a direct target of microRNA-193a-3p that promotes the multi-chemoresistance of the bladder cancer cell, PSEN1 acts as an important executor for the microRNA-193a-3p's positive impact on the multi-chemoresistance of bladder cancer, probably via its activating effect on DNA damage response pathway. In addition to the mechanistic insights, the key players in this microRNA-193a-3p/PSEN1 axis are likely the diagnostic and/or therapeutic targets for an effective chemotherapy of bladder cancer. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Aurora kinase A (AURKA) | [10] | |||
| Sensitive Disease | Multiple myeloma [ICD-11: 2A83.0] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Multiple myeloma [ICD-11: 2A83] | |||
| The Specified Disease | Myeloma | |||
| The Studied Tissue | Peripheral blood | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.27E-01 Fold-change: -1.02E-01 Z-score: -6.42E-01 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | NCI-H929 cells | Bone marrow | Homo sapiens (Human) | CVCL_1600 |
| U266 cells | Bone marrow | Homo sapiens (Human) | CVCL_0566 | |
| MM1S cells | Peripheral blood | Homo sapiens (Human) | CVCL_8792 | |
| OPM-2 cells | Peripheral blood | Homo sapiens (Human) | CVCL_1625 | |
| RPMI-8226 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0014 | |
| KMS11 cells | Peripheral blood | Homo sapiens (Human) | CVCL_2989 | |
| In Vivo Model | Mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Ectopic expression of miR137 strongly reduced the expression of AURkA and p-ATM/Chk2 in MM cells, and increased the expression of p53, and p21, overexpression of miR137 could reduce drug resistance and overcome chromosomal instability of the MM cells via affecting the apoptosis and RNA damage pathways. | |||
|
|
||||
| Key Molecule: hsa-mir-137 | [10] | |||
| Sensitive Disease | Multiple myeloma [ICD-11: 2A83.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | NCI-H929 cells | Bone marrow | Homo sapiens (Human) | CVCL_1600 |
| U266 cells | Bone marrow | Homo sapiens (Human) | CVCL_0566 | |
| MM1S cells | Peripheral blood | Homo sapiens (Human) | CVCL_8792 | |
| OPM-2 cells | Peripheral blood | Homo sapiens (Human) | CVCL_1625 | |
| RPMI-8226 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0014 | |
| KMS11 cells | Peripheral blood | Homo sapiens (Human) | CVCL_2989 | |
| In Vivo Model | Mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Ectopic expression of miR137 strongly reduced the expression of AURkA and p-ATM/Chk2 in MM cells, and increased the expression of p53, and p21, overexpression of miR137 could reduce drug resistance and overcome chromosomal instability of the MM cells via affecting the apoptosis and RNA damage pathways. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Cytochrome P450 family 1 subfamily B member1 (CYP1B1) | [11] | |||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | miR27b/CCNG1/p53 signaling pathway | Regulation | N.A. | |
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| SNU182 cells | Liver | Homo sapiens (Human) | CVCL_0090 | |
| SNU-739 cells | Liver | Homo sapiens (Human) | CVCL_5088 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | |||
| Mechanism Description | miR-27b synergizes with anticancer drugs througth enhancing anticancer drug-induced cell death which due to p53 activation and CYP1B1 suppression. | |||
|
|
||||
| Key Molecule: hsa-mir-27b | [11] | |||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | miR27b/CCNG1/p53 signaling pathway | Regulation | N.A. | |
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| SNU182 cells | Liver | Homo sapiens (Human) | CVCL_0090 | |
| SNU-739 cells | Liver | Homo sapiens (Human) | CVCL_5088 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | |||
| Mechanism Description | miR-27b synergizes with anticancer drugs througth enhancing anticancer drug-induced cell death which due to p53 activation and CYP1B1 suppression. | |||
|
|
||||
| Key Molecule: Cyclin-G1 (CCNG1) | [11] | |||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| miR27b/CCNG1/p53 signaling pathway | Regulation | N.A. | ||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| SNU182 cells | Liver | Homo sapiens (Human) | CVCL_0090 | |
| SNU-739 cells | Liver | Homo sapiens (Human) | CVCL_5088 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | |||
| Mechanism Description | miR-27b synergizes with anticancer drugs througth enhancing anticancer drug-induced cell death which due to p53 activation and CYP1B1 suppression. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: ATP-binding cassette sub-family G2 (ABCG2) | [8] | |||
| Resistant Disease | Sarcoma [ICD-11: 2C35.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SW-872 cells | Skin | Homo sapiens (Human) | CVCL_1730 |
| SW-1353 cells | Brain | Homo sapiens (Human) | CVCL_0543 | |
| TE-671 cells | Peripheral blood | Homo sapiens (Human) | CVCL_1756 | |
| SW-684 cells | Skin | Homo sapiens (Human) | CVCL_1726 | |
| SW-982 cells | Testicular | Homo sapiens (Human) | CVCL_1734 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | By investigating of important regulators of stem cell biology, real-time RT-PCR data showed an increased expression of c-Myc, beta-catenin, and SOX-2 in the ALDH1high population and a significant higher level of ABCG2. Statistical analysis of data demonstrated that ALDH1high cells of SW-982 and SW-1353 showed higher resistance to commonly used chemotherapeutic agents like doxorubicin, epirubicin, and cisplatin than ALDH1low cells. This study demonstrates that in different sarcoma cell lines, high ALDH1 activity can be used to identify a subpopulation of cells characterized by a significantly higher proliferation rate, increased colony forming, increased expression of ABC transporter genes and stemness markers compared to control cells. In addition, enhanced drug resistance was demonstrated. | |||
|
|
||||
| Key Molecule: Aldehyde dehydrogenase 1 family member A1 (ALDH1A1) | [8] | |||
| Resistant Disease | Sarcoma [ICD-11: 2C35.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SW-872 cells | Skin | Homo sapiens (Human) | CVCL_1730 |
| SW-1353 cells | Brain | Homo sapiens (Human) | CVCL_0543 | |
| TE-671 cells | Peripheral blood | Homo sapiens (Human) | CVCL_1756 | |
| SW-684 cells | Skin | Homo sapiens (Human) | CVCL_1726 | |
| SW-982 cells | Testicular | Homo sapiens (Human) | CVCL_1734 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | By investigating of important regulators of stem cell biology, real-time RT-PCR data showed an increased expression of c-Myc, beta-catenin, and SOX-2 in the ALDH1high population and a significant higher level of ABCG2. Statistical analysis of data demonstrated that ALDH1high cells of SW-982 and SW-1353 showed higher resistance to commonly used chemotherapeutic agents like doxorubicin, epirubicin, and cisplatin than ALDH1low cells. This study demonstrates that in different sarcoma cell lines, high ALDH1 activity can be used to identify a subpopulation of cells characterized by a significantly higher proliferation rate, increased colony forming, increased expression of ABC transporter genes and stemness markers compared to control cells. In addition, enhanced drug resistance was demonstrated. | |||
| Key Molecule: Myc proto-oncogene protein (MYC) | [8] | |||
| Resistant Disease | Sarcoma [ICD-11: 2C35.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SW-872 cells | Skin | Homo sapiens (Human) | CVCL_1730 |
| SW-1353 cells | Brain | Homo sapiens (Human) | CVCL_0543 | |
| TE-671 cells | Peripheral blood | Homo sapiens (Human) | CVCL_1756 | |
| SW-684 cells | Skin | Homo sapiens (Human) | CVCL_1726 | |
| SW-982 cells | Testicular | Homo sapiens (Human) | CVCL_1734 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | By investigating of important regulators of stem cell biology, real-time RT-PCR data showed an increased expression of c-Myc, beta-catenin, and SOX-2 in the ALDH1high population and a significant higher level of ABCG2. Statistical analysis of data demonstrated that ALDH1high cells of SW-982 and SW-1353 showed higher resistance to commonly used chemotherapeutic agents like doxorubicin, epirubicin, and cisplatin than ALDH1low cells. This study demonstrates that in different sarcoma cell lines, high ALDH1 activity can be used to identify a subpopulation of cells characterized by a significantly higher proliferation rate, increased colony forming, increased expression of ABC transporter genes and stemness markers compared to control cells. In addition, enhanced drug resistance was demonstrated. | |||
| Key Molecule: Transcription factor SOX-2 (SOX2) | [8] | |||
| Resistant Disease | Sarcoma [ICD-11: 2C35.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SW-872 cells | Skin | Homo sapiens (Human) | CVCL_1730 |
| SW-1353 cells | Brain | Homo sapiens (Human) | CVCL_0543 | |
| TE-671 cells | Peripheral blood | Homo sapiens (Human) | CVCL_1756 | |
| SW-684 cells | Skin | Homo sapiens (Human) | CVCL_1726 | |
| SW-982 cells | Testicular | Homo sapiens (Human) | CVCL_1734 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | By investigating of important regulators of stem cell biology, real-time RT-PCR data showed an increased expression of c-Myc, beta-catenin, and SOX-2 in the ALDH1high population and a significant higher level of ABCG2. Statistical analysis of data demonstrated that ALDH1high cells of SW-982 and SW-1353 showed higher resistance to commonly used chemotherapeutic agents like doxorubicin, epirubicin, and cisplatin than ALDH1low cells. This study demonstrates that in different sarcoma cell lines, high ALDH1 activity can be used to identify a subpopulation of cells characterized by a significantly higher proliferation rate, increased colony forming, increased expression of ABC transporter genes and stemness markers compared to control cells. In addition, enhanced drug resistance was demonstrated. | |||
| Key Molecule: Catenin beta interacting protein 1 (CTNNBIP1) | [8] | |||
| Resistant Disease | Sarcoma [ICD-11: 2C35.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SW-872 cells | Skin | Homo sapiens (Human) | CVCL_1730 |
| SW-1353 cells | Brain | Homo sapiens (Human) | CVCL_0543 | |
| TE-671 cells | Peripheral blood | Homo sapiens (Human) | CVCL_1756 | |
| SW-684 cells | Skin | Homo sapiens (Human) | CVCL_1726 | |
| SW-982 cells | Testicular | Homo sapiens (Human) | CVCL_1734 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | By investigating of important regulators of stem cell biology, real-time RT-PCR data showed an increased expression of c-Myc, beta-catenin, and SOX-2 in the ALDH1high population and a significant higher level of ABCG2. Statistical analysis of data demonstrated that ALDH1high cells of SW-982 and SW-1353 showed higher resistance to commonly used chemotherapeutic agents like doxorubicin, epirubicin, and cisplatin than ALDH1low cells. This study demonstrates that in different sarcoma cell lines, high ALDH1 activity can be used to identify a subpopulation of cells characterized by a significantly higher proliferation rate, increased colony forming, increased expression of ABC transporter genes and stemness markers compared to control cells. In addition, enhanced drug resistance was demonstrated. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-miR-129-5p | [1] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Activation | hsa05200 | |
| Cell migration | Activation | hsa04670 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 toxicity assay | |||
| Mechanism Description | NONHSAT101069 was upregulated in BC tissues and promoted epirubicin resistance, migration and invasion of BC cells via regulation of NONHSAT101069/miR-129-5p/Twist1 axis, highlighting its potential as an oncogene and a therapeutic biomarker for BC. | |||
| Key Molecule: Long non-protein coding RNA (NONHSAT101069) | [1] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Activation | hsa05200 | |
| Cell migration | Activation | hsa04670 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| SUM1315 cells | Breast | Homo sapiens (Human) | CVCL_5589 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR,Northern blot | |||
| Experiment for Drug Resistance |
CCK8 toxicity assay | |||
| Mechanism Description | Twist1 is a target protein of miR-129-5p and positively regulates by NONHSAT101069 in BC cells. and it stimulates epirubicin resistance, migration, invasion, and EMT progression of BC cells. | |||
| Key Molecule: H19, imprinted maternally expressed transcript (H19) | [12] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cullin4A/MDR1 signaling pathway | Regulation | N.A. | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Quantitative real-time RT-PCR | |||
| Experiment for Drug Resistance |
CellTiter AQueous One Solution Cell Proliferation Assay | |||
| Mechanism Description | LncRNA H19 is a major mediator of doxorubicin chemoresistance in breast cancer cells through a cullin4A-MDR1 pathway. H19 overexpression was contributed to cancer cell resistance to anthracyclines and paclitaxel as knockdown of H19 LncRNA by a specific H19 shRNA in Dox-resistant cells significantly improved the cell sensitivity to anthracyclines and paclitaxel. | |||
| Key Molecule: Bcl-2-interacting killer (BIK) | [7] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| MCF-7R cells | Breast | Homo sapiens (Human) | CVCL_Y493 | |
| ZR-75-1R cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Annexin V apoptosis assay; Fow cytometry analysis | |||
| Mechanism Description | LncRNA H19 attenuated cell apoptosis in response to PTX treatment by inhibiting transcription of pro-apoptotic genes BIk and NOXA. | |||
| Key Molecule: Phorbol-12-myristate-13-acetate-induced protein 1 (PMAIP1) | [7] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| MCF-7R cells | Breast | Homo sapiens (Human) | CVCL_Y493 | |
| ZR-75-1R cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Annexin V apoptosis assay; Fow cytometry analysis | |||
| Mechanism Description | LncRNA H19 attenuated cell apoptosis in response to PTX treatment by inhibiting transcription of pro-apoptotic genes BIk and NOXA. | |||
| Key Molecule: hsa-let-7a | [13] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Lower let-7a expression was associated with epirubicin resistance in primary breast tumors. Moreover, upregulation of let-7a expression sensitized resistant breast tumor cell lines to epirubicin by enhancing cellular apoptosis in vitro. Collectively, these findings indicate that lower expression of let-7a miRNA can induce chemoresistance in breast cancer by enhancing cellular apoptosis and suggest that let-7a may be used as a therapeutic target to modulate epirubicin-based chemotherapy resistance. | |||
| Key Molecule: hsa-mir-204 | [14] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell invasion | Activation | hsa05200 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Low miR-024 expression was enhancing chemotherapeutic resistance of breast cancer patients. | |||
| Key Molecule: hsa-mir-125b | [3] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | p53 signaling pathway | Inhibition | hsa04115 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| BT20 cells | Breast | Homo sapiens (Human) | CVCL_0178 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Trypan blue dye exclusion assay | |||
| Mechanism Description | E2F3, and in some settings E2F1, induce apoptosis through p53-dependent or -independent pathways, Overexpression of miR-125b in MCF-7 cells significantly down-regulated E2F3 protein level, overexpression of miR-125b caused a marked inhibition of anticancer drug activity and increased resistance in breast cancer cells in vitro. | |||
|
|
||||
| Key Molecule: 6-Phosphogluconate dehydrogenase (6PGD) | [15] | |||
| Metabolic Type | Glucose metabolism | |||
| Resistant Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Adrenergic signaling in cardiomyocytes | Activation | hsa04261 | |
| In Vitro Model | BT-549 cells | Breast | Homo sapiens (Human) | CVCL_1092 |
| MDA-MB-23 cells1 | Breast | Homo sapiens (Human) | CVCL_0062 | |
| Experiment for Molecule Alteration |
qRT-PCR; Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | In summary, this study investigated the important role of 6PGD in promoting TNBC progression and attenuating chemotherapy response efficacy of chemotherapy-resistant cells. Inhibition of 6PGD and epirubicin exerted synergistic effects on resistant cells, effectively increasing the sensitivity of resistant cells to chemotherapeutic agents through metabolic remodeling. Therefore, 6PGD might be a potential and important metabolic target in clinical applications such as reversing chemotherapy resistance in TNBC and tumor therapies. | |||
|
|
||||
| Key Molecule: Cyclin-G2 (CCNG2) | [16] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Exosomal microRNA miR1246 promotes cell proliferation, invasion and drug resistance by suppressing the expression level of CCNG2 in Breast Cancer. | |||
| Key Molecule: hsa-miR-1246 | [16] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Exosomal microRNA miR1246 promotes cell proliferation, invasion and drug resistance by suppressing the expression level of CCNG2 in Breast Cancer. | |||
|
|
||||
| Key Molecule: Twist-related protein 1 (TWST1) | [1] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Activation | hsa05200 | |
| Cell migration | Activation | hsa04670 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| SUM1315 cells | Breast | Homo sapiens (Human) | CVCL_5589 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR; luciferase reporter assay; Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 toxicity assay | |||
| Mechanism Description | Twist1 is a target protein of miR-129-5p and positively regulates by NONHSAT101069 in BC cells. and it stimulates epirubicin resistance, migration, invasion, and EMT progression of BC cells. | |||
| Key Molecule: Transcription factor E2F3 (E2F3) | [3] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | p53 signaling pathway | Inhibition | hsa04115 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| BT20 cells | Breast | Homo sapiens (Human) | CVCL_0178 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Trypan blue dye exclusion assay | |||
| Mechanism Description | E2F3, and in some settings E2F1, induce apoptosis through p53-dependent or -independent pathways, Overexpression of miR-125b in MCF-7 cells significantly down-regulated E2F3 protein level, overexpression of miR-125b caused a marked inhibition of anticancer drug activity and increased resistance in breast cancer cells in vitro. | |||
| Key Molecule: Fanconi anemia group D2 protein (FANCD2) | [17] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Missense mutation | p.G56V |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | ATF2/ATF3/ATF4 signaling pathway | Inhibition | hsa04915 | |
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Circulating-free DNA assay; Whole exome sequencing assay | |||
| Mechanism Description | Quantification of allele fractions in plasma identified increased representation of mutant alleles in association with emergence of therapy resistance. | |||
| Key Molecule: Hyaluronan mediated motility receptor (HMMR) | [18] | |||
| Resistant Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | RHAMM signaling pathway | Regulation | N.A. | |
| In Vitro Model | 231-EPIR cells | Breast | Homo sapiens (Human) | N.A. |
| M-EPIR cells | Breast | Homo sapiens (Human) | N.A. | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | We demonstrated that RHAMM immensely contributes to breast cancer chemoresistance by several mechanisms: promoting proliferation and migration, enrichment of breast cancer stemness, and induction of EMT. Also, we found that RHAMM served as a potent prognostic factor in breast cancer patients, especially in those who received chemotherapy. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Lactate dehydrogenase A (LDHA) | [5] | |||
| Resistant Disease | Prostate cancer [ICD-11: 2C82.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Mechanism Description | 7928 genes were identified as genes related to tumor progression and metastasis. Of these, 7 genes were found to be associated with PCa prognosis. The scRNA-seq and TCGA data showed that the expression of LDHA was higher in tumors and associated with poor prognosis of PCa. In addition, upregulation of LDHA in PCa cells induces osteoclast differentiation. Additionally, high LDHA expression was associated with resistance to Epirubicin, Elliptinium acetate, and doxorubicin. Cellular experiments demonstrated that LDHA knockdown inhibited doxorubicin resistance in PCa cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Cytochrome P450 family 1 subfamily B member1 (CYP1B1) | [11] | |||
| Sensitive Disease | Kidney cancer [ICD-11: 2C90.1] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | miR27b/CCNG1/p53 signaling pathway | Regulation | N.A. | |
| In Vitro Model | 769-P cells | Kidney | Homo sapiens (Human) | CVCL_1050 |
| 786-O cells | Kidney | Homo sapiens (Human) | CVCL_1051 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | |||
| Mechanism Description | miR-27b synergizes with anticancer drugs througth enhancing anticancer drug-induced cell death which due to p53 activation and CYP1B1 suppression. | |||
|
|
||||
| Key Molecule: hsa-mir-27b | [11] | |||
| Sensitive Disease | Kidney cancer [ICD-11: 2C90.1] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | miR27b/CCNG1/p53 signaling pathway | Regulation | N.A. | |
| In Vitro Model | 769-P cells | Kidney | Homo sapiens (Human) | CVCL_1050 |
| 786-O cells | Kidney | Homo sapiens (Human) | CVCL_1051 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | |||
| Mechanism Description | miR-27b synergizes with anticancer drugs througth enhancing anticancer drug-induced cell death which due to p53 activation and CYP1B1 suppression. | |||
|
|
||||
| Key Molecule: Cyclin-G1 (CCNG1) | [11] | |||
| Sensitive Disease | Kidney cancer [ICD-11: 2C90.1] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| miR27b/CCNG1/p53 signaling pathway | Regulation | N.A. | ||
| In Vitro Model | 769-P cells | Kidney | Homo sapiens (Human) | CVCL_1050 |
| 786-O cells | Kidney | Homo sapiens (Human) | CVCL_1051 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | |||
| Mechanism Description | miR-27b synergizes with anticancer drugs througth enhancing anticancer drug-induced cell death which due to p53 activation and CYP1B1 suppression. | |||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
