Molecule Information
General Information of the Molecule (ID: Mol00300)
| Name |
Cytochrome P450 family 1 subfamily B member1 (CYP1B1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
CYPIB1; Hydroperoxy icosatetraenoate dehydratase
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
CYP1B1
|
||||
| Gene ID | |||||
| Location |
chr2:38066973-38109902[-]
|
||||
| Sequence |
MGTSLSPNDPWPLNPLSIQQTTLLLLLSVLATVHVGQRLLRQRRRQLRSAPPGPFAWPLI
GNAAAVGQAAHLSFARLARRYGDVFQIRLGSCPIVVLNGERAIHQALVQQGSAFADRPAF ASFRVVSGGRSMAFGHYSEHWKVQRRAAHSMMRNFFTRQPRSRQVLEGHVLSEARELVAL LVRGSADGAFLDPRPLTVVAVANVMSAVCFGCRYSHDDPEFRELLSHNEEFGRTVGAGSL VDVMPWLQYFPNPVRTVFREFEQLNRNFSNFILDKFLRHCESLRPGAAPRDMMDAFILSA EKKAAGDSHGGGARLDLENVPATITDIFGASQDTLSTALQWLLLLFTRYPDVQTRVQAEL DQVVGRDRLPCMGDQPNLPYVLAFLYEAMRFSSFVPVTIPHATTANTSVLGYHIPKDTVV FVNQWSVNHDPLKWPNPENFDPARFLDKDGLINKDLTSRVMIFSVGKRRCIGEELSKMQL FLFISILAHQCDFRANPNEPAKMNFSYGLTIKPKSFKVNVTLRESMELLDSAVQNLQAKE TCQ Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
A cytochrome P450 monooxygenase involved in the metabolism of various endogenous substrates, including fatty acids, steroid hormones and vitamins. Mechanistically, uses molecular oxygen inserting one oxygen atom into a substrate, and reducing the second into a water molecule, with two electrons provided by NADPH via cytochrome P450 reductase (NADPH--hemoprotein reductase). Exhibits catalytic activity for the formation of hydroxyestrogens from estrone (E1) and 17beta-estradiol (E2), namely 2- and 4-hydroxy E1 and E2. Displays a predominant hydroxylase activity toward E2 at the C-4 position. Metabolizes testosterone and progesterone to B or D ring hydroxylated metabolites. May act as a major enzyme for all-trans retinoic acid biosynthesis in extrahepatic tissues. Catalyzes two successive oxidative transformation of all-trans retinol to all-trans retinal and then to the active form all-trans retinoic acid. Catalyzes the epoxidation of double bonds of certain PUFA. Converts arachidonic acid toward epoxyeicosatrienoic acid (EpETrE) regioisomers, 8,9-, 11,12-, and 14,15- EpETrE, that function as lipid mediators in the vascular system. Additionally, displays dehydratase activity toward oxygenated eicosanoids hydroperoxyeicosatetraenoates (HpETEs). This activity is independent of cytochrome P450 reductase, NADPH, and O2. Also involved in the oxidative metabolism of xenobiotics, particularly converting polycyclic aromatic hydrocarbons and heterocyclic aryl amines procarcinogens to DNA-damaging products. Plays an important role in retinal vascular development. Under hyperoxic O2 conditions, promotes retinal angiogenesis and capillary morphogenesis, likely by metabolizing the oxygenated products generated during the oxidative stress. Also, contributes to oxidative homeostasis and ultrastructural organization and function of trabecular meshwork tissue through modulation of POSTN expression.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
5 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Kidney cancer [ICD-11: 2C90.1] | [1] | |||
| Sensitive Disease | Kidney cancer [ICD-11: 2C90.1] | |||
| Sensitive Drug | Gefitinib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Kidney cancer [ICD-11: 2C90] | |||
| The Specified Disease | Renal cancer | |||
| The Studied Tissue | Kidney | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.73E-03 Fold-change: -1.49E-01 Z-score: -3.05E+00 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | miR27b/CCNG1/p53 signaling pathway | Regulation | N.A. | |
| In Vitro Model | 769-P cells | Kidney | Homo sapiens (Human) | CVCL_1050 |
| 786-O cells | Kidney | Homo sapiens (Human) | CVCL_1051 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | |||
| Mechanism Description | miR-27b synergizes with anticancer drugs througth enhancing anticancer drug-induced cell death which due to p53 activation and CYP1B1 suppression. | |||
| Disease Class: Liver cancer [ICD-11: 2C12.6] | [1] | |||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Sensitive Drug | Gefitinib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | miR27b/CCNG1/p53 signaling pathway | Regulation | N.A. | |
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| SNU182 cells | Liver | Homo sapiens (Human) | CVCL_0090 | |
| SNU-739 cells | Liver | Homo sapiens (Human) | CVCL_5088 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | |||
| Mechanism Description | miR-27b synergizes with anticancer drugs througth enhancing anticancer drug-induced cell death which due to p53 activation and CYP1B1 suppression. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Liver cancer [ICD-11: 2C12.6] | [1] | |||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | miR27b/CCNG1/p53 signaling pathway | Regulation | N.A. | |
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| SNU182 cells | Liver | Homo sapiens (Human) | CVCL_0090 | |
| SNU-739 cells | Liver | Homo sapiens (Human) | CVCL_5088 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | |||
| Mechanism Description | miR-27b synergizes with anticancer drugs througth enhancing anticancer drug-induced cell death which due to p53 activation and CYP1B1 suppression. | |||
| Disease Class: Kidney cancer [ICD-11: 2C90.1] | [1] | |||
| Sensitive Disease | Kidney cancer [ICD-11: 2C90.1] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | miR27b/CCNG1/p53 signaling pathway | Regulation | N.A. | |
| In Vitro Model | 769-P cells | Kidney | Homo sapiens (Human) | CVCL_1050 |
| 786-O cells | Kidney | Homo sapiens (Human) | CVCL_1051 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | |||
| Mechanism Description | miR-27b synergizes with anticancer drugs througth enhancing anticancer drug-induced cell death which due to p53 activation and CYP1B1 suppression. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Liver cancer [ICD-11: 2C12.6] | [1] | |||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Sensitive Drug | Epirubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | miR27b/CCNG1/p53 signaling pathway | Regulation | N.A. | |
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| SNU182 cells | Liver | Homo sapiens (Human) | CVCL_0090 | |
| SNU-739 cells | Liver | Homo sapiens (Human) | CVCL_5088 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | |||
| Mechanism Description | miR-27b synergizes with anticancer drugs througth enhancing anticancer drug-induced cell death which due to p53 activation and CYP1B1 suppression. | |||
| Disease Class: Kidney cancer [ICD-11: 2C90.1] | [1] | |||
| Sensitive Disease | Kidney cancer [ICD-11: 2C90.1] | |||
| Sensitive Drug | Epirubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | miR27b/CCNG1/p53 signaling pathway | Regulation | N.A. | |
| In Vitro Model | 769-P cells | Kidney | Homo sapiens (Human) | CVCL_1050 |
| 786-O cells | Kidney | Homo sapiens (Human) | CVCL_1051 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | |||
| Mechanism Description | miR-27b synergizes with anticancer drugs througth enhancing anticancer drug-induced cell death which due to p53 activation and CYP1B1 suppression. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Liver cancer [ICD-11: 2C12.6] | [1] | |||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Sensitive Drug | Etoposide | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | miR27b/CCNG1/p53 signaling pathway | Regulation | N.A. | |
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| SNU182 cells | Liver | Homo sapiens (Human) | CVCL_0090 | |
| SNU-739 cells | Liver | Homo sapiens (Human) | CVCL_5088 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | |||
| Mechanism Description | miR-27b synergizes with anticancer drugs througth enhancing anticancer drug-induced cell death which due to p53 activation and CYP1B1 suppression. | |||
| Disease Class: Kidney cancer [ICD-11: 2C90.1] | [1] | |||
| Sensitive Disease | Kidney cancer [ICD-11: 2C90.1] | |||
| Sensitive Drug | Etoposide | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | miR27b/CCNG1/p53 signaling pathway | Regulation | N.A. | |
| In Vitro Model | 769-P cells | Kidney | Homo sapiens (Human) | CVCL_1050 |
| 786-O cells | Kidney | Homo sapiens (Human) | CVCL_1051 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | |||
| Mechanism Description | miR-27b synergizes with anticancer drugs througth enhancing anticancer drug-induced cell death which due to p53 activation and CYP1B1 suppression. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Kidney cancer [ICD-11: 2C90.1] | [1] | |||
| Sensitive Disease | Kidney cancer [ICD-11: 2C90.1] | |||
| Sensitive Drug | Sorafenib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | miR27b/CCNG1/p53 signaling pathway | Regulation | N.A. | |
| In Vitro Model | 769-P cells | Kidney | Homo sapiens (Human) | CVCL_1050 |
| 786-O cells | Kidney | Homo sapiens (Human) | CVCL_1051 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | |||
| Mechanism Description | miR-27b synergizes with anticancer drugs througth enhancing anticancer drug-induced cell death which due to p53 activation and CYP1B1 suppression. | |||
| Disease Class: Liver cancer [ICD-11: 2C12.6] | [1] | |||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Sensitive Drug | Sorafenib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | miR27b/CCNG1/p53 signaling pathway | Regulation | N.A. | |
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| SNU182 cells | Liver | Homo sapiens (Human) | CVCL_0090 | |
| SNU-739 cells | Liver | Homo sapiens (Human) | CVCL_5088 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | |||
| Mechanism Description | miR-27b synergizes with anticancer drugs througth enhancing anticancer drug-induced cell death which due to p53 activation and CYP1B1 suppression. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Liver | |
| The Specified Disease | Liver cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.27E-01; Fold-change: 2.43E-01; Z-score: 2.46E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 4.90E-02; Fold-change: -1.73E-01; Z-score: -2.73E-01 | |
| The Expression Level of Disease Section Compare with the Other Disease Section | p-value: 7.82E-01; Fold-change: 5.17E-02; Z-score: 9.74E-02 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
Molecule expression in tissue other than the diseased tissue of patients
|
||
| Disease-specific Molecule Abundances |
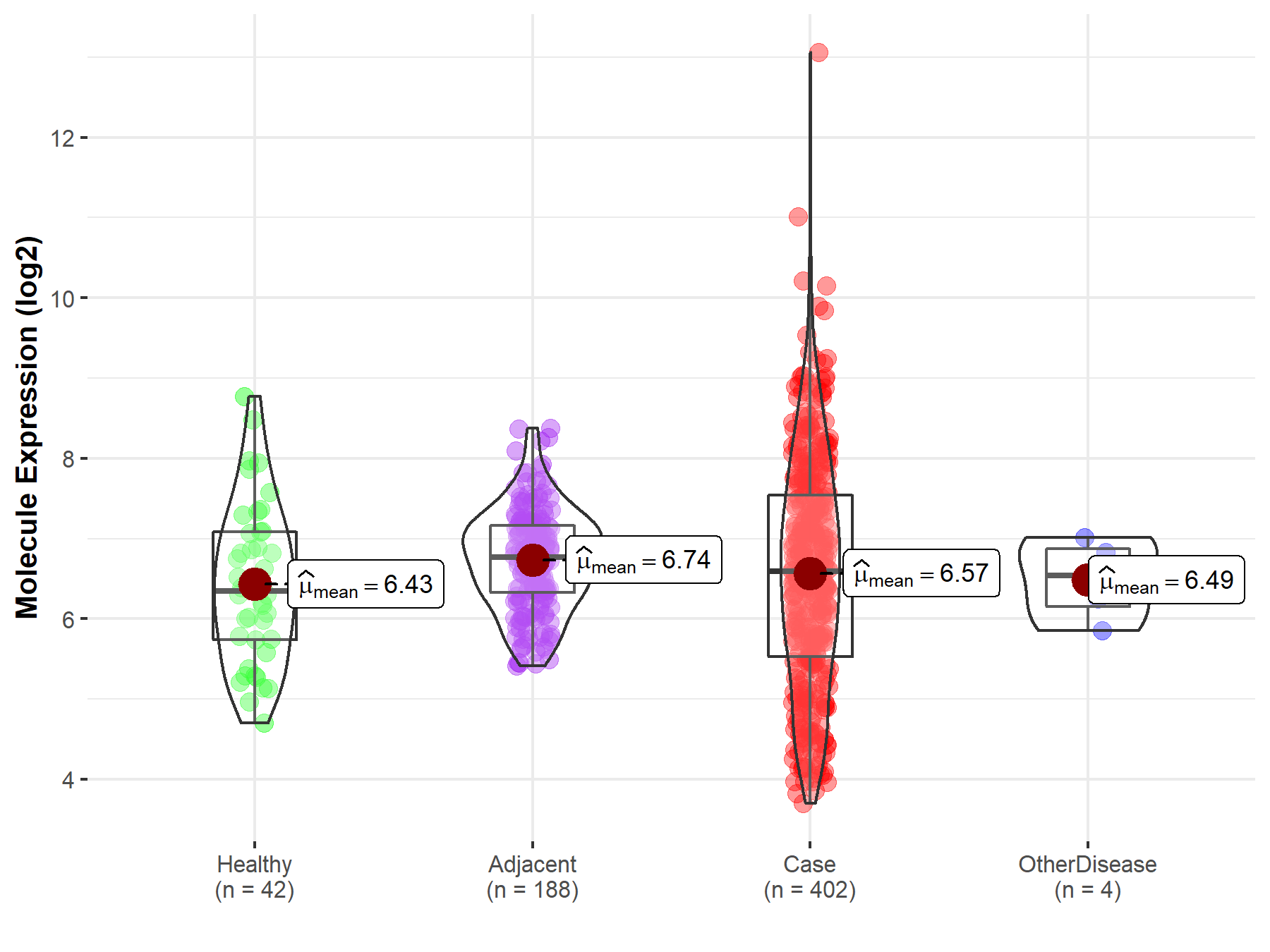
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Kidney | |
| The Specified Disease | Kidney cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.73E-03; Fold-change: -1.17E+00; Z-score: -1.30E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.39E-01; Fold-change: -2.46E-01; Z-score: -3.48E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

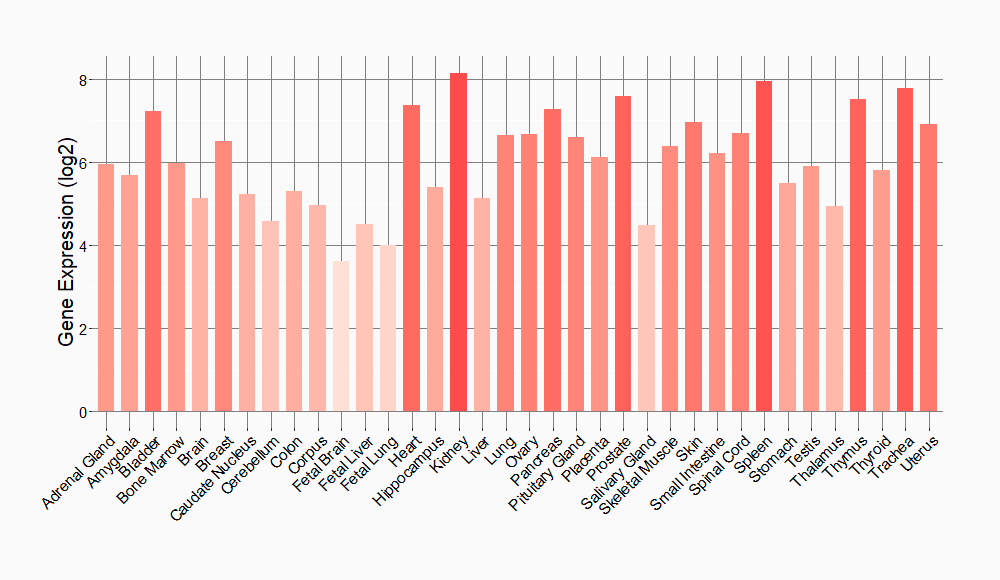
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
