Molecule Information
General Information of the Molecule (ID: Mol01207)
| Name |
H19, imprinted maternally expressed transcript (H19)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
H19
Click to Show/Hide
|
||||
| Molecule Type |
LncRNA
|
||||
| Gene Name |
psiTPTE22
|
||||
| Gene ID | |||||
| Location |
chr11:1995171-2001470[-]
|
||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
19 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [1] | |||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Gastric cancer [ICD-11: 2B72] | |||
| The Specified Disease | Stomach adenocarcinoma | |||
| The Studied Tissue | Stomach | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.86E-01 Fold-change: 6.91E-01 Z-score: 1.32E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | FADD/Caspase 8/Caspase 3 signaling pathway | Regulation | N.A. | |
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| GES-1 cells | Gastric | Homo sapiens (Human) | CVCL_EQ22 | |
| SGC-7901/DDP cells | Gastric | Homo sapiens (Human) | CVCL_0520 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometric analysis; Colony formation assay; Colony formation assay | |||
| Mechanism Description | Long Noncoding RNA H19/miR675 Axis Promotes Gastric Cancer via FADD/Caspase 8/Caspase 3 signaling Pathway. H19/miR675 targets FADD and inhibits caspase 8/caspase 3, H19 inhibits the expression of FADD through miR675 targeting. | |||
| Disease Class: Seminoma [ICD-11: 2C80.3] | [3] | |||
| Resistant Disease | Seminoma [ICD-11: 2C80.3] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Testicular cancer [ICD-11: 2C80] | |||
| The Specified Disease | Testicular germ cell tumor | |||
| The Studied Tissue | Testis | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.09E-08 Fold-change: 3.91E+00 Z-score: 6.05E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| PI3K/AKT/mTOR signaling pathway | Regulation | N.A. | ||
| In Vitro Model | TCam-2 cells | Testicle | Homo sapiens (Human) | CVCL_T012 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Long non-coding RNA H19 promotes TDRG1 expression and cisplatin resistance by sequestering miRNA-106b-5p in seminoma. | |||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [9] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| MCF-7R cells | Breast | Homo sapiens (Human) | CVCL_Y493 | |
| ZR-75-1R cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Annexin V apoptosis assay; Fow cytometry analysis | |||
| Mechanism Description | LncRNA H19 attenuated cell apoptosis in response to PTX treatment by inhibiting transcription of pro-apoptotic genes BIk and NOXA. | |||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [10] | |||
| Resistant Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Activation | hsa05200 | |
| In Vitro Model | A2780-DR cells | Ovary | Homo sapiens (Human) | CVCL_EG64 |
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
Clonogenic assay | |||
| Mechanism Description | The Essential Role of H19 Contributing to Cisplatin Resistance by Regulating Glutathione Metabolism in High-Grade Serous Ovarian Cancer.Additionally, we verified that different H19 expression levels in HGSC tissues showed strong correlation with cancer recurrence. H19 knockdown in A2780-DR cells resulted in recovery of cisplatin sensitivity in vitro and in vivo. Quantitative proteomics analysis indicated that six NRF2-targeted proteins, including NQO1, GSR, G6PD, GCLC, GCLM and GSTP1 involved in the glutathione metabolism pathway, were reduced in H19-knockdown cells. Furthermore, H19-knockdown cells were markedly more sensitive to hydrogen-peroxide treatment and exhibited lower glutathione levels. Our results reveal a previously unknown link between H19 and glutathione metabolism in the regulation of cancer-drug resistance. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [10] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | A2780-DR cells | Ovary | Homo sapiens (Human) | CVCL_EG64 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Clonogenic assay | |||
| Mechanism Description | H19 overexpression contributes to cisplatin resistance. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colon cancer [ICD-11: 2B90.1] | [2] | |||
| Resistant Disease | Colon cancer [ICD-11: 2B90.1] | |||
| Resistant Drug | Calcitriol | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Colon cancer [ICD-11: 2B90] | |||
| The Specified Disease | Colon adenocarcinoma | |||
| The Studied Tissue | Colon | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.23E-01 Fold-change: 4.47E-01 Z-score: 9.89E-01 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Wnt/Beta-catenin signaling pathway | Inhibition | hsa04310 | |
| In Vitro Model | DLD1 cells | Colon | Homo sapiens (Human) | CVCL_0248 |
| HT-29 cells | Colon | Homo sapiens (Human) | CVCL_0320 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | H19 overexpression induces resistance to 1,25(OH)2D3 by inhibiting the expression of VDR Through miR675-5p in colon cancer cells. vdr signaling was able to attenuate the proliferation and migration of colon cancer cells via multiple mechanisms including inhibiting Wnt/beta-catenin pathway, VDR signaling inhibits the expression of h19 by regulating the C-Myc/mad-1 Network. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Malignant glioma [ICD-11: 2A00.2] | [4] | |||
| Resistant Disease | Malignant glioma [ICD-11: 2A00.2] | |||
| Resistant Drug | Temozolomide | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Brain cancer [ICD-11: 2A00] | |||
| The Specified Disease | Glioblastoma multiforme | |||
| The Studied Tissue | Brain | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.56E-04 Fold-change: 2.67E+00 Z-score: 3.51E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| U87 cells | Brain | Homo sapiens (Human) | CVCL_0022 | |
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Knockdown of long noncoding RNA H19 sensitizes human glioma cells to temozolomide therapy.the expression level of H19 transcripts was increased compared to wild-type or nonresistant clones.Furthermore, the reduced expression of H19 altered major drug resistance genes, such as ABCB1 (MDR1), ABCC (MRP), and ABCG2 (BCRP), both at the mRNA and protein levels. Taken together, these findings suggest that H19 plays an important role in the development of TMZ resistance, and may represent a novel therapeutic target for TMZ-resistant gliomas.Our results suggested that knockdown of H19 significantly downregulated the expression of these drug-resistant genes, both at the mRNA (P<0.001 vs respective control siRNA) and protein levels. These data confirm that the H19-induced TMZ resistance is in part mediated by MDR, MRP, and ABCG2. | |||
|
|
||||
| Disease Class: Malignant glioma [ICD-11: 2A00.2] | [18] | |||
| Resistant Disease | Malignant glioma [ICD-11: 2A00.2] | |||
| Resistant Drug | Temozolomide | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell colony | Activation | hsa05200 | ||
| Cell viability | Activation | hsa05200 | ||
| Wnt/beta-catenin signaling pathway | Activation | hsa04310 | ||
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| M059J cells | Brain | Homo sapiens (Human) | CVCL_0400 | |
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Silencing of H19 decreases chemoresistance through suppressing EMT via the Wnt/beta-Catenin pathway. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lung small cell carcinoma [ICD-11: 2C25.2] | [5] | |||
| Resistant Disease | Lung small cell carcinoma [ICD-11: 2C25.2] | |||
| Resistant Drug | Etoposide | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | |||
| The Specified Disease | Lung adenocarcinoma | |||
| The Studied Tissue | Lung | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.58E-05 Fold-change: 2.58E+00 Z-score: 4.17E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/AdrVp cells | Breast | Homo sapiens (Human) | CVCL_4Y46 | |
| Experiment for Molecule Alteration |
RT-PCR; Northern blotting analysis | |||
| Experiment for Drug Resistance |
Clonogenic assay | |||
| Mechanism Description | The mRNA of the H19 gene is overexpressed in MCF-7/AdrVp cells relative toparental MCF-7 cells or drug-sensitive MCF-7/AdrVp revertant cells. H19is an imprinted gene with an important role in fetal differentiation, as well as a postulated function as a tumor suppressor gene. Another p95-over-expressing multidrug-resistant cell line, human lung carcinoma NCI-H1688, also displays high levels of 1119 mRNA. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hepatocellular carcinoma [ICD-11: 2C12.2] | [6] | |||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Resistant Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Liver cancer [ICD-11: 2C12] | |||
| The Specified Disease | Liver hepatocellular carcinoma | |||
| The Studied Tissue | Liver | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.95E-05 Fold-change: 2.36E+00 Z-score: 4.33E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Antisense H19 oligonucleotides transfection induced a marked increase in the percentage of MDR1 promoter methylation and decrease in MDR1 expression in R-HepG2 cells. Thus, the H19 gene is believed to induce P-glycoprotein expression and MDR1-associated drug resistance at least in liver cancer cells through regulation of MDR1 promoter methylation. | |||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [11] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cullin4A/MDR1 signaling pathway | Regulation | N.A. | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Quantitative real-time RT-PCR | |||
| Experiment for Drug Resistance |
CellTiter AQueous One Solution Cell Proliferation Assay | |||
| Mechanism Description | LncRNA H19 is a major mediator of doxorubicin chemoresistance in breast cancer cells through a cullin4A-MDR1 pathway. H19 overexpression was contributed to cancer cell resistance to anthracyclines and paclitaxel as knockdown of H19 LncRNA by a specific H19 shRNA in Dox-resistant cells significantly improved the cell sensitivity to anthracyclines and paclitaxel. | |||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [5] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/AdrVp cells | Breast | Homo sapiens (Human) | CVCL_4Y46 | |
| Experiment for Molecule Alteration |
RT-PCR; Northern blotting analysis | |||
| Experiment for Drug Resistance |
Clonogenic assay | |||
| Mechanism Description | The mRNA of the H19 gene is overexpressed in MCF-7/AdrVp cells relative toparental MCF-7 cells or drug-sensitive MCF-7/AdrVp revertant cells. H19is an imprinted gene with an important role in fetal differentiation, as well as a postulated function as a tumor suppressor gene. Another p95-over-expressing multidrug-resistant cell line, human lung carcinoma NCI-H1688, also displays high levels of 1119 mRNA. | |||
| Disease Class: Leukemia [ICD-11: 2B33.6] | [5] | |||
| Resistant Disease | Leukemia [ICD-11: 2B33.6] | |||
| Resistant Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/AdrVp cells | Breast | Homo sapiens (Human) | CVCL_4Y46 | |
| Experiment for Molecule Alteration |
RT-PCR; Northern blotting analysis | |||
| Experiment for Drug Resistance |
Clonogenic assay | |||
| Mechanism Description | The mRNA of the H19 gene is overexpressed in MCF-7/AdrVp cells relative toparental MCF-7 cells or drug-sensitive MCF-7/AdrVp revertant cells. H19is an imprinted gene with an important role in fetal differentiation, as well as a postulated function as a tumor suppressor gene. Another p95-over-expressing multidrug-resistant cell line, human lung carcinoma NCI-H1688, also displays high levels of 1119 mRNA. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hepatic fibrosis [ICD-11: DB93.0] | [7] | |||
| Resistant Disease | Hepatic fibrosis [ICD-11: DB93.0] | |||
| Resistant Drug | Artenimol | |||
| Molecule Alteration | Up-regulation | Interaction |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | HSC cells | N.A. | N.A. | N.A. |
| Experiment for Molecule Alteration |
Knockdown assay | |||
| Mechanism Description | LncRNA-H19 induces hepatic stellate cell activation via upregulating alcohol dehydrogenase III-mediated retinoic acid signals. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Multiple myeloma [ICD-11: 2A83.0] | [8] | |||
| Resistant Disease | Multiple myeloma [ICD-11: 2A83.0] | |||
| Resistant Drug | Bortezomib | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | 8226 cells | Bone marrow | Homo sapiens (Human) | CVCL_0014 |
| NCI-H929 cells | Bone marrow | Homo sapiens (Human) | CVCL_1600 | |
| U266 cells | Bone marrow | Homo sapiens (Human) | CVCL_0566 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | LncRNA H19 overexpression induces bortezomib resistance in multiple myeloma by targeting MCL-1 via miR-29b-3p. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [11] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Epirubicin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cullin4A/MDR1 signaling pathway | Regulation | N.A. | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Quantitative real-time RT-PCR | |||
| Experiment for Drug Resistance |
CellTiter AQueous One Solution Cell Proliferation Assay | |||
| Mechanism Description | LncRNA H19 is a major mediator of doxorubicin chemoresistance in breast cancer cells through a cullin4A-MDR1 pathway. H19 overexpression was contributed to cancer cell resistance to anthracyclines and paclitaxel as knockdown of H19 LncRNA by a specific H19 shRNA in Dox-resistant cells significantly improved the cell sensitivity to anthracyclines and paclitaxel. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: ER positive breast cancer [ICD-11: 2C60.6] | [12] | |||
| Sensitive Disease | ER positive breast cancer [ICD-11: 2C60.6] | |||
| Sensitive Drug | Fulvestrant | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| LCC2 cells | Breast | Homo sapiens (Human) | CVCL_DP51 | |
| LCC9 cells | Breast | Homo sapiens (Human) | CVCL_DP52 | |
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
WST-8 assay | |||
| Mechanism Description | H19 plays a central role in maintaining endocrine therapy resistance by modulating ERalpha expression in these cells. Moreover, decreasing H19 levels using pharmacological inhibitors, that inhibit pathways regulating H19 expression in the ETR cells, helps overcome Tamoxifen and Fulvestrant-resistance. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [13] | |||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Sensitive Drug | Gefitinib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | HCC827 cells | Lung | Homo sapiens (Human) | CVCL_2063 |
| HCC4006 cells | Lung | Homo sapiens (Human) | CVCL_1269 | |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay; TUNEL assay | |||
| Mechanism Description | Knockdown of H19 by siRNA transfection can significantly reduce the expression of N-cadherin, as well as increase E-cadherin and vimentin level, which improved tamoxifen sensitivity in tamoxifen-resistant breast cancer cells. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Myocardial ischemia [ICD-11: BA6Z.0] | [7] | |||
| Resistant Disease | Myocardial ischemia [ICD-11: BA6Z.0] | |||
| Resistant Drug | Hydrogen peroxide | |||
| Molecule Alteration | Up-regulation | Interaction |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | H9C2 cells | Ovary | Cricetulus griseus (Chinese hamster) | CVCL_A0TS |
| In Vivo Model | Rat model of heart IP and I/R injury | Rattus norvegicus | ||
| Experiment for Molecule Alteration |
Knockdown assay; Overexpression assay; qRT-PCR; Western bloting analysis; RIP experiments assay | |||
| Experiment for Drug Resistance |
CCK8 assay; caspase-3 activity assay | |||
| Mechanism Description | LncRNA H19 is involved in myocardial IP via increasing the stability of nucleolin protein and LncRNA H19 may represent a potential therapeutic target for the treatment of the myocardial injury. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [14] | |||
| Resistant Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Resistant Drug | Methotrexate | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Wnt/Beta-catenin signaling pathway | Activation | hsa04310 | |
| In Vitro Model | HT-29 cells | Colon | Homo sapiens (Human) | CVCL_0320 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | H19 mediates methotrexate resistance in colorectal cancer through activating Wnt/beta-catenin pathway. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [15] | |||
| Resistant Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Resistant Drug | Oxaliplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Beta-catenin signaling pathway | Activation | hsa04520 | |
| Cell apoptosis | Activation | hsa04210 | ||
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | SW480 cells | Colon | Homo sapiens (Human) | CVCL_0546 |
| HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |
| In Vivo Model | NOD/SCID mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | CAF-derived exosomes transfer LncRNA H19 to colorectal cancer cells and H19 activated the beta-catenin pathway via acting as a competing endogenous RNA sponge for miR-141, while miR-141 inhibited the stemness of CRC cells. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [11] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cullin4A/MDR1 signaling pathway | Regulation | N.A. | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Quantitative real-time RT-PCR | |||
| Experiment for Drug Resistance |
CellTiter AQueous One Solution Cell Proliferation Assay | |||
| Mechanism Description | LncRNA H19 is a major mediator of doxorubicin chemoresistance in breast cancer cells through a cullin4A-MDR1 pathway. H19 overexpression was contributed to cancer cell resistance to anthracyclines and paclitaxel as knockdown of H19 LncRNA by a specific H19 shRNA in Dox-resistant cells significantly improved the cell sensitivity to anthracyclines and paclitaxel. | |||
| Disease Class: ER positive breast cancer [ICD-11: 2C60.6] | [9] | |||
| Resistant Disease | ER positive breast cancer [ICD-11: 2C60.6] | |||
| Resistant Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| MCF-7R cells | Breast | Homo sapiens (Human) | CVCL_Y493 | |
| ZR-75-1R cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Annexin V apoptosis assay; Fow cytometry analysis | |||
| Mechanism Description | LncRNA H19 was identified as a powerful factor associated with paclitaxel (PTX) resistance in ERalpha-positive breast cancer cells, but not in ERalpha-negative breast cancer cells. LncRNA H19 was one of the downstream target molecules of ERalpha. Altered ERalpha expression may therefore change H19 levels to modulate the apoptosis response to chemotherapy in breast cancer cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [16] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | AKT signaling pathway | Inhibition | hsa04151 | |
| Cell apoptosis | Activation | hsa04210 | ||
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | |
| MDA-MB-157 cells | Breast | Homo sapiens (Human) | CVCL_0618 | |
| In Vivo Model | BALB/c nude mouse xenograft mode | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Knockdown of LncRNA H19 restores chemo-sensitivity in paclitaxel-resistant triple-negative breast cancer through triggering apoptosis and regulating Akt signaling pathway. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [11] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Pirarubicin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cullin4A/MDR1 signaling pathway | Regulation | N.A. | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Quantitative real-time RT-PCR | |||
| Experiment for Drug Resistance |
CellTiter AQueous One Solution Cell Proliferation Assay | |||
| Mechanism Description | LncRNA H19 is a major mediator of doxorubicin chemoresistance in breast cancer cells through a cullin4A-MDR1 pathway. H19 overexpression was contributed to cancer cell resistance to anthracyclines and paclitaxel as knockdown of H19 LncRNA by a specific H19 shRNA in Dox-resistant cells significantly improved the cell sensitivity to anthracyclines and paclitaxel. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: ER positive breast cancer [ICD-11: 2C60.6] | [12] | |||
| Sensitive Disease | ER positive breast cancer [ICD-11: 2C60.6] | |||
| Sensitive Drug | Tamoxifen | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| LCC2 cells | Breast | Homo sapiens (Human) | CVCL_DP51 | |
| LCC9 cells | Breast | Homo sapiens (Human) | CVCL_DP52 | |
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
WST-8 assay | |||
| Mechanism Description | H19 plays a central role in maintaining endocrine therapy resistance by modulating ERalpha expression in these cells. Moreover, decreasing H19 levels using pharmacological inhibitors, that inhibit pathways regulating H19 expression in the ETR cells, helps overcome Tamoxifen and Fulvestrant-resistance. | |||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [17] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Tamoxifen | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell viability | Inhibition | hsa05200 | ||
| Wnt signaling pathway | Inhibition | hsa04310 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | Knockdown of H19 by siRNA transfection can significantly reduce the expression of N-cadherin, as well as increase E-cadherin and vimentin level, which improved tamoxifen sensitivity in tamoxifen-resistant breast cancer cells. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [19] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Trastuzumab | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Higher expression of H19 might lead to trastuzumab resistance in HER2-positive breast cancer patients. High H19 expression was associated with a worse clinical prognosis and a lower PFS. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [5] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Verapamil | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/AdrVp cells | Breast | Homo sapiens (Human) | CVCL_4Y46 | |
| Experiment for Molecule Alteration |
RT-PCR; Northern blotting analysis | |||
| Experiment for Drug Resistance |
Clonogenic assay | |||
| Mechanism Description | The mRNA of the H19 gene is overexpressed in MCF-7/AdrVp cells relative toparental MCF-7 cells or drug-sensitive MCF-7/AdrVp revertant cells. H19is an imprinted gene with an important role in fetal differentiation, as well as a postulated function as a tumor suppressor gene. Another p95-over-expressing multidrug-resistant cell line, human lung carcinoma NCI-H1688, also displays high levels of 1119 mRNA. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Leukemia [ICD-11: 2B33.6] | [5] | |||
| Resistant Disease | Leukemia [ICD-11: 2B33.6] | |||
| Resistant Drug | Vincristine | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/AdrVp cells | Breast | Homo sapiens (Human) | CVCL_4Y46 | |
| Experiment for Molecule Alteration |
RT-PCR; Northern blotting analysis | |||
| Experiment for Drug Resistance |
Clonogenic assay | |||
| Mechanism Description | The mRNA of the H19 gene is overexpressed in MCF-7/AdrVp cells relative toparental MCF-7 cells or drug-sensitive MCF-7/AdrVp revertant cells. H19is an imprinted gene with an important role in fetal differentiation, as well as a postulated function as a tumor suppressor gene. Another p95-over-expressing multidrug-resistant cell line, human lung carcinoma NCI-H1688, also displays high levels of 1119 mRNA. | |||
Investigative Drug(s)
7 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [7] | |||
| Resistant Disease | Melanoma [ICD-11: 2C30.0] | |||
| Resistant Drug | Cisplatinum | |||
| Molecule Alteration | Up-regulation | Interaction |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 |
| M8 cells | Skin | Homo sapiens (Human) | N.A. | |
| WM35 cells | Skin | Homo sapiens (Human) | CVCL_0580 | |
| SK-MEL-2 cells | Skin | Homo sapiens (Human) | CVCL_0069 | |
| A2508 cells | N.A. | N.A. | N.A. | |
| Experiment for Molecule Alteration |
Knockdown assay; qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Down-regulation of LncRNA H19 sensitizes melanoma cells to cisplatin by regulating the miR-18b/IGF1 axis. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [20] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Huaier | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell metastasis | Activation | hsa05205 | ||
| Cell proliferation | Activation | hsa05200 | ||
| Cell viability | Activation | hsa05200 | ||
| Tumorigenesis | Activation | hsa05206 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| Experiment for Molecule Alteration |
qPCR; Microarray assay | |||
| Experiment for Drug Resistance |
Flow cytometry assay; MTT assay | |||
| Mechanism Description | CBL was a direct target of miR675-5p, overexpression miR675-5p decreased the expression of CBL. H19/miR675-5p increases cell proliferation by regulating CBL. Huaier extract reduced the expression of H19 and miR675-5p, thereby inhibiting breast cancer progression through increasing CBL. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [20] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Huaier extract | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | LncRH19/MiR675-5p signaling pathway | Regulation | N.A. | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Flow cytometry assay; MTT assay | |||
| Mechanism Description | miR675-5p was identified as a mature product of H19. The overexpression of H19 or miR675-5p reverses the tumor-inhibitory effects of Huaier extract. However, knocking down H19 or miR675-5p sensitizes breast cancer cells to Huaier extract. Huaier extract inhibits breast cancer cell proliferation and induces apoptosis via the LncRNA H19/miR675-5p/CBL signaling pathway. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Sepsis [ICD-11: 1G40.0] | [7] | |||
| Resistant Disease | Sepsis [ICD-11: 1G40.0] | |||
| Resistant Drug | Lipopolysaccharide | |||
| Molecule Alteration | Down-regulation | Interaction |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | UL-1 cells | Ascites | Canis lupus familiaris (Dog) | CVCL_L421 |
| In Vivo Model | Male BALB/c nude mice model | Mus musculus | ||
| Experiment for Molecule Alteration |
Overexpression assay | |||
| Mechanism Description | LncRNA H19 functions as an Aquaporin 1 competitive endogenous RNA to regulate microRNA-874 expression in LPS sepsis. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hyperglycemia [ICD-11: MA18.0] | [7] | |||
| Resistant Disease | Hyperglycemia [ICD-11: MA18.0] | |||
| Resistant Drug | Mangostin | |||
| Molecule Alteration | Down-regulation | Interaction |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HUVEC-12 cells | Endothelium | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
RNA-seq assay | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | H19 exerted its function via the modulation of H19/miR-140/HE4 in hyperglycemia with alpha-Mangostin. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Nonalcoholic fatty liver disease [ICD-11: DB92.0] | [7] | |||
| Resistant Disease | Nonalcoholic fatty liver disease [ICD-11: DB92.0] | |||
| Resistant Drug | Nonesterified | |||
| Molecule Alteration | Up-regulation | Interaction |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| In Vivo Model | C57BL/6 mice model | Mus musculus | ||
| Experiment for Molecule Alteration |
IHC staining assay; ELISA assay; Luciferase assay; RNA pull down assay; qRT-PCR; Western bloting analysis; Overexpression assay; Knockdown assay | |||
| Mechanism Description | LncRNA-H19 promotes hepatic lipogenesis by directly regulating miR-130a/PPAR gama axis in nonalcoholic fatty liver disease. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Duchenne muscular dystrophy [ICD-11: 8C70.1] | [7] | |||
| Resistant Disease | Duchenne muscular dystrophy [ICD-11: 8C70.1] | |||
| Resistant Drug | Preimplantation factor | |||
| Molecule Alteration | Up-regulation | Interaction |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | C2C12 cells | Skeletal muscle | Homo sapiens (Human) | CVCL_0188 |
| In Vivo Model | Mdx mice model | Mus musculus | ||
| Mechanism Description | SPIF promotes myoblast differentiation and utrophin expression while inhibiting fibrosis in Duchenne muscular dystrophy via the H19/miR-675/let-7 and miR-21 pathways. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Brain | |
| The Specified Disease | Brain lower grade glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.97E-01; Fold-change: -5.55E-03 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
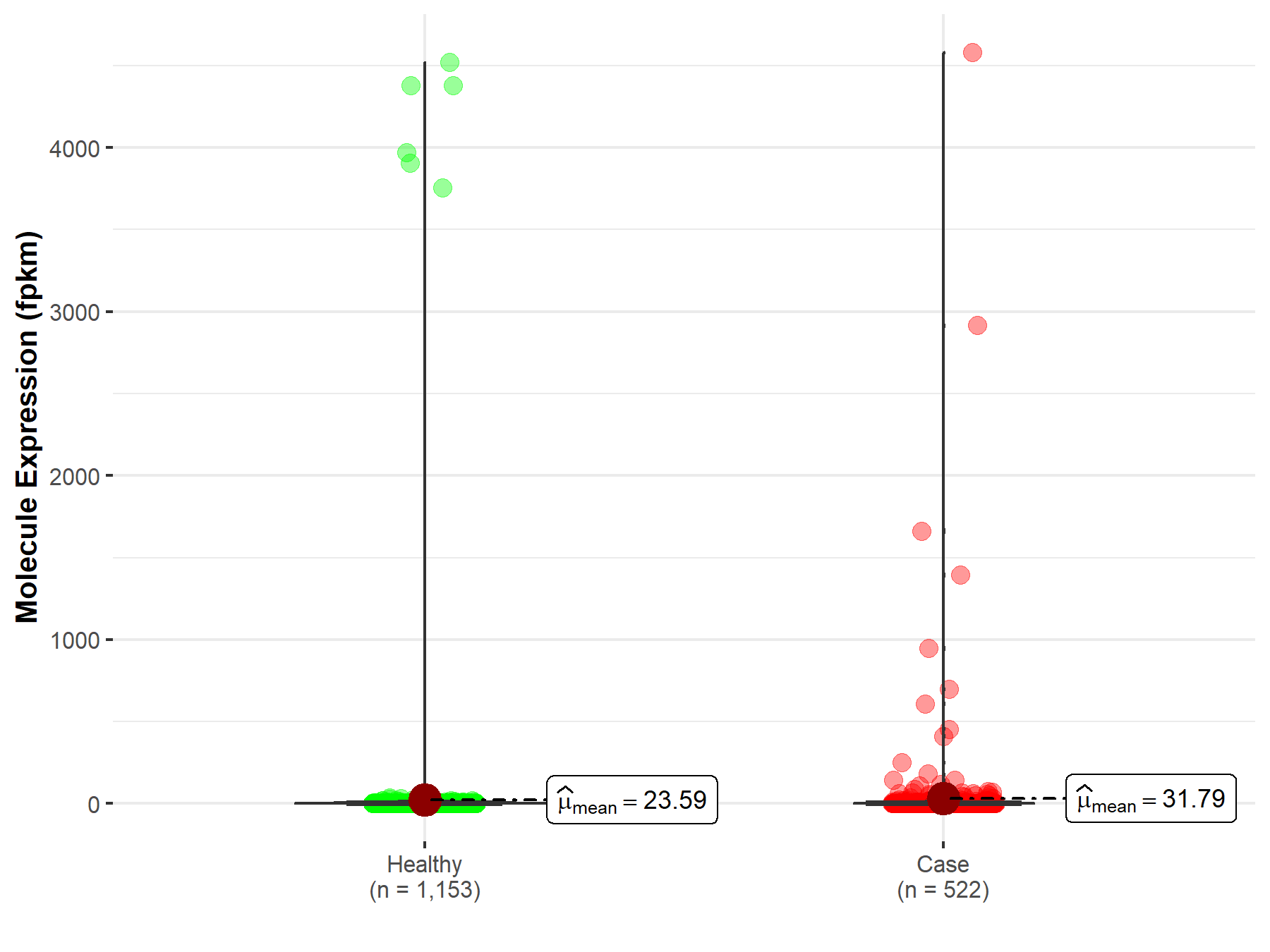
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brain | |
| The Specified Disease | Glioblastoma multiforme | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.81E-131; Fold-change: -5.10E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
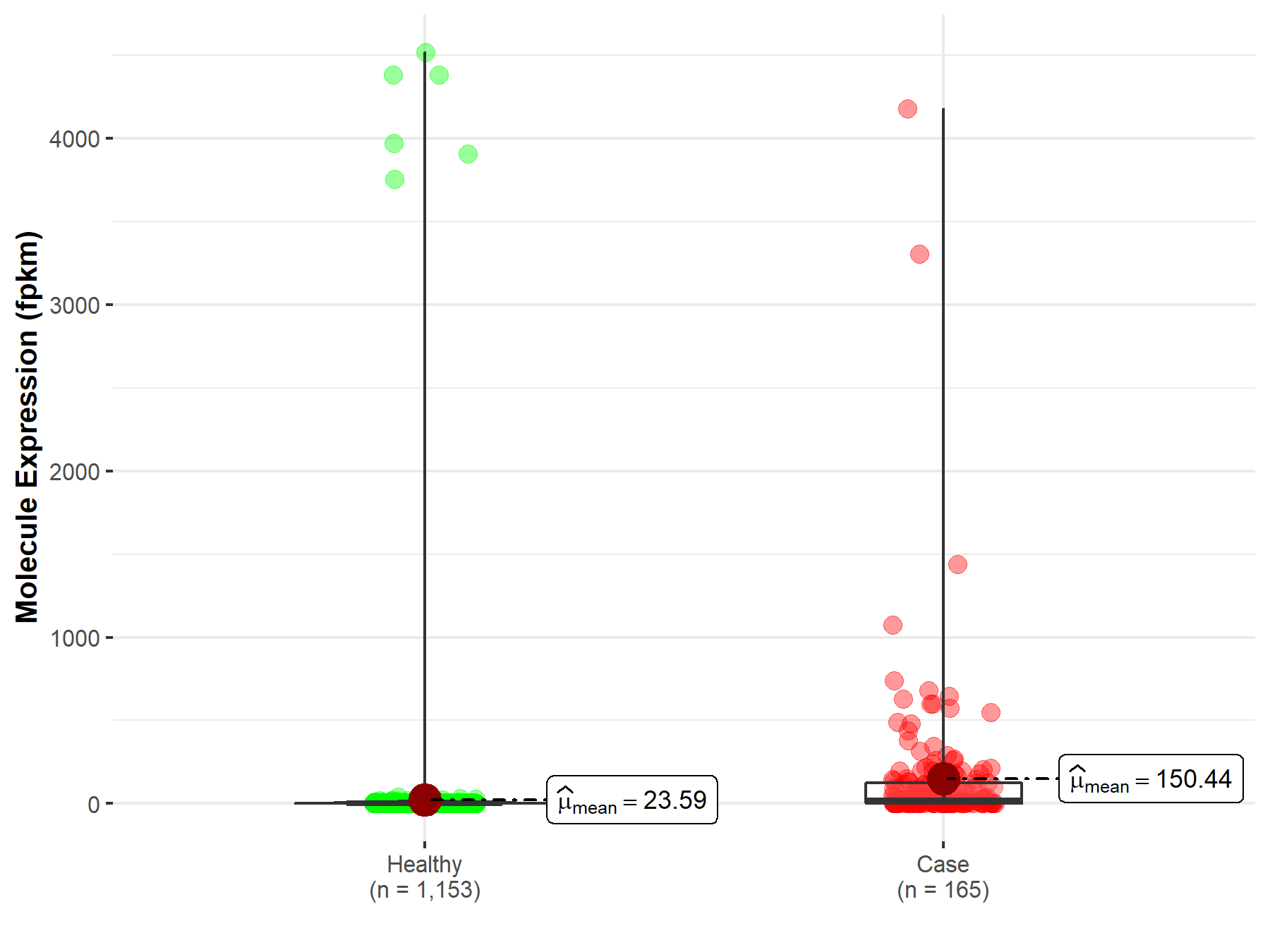
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Stomach | |
| The Specified Disease | Stomach adenocarcinoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.10E-06; Fold-change: -8.69E-02 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
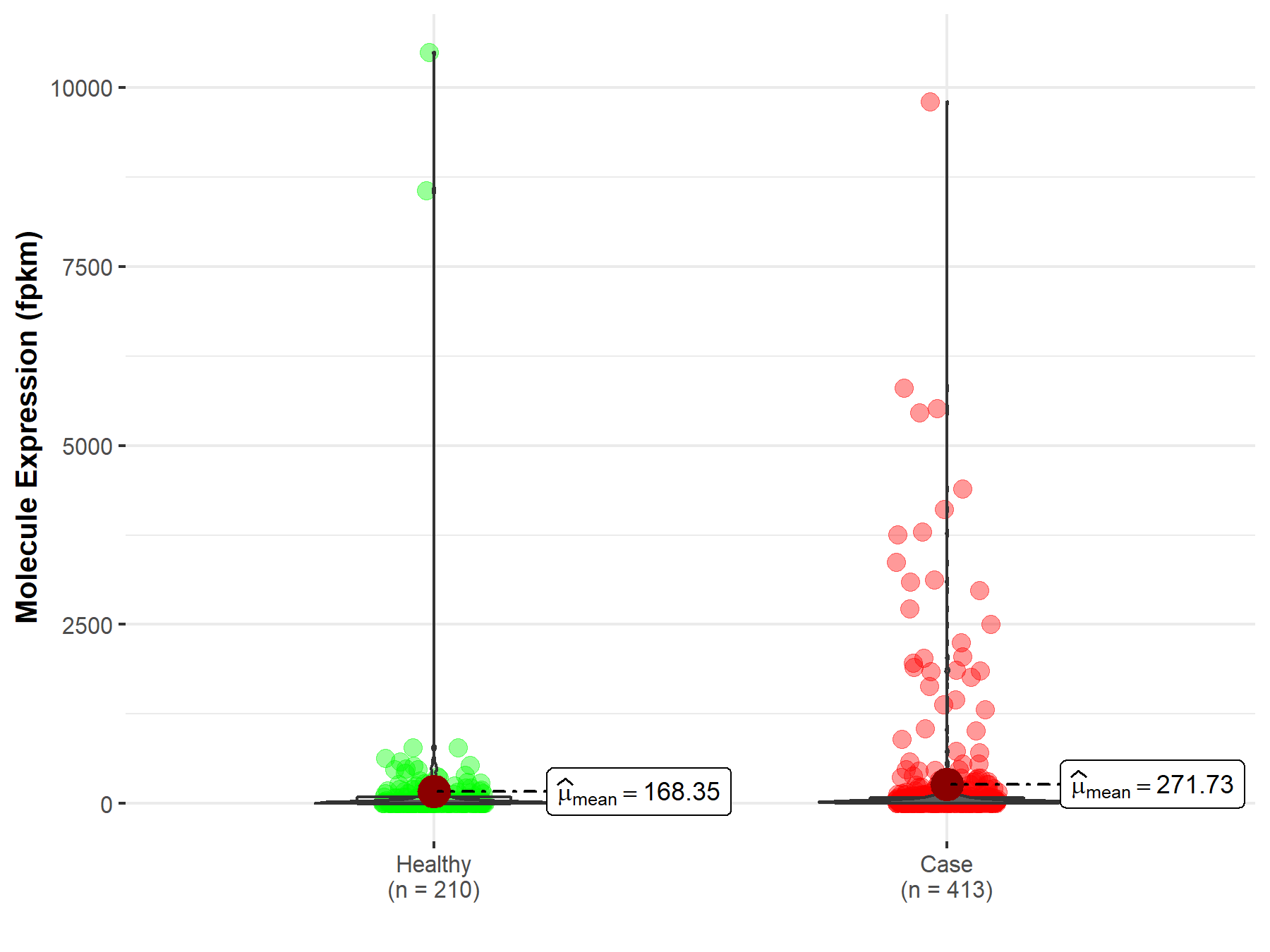
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Colon | |
| The Specified Disease | Colon adenocarcinoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.71E-02; Fold-change: -3.00E-02 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
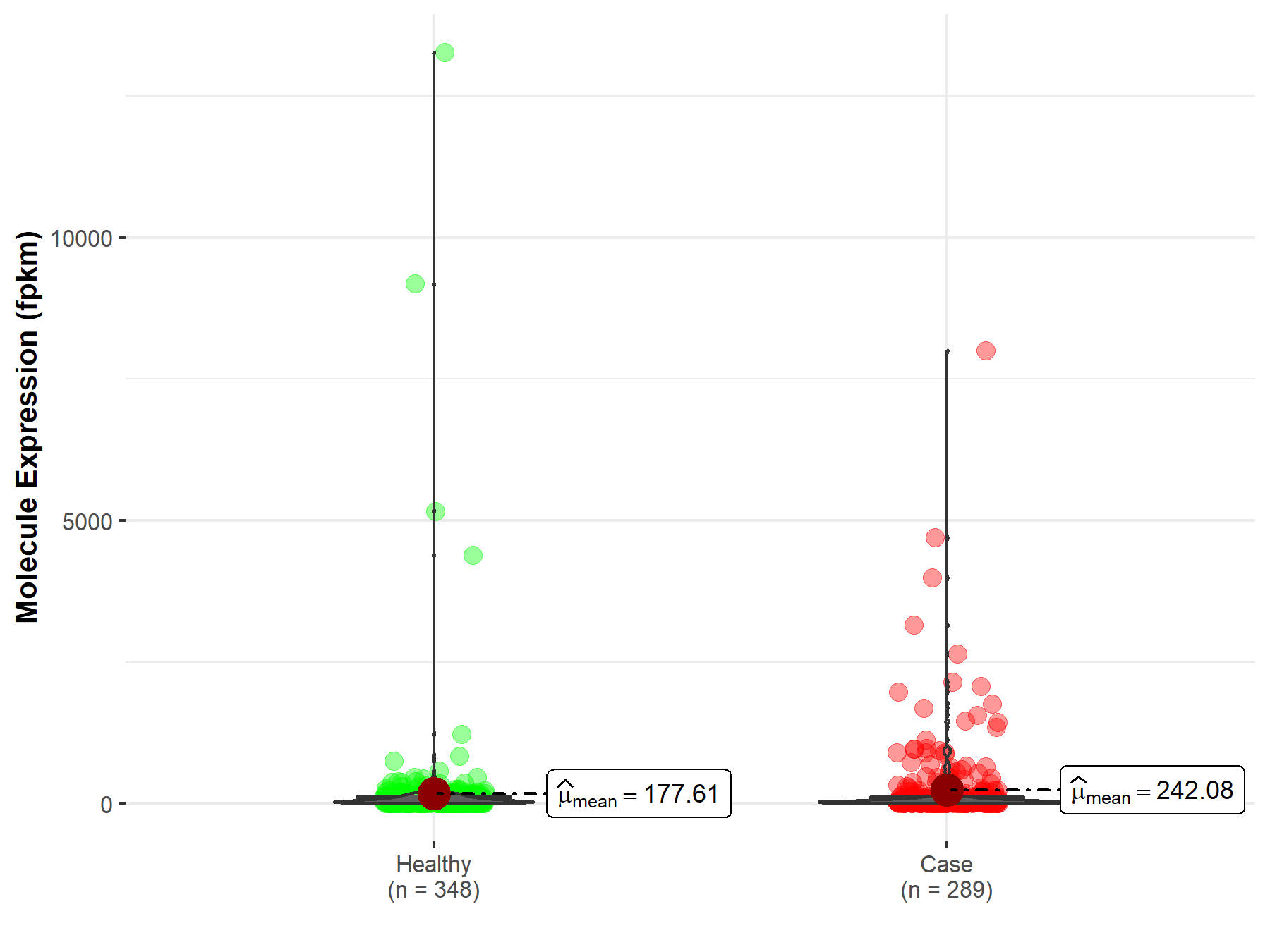
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Rectum | |
| The Specified Disease | Rectum adenocarcinoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.83E-03; Fold-change: -2.19E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
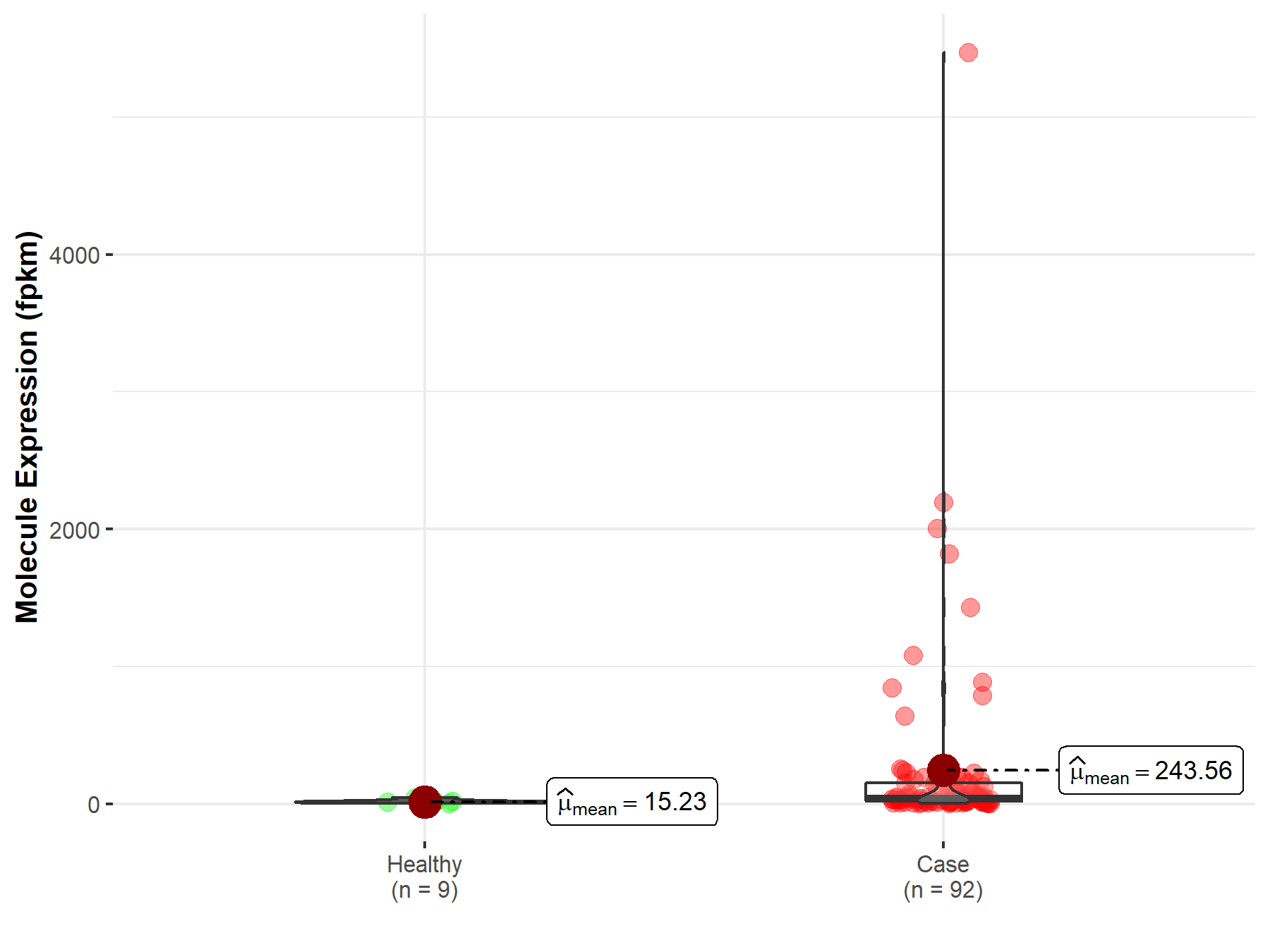
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Bile duct | |
| The Specified Disease | Cholangiocarcinoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.97E-03; Fold-change: 1.68E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
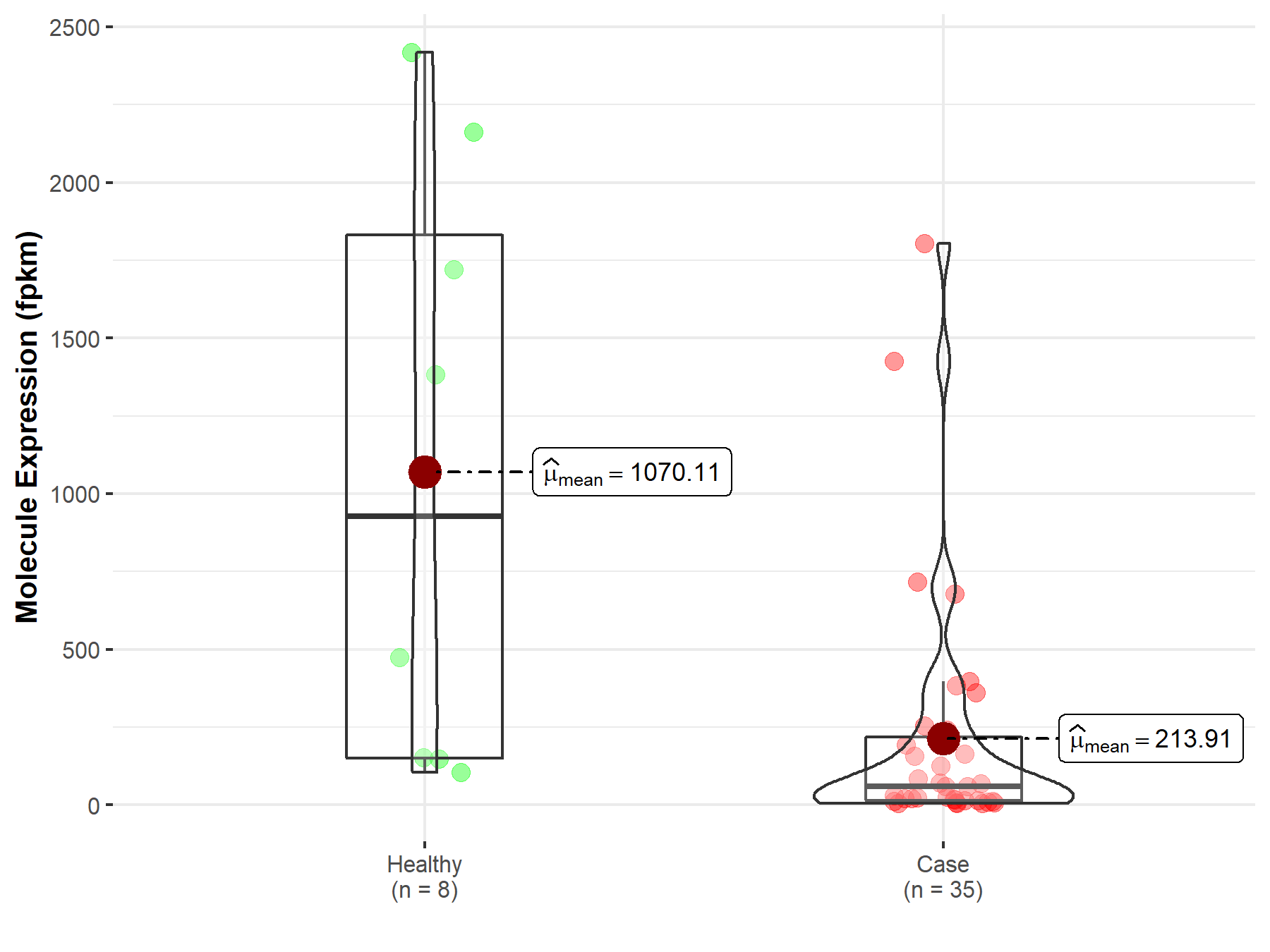
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Liver | |
| The Specified Disease | Liver hepatocellular carcinoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.03E-03; Fold-change: 4.86E-02 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
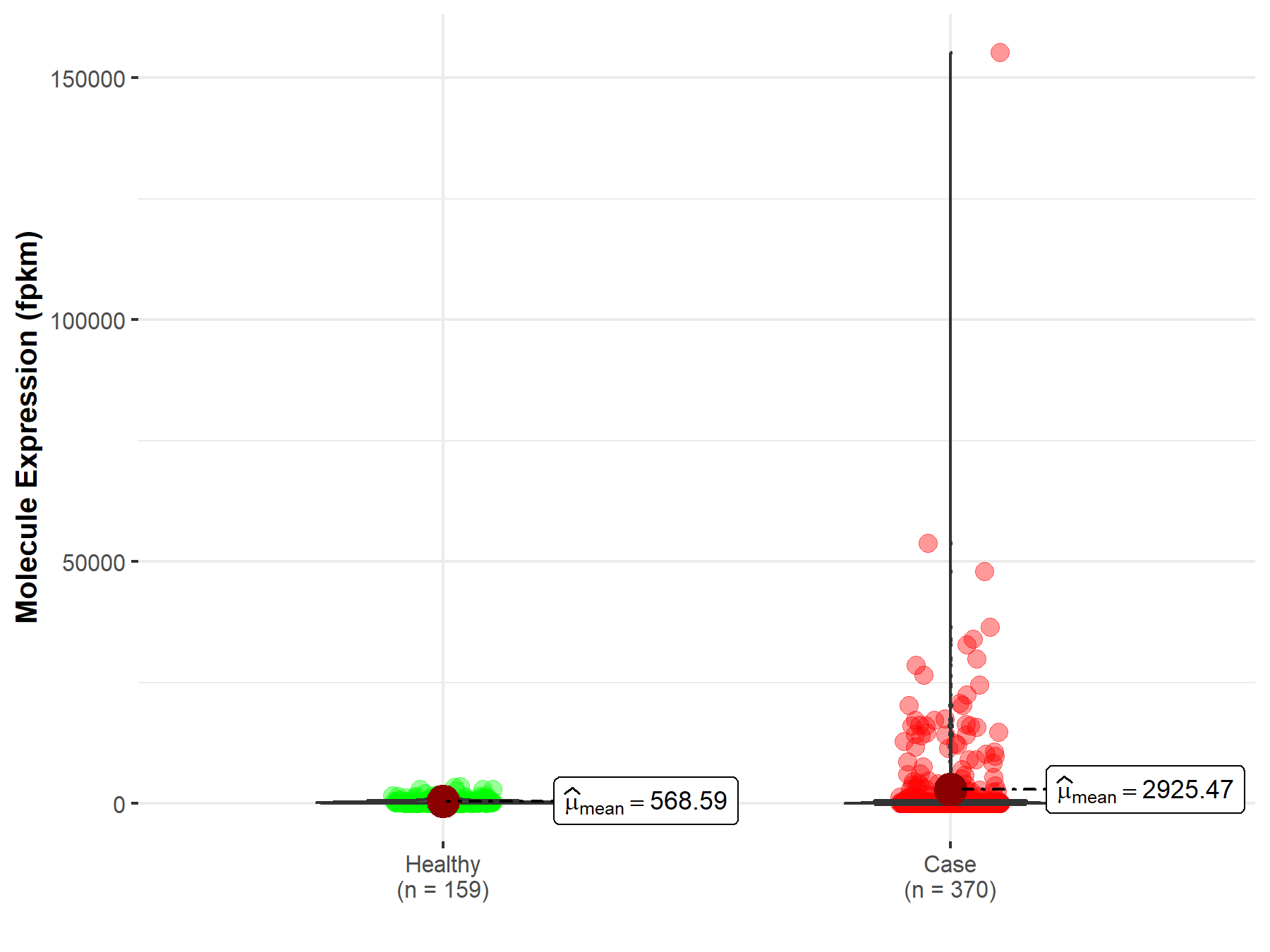
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Lung | |
| The Specified Disease | Lung adenocarcinoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.10E-01; Fold-change: 2.90E-03 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
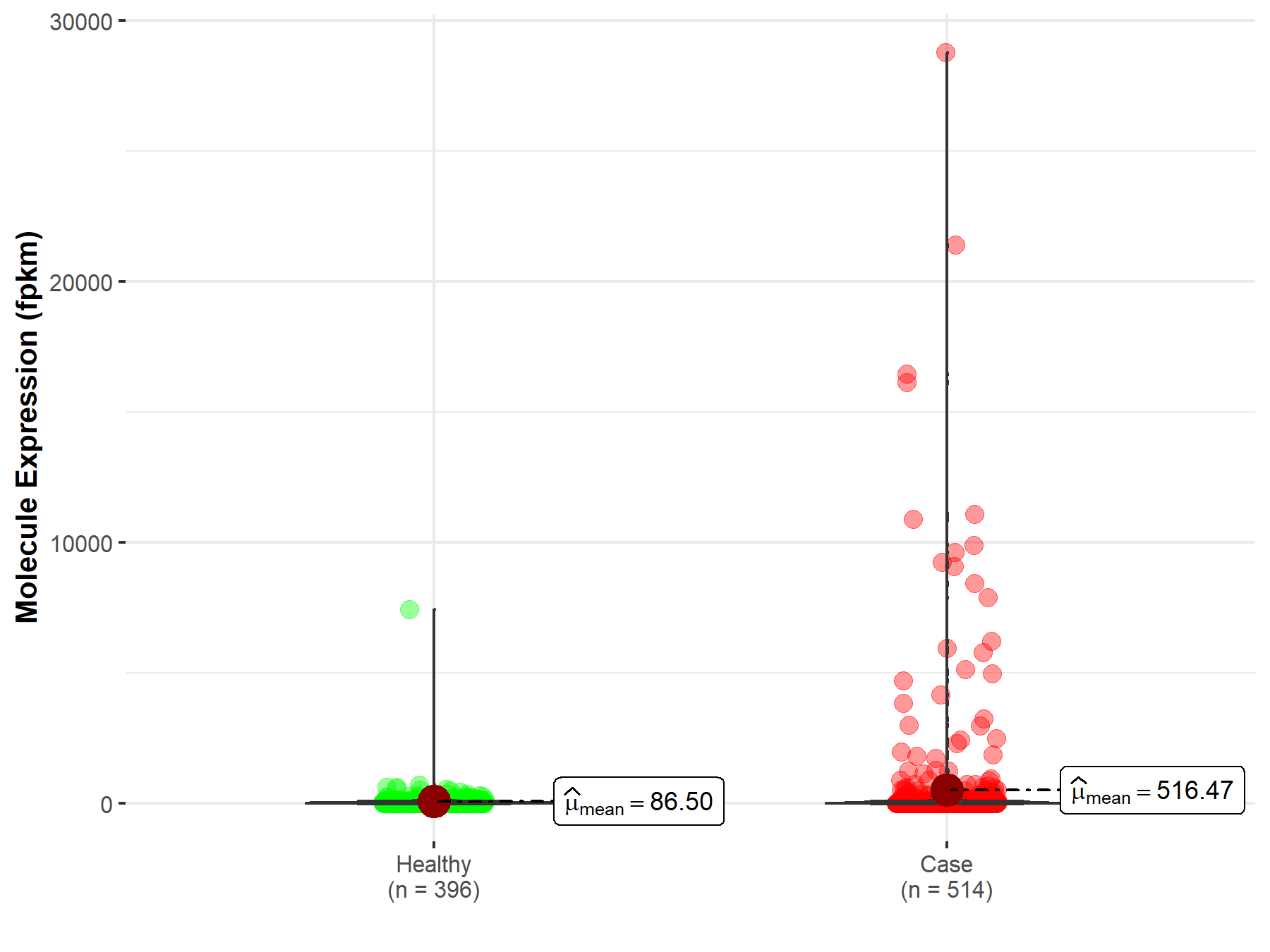
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Lung | |
| The Specified Disease | Lung squamous cell carcinoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.14E-04; Fold-change: -4.59E-02 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
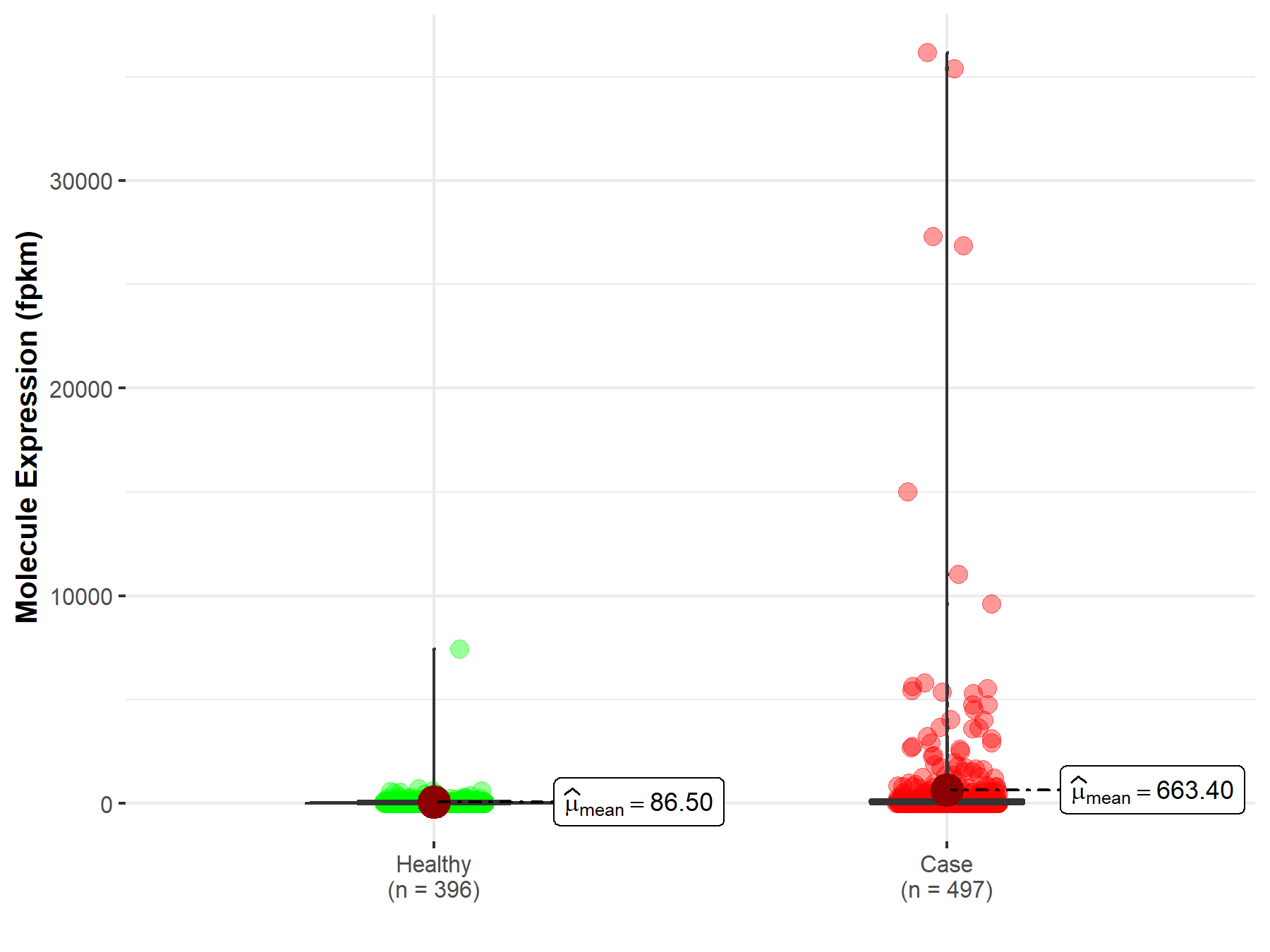
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Skin | |
| The Specified Disease | Skin cutaneous melanoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.26E-40; Fold-change: -1.24E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Ovary | |
| The Specified Disease | Ovarian serous cystadenocarcinoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.40E-01; Fold-change: 1.48E-03 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
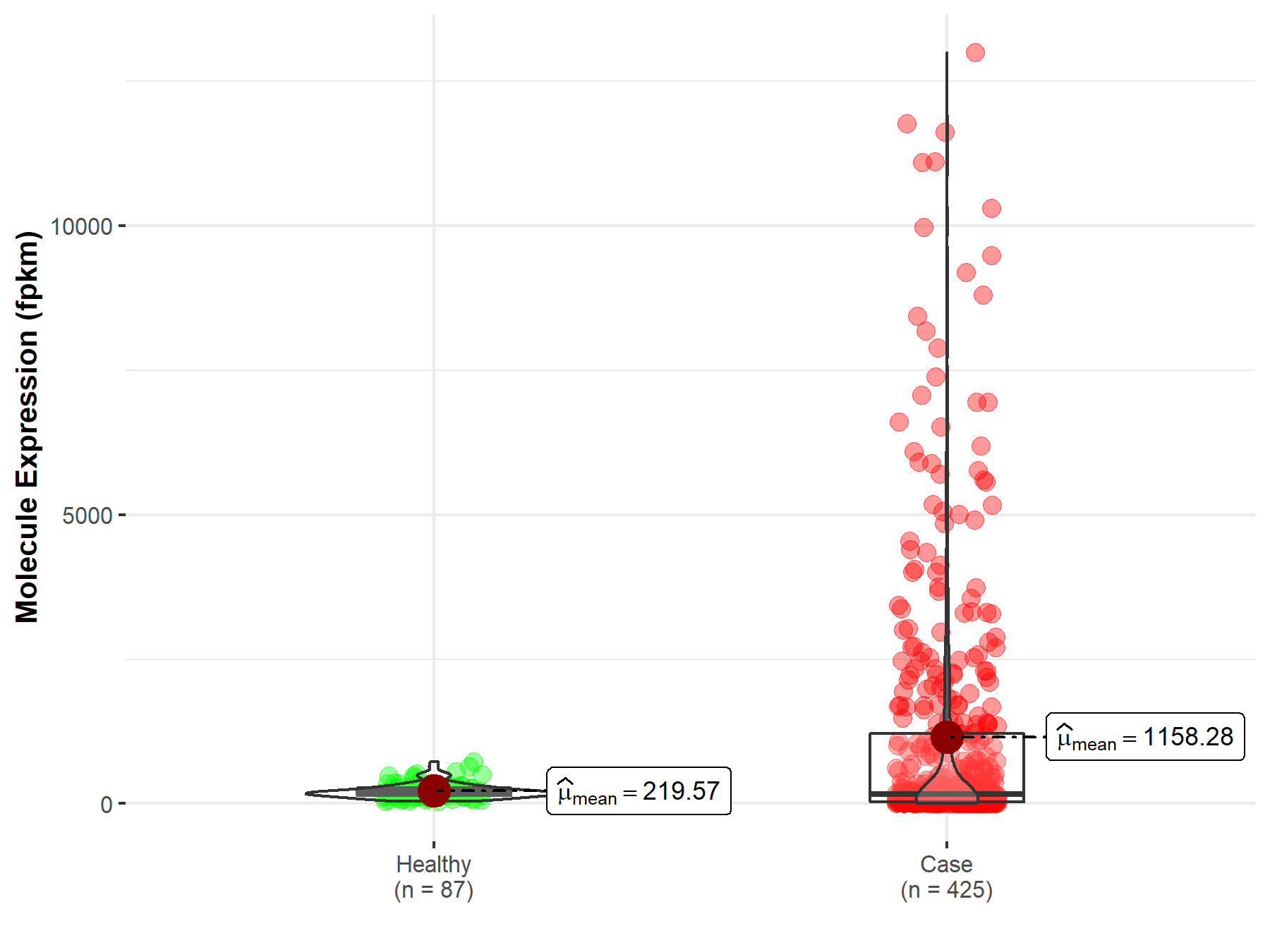
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Testis | |
| The Specified Disease | Testicular germ cell tumor | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.17E-17; Fold-change: -1.19E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
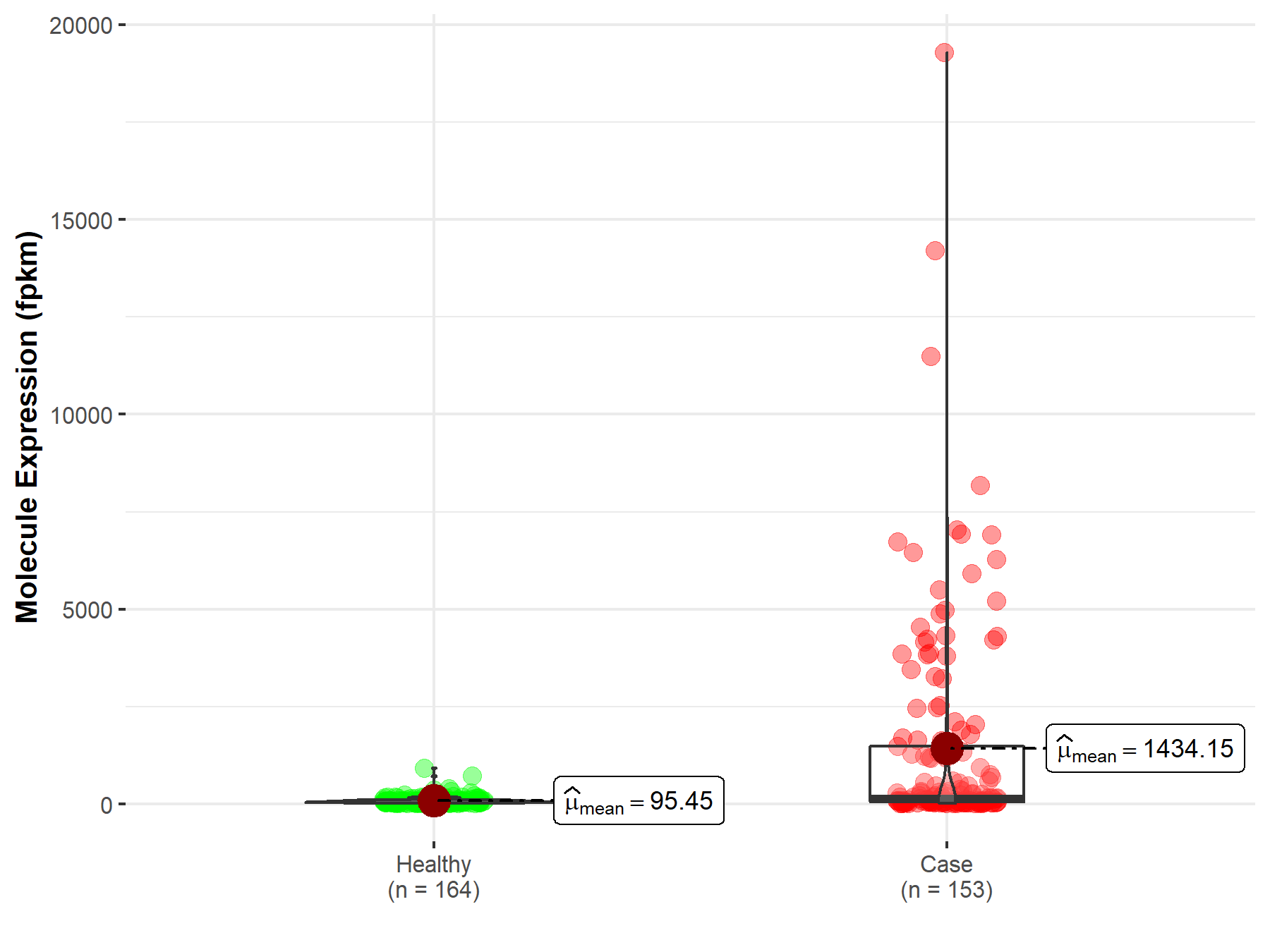
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

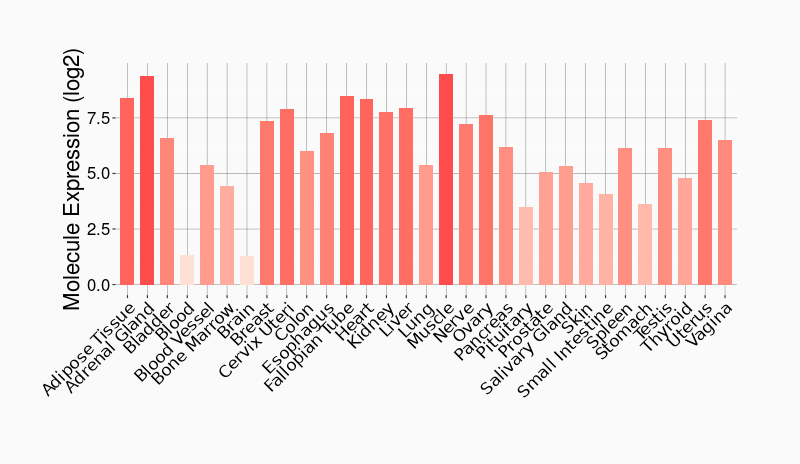
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
