Molecule Information
General Information of the Molecule (ID: Mol00022)
| Name |
Aurora kinase A (AURKA)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Aurora 2; Aurora/IPL1-related kinase 1; ARK-1; Aurora-related kinase 1; hARK1; Breast tumor-amplified kinase; Serine/threonine-protein kinase 15; Serine/threonine-protein kinase 6; Serine/threonine-protein kinase aurora-A; AIK; AIRK1; ARK1; AURA; AYK1; BTAK; IAK1; STK15; STK6
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
AURKA
|
||||
| Gene ID | |||||
| Location |
chr20:56369389-56392337[-]
|
||||
| Sequence |
MDRSKENCISGPVKATAPVGGPKRVLVTQQFPCQNPLPVNSGQAQRVLCPSNSSQRIPLQ
AQKLVSSHKPVQNQKQKQLQATSVPHPVSRPLNNTQKSKQPLPSAPENNPEEELASKQKN EESKKRQWALEDFEIGRPLGKGKFGNVYLAREKQSKFILALKVLFKAQLEKAGVEHQLRR EVEIQSHLRHPNILRLYGYFHDATRVYLILEYAPLGTVYRELQKLSKFDEQRTATYITEL ANALSYCHSKRVIHRDIKPENLLLGSAGELKIADFGWSVHAPSSRRTTLCGTLDYLPPEM IEGRMHDEKVDLWSLGVLCYEFLVGKPPFEANTYQETYKRISRVEFTFPDFVTEGARDLI SRLLKHNPSQRPMLREVLEHPWITANSSKPSNCQNKESASKQS Click to Show/Hide
|
||||
| Function |
Mitotic serine/threonine kinase that contributes to the regulation of cell cycle progression. Associates with the centrosome and the spindle microtubules during mitosis and plays a critical role in various mitotic events including the establishment of mitotic spindle, centrosome duplication, centrosome separation as well as maturation, chromosomal alignment, spindle assembly checkpoint, and cytokinesis. Required for normal spindle positioning during mitosis and for the localization of NUMA1 and DCTN1 to the cell cortex during metaphase. Required for initial activation of CDK1 at centrosomes. Phosphorylates numerous target proteins, including ARHGEF2, BORA, BRCA1, CDC25B, DLGP5, HDAC6, KIF2A, LATS2, NDEL1, PARD3, PPP1R2, PLK1, RASSF1, TACC3, p53/TP53 and TPX2. Regulates KIF2A tubulin depolymerase activity. Important for microtubule formation and/or stabilization. Required for normal axon formation. Plays a role in microtubule remodeling during neurite extension. Also acts as a key regulatory component of the p53/TP53 pathway, and particularly the checkpoint-response pathways critical for oncogenic transformation of cells, by phosphorylating and destabilizing p53/TP53. Phosphorylates its own inhibitors, the protein phosphatase type 1 (PP1) isoforms, to inhibit their activity. Necessary for proper cilia disassembly prior to mitosis. Regulates protein levels of the anti-apoptosis protein BIRC5 by suppressing the expression of the SCF(FBXL7) E3 ubiquitin-protein ligase substrate adapter FBXL7 through the phosphorylation of the transcription factor FOXP1.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
3 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Glioblastoma [ICD-11: 2A00.02] | [1] | |||
| Sensitive Disease | Glioblastoma [ICD-11: 2A00.02] | |||
| Sensitive Drug | Temozolomide | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Brain cancer [ICD-11: 2A00] | |||
| The Specified Disease | Glioblastoma | |||
| The Studied Tissue | Nervous tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.24E-03 Fold-change: -1.17E+00 Z-score: -4.08E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | LN229 cells | Brain | Homo sapiens (Human) | CVCL_0393 |
| A172 cells | Brain | Homo sapiens (Human) | CVCL_0131 | |
| T98G cells | Brain | Homo sapiens (Human) | CVCL_0556 | |
| M059J cells | Brain | Homo sapiens (Human) | CVCL_0400 | |
| M059k cells | Brain | Homo sapiens (Human) | CVCL_0401 | |
| U-87 MG cells | Brain | Homo sapiens (Human) | CVCL_0022 | |
| U118 MG cells | Brain | Homo sapiens (Human) | CVCL_0633 | |
| U138-MG cells | Brain | Homo sapiens (Human) | CVCL_0020 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Dual luciferase assay; Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | miR124 suppresses glioblastoma growth and potentiates chemosensitivity by inhibiting AURkA. Re-expression of AURkA rescued miR124-mediated growth suppression. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Multiple myeloma [ICD-11: 2A83.0] | [2] | |||
| Sensitive Disease | Multiple myeloma [ICD-11: 2A83.0] | |||
| Sensitive Drug | Epirubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Multiple myeloma [ICD-11: 2A83] | |||
| The Specified Disease | Myeloma | |||
| The Studied Tissue | Peripheral blood | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.27E-01 Fold-change: -1.02E-01 Z-score: -6.42E-01 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | NCI-H929 cells | Bone marrow | Homo sapiens (Human) | CVCL_1600 |
| U266 cells | Bone marrow | Homo sapiens (Human) | CVCL_0566 | |
| MM1S cells | Peripheral blood | Homo sapiens (Human) | CVCL_8792 | |
| OPM-2 cells | Peripheral blood | Homo sapiens (Human) | CVCL_1625 | |
| RPMI-8226 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0014 | |
| KMS11 cells | Peripheral blood | Homo sapiens (Human) | CVCL_2989 | |
| In Vivo Model | Mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Ectopic expression of miR137 strongly reduced the expression of AURkA and p-ATM/Chk2 in MM cells, and increased the expression of p53, and p21, overexpression of miR137 could reduce drug resistance and overcome chromosomal instability of the MM cells via affecting the apoptosis and RNA damage pathways. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Multiple myeloma [ICD-11: 2A83.0] | [2] | |||
| Sensitive Disease | Multiple myeloma [ICD-11: 2A83.0] | |||
| Sensitive Drug | Bortezomib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | NCI-H929 cells | Bone marrow | Homo sapiens (Human) | CVCL_1600 |
| U266 cells | Bone marrow | Homo sapiens (Human) | CVCL_0566 | |
| MM1S cells | Peripheral blood | Homo sapiens (Human) | CVCL_8792 | |
| OPM-2 cells | Peripheral blood | Homo sapiens (Human) | CVCL_1625 | |
| RPMI-8226 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0014 | |
| KMS11 cells | Peripheral blood | Homo sapiens (Human) | CVCL_2989 | |
| In Vivo Model | Mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Epigenetic silencing of miR137 induces drug resistance and chromosomal instability by targeting AURkA in multiple myeloma. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Nervous tissue | |
| The Specified Disease | Brain cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.56E-166; Fold-change: 1.56E+00; Z-score: 2.62E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.34E-03; Fold-change: 1.33E+00; Z-score: 5.30E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
|
Click to View the Clearer Original Diagram |
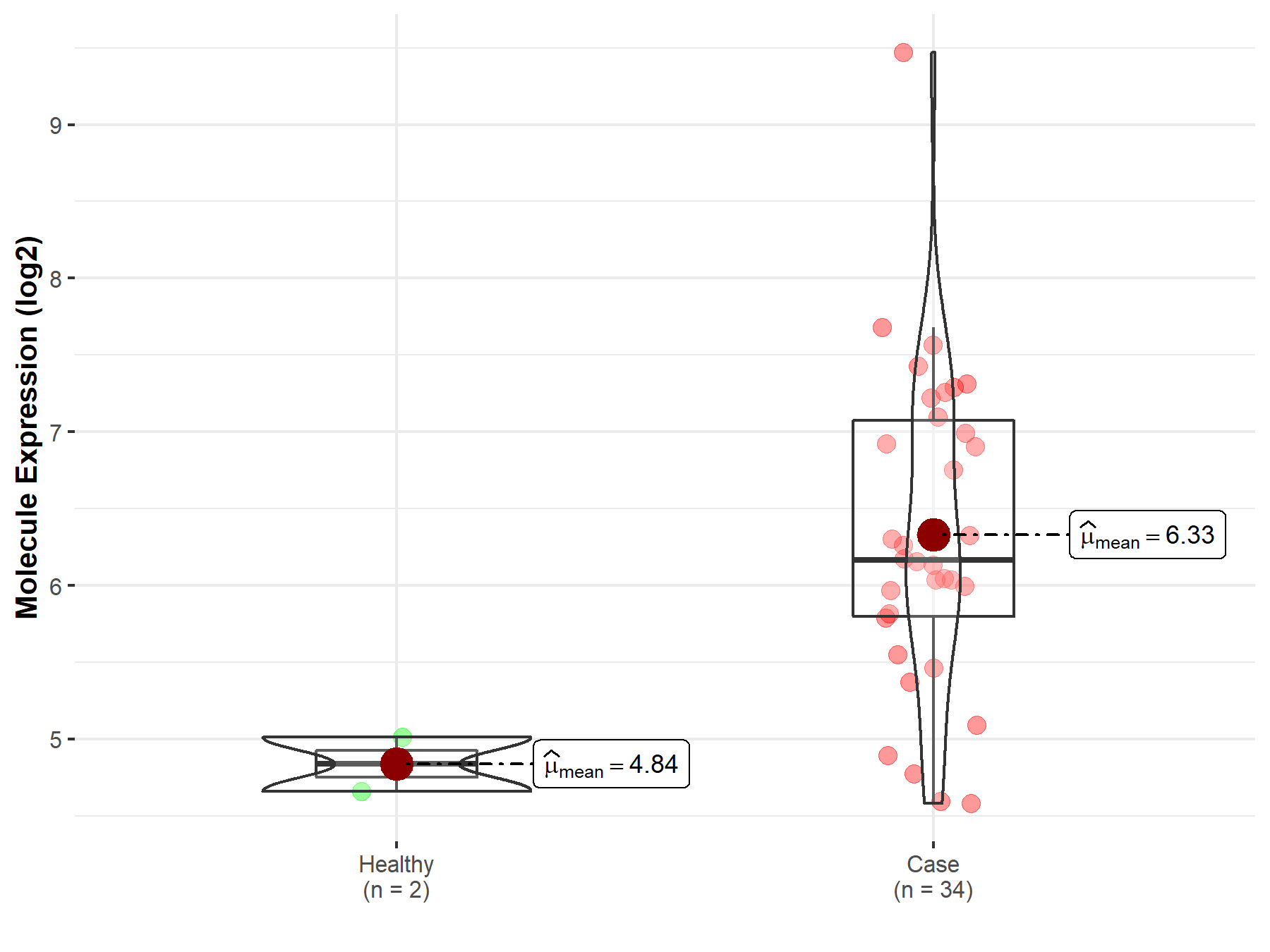
| The Studied Tissue | White matter | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.06E-04; Fold-change: 1.11E+00; Z-score: 2.09E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
|
Click to View the Clearer Original Diagram |
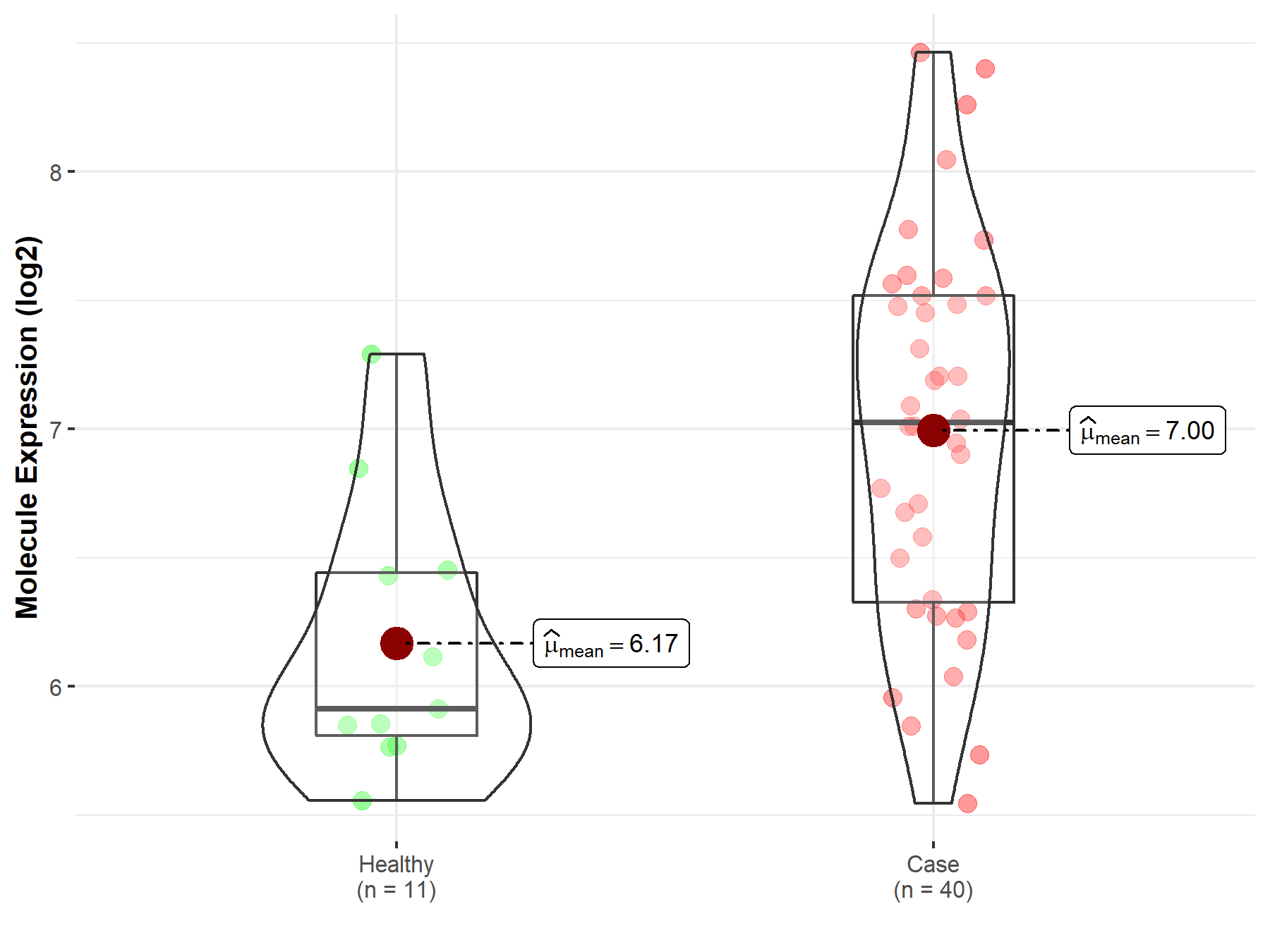
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Neuroectodermal tumor | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.53E-17; Fold-change: 3.12E+00; Z-score: 9.55E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
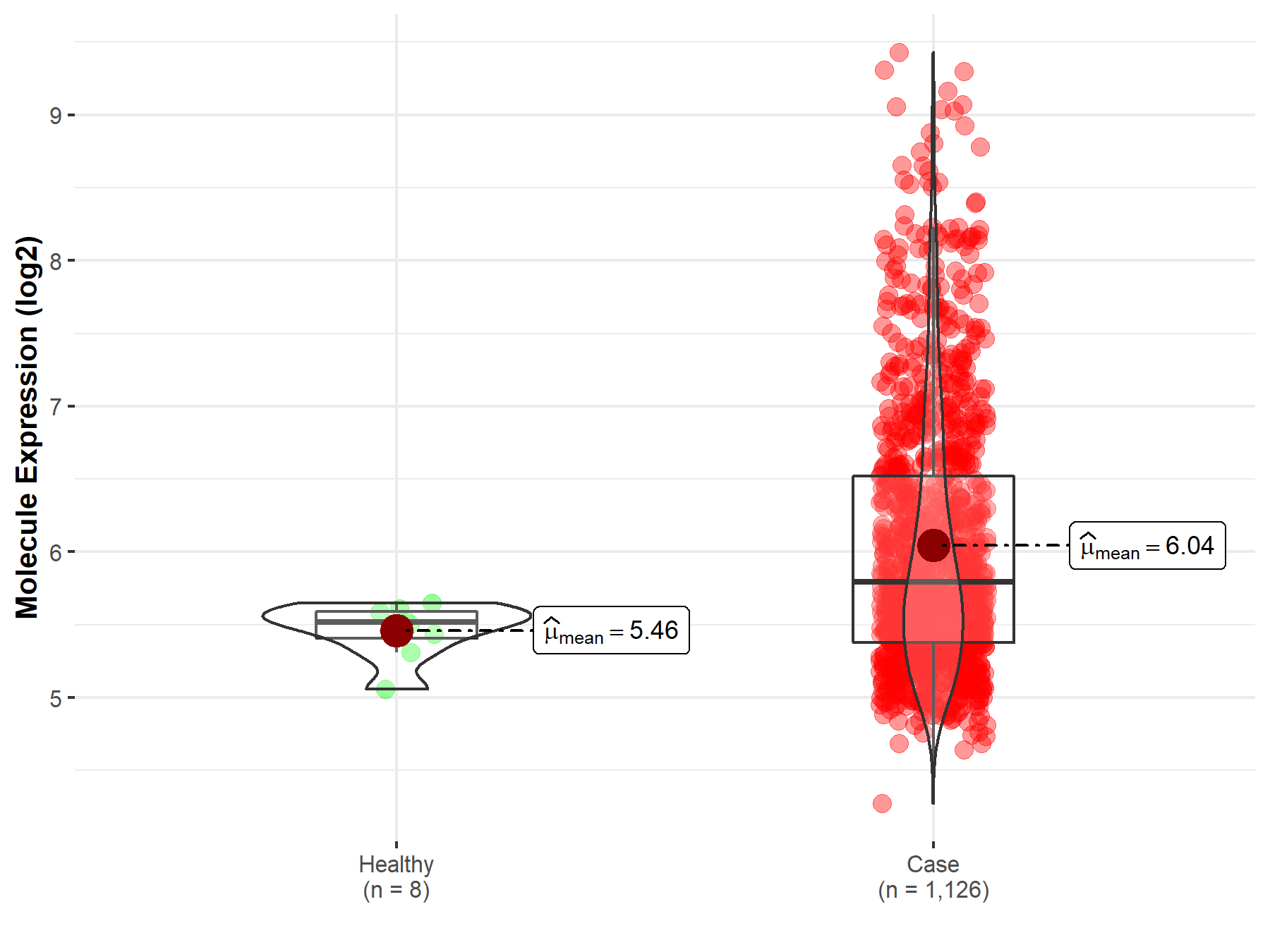
| The Studied Tissue | Bone marrow | |
| The Specified Disease | Multiple myeloma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.17E-05; Fold-change: 2.72E-01; Z-score: 1.39E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
|
Click to View the Clearer Original Diagram |
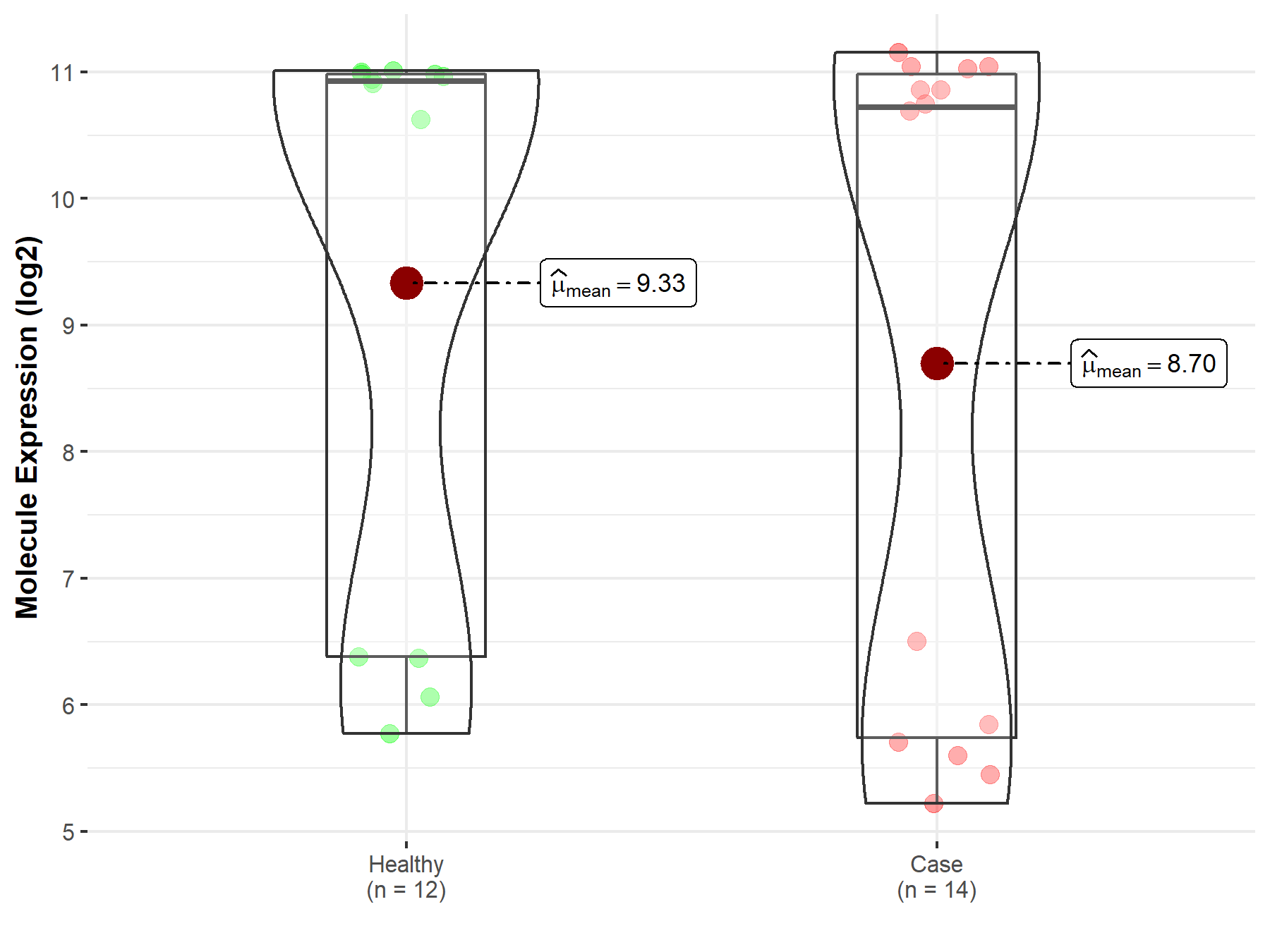
| The Studied Tissue | Peripheral blood | |
| The Specified Disease | Multiple myeloma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.27E-01; Fold-change: -2.02E-01; Z-score: -8.57E-02 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals


|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
