Molecule Information
General Information of the Molecule (ID: Mol00034)
| Name |
Serine/threonine-protein kinase B-raf (BRAF)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Proto-oncogene B-Raf; p94; v-Raf murine sarcoma viral oncogene homolog B1; BRAF1; RAFB1
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
BRAF
|
||||
| Gene ID | |||||
| Location |
chr7:140719327-140924929[-]
|
||||
| Sequence |
MAALSGGGGGGAEPGQALFNGDMEPEAGAGAGAAASSAADPAIPEEVWNIKQMIKLTQEH
IEALLDKFGGEHNPPSIYLEAYEEYTSKLDALQQREQQLLESLGNGTDFSVSSSASMDTV TSSSSSSLSVLPSSLSVFQNPTDVARSNPKSPQKPIVRVFLPNKQRTVVPARCGVTVRDS LKKALMMRGLIPECCAVYRIQDGEKKPIGWDTDISWLTGEELHVEVLENVPLTTHNFVRK TFFTLAFCDFCRKLLFQGFRCQTCGYKFHQRCSTEVPLMCVNYDQLDLLFVSKFFEHHPI PQEEASLAETALTSGSSPSAPASDSIGPQILTSPSPSKSIPIPQPFRPADEDHRNQFGQR DRSSSAPNVHINTIEPVNIDDLIRDQGFRGDGGSTTGLSATPPASLPGSLTNVKALQKSP GPQRERKSSSSSEDRNRMKTLGRRDSSDDWEIPDGQITVGQRIGSGSFGTVYKGKWHGDV AVKMLNVTAPTPQQLQAFKNEVGVLRKTRHVNILLFMGYSTKPQLAIVTQWCEGSSLYHH LHIIETKFEMIKLIDIARQTAQGMDYLHAKSIIHRDLKSNNIFLHEDLTVKIGDFGLATV KSRWSGSHQFEQLSGSILWMAPEVIRMQDKNPYSFQSDVYAFGIVLYELMTGQLPYSNIN NRDQIIFMVGRGYLSPDLSKVRSNCPKAMKRLMAECLKKKRDERPLFPQILASIELLARS LPKIHRSASEPSLNRAGFQTEDFSLYACASPKTPIQAGGYGAFPVH Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Protein kinase involved in the transduction of mitogenic signals from the cell membrane to the nucleus (Probable). Phosphorylates MAP2K1, and thereby activates the MAP kinase signal transduction pathway. May play a role in the postsynaptic responses of hippocampal neurons.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
13 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [2] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | Binimetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Cutaneous melanoma tissue | N.A. | |||||||||||
| Mechanism Description | The missense mutation p.V600E (c.1799T>A) in gene BRAF cause the sensitivity of Binimetinib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [3] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | Binimetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.L597S (c.1789_1790delCTinsTC) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | MAPK signaling pathway | Inhibition | hsa04010 | ||||||||||
| In Vitro Model | Skin sample | N.A. | |||||||||||
| In Vivo Model | Mouse PDX model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
Crystal violet staining assay | ||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [2] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | Binimetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600X (c.1798_1800) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Cutaneous melanoma tissue | N.A. | |||||||||||
| Mechanism Description | The missense mutation p.V600X (c.1798_1800) in gene BRAF cause the sensitivity of Binimetinib by unusual activation of pro-survival pathway | ||||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [4] | ||||||||||||
| Resistant Disease | Colorectal cancer [ICD-11: 2B91.1] | ||||||||||||
| Resistant Drug | Cetuximab | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Colon cells | Colon | Homo sapiens (Human) | N.A. | |||||||||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Liquid biopsy assay | ||||||||||||
| Mechanism Description | Mechanisms of resistance to EGFR blockade include the emergence of kRAS, NRAS and EGFR extracellular domain mutations as well as HER2/MET alterations. | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [5] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Sensitive Drug | Dabrafenib/Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [6] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | Dabrafenib/Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: Thyroid gland cancer [ICD-11: 2D10.0] | [7] | ||||||||||||
| Sensitive Disease | Thyroid gland cancer [ICD-11: 2D10.0] | ||||||||||||
| Sensitive Drug | Dabrafenib/Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: FGFR-tacc positive glioblastoma [ICD-11: 2A00.01] | [8] | ||||||||||||
| Sensitive Disease | FGFR-tacc positive glioblastoma [ICD-11: 2A00.01] | ||||||||||||
| Sensitive Drug | Dabrafenib/Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: Salivary gland adenoid cystic carcinoma [ICD-11: 2E60.0] | [9] | ||||||||||||
| Sensitive Disease | Salivary gland adenoid cystic carcinoma [ICD-11: 2E60.0] | ||||||||||||
| Sensitive Drug | Dabrafenib/Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Human laryngeal cells isolates | N.A. | |||||||||||
| Experiment for Molecule Alteration |
ctDNA sequencing assay | ||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [10] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | Dabrafenib/Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [11] | ||||||||||||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | ||||||||||||
| Sensitive Drug | Dabrafenib/Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: Cholangiocarcinoma [ICD-11: 2C12.0] | [12] | ||||||||||||
| Sensitive Disease | Cholangiocarcinoma [ICD-11: 2C12.0] | ||||||||||||
| Sensitive Drug | Dabrafenib/Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [13] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Sensitive Drug | Dabrafenib/Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600X (c.1798_1800) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [14] | ||||||||||||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | ||||||||||||
| Sensitive Drug | Dabrafenib/Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600X (c.1798_1800) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Colorectum | N.A. | |||||||||||
| In Vivo Model | Patient-Derived xenograft mouse model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Reverse-phase protein array (RPPA) analysis; Targeted next-generation sequencing (NGS) assay | ||||||||||||
| Experiment for Drug Resistance |
Immunohistochemistry assay | ||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [10] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | Dabrafenib/Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600K (c.1798_1799delGTinsAA) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [13] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Sensitive Drug | Dabrafenib/Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600K (c.1798_1799delGTinsAA) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [14] | ||||||||||||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | ||||||||||||
| Sensitive Drug | Dabrafenib/Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600X (c.1798_1799) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Colorectum | N.A. | |||||||||||
| In Vivo Model | Patient-Derived xenograft mouse model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Reverse-phase protein array (RPPA) analysis; Targeted next-generation sequencing (NGS) assay | ||||||||||||
| Experiment for Drug Resistance |
Immunohistochemistry assay | ||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [15] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | Dabrafenib/Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600X (c.1798_1799) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
|
|
|||||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [16] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Sensitive Drug | Dabrafenib/Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | MAPK signaling pathway | Inhibition | hsa04010 | ||||||||||
| Disease Class: Metastatic neuroendocrine carcinoma [ICD-11: 2D4Y.0] | [17] | ||||||||||||
| Sensitive Disease | Metastatic neuroendocrine carcinoma [ICD-11: 2D4Y.0] | ||||||||||||
| Sensitive Drug | Dabrafenib/Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1406G>C) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: Pleomorphic xanthoastrocytoma [ICD-11: 2A00.0Y] | [8] | ||||||||||||
| Sensitive Disease | Pleomorphic xanthoastrocytoma [ICD-11: 2A00.0Y] | ||||||||||||
| Sensitive Drug | Dabrafenib/Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | ERK signaling pathway | Inhibition | hsa04210 | ||||||||||
| In Vitro Model | Brain | N.A. | |||||||||||
| Disease Class: Metastatic neuroendocrine carcinoma [ICD-11: 2D4Y.0] | [17] | ||||||||||||
| Sensitive Disease | Metastatic neuroendocrine carcinoma [ICD-11: 2D4Y.0] | ||||||||||||
| Sensitive Drug | Dabrafenib/Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: FGFR-tacc positive glioblastoma [ICD-11: 2A00.01] | [18] | ||||||||||||
| Sensitive Disease | FGFR-tacc positive glioblastoma [ICD-11: 2A00.01] | ||||||||||||
| Sensitive Drug | Dabrafenib/Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600D (c.1799_1800delTGinsAC) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | BRAF/MEK/MAPK signaling pathway | Inhibition | hsa04010 | ||||||||||
| In Vitro Model | Brain | N.A. | |||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [19] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Sensitive Drug | Dabrafenib/Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.G469A (c.1406G>C) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | ERK signaling pathway | Inhibition | hsa04210 | ||||||||||
| In Vitro Model | HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 | |||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Promega assay | ||||||||||||
| Mechanism Description | The ERK pathway is the major deregulated pathway associated with BRAF-mutated cancers. ERK pathway inhibition has been shown to have anti-proliferative effects in cells harboring both kinase-activating and impairing BRAF mutations. The combination of Trametinib and Dabrafenib leads to more prolonged ERK inhibition and has anti-proliferative and pro-apoptotic effects in cells harboring both types of non-V600 BRAF mutations. | ||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [20] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | Dabrafenib/Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600X (c.1798_1800) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | MAPK signaling pathway | Inhibition | hsa04010 | ||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: EGFR-mutant non-small cell lung cancer [ICD-11: 2C25.7] | [21] | |||
| Resistant Disease | EGFR-mutant non-small cell lung cancer [ICD-11: 2C25.7] | |||
| Resistant Drug | Erlotinib | |||
| Molecule Alteration | Mutation | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Experiment for Molecule Alteration |
Low throughput experiment assay | |||
| Experiment for Drug Resistance |
Progression-free survival assay | |||
| Mechanism Description | Acquired resistance can occur through failure of drug delivery to the target, as in isolated central nervous system (CNS) progression, or by selection of biological variants during TkI exposure. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [22] | |||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Sensitive Drug | Fluorouracil | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT asssay; Innocyte invasion assay | |||
| Mechanism Description | microRNA-224 was differentially expressed in dysplastic colorectal disease and in isogeneic kRAS WT and mutant HCT116 cells. Antagomir-mediated miR-224 silencing in HCT116 kRAS WT cells phenocopied kRAS mutation, increased kRAS activity and ERk and AkT phosphorylation. 5-FU chemosensitivity was significantly increased in miR-224 knockdown cells, and in NIH3T3 cells expressing kRAS and BRAF mutant proteins. Bioinformatics analysis of predicted miR-224 target genes predicted altered cell proliferation, invasion and epithelial-mesenchymal transition (EMT) phenotypes that were experimentally confirmed in miR-224 knockdown cells. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: EGFR-mutant non-small cell lung cancer [ICD-11: 2C25.7] | [21] | |||
| Resistant Disease | EGFR-mutant non-small cell lung cancer [ICD-11: 2C25.7] | |||
| Resistant Drug | Gefitinib | |||
| Molecule Alteration | Mutation | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Experiment for Molecule Alteration |
Low throughput experiment assay | |||
| Experiment for Drug Resistance |
Progression-free survival assay | |||
| Mechanism Description | Acquired resistance can occur through failure of drug delivery to the target, as in isolated central nervous system (CNS) progression, or by selection of biological variants during TkI exposure. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Gastrointestinal stromal cancer [ICD-11: 2B5B.1] | [23] | ||||||||||||
| Resistant Disease | Gastrointestinal stromal cancer [ICD-11: 2B5B.1] | ||||||||||||
| Resistant Drug | Imatinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | RAS/RAF/Mek/ERK signaling pathway | Activation | hsa04010 | ||||||||||
| Experiment for Molecule Alteration |
Direct sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
High-performance liquid chromatography screening assay | ||||||||||||
| Mechanism Description | This finding, in combination with the loss of kIT expression, suggests the possibility of activation of RAS-RAF-MEk-ERk pathways driven by a kIT-independent oncogenic mechanism. Most mutations lie within the kinase domain with a single nucleotide substitution at position 1799 in exon 15, leading to the V600E amino-acid substitution (98 %). | ||||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [24] | ||||||||||||
| Resistant Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Resistant Drug | Nivolumab | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [24] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Nivolumab | |||
| Molecule Alteration | Missense mutation | p.V600K (c.1798_1799delGTinsAA) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [4] | ||||||||||||
| Resistant Disease | Colorectal cancer [ICD-11: 2B91.1] | ||||||||||||
| Resistant Drug | Panitumumab | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Colon cells | Colon | Homo sapiens (Human) | N.A. | |||||||||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Liquid biopsy assay | ||||||||||||
| Mechanism Description | Mechanisms of resistance to EGFR blockade include the emergence of kRAS, NRAS and EGFR extracellular domain mutations as well as HER2/MET alterations. | ||||||||||||
| Disease Class: Metastatic colorectal cancer [ICD-11: 2D85.0] | [25] | ||||||||||||
| Resistant Disease | Metastatic colorectal cancer [ICD-11: 2D85.0] | ||||||||||||
| Resistant Drug | Panitumumab | ||||||||||||
| Molecule Alteration | Mutation | . |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | RAS/MEk/ERK signaling pathway | Inhibition | hsa04010 | ||||||||||
| Experiment for Molecule Alteration |
Sanger sequencing assay; Next-generation sequencing assay | ||||||||||||
| Mechanism Description | Mutations in kRAS, NRAS, and BRAF and amplification of ERBB2 and MET drive primary (de novo) resistance to anti-EGFR treatment. | ||||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | [26] | |||
| Resistant Disease | Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | |||
| Resistant Drug | Regorafenib | |||
| Molecule Alteration | Other | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Metastatic GI stromal tumor tissue | N.A. | ||
| Mechanism Description | The mutation in gene BRAF cause the resistance of Regorafenib by unusual activation of pro-survival pathway. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | [27] | |||
| Resistant Disease | Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | |||
| Resistant Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.K601E (c.1801A>G) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 |
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |
| NIH-3T3 cells | Embryo | Mus musculus (Mouse) | CVCL_0594 | |
| NT-3 cells | Lymph node | Homo sapiens (Human) | CVCL_VG81 | |
| Experiment for Drug Resistance |
BioRad TC20 automated cell counter assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [28] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.L597Q (c.1790T>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [29] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.K601E (c.1801A>G) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Melanoma thyroid metastasis | N.A. | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Mechanism Description | The missense mutation p.K601E (c.1801A>G) in gene BRAF cause the sensitivity of Trametinib by unusual activation of pro-survival pathway | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [30] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.K601E (c.1801A>G) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [30] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.L597R (c.1790T>G) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Experiment for Molecule Alteration |
Whole-gene resequencing assay | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [30] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.V600R (c.1798_1799delGTinsAG) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [31] | |||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.G466V (c.1397G>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | ERK signaling pathway | Activation | hsa04210 | |
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |
| HT-29 cells | Colon | Homo sapiens (Human) | CVCL_0320 | |
| H1650 cells | Pleural effusion | Homo sapiens (Human) | CVCL_4V01 | |
| HTB-56 cells | Pleural effusion | Homo sapiens (Human) | CVCL_0236 | |
| HTB-38 cells | Colon | Homo sapiens (Human) | CVCL_0320 | |
| HTB-183 cells | Lymph node | Homo sapiens (Human) | CVCL_1577 | |
| H661 cells | Lymph node | Homo sapiens (Human) | CVCL_1577 | |
| H508 cells | Abdominal wall | Homo sapiens (Human) | CVCL_1564 | |
| H2405 cells | Lung | Homo sapiens (Human) | CVCL_1551 | |
| H1666 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1485 | |
| H1395 cells | Lung | Homo sapiens (Human) | CVCL_1467 | |
| CRL-5944 cells | Ascites | Homo sapiens (Human) | CVCL_1551 | |
| CRL-5885 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1485 | |
| CRL-5883 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1483 | |
| CRL-5868 cells | Lung | Homo sapiens (Human) | CVCL_1467 | |
| CRL-5803 cells | Lymph node | Homo sapiens (Human) | CVCL_0060 | |
| CCL-253 cells | Abdominal wall | Homo sapiens (Human) | CVCL_1564 | |
| CCL-185 cells | Bowel | Homo sapiens (Human) | CVCL_0023 | |
| Calu-6 cells | Lung | Homo sapiens (Human) | CVCL_0236 | |
| In Vivo Model | NSG mouse PDX model | Mus musculus | ||
| Experiment for Drug Resistance |
Promega assay | |||
| Mechanism Description | Researchers defined three distinct functional classes of BRAF mutants in human tumours. The mutants activate ERK signalling by different mechanisms that dictate their sensitivity to therapeutic inhibitors of the pathway. | |||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [31] | |||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.G596R (c.1786G>C) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | ERK signaling pathway | Activation | hsa04210 | |
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |
| HT-29 cells | Colon | Homo sapiens (Human) | CVCL_0320 | |
| H1650 cells | Pleural effusion | Homo sapiens (Human) | CVCL_4V01 | |
| HTB-56 cells | Pleural effusion | Homo sapiens (Human) | CVCL_0236 | |
| HTB-38 cells | Colon | Homo sapiens (Human) | CVCL_0320 | |
| HTB-183 cells | Lymph node | Homo sapiens (Human) | CVCL_1577 | |
| H661 cells | Lymph node | Homo sapiens (Human) | CVCL_1577 | |
| H508 cells | Abdominal wall | Homo sapiens (Human) | CVCL_1564 | |
| H2405 cells | Lung | Homo sapiens (Human) | CVCL_1551 | |
| H1666 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1485 | |
| H1395 cells | Lung | Homo sapiens (Human) | CVCL_1467 | |
| CRL-5944 cells | Ascites | Homo sapiens (Human) | CVCL_1551 | |
| CRL-5885 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1485 | |
| CRL-5883 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1483 | |
| CRL-5868 cells | Lung | Homo sapiens (Human) | CVCL_1467 | |
| CRL-5803 cells | Lymph node | Homo sapiens (Human) | CVCL_0060 | |
| CCL-253 cells | Abdominal wall | Homo sapiens (Human) | CVCL_1564 | |
| CCL-185 cells | Bowel | Homo sapiens (Human) | CVCL_0023 | |
| Calu-6 cells | Lung | Homo sapiens (Human) | CVCL_0236 | |
| In Vivo Model | NSG mouse PDX model | Mus musculus | ||
| Experiment for Drug Resistance |
Promega assay | |||
| Mechanism Description | Researchers defined three distinct functional classes of BRAF mutants in human tumours. The mutants activate ERK signalling by different mechanisms that dictate their sensitivity to therapeutic inhibitors of the pathway. | |||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [31] | |||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.G466V (c.1397G>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | ERK signaling pathway | Activation | hsa04210 | |
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |
| HT-29 cells | Colon | Homo sapiens (Human) | CVCL_0320 | |
| H1650 cells | Pleural effusion | Homo sapiens (Human) | CVCL_4V01 | |
| HTB-56 cells | Pleural effusion | Homo sapiens (Human) | CVCL_0236 | |
| HTB-38 cells | Colon | Homo sapiens (Human) | CVCL_0320 | |
| HTB-183 cells | Lymph node | Homo sapiens (Human) | CVCL_1577 | |
| H661 cells | Lymph node | Homo sapiens (Human) | CVCL_1577 | |
| H508 cells | Abdominal wall | Homo sapiens (Human) | CVCL_1564 | |
| H2405 cells | Lung | Homo sapiens (Human) | CVCL_1551 | |
| H1666 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1485 | |
| H1395 cells | Lung | Homo sapiens (Human) | CVCL_1467 | |
| CRL-5944 cells | Ascites | Homo sapiens (Human) | CVCL_1551 | |
| CRL-5885 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1485 | |
| CRL-5883 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1483 | |
| CRL-5868 cells | Lung | Homo sapiens (Human) | CVCL_1467 | |
| CRL-5803 cells | Lymph node | Homo sapiens (Human) | CVCL_0060 | |
| CCL-253 cells | Abdominal wall | Homo sapiens (Human) | CVCL_1564 | |
| CCL-185 cells | Bowel | Homo sapiens (Human) | CVCL_0023 | |
| Calu-6 cells | Lung | Homo sapiens (Human) | CVCL_0236 | |
| In Vivo Model | NSG mouse PDX model | Mus musculus | ||
| Experiment for Drug Resistance |
Promega assay | |||
| Mechanism Description | Researchers defined three distinct functional classes of BRAF mutants in human tumours. The mutants activate ERK signalling by different mechanisms that dictate their sensitivity to therapeutic inhibitors of the pathway. | |||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [32] | |||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Complex-indel | p.L485_P490delinsY (c.1453_1470delinsTAT) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |
| BxPC-3 cells | Pancreas | Homo sapiens (Human) | CVCL_0186 | |
| NIH 3T3 cells | Colon | Homo sapiens (Human) | CVCL_0594 | |
| HEK 293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |
| OV-90 cells | Ascites | Homo sapiens (Human) | CVCL_3768 | |
| H2405 cells | Lung | Homo sapiens (Human) | CVCL_1551 | |
| In Vivo Model | NIH nude rat xenograft model | Rattus norvegicus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay; Colony transformation assay; Cell-cycle analysis; BrdUrd incorporation assay | |||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [32] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | IF-deletion | p.N486_P490delNVTAP (c.1457_1471del15) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |
| BxPC-3 cells | Pancreas | Homo sapiens (Human) | CVCL_0186 | |
| NIH 3T3 cells | Colon | Homo sapiens (Human) | CVCL_0594 | |
| HEK 293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |
| OV-90 cells | Ascites | Homo sapiens (Human) | CVCL_3768 | |
| H2405 cells | Lung | Homo sapiens (Human) | CVCL_1551 | |
| In Vivo Model | NIH nude rat xenograft model | Rattus norvegicus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay; Colony transformation assay; Cell-cycle analysis; BrdUrd incorporation assay | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [33] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.L597V (c.1789C>G) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Skin | N.A. | ||
| Experiment for Molecule Alteration |
Tumour genotyping assay | |||
| Mechanism Description | The missense mutation p.L597V (c.1789C>G) in gene BRAF cause the sensitivity of Trametinib by unusual activation of pro-survival pathway | |||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [29] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.L597S (c.1789_1790delCTinsTC) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Melanoma thyroid metastasis | N.A. | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Mechanism Description | The missense mutation p.L597S (c.1789_1790delCTinsTC) in gene BRAF cause the sensitivity of Trametinib by unusual activation of pro-survival pathway | |||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [29] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.L597R (c.1790T>G) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Melanoma thyroid metastasis | N.A. | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Mechanism Description | The missense mutation p.L597R (c.1790T>G) in gene BRAF cause the sensitivity of Trametinib by unusual activation of pro-survival pathway | |||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [29] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.K601E (c.1801A>G) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Melanoma thyroid metastasis | N.A. | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Mechanism Description | The missense mutation p.K601E (c.1801A>G) in gene BRAF cause the sensitivity of Trametinib by unusual activation of pro-survival pathway | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [29] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.L597S (c.1789_1790delCTinsTC) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Melanoma thyroid metastasis | N.A. | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Mechanism Description | The missense mutation p.L597S (c.1789_1790delCTinsTC) in gene BRAF cause the sensitivity of Trametinib by unusual activation of pro-survival pathway | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [31] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.D594G (c.1781A>G) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | ERK signaling pathway | Activation | hsa04210 | |
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |
| HT-29 cells | Colon | Homo sapiens (Human) | CVCL_0320 | |
| H1650 cells | Pleural effusion | Homo sapiens (Human) | CVCL_4V01 | |
| HTB-56 cells | Pleural effusion | Homo sapiens (Human) | CVCL_0236 | |
| HTB-38 cells | Colon | Homo sapiens (Human) | CVCL_0320 | |
| HTB-183 cells | Lymph node | Homo sapiens (Human) | CVCL_1577 | |
| H661 cells | Lymph node | Homo sapiens (Human) | CVCL_1577 | |
| H508 cells | Abdominal wall | Homo sapiens (Human) | CVCL_1564 | |
| H2405 cells | Lung | Homo sapiens (Human) | CVCL_1551 | |
| H1666 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1485 | |
| H1395 cells | Lung | Homo sapiens (Human) | CVCL_1467 | |
| CRL-5944 cells | Ascites | Homo sapiens (Human) | CVCL_1551 | |
| CRL-5885 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1485 | |
| CRL-5883 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1483 | |
| CRL-5868 cells | Lung | Homo sapiens (Human) | CVCL_1467 | |
| CRL-5803 cells | Lymph node | Homo sapiens (Human) | CVCL_0060 | |
| CCL-253 cells | Abdominal wall | Homo sapiens (Human) | CVCL_1564 | |
| CCL-185 cells | Bowel | Homo sapiens (Human) | CVCL_0023 | |
| Calu-6 cells | Lung | Homo sapiens (Human) | CVCL_0236 | |
| In Vivo Model | NSG mouse PDX model | Mus musculus | ||
| Experiment for Drug Resistance |
Promega assay | |||
| Mechanism Description | Researchers defined three distinct functional classes of BRAF mutants in human tumours. The mutants activate ERK signalling by different mechanisms that dictate their sensitivity to therapeutic inhibitors of the pathway. | |||
| Disease Class: Bladder cancer [ICD-11: 2C94.0] | [34] | |||
| Sensitive Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Other | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [3] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.L597S (c.1789_1790delCTinsTC) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | MAPK signaling pathway | Inhibition | hsa04010 | |
| In Vitro Model | Skin sample | N.A. | ||
| In Vivo Model | Mouse PDX model | Mus musculus | ||
| Experiment for Drug Resistance |
Crystal violet staining assay | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [35] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.D594V (c.1781A>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 |
| Experiment for Molecule Alteration |
Immunoblotting analysis | |||
| Mechanism Description | The missense mutation p.D594V (c.1781A>T) in gene BRAF cause the sensitivity of Trametinib by unusual activation of pro-survival pathway | |||
| Disease Class: Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | [32] | |||
| Sensitive Disease | Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Complex-indel | p.V487_P492delinsA (c.1460_1474del15) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |
| BxPC-3 cells | Pancreas | Homo sapiens (Human) | CVCL_0186 | |
| NIH 3T3 cells | Colon | Homo sapiens (Human) | CVCL_0594 | |
| HEK 293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |
| OV-90 cells | Ascites | Homo sapiens (Human) | CVCL_3768 | |
| H2405 cells | Lung | Homo sapiens (Human) | CVCL_1551 | |
| In Vivo Model | NIH nude rat xenograft model | Rattus norvegicus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay; Colony transformation assay; Cell-cycle analysis; BrdUrd incorporation assay | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [36] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Synonymous | p.K601K (c.1803A>G) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [3] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Synonymous | p.L597L (c.1791A>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [30] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.K601R (c.1802A>G) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [33] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.L597R (c.1790T>G) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | 293H cells | Fetal kidney | Homo sapiens (Human) | CVCL_ZK99 |
| Experiment for Molecule Alteration |
Whole genome sequencing assay | |||
| Disease Class: Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | [37] | |||
| Sensitive Disease | Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | IF-deletion | p.N486_P490delNVTAP (c.1457_1471del15) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Blood sample | N.A. | ||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [38] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.F247L (c.739T>C) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | MAPK signaling pathway | Activation | hsa04010 | |
| In Vitro Model | MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| HMLER cells | Breast | Homo sapiens (Human) | CVCL_DG85 | |
| In Vivo Model | Athymic mouse PDX model | Mus musculus | ||
| Experiment for Molecule Alteration |
Immunoblotting assay; Immunofluorescence assay; qPCR | |||
| Experiment for Drug Resistance |
Promega assay | |||
| Mechanism Description | Reseachers identified an oncogenic mutation, F247L, whose expression robustly activated the MAPK pathway and sensitized cells to BRAF and MEK inhibitors. | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [6] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.V600K (c.1798_1799delGTinsAA) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [39] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | Trametinib/Dabrafenib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600X (c.1798_1799) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Skin | N.A. | |||||||||||
| Experiment for Drug Resistance |
Tumor evaluation assay | ||||||||||||
|
|
|||||||||||||
| Disease Class: Thyroid gland cancer [ICD-11: 2D10.0] | [40] | ||||||||||||
| Sensitive Disease | Thyroid gland cancer [ICD-11: 2D10.0] | ||||||||||||
| Sensitive Drug | Trametinib/Dabrafenib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | MAPK signaling pathway | Inhibition | hsa04010 | ||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Papillary thyroid carcinoma [ICD-11: 2D10.1] | [41] | ||||||||||||
| Resistant Disease | Papillary thyroid carcinoma [ICD-11: 2D10.1] | ||||||||||||
| Resistant Drug | Vemurafenib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
Low throughput experiment assay | ||||||||||||
| Mechanism Description | BRAFV600E is the most common mutation in PTC, occurring in about 60% of PTC tumors, and has been described as a clonal event since it occurs in the majority of tumor cells. BRAFV600E PTC exhibits primary resistance to RAI treatment, higher rates of tumor recurrence and metastases, and lower survival rates. Remarkably, the BRAFV600E mutation not only promotes thyroid tumor cell proliferation, adhesion, migration and invasion, but also up-regulates epigenetic pathways that silence expression of the sodium/iodide symporter. This blocks iodide uptake, which may be one cause of primary resistance to RAI. Present in other cancers, including 40-70% of malignant melanomas and 10% of colorectal cancers, BRAFV600E positive tumors provide one important case study for the evolution of drug resistance. | ||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [42] | ||||||||||||
| Resistant Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Resistant Drug | Vemurafenib | ||||||||||||
| Molecule Alteration | Structural variation | Copy number gain |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
Whole-exome sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Multivariate analysis of overall or disease-free survival assay | ||||||||||||
| Mechanism Description | Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. | ||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [43] | ||||||||||||
| Resistant Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Resistant Drug | Vemurafenib | ||||||||||||
| Molecule Alteration | Expression | Down-regulation |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | ERK1/2/MEK activation signaling pathway|hsa04210) | Regulation | N.A. | ||||||||||
| MAPK signaling pathway | Activation | hsa04010 | |||||||||||
| PI3K signaling pathway | Activation | hsa04151 | |||||||||||
| RAS signaling pathway | Activation | hsa04014 | |||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| Experiment for Molecule Alteration |
Western blot analysis; GTPase assay | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Mechanism Description | miR204-5p and miR211-5p contribute to BRAF inhibitor resistance in melanoma. MTT assays revealed a moderate but consistent increase in resistance to VMF in cells overexpressing miR211-5p or miR204-5p. Joint overexpression of miR204-5p and miR211-5p durably stimulated Ras and MAPk upregulation. Resistance to BRAFi in melanoma involves genetic alterations that lead to reactivation of the MAPk pathway or activation of PI3-k/AkT signalling. | ||||||||||||
Clinical Trial Drug(s)
18 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Lymphatic system cancer [ICD-11: 2E81.1] | [44] | ||||||||||||
| Sensitive Disease | Lymphatic system cancer [ICD-11: 2E81.1] | ||||||||||||
| Sensitive Drug | Cobimetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |||||||||
| Experiment for Molecule Alteration |
Whole exome sequencing; Targeted Exon Sequencing | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay; IC50 assay | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [45] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | Cobimetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Mechanism Description | The missense mutation p.V600E (c.1799T>A) in gene BRAF cause the sensitivity of Cobimetinib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [44] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | Cobimetinib | ||||||||||||
| Molecule Alteration | Complex-indel | p.N486_T491delinsK (c.1458_1472del15) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |||||||||
| Experiment for Molecule Alteration |
Whole exome sequencing; Targeted Exon Sequencing | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay; IC50 assay | ||||||||||||
| Disease Class: Lymphatic system cancer [ICD-11: 2E81.1] | [44] | ||||||||||||
| Sensitive Disease | Lymphatic system cancer [ICD-11: 2E81.1] | ||||||||||||
| Sensitive Drug | Cobimetinib | ||||||||||||
| Molecule Alteration | Complex-indel | p.N486_T491delinsK (c.1458_1472del15) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |||||||||
| Experiment for Molecule Alteration |
Whole exome sequencing; Targeted Exon Sequencing | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay; IC50 assay | ||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [3] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | Cobimetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.L597S (c.1789_1790delCTinsTC) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | MAPK signaling pathway | Inhibition | hsa04010 | ||||||||||
| In Vitro Model | Skin sample | N.A. | |||||||||||
| In Vivo Model | Mouse PDX model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
Crystal violet staining assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [46] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | Ganetespib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
WST-1 cell proliferation assay | ||||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [47] | |||
| Resistant Disease | Melanoma [ICD-11: 2C30.0] | |||
| Resistant Drug | Refametinib | |||
| Molecule Alteration | Missense mutation | p.V600X (c.1798_1799) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Mechanism Description | The missense mutation p.V600X (c.1798_1799) in gene BRAF cause the resistance of Refametinib by aberration of the drug's therapeutic target | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [48] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | Refametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |||||||||
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | ||||||||||
| HT-29 cells | Colon | Homo sapiens (Human) | CVCL_0320 | ||||||||||
| A431 cells | Skin | Homo sapiens (Human) | CVCL_0037 | ||||||||||
| COLO205 cells | Colon | Homo sapiens (Human) | CVCL_F402 | ||||||||||
| BxPc3 cells | Pancreas | Homo sapiens (Human) | CVCL_0186 | ||||||||||
| SkMEL28 cells | Skin | Homo sapiens (Human) | CVCL_0526 | ||||||||||
| In Vivo Model | Female athymic nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Biochemical kinase assays | ||||||||||||
| Experiment for Drug Resistance |
CellTiter 96 Aqueous One assay | ||||||||||||
| Mechanism Description | The missense mutation p.V600E (c.1799T>A) in gene BRAF cause the sensitivity of Refametinib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [48] | ||||||||||||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | ||||||||||||
| Sensitive Drug | Refametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |||||||||
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | ||||||||||
| HT-29 cells | Colon | Homo sapiens (Human) | CVCL_0320 | ||||||||||
| A431 cells | Skin | Homo sapiens (Human) | CVCL_0037 | ||||||||||
| COLO205 cells | Colon | Homo sapiens (Human) | CVCL_F402 | ||||||||||
| BxPc3 cells | Pancreas | Homo sapiens (Human) | CVCL_0186 | ||||||||||
| SkMEL28 cells | Skin | Homo sapiens (Human) | CVCL_0526 | ||||||||||
| In Vivo Model | Female athymic nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Biochemical kinase assays | ||||||||||||
| Experiment for Drug Resistance |
CellTiter 96 Aqueous One assay | ||||||||||||
| Mechanism Description | The missense mutation p.V600E (c.1799T>A) in gene BRAF cause the sensitivity of Refametinib by unusual activation of pro-survival pathway | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: FGFR-tacc positive glioblastoma [ICD-11: 2A00.01] | [49] | ||||||||||||
| Sensitive Disease | FGFR-tacc positive glioblastoma [ICD-11: 2A00.01] | ||||||||||||
| Sensitive Drug | Selumetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: FGFR-tacc positive glioblastoma [ICD-11: 2A00.01] | [49] | ||||||||||||
| Sensitive Disease | FGFR-tacc positive glioblastoma [ICD-11: 2A00.01] | ||||||||||||
| Sensitive Drug | Selumetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: Thyroid gland cancer [ICD-11: 2D10.0] | [50] | ||||||||||||
| Sensitive Disease | Thyroid gland cancer [ICD-11: 2D10.0] | ||||||||||||
| Sensitive Drug | Selumetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | BRAF/MEK/MAPK signaling pathway | Inhibition | hsa04010 | ||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| BCPAP cells | Thyroid | Homo sapiens (Human) | CVCL_0153 | ||||||||||
| SW1736 cells | Thyroid | Homo sapiens (Human) | CVCL_3883 | ||||||||||
| C643 cells | Thyroid gland | Homo sapiens (Human) | CVCL_5969 | ||||||||||
| CAL62 cells | Thyroid gland | Homo sapiens (Human) | CVCL_1112 | ||||||||||
| In Vivo Model | Athymic nude mouse PDX xenografts model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting assay; Immunoprecipitation assy | ||||||||||||
| Experiment for Drug Resistance |
SRB staining assay; Promega assay | ||||||||||||
| Mechanism Description | Activation of the Mitogen Activated Protein (MAP) Kinase pathway was increased in all four of the dasatinib-resistant cell lines, likely due to B-Raf and c-Raf dimerization. Furthermore, MAP2K1/MAP2K2 (MEK1/2) inhibition restored sensitivity in all four of the dasatinib-resistant cell lines, and overcome acquired resistance to dasatinib in the RAS-mutant Cal62 cell line, in vivo. Together, these studies demonstrate that acquisition of the c-Src gatekeeper mutation and MAP Kinase pathway signaling play important roles in promoting resistance to the Src inhibitor, dasatinib. | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [6] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | Vemurafenib/Cobimetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [10] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | Vemurafenib/Cobimetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [51] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | Vemurafenib/Cobimetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600X (c.1798_1799) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [6] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | Vemurafenib/Cobimetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600K (c.1798_1799delGTinsAA) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [52] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | E6201 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| G-361 cells | Skin | Homo sapiens (Human) | CVCL_1220 | ||||||||||
| MDA-MB-435s cells | Breast | Homo sapiens (Human) | CVCL_0622 | ||||||||||
| SEKI cells | Skin | Homo sapiens (Human) | CVCL_3162 | ||||||||||
| HMV-1 cells | Uterus | Homo sapiens (Human) | CVCL_8233 | ||||||||||
| HMCB cells | Skin | Homo sapiens (Human) | CVCL_3317 | ||||||||||
| CHL-1 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1122 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; Immunohistochemistry assay | ||||||||||||
| Experiment for Drug Resistance |
WST-8 assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [53] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | Pictilisib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
ICH assay | ||||||||||||
| Experiment for Drug Resistance |
Plasma level assay; Electrochemiluminescense assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [54] | ||||||||||||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | ||||||||||||
| Sensitive Drug | Pimasertib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HT29 Cells | Colon | Homo sapiens (Human) | CVCL_A8EZ | |||||||||
| H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | ||||||||||
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | ||||||||||
| LOVO cells | Colon | Homo sapiens (Human) | CVCL_0399 | ||||||||||
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | ||||||||||
| HCT15 cells | Colon | Homo sapiens (Human) | CVCL_0292 | ||||||||||
| COLO205 cells | Colon | Homo sapiens (Human) | CVCL_F402 | ||||||||||
| In Vivo Model | Female balb/c athymic (nu+/nu+) mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Mechanism Description | The missense mutation p.V600E (c.1799T>A) in gene BRAF cause the sensitivity of Pimasertib by aberration of the drug's therapeutic target | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [55] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Sensitive Drug | PLX8394 | ||||||||||||
| Molecule Alteration | Missense mutation | p.G469A (c.1406G>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HCC364 cells | Lung | Homo sapiens (Human) | CVCL_5134 | |||||||||
| H2405 cells | Lung | Homo sapiens (Human) | CVCL_1551 | ||||||||||
| H2087 cells | Lymph node | Homo sapiens (Human) | CVCL_1524 | ||||||||||
| H1755 cells | Liver | Homo sapiens (Human) | CVCL_1492 | ||||||||||
| H1666 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1485 | ||||||||||
| H1437 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1472 | ||||||||||
| H1395 cells | Lung | Homo sapiens (Human) | CVCL_1467 | ||||||||||
| CAL-12T cells | Lung | Homo sapiens (Human) | CVCL_1105 | ||||||||||
| In Vivo Model | SCID beige mouse PDX model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Promega assay | ||||||||||||
| Mechanism Description | PLX8394 was effective against treatment-naive BRAF-mutant LAs and those with acquired vemurafenib resistance caused by an alternatively spliced, truncated BRAFV600E that promotes vemurafenib-insensitive MAPK pathway signaling. Acquired PLX8394 resistance occurs via EGFR-mediated RAS-mTOR signaling and is prevented by upfront combination therapy with PLX8394 and either an EGFR or mTOR inhibitor. | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [56] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | PLX8394 | ||||||||||||
| Molecule Alteration | Synonymous | p.K601K (c.1803A>G) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [56] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | PLX8394 | ||||||||||||
| Molecule Alteration | Synonymous | p.G464G (c.1392A>T) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [56] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | PLX8394 | ||||||||||||
| Molecule Alteration | Missense mutation | p.G469R (c.1405G>A) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [56] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | PLX8394 | ||||||||||||
| Molecule Alteration | Missense mutation | p.G469A (c.1406G>C) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [56] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | PLX8394 | ||||||||||||
| Molecule Alteration | Missense mutation | p.G469V (c.1406G>T) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [56] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | PLX8394 | ||||||||||||
| Molecule Alteration | Synonymous | p.L597L (c.1791A>T) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [55] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Sensitive Drug | PLX8394 | ||||||||||||
| Molecule Alteration | Missense mutation | p.G466V (c.1397G>T) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HCC364 cells | Lung | Homo sapiens (Human) | CVCL_5134 | |||||||||
| H2405 cells | Lung | Homo sapiens (Human) | CVCL_1551 | ||||||||||
| H2087 cells | Lymph node | Homo sapiens (Human) | CVCL_1524 | ||||||||||
| H1755 cells | Liver | Homo sapiens (Human) | CVCL_1492 | ||||||||||
| H1666 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1485 | ||||||||||
| H1437 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1472 | ||||||||||
| H1395 cells | Lung | Homo sapiens (Human) | CVCL_1467 | ||||||||||
| CAL-12T cells | Lung | Homo sapiens (Human) | CVCL_1105 | ||||||||||
| In Vivo Model | SCID beige mouse PDX model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Promega assay | ||||||||||||
| Mechanism Description | PLX8394 was effective against treatment-naive BRAF-mutant LAs and those with acquired vemurafenib resistance caused by an alternatively spliced, truncated BRAFV600E that promotes vemurafenib-insensitive MAPK pathway signaling. Acquired PLX8394 resistance occurs via EGFR-mediated RAS-mTOR signaling and is prevented by upfront combination therapy with PLX8394 and either an EGFR or mTOR inhibitor. | ||||||||||||
|
|
|||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [57] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | PLX8394 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | PRT cells | Brain | Homo sapiens (Human) | CVCL_7207 | |||||||||
| 1205Lu cells | Skin | Homo sapiens (Human) | CVCL_5239 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTT assay; AlamarBlue assay; Colony growth assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [58] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | RAF-265 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| In Vivo Model | Nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
BRAF kinase assay | ||||||||||||
| Experiment for Drug Resistance |
Flow cytometry assay | ||||||||||||
| Mechanism Description | The missense mutation p.V600E (c.1799T>A) in gene BRAF cause the sensitivity of RAF-265 by aberration of the drug's therapeutic target | ||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [59] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | RAF-265 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Skin sample | N.A. | |||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Thyroid gland cancer [ICD-11: 2D10.0] | [60] | ||||||||||||
| Sensitive Disease | Thyroid gland cancer [ICD-11: 2D10.0] | ||||||||||||
| Sensitive Drug | RO-5126766 free base | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | ERK signaling pathway | Inhibition | hsa04210 | ||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [61] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | RO-5126766 free base | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| NCI-N87 cells | Gastric | Homo sapiens (Human) | CVCL_1603 | ||||||||||
| MkN-45 cells | Gastric | Homo sapiens (Human) | CVCL_0434 | ||||||||||
| NCI-H716 cells | Colon | Homo sapiens (Human) | CVCL_1581 | ||||||||||
| SNU-16 cells | Gastric | Homo sapiens (Human) | CVCL_0076 | ||||||||||
| NCI-H520 cells | Lung | Homo sapiens (Human) | CVCL_1566 | ||||||||||
| RT-4 cells | Urinary bladder | Homo sapiens (Human) | CVCL_0036 | ||||||||||
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | ||||||||||
| NCI-N87 cells | Gastric | Homo sapiens (Human) | CVCL_1603 | ||||||||||
| NCI-H716 cells | Colon | Homo sapiens (Human) | CVCL_1581 | ||||||||||
| KATO-3 cells | Gastric | Homo sapiens (Human) | CVCL_0371 | ||||||||||
| SNU-16 cells | Gastric | Homo sapiens (Human) | CVCL_0076 | ||||||||||
| NCI-H520 cells | Lung | Homo sapiens (Human) | CVCL_1566 | ||||||||||
| RT-4 cells | Urinary bladder | Homo sapiens (Human) | CVCL_0036 | ||||||||||
| UM-UC-14 cells | Kidney | Homo sapiens (Human) | CVCL_2747 | ||||||||||
| SUM-52PE cells | Pleural effusion | Homo sapiens (Human) | CVCL_3425 | ||||||||||
| NCI-H1581 cells | Lung | Homo sapiens (Human) | CVCL_1479 | ||||||||||
| MFE296 cells | Endometrium | Homo sapiens (Human) | CVCL_1406 | ||||||||||
| MFE280 cells | Endometrium | Homo sapiens (Human) | CVCL_1405 | ||||||||||
| KMS-11 cells | Pleural effusion | Homo sapiens (Human) | CVCL_2989 | ||||||||||
| HSC-39 cells | Ascites | Homo sapiens (Human) | CVCL_A385 | ||||||||||
| DMS-114 cells | Lung | Homo sapiens (Human) | CVCL_1174 | ||||||||||
| AN3 CA cells | Endometrium | Homo sapiens (Human) | CVCL_0028 | ||||||||||
| UM-UC-14 cells | Kidney | Homo sapiens (Human) | CVCL_2747 | ||||||||||
| KATO-III cells | Pleural effusion | Homo sapiens (Human) | CVCL_0371 | ||||||||||
| AN3 CA cells | Endometrium | Homo sapiens (Human) | CVCL_0028 | ||||||||||
| Experiment for Molecule Alteration |
Microarray assay; Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CCK-8 assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: FGFR-tacc positive glioblastoma [ICD-11: 2A00.01] | [62] | ||||||||||||
| Sensitive Disease | FGFR-tacc positive glioblastoma [ICD-11: 2A00.01] | ||||||||||||
| Sensitive Drug | Ulixertinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | MAPK signaling pathway | Inhibition | hsa04010 | ||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [62] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Sensitive Drug | Ulixertinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | MAPK signaling pathway | Inhibition | hsa04010 | ||||||||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [63] | ||||||||||||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | ||||||||||||
| Sensitive Drug | Ulixertinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | MAPK signaling pathway | Inhibition | hsa04010 | ||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | ||||||||||
| RkO cells | Colon | Homo sapiens (Human) | CVCL_0504 | ||||||||||
| G-361 cells | Skin | Homo sapiens (Human) | CVCL_1220 | ||||||||||
| HT-29 cells | Colon | Homo sapiens (Human) | CVCL_0320 | ||||||||||
| SW48 cells | Colon | Homo sapiens (Human) | CVCL_1724 | ||||||||||
| COLO205 cells | Colon | Homo sapiens (Human) | CVCL_F402 | ||||||||||
| AN3CA cells | Ovary | Homo sapiens (Human) | CVCL_0028 | ||||||||||
| MIA PaCa-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0428 | ||||||||||
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | ||||||||||
| In Vivo Model | Athymic nude mouse PDX model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
Standard coupled-enzyme assay | ||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [63] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | Ulixertinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | MAPK signaling pathway | Inhibition | hsa04010 | ||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | ||||||||||
| RkO cells | Colon | Homo sapiens (Human) | CVCL_0504 | ||||||||||
| G-361 cells | Skin | Homo sapiens (Human) | CVCL_1220 | ||||||||||
| HT-29 cells | Colon | Homo sapiens (Human) | CVCL_0320 | ||||||||||
| SW48 cells | Colon | Homo sapiens (Human) | CVCL_1724 | ||||||||||
| COLO205 cells | Colon | Homo sapiens (Human) | CVCL_F402 | ||||||||||
| AN3CA cells | Ovary | Homo sapiens (Human) | CVCL_0028 | ||||||||||
| MIA PaCa-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0428 | ||||||||||
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | ||||||||||
| In Vivo Model | Athymic nude mouse PDX model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
Standard coupled-enzyme assay | ||||||||||||
| Disease Class: Head and neck cancer [ICD-11: 2D42.0] | [62] | ||||||||||||
| Sensitive Disease | Head and neck cancer [ICD-11: 2D42.0] | ||||||||||||
| Sensitive Drug | Ulixertinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.G469A (c.1406G>C) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | MAPK signaling pathway | Inhibition | hsa04010 | ||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [62] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Sensitive Drug | Ulixertinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.L597Q (c.1790T>A) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | MAPK signaling pathway | Inhibition | hsa04010 | ||||||||||
| Disease Class: Gallbladder cancer [ICD-11: 2C13.0] | [62] | ||||||||||||
| Sensitive Disease | Gallbladder cancer [ICD-11: 2C13.0] | ||||||||||||
| Sensitive Drug | Ulixertinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.L485W (c.1454T>G) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: Small intestine carcinoma [ICD-11: 2B80.20] | [62] | ||||||||||||
| Sensitive Disease | Small intestine carcinoma [ICD-11: 2B80.20] | ||||||||||||
| Sensitive Drug | Ulixertinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.G469A (c.1406G>C) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | MAPK signaling pathway | Inhibition | hsa04010 | ||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Colon cancer [ICD-11: 2B90.1] | [64] | ||||||||||||
| Sensitive Disease | Colon cancer [ICD-11: 2B90.1] | ||||||||||||
| Sensitive Drug | Agerafenib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| Mechanism Description | The missense mutation p.V600E (c.1799T>A) in gene BRAF cause the sensitivity of Agerafenib by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [64] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | Agerafenib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| Mechanism Description | The missense mutation p.V600E (c.1799T>A) in gene BRAF cause the sensitivity of Agerafenib by aberration of the drug's therapeutic target | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [65] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | Lifirafenib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HT29 Cells | Colon | Homo sapiens (Human) | CVCL_A8EZ | |||||||||
| SW620 cells | Colon | Homo sapiens (Human) | CVCL_0547 | ||||||||||
| HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | ||||||||||
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | ||||||||||
| HCC827 cells | Lung | Homo sapiens (Human) | CVCL_2063 | ||||||||||
| A431 cells | Skin | Homo sapiens (Human) | CVCL_0037 | ||||||||||
| COLO205 cells | Colon | Homo sapiens (Human) | CVCL_F402 | ||||||||||
| WiDR cells | Colon | Homo sapiens (Human) | CVCL_2760 | ||||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| SkMEL28 cells | Skin | Homo sapiens (Human) | CVCL_0526 | ||||||||||
| In Vivo Model | Female NOD/SCID and BALB/c nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay; Tumor volume measurement assay | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [65] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | Lifirafenib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HT29 Cells | Colon | Homo sapiens (Human) | CVCL_A8EZ | |||||||||
| SW620 cells | Colon | Homo sapiens (Human) | CVCL_0547 | ||||||||||
| HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | ||||||||||
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | ||||||||||
| HCC827 cells | Lung | Homo sapiens (Human) | CVCL_2063 | ||||||||||
| A431 cells | Skin | Homo sapiens (Human) | CVCL_0037 | ||||||||||
| COLO205 cells | Colon | Homo sapiens (Human) | CVCL_F402 | ||||||||||
| WiDR cells | Colon | Homo sapiens (Human) | CVCL_2760 | ||||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| SkMEL28 cells | Skin | Homo sapiens (Human) | CVCL_0526 | ||||||||||
| In Vivo Model | Female NOD/SCID and BALB/c nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay; Tumor volume measurement assay | ||||||||||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [65] | ||||||||||||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | ||||||||||||
| Sensitive Drug | Lifirafenib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HT29 Cells | Colon | Homo sapiens (Human) | CVCL_A8EZ | |||||||||
| SW620 cells | Colon | Homo sapiens (Human) | CVCL_0547 | ||||||||||
| HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | ||||||||||
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | ||||||||||
| HCC827 cells | Lung | Homo sapiens (Human) | CVCL_2063 | ||||||||||
| A431 cells | Skin | Homo sapiens (Human) | CVCL_0037 | ||||||||||
| COLO205 cells | Colon | Homo sapiens (Human) | CVCL_F402 | ||||||||||
| WiDR cells | Colon | Homo sapiens (Human) | CVCL_2760 | ||||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| SkMEL28 cells | Skin | Homo sapiens (Human) | CVCL_0526 | ||||||||||
| In Vivo Model | Female NOD/SCID and BALB/c nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay; Tumor volume measurement assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [66] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | BI-847325 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| WM793 cells | N.A. | Homo sapiens (Human) | CVCL_8787/CVCL_5414 | ||||||||||
| WM39 cells | Skin | Homo sapiens (Human) | CVCL_2240 | ||||||||||
| WM164 cells | Skin | Homo sapiens (Human) | CVCL_7928 | ||||||||||
| RPMI-7951 cells | Lymph node | Homo sapiens (Human) | CVCL_1666 | ||||||||||
| 1205Lu cells | Skin | Homo sapiens (Human) | CVCL_5239 | ||||||||||
| In Vivo Model | BALB SCID mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; qPCR; Proteasome-Glo Chymotrypsin-like cell-based assay | ||||||||||||
| Experiment for Drug Resistance |
Alamar blue assay; Colony formation assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Hematologic Cancer [ICD-11: MG24.Y] | [32] | ||||||||||||
| Sensitive Disease | Hematologic Cancer [ICD-11: MG24.Y] | ||||||||||||
| Sensitive Drug | LY3009120 | ||||||||||||
| Molecule Alteration | Missense mutation | p.K601N (c.1803A>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | ||||||||||
| BxPC-3 cells | Pancreas | Homo sapiens (Human) | CVCL_0186 | ||||||||||
| NIH 3T3 cells | Colon | Homo sapiens (Human) | CVCL_0594 | ||||||||||
| HEK 293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | ||||||||||
| OV-90 cells | Ascites | Homo sapiens (Human) | CVCL_3768 | ||||||||||
| H2405 cells | Lung | Homo sapiens (Human) | CVCL_1551 | ||||||||||
| In Vivo Model | NIH nude rat xenograft model | Rattus norvegicus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay; Colony transformation assay; Cell-cycle analysis; BrdUrd incorporation assay | ||||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [32] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Sensitive Drug | LY3009120 | ||||||||||||
| Molecule Alteration | Complex-indel | p.L485_P490delinsY (c.1453_1470delinsTAT) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | ||||||||||
| BxPC-3 cells | Pancreas | Homo sapiens (Human) | CVCL_0186 | ||||||||||
| NIH 3T3 cells | Colon | Homo sapiens (Human) | CVCL_0594 | ||||||||||
| HEK 293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | ||||||||||
| OV-90 cells | Ascites | Homo sapiens (Human) | CVCL_3768 | ||||||||||
| H2405 cells | Lung | Homo sapiens (Human) | CVCL_1551 | ||||||||||
| In Vivo Model | NIH nude rat xenograft model | Rattus norvegicus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay; Colony transformation assay; Cell-cycle analysis; BrdUrd incorporation assay | ||||||||||||
| Disease Class: Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | [32] | ||||||||||||
| Sensitive Disease | Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | ||||||||||||
| Sensitive Drug | LY3009120 | ||||||||||||
| Molecule Alteration | Complex-indel | p.V487_P492delinsA (c.1460_1474del15) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | ||||||||||
| BxPC-3 cells | Pancreas | Homo sapiens (Human) | CVCL_0186 | ||||||||||
| NIH 3T3 cells | Colon | Homo sapiens (Human) | CVCL_0594 | ||||||||||
| HEK 293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | ||||||||||
| OV-90 cells | Ascites | Homo sapiens (Human) | CVCL_3768 | ||||||||||
| H2405 cells | Lung | Homo sapiens (Human) | CVCL_1551 | ||||||||||
| In Vivo Model | NIH nude rat xenograft model | Rattus norvegicus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay; Colony transformation assay; Cell-cycle analysis; BrdUrd incorporation assay | ||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [67] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | LY3009120 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| Sk-Mel28 cells | Skin | Homo sapiens (Human) | CVCL_0526 | ||||||||||
| A2058 cells | Skin | Homo sapiens (Human) | CVCL_1059 | ||||||||||
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | ||||||||||
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | ||||||||||
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | ||||||||||
| WM2664 cells | Skin | Homo sapiens (Human) | CVCL_2765 | ||||||||||
| SkMEL 30 cells | Skin | Homo sapiens (Human) | CVCL_0039 | ||||||||||
| SkMEL 2 cells | Skin | Homo sapiens (Human) | CVCL_0069 | ||||||||||
| SH4 cells | Skin | Mus musculus (Mouse) | CVCL_7702 | ||||||||||
| MEXF-535 cells | Skin | Homo sapiens (Human) | N.A. | ||||||||||
| MEXF-1792 cells | Skin | Homo sapiens (Human) | N.A. | ||||||||||
| MEXF-1341 cells | Skin | Homo sapiens (Human) | N.A. | ||||||||||
| M14 cells | Hypodermis | Homo sapiens (Human) | CVCL_1395 | ||||||||||
| GAK cells | Lnguinal lymph node | Homo sapiens (Human) | CVCL_1225 | ||||||||||
| Colo829 cells | Skin | Homo sapiens (Human) | CVCL_1137 | ||||||||||
| In Vivo Model | Female NIH nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; Crystallization assay; X-ray data collection and structure determination assay | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay; Enzymatic kinase assay | ||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [3] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | LY3009120 | ||||||||||||
| Molecule Alteration | Missense mutation | p.L597S (c.1789_1790delCTinsTC) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | MAPK signaling pathway | Inhibition | hsa04010 | ||||||||||
| In Vitro Model | Skin sample | N.A. | |||||||||||
| In Vivo Model | Mouse PDX model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
Crystal violet staining assay | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [32] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | LY3009120 | ||||||||||||
| Molecule Alteration | IF-deletion | p.L485_P490delLNVTAP (c.1453_1470del18) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | ||||||||||
| BxPC-3 cells | Pancreas | Homo sapiens (Human) | CVCL_0186 | ||||||||||
| NIH 3T3 cells | Colon | Homo sapiens (Human) | CVCL_0594 | ||||||||||
| HEK 293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | ||||||||||
| OV-90 cells | Ascites | Homo sapiens (Human) | CVCL_3768 | ||||||||||
| H2405 cells | Lung | Homo sapiens (Human) | CVCL_1551 | ||||||||||
| In Vivo Model | NIH nude rat xenograft model | Rattus norvegicus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay; Colony transformation assay; Cell-cycle analysis; BrdUrd incorporation assay | ||||||||||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [32] | ||||||||||||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | ||||||||||||
| Sensitive Drug | LY3009120 | ||||||||||||
| Molecule Alteration | IF-deletion | p.N486_P490delNVTAP (c.1457_1471del15) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | ||||||||||
| BxPC-3 cells | Pancreas | Homo sapiens (Human) | CVCL_0186 | ||||||||||
| NIH 3T3 cells | Colon | Homo sapiens (Human) | CVCL_0594 | ||||||||||
| HEK 293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | ||||||||||
| OV-90 cells | Ascites | Homo sapiens (Human) | CVCL_3768 | ||||||||||
| H2405 cells | Lung | Homo sapiens (Human) | CVCL_1551 | ||||||||||
| In Vivo Model | NIH nude rat xenograft model | Rattus norvegicus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay; Colony transformation assay; Cell-cycle analysis; BrdUrd incorporation assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [68] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | Omipalisib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay | ||||||||||||
| Mechanism Description | The missense mutation p.V600E (c.1799T>A) in gene BRAF cause the sensitivity of Omipalisib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [68] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | Omipalisib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600K (c.1798_1799delGTinsAA) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay | ||||||||||||
| Mechanism Description | The missense mutation p.V600K (c.1798_1799delGTinsAA) in gene BRAF cause the sensitivity of Omipalisib by unusual activation of pro-survival pathway | ||||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: FGFR-tacc positive glioblastoma [ICD-11: 2A00.01] | [69] | ||||||||||||
| Resistant Disease | FGFR-tacc positive glioblastoma [ICD-11: 2A00.01] | ||||||||||||
| Resistant Drug | PLX4720 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | GBM cells | Brain | Homo sapiens (Human) | N.A. | |||||||||
| In Vivo Model | Athymic mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
qPCR | ||||||||||||
| Experiment for Drug Resistance |
Alamar blue proliferation assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [70] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | PLX4720 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | ||||||||||||
| Mechanism Description | The missense mutation p.V600E (c.1799T>A) in gene BRAF cause the sensitivity of PLX4720 by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: FGFR-tacc positive glioblastoma [ICD-11: 2A00.01] | [71] | ||||||||||||
| Sensitive Disease | FGFR-tacc positive glioblastoma [ICD-11: 2A00.01] | ||||||||||||
| Sensitive Drug | PLX4720 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | LN229 cells | Brain | Homo sapiens (Human) | CVCL_0393 | |||||||||
| A172 cells | Brain | Homo sapiens (Human) | CVCL_0131 | ||||||||||
| U87 cells | Brain | Homo sapiens (Human) | CVCL_0022 | ||||||||||
| SNB19 cells | Brain | Homo sapiens (Human) | CVCL_0535 | ||||||||||
| U138 cells | Brain | Homo sapiens (Human) | CVCL_0020 | ||||||||||
| T98G cells | Brain | Homo sapiens (Human) | CVCL_0556 | ||||||||||
| HS683 cells | Brain | Homo sapiens (Human) | CVCL_0844 | ||||||||||
| DBTRG-05MG cells | Brain | Homo sapiens (Human) | CVCL_1169 | ||||||||||
| NMC-G1 cells | Brain | Homo sapiens (Human) | CVCL_1608 | ||||||||||
| MO59J cells | Brain | Homo sapiens (Human) | CVCL_0400 | ||||||||||
| LN405 cells | Brain | Homo sapiens (Human) | CVCL_1378 | ||||||||||
| LN172 cells | N.A. | N.A. | N.A. | ||||||||||
| KG1c cells | Brain | Homo sapiens (Human) | CVCL_2971 | ||||||||||
| H4 cells | Brain | Homo sapiens (Human) | CVCL_1239 | ||||||||||
| GMS10 cells | Brain | Homo sapiens (Human) | CVCL_1233 | ||||||||||
| GAMG cells | Brain | Homo sapiens (Human) | CVCL_1226 | ||||||||||
| CCF-STTG1 cells | Brain | Homo sapiens (Human) | CVCL_1118 | ||||||||||
| AM-38 cells | Brain | Homo sapiens (Human) | CVCL_1070 | ||||||||||
| 8MGBA cells | Brain | Homo sapiens (Human) | CVCL_1052 | ||||||||||
| 42MGBA cells | Brain | Homo sapiens (Human) | CVCL_1798 | ||||||||||
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CCK-8 assay | ||||||||||||
| Mechanism Description | PLX4720 suppresses MEK-ERK phosphorylation and cell proliferation in MA cells containing BRAFV600E mutation. | ||||||||||||
| Disease Class: Colon cancer [ICD-11: 2B90.1] | [70] | ||||||||||||
| Sensitive Disease | Colon cancer [ICD-11: 2B90.1] | ||||||||||||
| Sensitive Drug | PLX4720 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | ||||||||||||
| Mechanism Description | The missense mutation p.V600E (c.1799T>A) in gene BRAF cause the sensitivity of PLX4720 by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [58] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | PLX4720 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| In Vivo Model | Nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
BRAF kinase assay | ||||||||||||
| Experiment for Drug Resistance |
Flow cytometry assay | ||||||||||||
| Mechanism Description | The missense mutation p.V600E (c.1799T>A) in gene BRAF cause the sensitivity of PLX4720 by aberration of the drug's therapeutic target | ||||||||||||
Discontinued Drug(s)
1 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [61] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | RO4987655 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| NCI-N87 cells | Gastric | Homo sapiens (Human) | CVCL_1603 | ||||||||||
| MkN-45 cells | Gastric | Homo sapiens (Human) | CVCL_0434 | ||||||||||
| NCI-H716 cells | Colon | Homo sapiens (Human) | CVCL_1581 | ||||||||||
| SNU-16 cells | Gastric | Homo sapiens (Human) | CVCL_0076 | ||||||||||
| NCI-H520 cells | Lung | Homo sapiens (Human) | CVCL_1566 | ||||||||||
| RT-4 cells | Urinary bladder | Homo sapiens (Human) | CVCL_0036 | ||||||||||
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | ||||||||||
| NCI-N87 cells | Gastric | Homo sapiens (Human) | CVCL_1603 | ||||||||||
| NCI-H716 cells | Colon | Homo sapiens (Human) | CVCL_1581 | ||||||||||
| KATO-3 cells | Gastric | Homo sapiens (Human) | CVCL_0371 | ||||||||||
| SNU-16 cells | Gastric | Homo sapiens (Human) | CVCL_0076 | ||||||||||
| NCI-H520 cells | Lung | Homo sapiens (Human) | CVCL_1566 | ||||||||||
| RT-4 cells | Urinary bladder | Homo sapiens (Human) | CVCL_0036 | ||||||||||
| UM-UC-14 cells | Kidney | Homo sapiens (Human) | CVCL_2747 | ||||||||||
| SUM-52PE cells | Pleural effusion | Homo sapiens (Human) | CVCL_3425 | ||||||||||
| NCI-H1581 cells | Lung | Homo sapiens (Human) | CVCL_1479 | ||||||||||
| MFE296 cells | Endometrium | Homo sapiens (Human) | CVCL_1406 | ||||||||||
| MFE280 cells | Endometrium | Homo sapiens (Human) | CVCL_1405 | ||||||||||
| KMS-11 cells | Pleural effusion | Homo sapiens (Human) | CVCL_2989 | ||||||||||
| HSC-39 cells | Ascites | Homo sapiens (Human) | CVCL_A385 | ||||||||||
| DMS-114 cells | Lung | Homo sapiens (Human) | CVCL_1174 | ||||||||||
| AN3 CA cells | Endometrium | Homo sapiens (Human) | CVCL_0028 | ||||||||||
| UM-UC-14 cells | Kidney | Homo sapiens (Human) | CVCL_2747 | ||||||||||
| KATO-III cells | Pleural effusion | Homo sapiens (Human) | CVCL_0371 | ||||||||||
| AN3 CA cells | Endometrium | Homo sapiens (Human) | CVCL_0028 | ||||||||||
| Experiment for Molecule Alteration |
Microarray assay; Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CCK-8 assay | ||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [72] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | RO4987655 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600X (c.1798_1799) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Skin sample | N.A. | |||||||||||
| Experiment for Molecule Alteration |
IHC assay; Tumor-DNA mutation analysis | ||||||||||||
| Experiment for Drug Resistance |
Pharmacokinetics analysis; Tumor biopsies analysis | ||||||||||||
Preclinical Drug(s)
14 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [73] | ||||||||||||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | ||||||||||||
| Sensitive Drug | Alpelisib/Cetuximab/Encorafenib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | MAPK signaling pathway | Inhibition | hsa04010 | ||||||||||
| In Vitro Model | WiDR cells | Colon | Homo sapiens (Human) | CVCL_2760 | |||||||||
| VACO432 cells | Colon | Homo sapiens (Human) | CVCL_5402 | ||||||||||
| HROC87 cells | Colon | Homo sapiens (Human) | CVCL_S854 | ||||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay; Western blot analysis; chemiluminescent detection assay | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent assay; CellTox green assay | ||||||||||||
| Mechanism Description | Resistant cells escaped drug treatments through one or more mechanisms leading to biochemical reactivation of the MAPK signaling pathway. | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: FGFR-tacc positive glioblastoma [ICD-11: 2A00.01] | [69] | ||||||||||||
| Sensitive Disease | FGFR-tacc positive glioblastoma [ICD-11: 2A00.01] | ||||||||||||
| Sensitive Drug | BI-2536 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | GBM cells | Brain | Homo sapiens (Human) | N.A. | |||||||||
| In Vivo Model | Athymic mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
qPCR | ||||||||||||
| Experiment for Drug Resistance |
Alamar blue proliferation assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [74] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | BI-69A11 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [75] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Buparlisib/Vemurafenib | |||
| Molecule Alteration | Missense mutation | p.V600X (c.1798_1800) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 |
| Melanoma cells | Skin | Homo sapiens (Human) | N.A. | |
| WM cells | N.A. | Homo sapiens (Human) | N.A. | |
| SK cells | Brain | Homo sapiens (Human) | N.A. | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay; Fluorescent microscopy assay; Apoptosis analysis | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [76] | ||||||||||||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | ||||||||||||
| Sensitive Drug | CCT196969 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| In Vivo Model | Female nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay; IC50 assay | ||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [77] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | CCT196969 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600D (c.1799_1800delTGinsAC) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [76] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | CCT196969 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| In Vivo Model | Female xenograft nude mouse model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; Sanger Sequencing | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay; Tumor volume measurement assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [76] | ||||||||||||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | ||||||||||||
| Sensitive Drug | CCT241161 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| In Vivo Model | Female nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay; IC50 assay | ||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [77] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | CCT241161 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600D (c.1799_1800delTGinsAC) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [76] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | CCT241161 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| In Vivo Model | Female xenograft nude mouse model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; Sanger Sequencing | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay; Tumor volume measurement assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [78] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | Cediranib/PLX4720 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| WM451 cells | Skin | Homo sapiens (Human) | CVCL_6357 | ||||||||||
| In Vivo Model | Nu/Nu(ISTMel1) mouse xenograft model; NSG (RPMI7951) female mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; Immunohistochemistry assay | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [73] | ||||||||||||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | ||||||||||||
| Sensitive Drug | Cetuximab/Selumetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | MAPK signaling pathway | Inhibition | hsa04010 | ||||||||||
| In Vitro Model | WiDR cells | Colon | Homo sapiens (Human) | CVCL_2760 | |||||||||
| VACO432 cells | Colon | Homo sapiens (Human) | CVCL_5402 | ||||||||||
| HROC87 cells | Colon | Homo sapiens (Human) | CVCL_S854 | ||||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay; Western blot analysis; chemiluminescent detection assay | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent assay; CellTox green assay | ||||||||||||
| Mechanism Description | Resistant cells escaped drug treatments through one or more mechanisms leading to biochemical reactivation of the MAPK signaling pathway. | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Thyroid gland cancer [ICD-11: 2D10.0] | [79] | ||||||||||||
| Sensitive Disease | Thyroid gland cancer [ICD-11: 2D10.0] | ||||||||||||
| Sensitive Drug | CLM3 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | 8305C cells | Thyroid | Homo sapiens (Human) | CVCL_1053 | |||||||||
| In Vivo Model | Male CD nu/nu mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Human cyclin D1 ELISA assay | ||||||||||||
| Experiment for Drug Resistance |
WST-1 assay; Cell counting assay; Hoechst uptake assay; Annexin V binding assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Thyroid gland cancer [ICD-11: 2D10.0] | [50] | ||||||||||||
| Sensitive Disease | Thyroid gland cancer [ICD-11: 2D10.0] | ||||||||||||
| Sensitive Drug | Dasatinib/SCH772984 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | BRAF/MEK/MAPK signaling pathway | Inhibition | hsa04010 | ||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| BCPAP cells | Thyroid | Homo sapiens (Human) | CVCL_0153 | ||||||||||
| SW1736 cells | Thyroid | Homo sapiens (Human) | CVCL_3883 | ||||||||||
| C643 cells | Thyroid gland | Homo sapiens (Human) | CVCL_5969 | ||||||||||
| CAL62 cells | Thyroid gland | Homo sapiens (Human) | CVCL_1112 | ||||||||||
| In Vivo Model | Athymic nude mouse PDX xenografts model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting assay; Immunoprecipitation assy | ||||||||||||
| Experiment for Drug Resistance |
SRB staining assay; Promega assay | ||||||||||||
| Mechanism Description | Activation of the Mitogen Activated Protein (MAP) Kinase pathway was increased in all four of the dasatinib-resistant cell lines, likely due to B-Raf and c-Raf dimerization. Furthermore, MAP2K1/MAP2K2 (MEK1/2) inhibition restored sensitivity in all four of the dasatinib-resistant cell lines, and overcome acquired resistance to dasatinib in the RAS-mutant Cal62 cell line, in vivo. Together, these studies demonstrate that acquisition of the c-Src gatekeeper mutation and MAP Kinase pathway signaling play important roles in promoting resistance to the Src inhibitor, dasatinib. | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Thyroid gland cancer [ICD-11: 2D10.0] | [50] | ||||||||||||
| Sensitive Disease | Thyroid gland cancer [ICD-11: 2D10.0] | ||||||||||||
| Sensitive Drug | Dasatinib/Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | BRAF/MEK/MAPK signaling pathway | Inhibition | hsa04010 | ||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| BCPAP cells | Thyroid | Homo sapiens (Human) | CVCL_0153 | ||||||||||
| SW1736 cells | Thyroid | Homo sapiens (Human) | CVCL_3883 | ||||||||||
| C643 cells | Thyroid gland | Homo sapiens (Human) | CVCL_5969 | ||||||||||
| CAL62 cells | Thyroid gland | Homo sapiens (Human) | CVCL_1112 | ||||||||||
| In Vivo Model | Athymic nude mouse PDX xenografts model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting assay; Immunoprecipitation assy | ||||||||||||
| Experiment for Drug Resistance |
SRB staining assay; Promega assay | ||||||||||||
| Mechanism Description | Activation of the Mitogen Activated Protein (MAP) Kinase pathway was increased in all four of the dasatinib-resistant cell lines, likely due to B-Raf and c-Raf dimerization. Furthermore, MAP2K1/MAP2K2 (MEK1/2) inhibition restored sensitivity in all four of the dasatinib-resistant cell lines, and overcome acquired resistance to dasatinib in the RAS-mutant Cal62 cell line, in vivo. Together, these studies demonstrate that acquisition of the c-Src gatekeeper mutation and MAP Kinase pathway signaling play important roles in promoting resistance to the Src inhibitor, dasatinib. | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [80] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | DEL-22379 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | ||||||||||
| In Vivo Model | Female athymic nu/nu mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Caspase-Glo 3/7 luminogenic assay | ||||||||||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [80] | ||||||||||||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | ||||||||||||
| Sensitive Drug | DEL-22379 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | ||||||||||
| In Vivo Model | Female athymic nu/nu mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Caspase-Glo 3/7 luminogenic assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [81] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | DETD-35 | ||||||||||||
| Molecule Alteration | Mutation | . |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A375-R cells | Skin | Homo sapiens (Human) | CVCL_6234 | |||||||||
| Experiment for Drug Resistance |
Cell viability assay | ||||||||||||
| Mechanism Description | Acquired resistance to?vemurafenib?(PLX4032) is a thorny issue in BRAFV600E?mutant?melanoma?therapy. Ferroptotic?programmed cell death?is a potential strategy for combating therapy-resistant cancers. This study uncovers the adaptation and abnormal upregulation of PUFAs and bioactive?oxylipin?metabolism in PLX4032 resistant melanoma cells. Phyto-sesquiterpene lactone, DET, and its derivative, DETD-35, induced?lipid?ROS?accumulation and triggered?ferroptotic cell death?in PLX4032 sensitive (A375) and resistant (A375-R) BRAFV600E?melanoma cells by reprogramming?glutathione?and primary metabolisms, lipid/oxylipin metabolism, and causing mitochondrial damage in which DETD-35 showed superior efficiency to DET. | ||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [82] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | DETD-35 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | MEK/ERK signaling pathway | Inhibition | hsa04011 | ||||||||||
| AKT signaling pathway | Inhibition | hsa04151 | |||||||||||
| STAT3 signaling pathway | Inhibition | hsa04550 | |||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| MeWo cells | Skin | Homo sapiens (Human) | CVCL_0445 | ||||||||||
| A2058 cells | Skin | Homo sapiens (Human) | CVCL_1059 | ||||||||||
| B16-F10 cells | Skin | Mus musculus (Mouse) | CVCL_0159 | ||||||||||
| SkMEL2 cells | Skin | Homo sapiens (Human) | CVCL_0069 | ||||||||||
| BRAF cells | N.A. | Homo sapiens (Human) | N.A. | ||||||||||
| In Vivo Model | NSG mouse PDX model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; Immunohistochemistry analysis | ||||||||||||
| Experiment for Drug Resistance |
MTT assay; Crystal violet staining assay; FACS assay | ||||||||||||
| Mechanism Description | DETD-35 overcame acquired vemurafenib resistance at least in part through deregulating MEK-ERK, Akt, and STAT3 signaling pathways and promoting apoptosis of cancer cells. | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [83] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | EBI-907 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V600E (c.1799T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.55 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
420
|
M
M
D
D
R
R
G
G
S
S
H
H
H
H
H
H
H
H
H
H
430
|
H
H
G
G
S
S
E
E
D
D
R
R
N
N
R
R
M
M
K
K
440
|
T
T
L
L
G
G
R
R
R
R
D
D
S
S
S
S
D
D
D
D
450
|
W
W
E
E
I
I
P
P
D
D
G
G
Q
Q
I
I
T
T
V
V
460
|
G
G
Q
Q
R
R
I
I
G
G
S
S
G
G
S
S
F
F
G
G
470
|
T
T
V
V
Y
Y
K
K
G
G
K
K
W
W
H
H
G
G
D
D
480
|
V
V
A
A
V
V
K
K
M
M
L
L
N
N
V
V
T
T
A
A
490
|
P
P
T
T
P
P
Q
Q
Q
Q
L
L
Q
Q
A
A
F
F
K
K
500
|
N
N
E
E
V
V
G
G
V
V
L
L
R
R
K
K
T
T
R
R
510
|
H
H
V
V
N
N
I
I
L
L
L
L
F
F
M
M
G
G
Y
Y
520
|
S
S
T
T
K
K
P
P
Q
Q
L
L
A
A
I
I
V
V
T
T
530
|
Q
Q
W
W
C
C
E
E
G
G
S
S
S
S
L
L
Y
Y
H
H
540
|
H
H
L
L
H
H
I
I
I
I
E
E
T
T
K
K
F
F
E
E
550
|
M
M
I
I
K
K
L
L
I
I
D
D
I
I
A
A
R
R
Q
Q
560
|
T
T
A
A
Q
Q
G
G
M
M
D
D
Y
Y
L
L
H
H
A
A
570
|
K
K
S
S
I
I
I
I
H
H
R
R
D
D
L
L
K
K
S
S
580
|
N
N
N
N
I
I
F
F
L
L
H
H
E
E
D
D
L
L
T
T
590
|
V
V
K
K
I
I
G
G
D
D
F
F
G
G
L
L
A
A
T
T
600
|
V
E
K
K
S
S
R
R
W
W
S
S
G
G
S
S
H
H
Q
Q
610
|
F
F
E
E
Q
Q
L
L
S
S
G
G
S
S
I
I
L
L
W
W
620
|
M
M
A
A
P
P
E
E
V
V
I
I
R
R
M
M
Q
Q
D
D
630
|
K
K
N
N
P
P
Y
Y
S
S
F
F
Q
Q
S
S
D
D
V
V
640
|
Y
Y
A
A
F
F
G
G
I
I
V
V
L
L
Y
Y
E
E
L
L
650
|
M
M
T
T
G
G
Q
Q
L
L
P
P
Y
Y
S
S
N
N
I
I
660
|
N
N
N
N
R
R
D
D
Q
Q
I
I
I
I
F
F
M
M
V
V
670
|
G
G
R
R
G
G
Y
Y
L
L
S
S
P
P
D
D
L
L
S
S
680
|
K
K
V
V
R
R
S
S
N
N
C
C
P
P
K
K
A
A
M
M
690
|
K
K
R
R
L
L
M
M
A
A
E
E
C
C
L
L
K
K
K
K
700
|
K
K
R
R
D
D
E
E
R
R
P
P
L
L
F
F
P
P
Q
Q
710
|
I
I
L
L
A
A
S
S
I
I
E
E
L
L
L
L
A
A
R
R
720
|
S
S
L
L
P
P
K
K
I
I
H
H
R
R
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| SW-480 cells | Colon | Homo sapiens (Human) | CVCL_0546 | ||||||||||
| Colo-205 cells | Ascites | Homo sapiens (Human) | CVCL_0218 | ||||||||||
| Calu-6 cells | Lung | Homo sapiens (Human) | CVCL_0236 | ||||||||||
| In Vivo Model | Nu/Nu Colo-205 xenograft mouse model; Nu/Nu A357 xenograft mouse model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo luminescent cell viability assay | ||||||||||||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Nervous tissue | |
| The Specified Disease | Brain cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.57E-67; Fold-change: 5.39E-01; Z-score: 1.14E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.65E-01; Fold-change: 5.98E-01; Z-score: 5.07E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
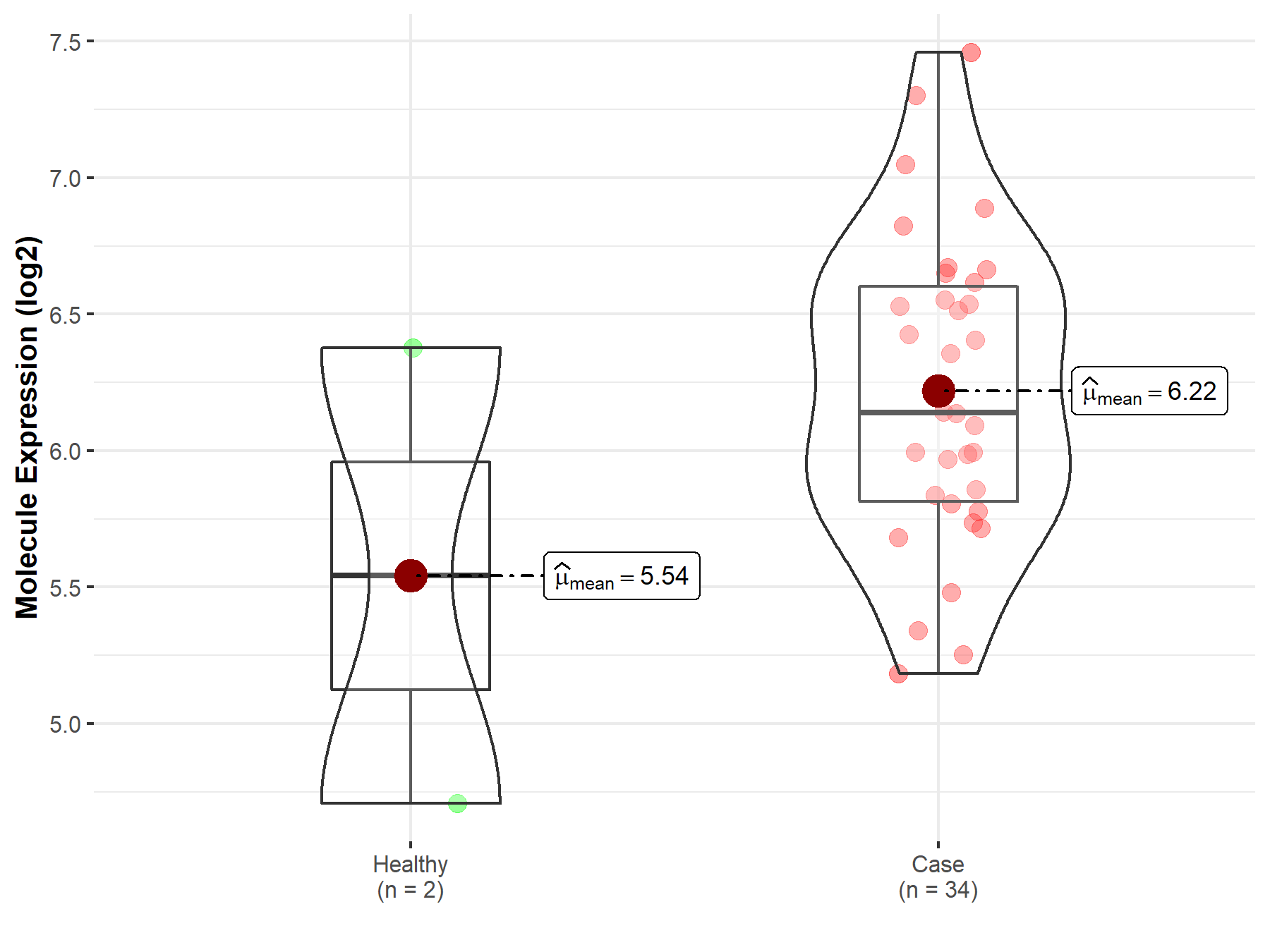
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | White matter | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.08E-03; Fold-change: 4.28E-01; Z-score: 1.30E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
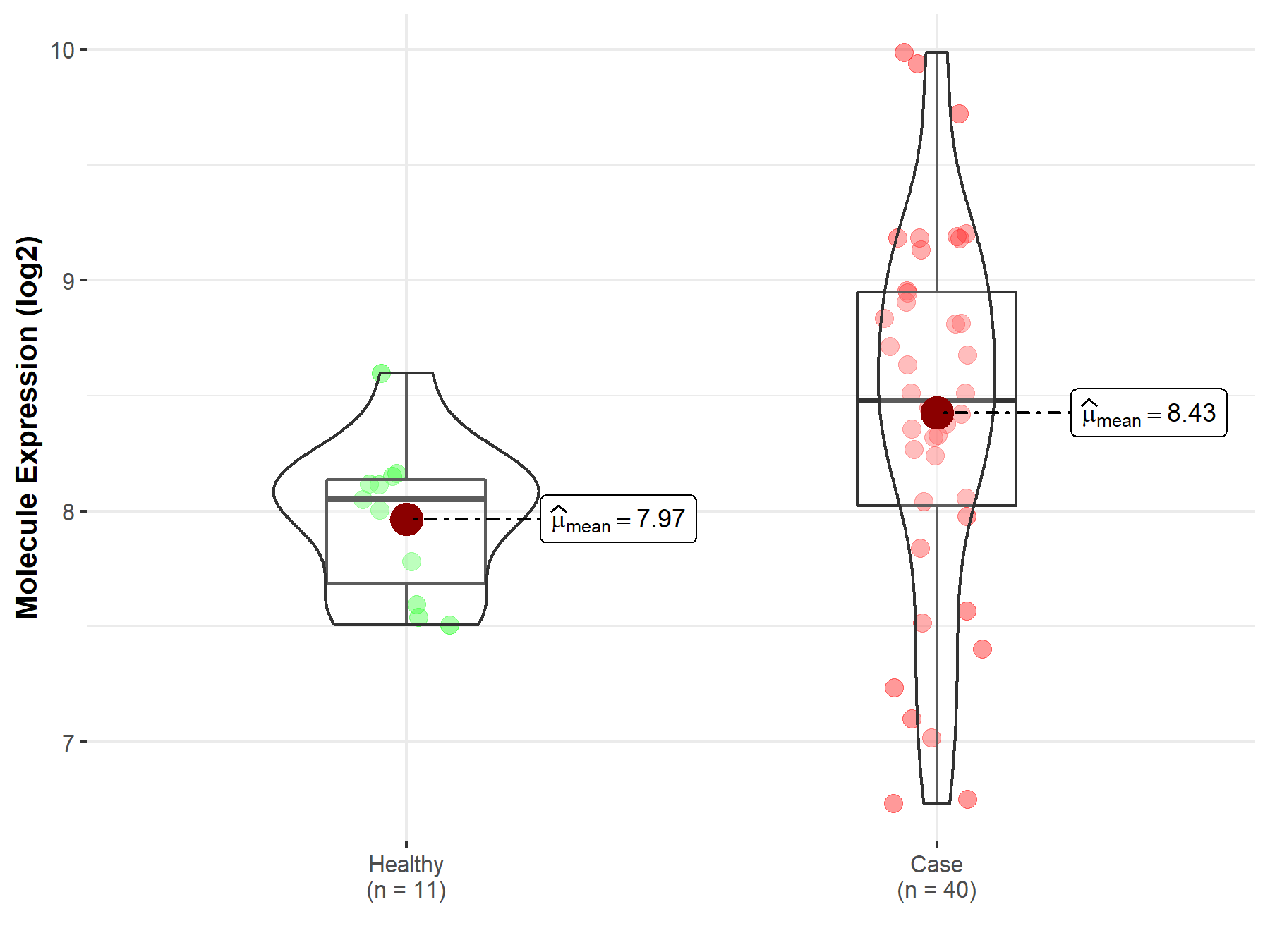
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Neuroectodermal tumor | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.11E-05; Fold-change: 1.65E+00; Z-score: 2.77E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Gastric tissue | |
| The Specified Disease | Gastric cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.50E-01; Fold-change: -1.65E-01; Z-score: -9.71E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 7.01E-02; Fold-change: -2.60E-01; Z-score: -6.67E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
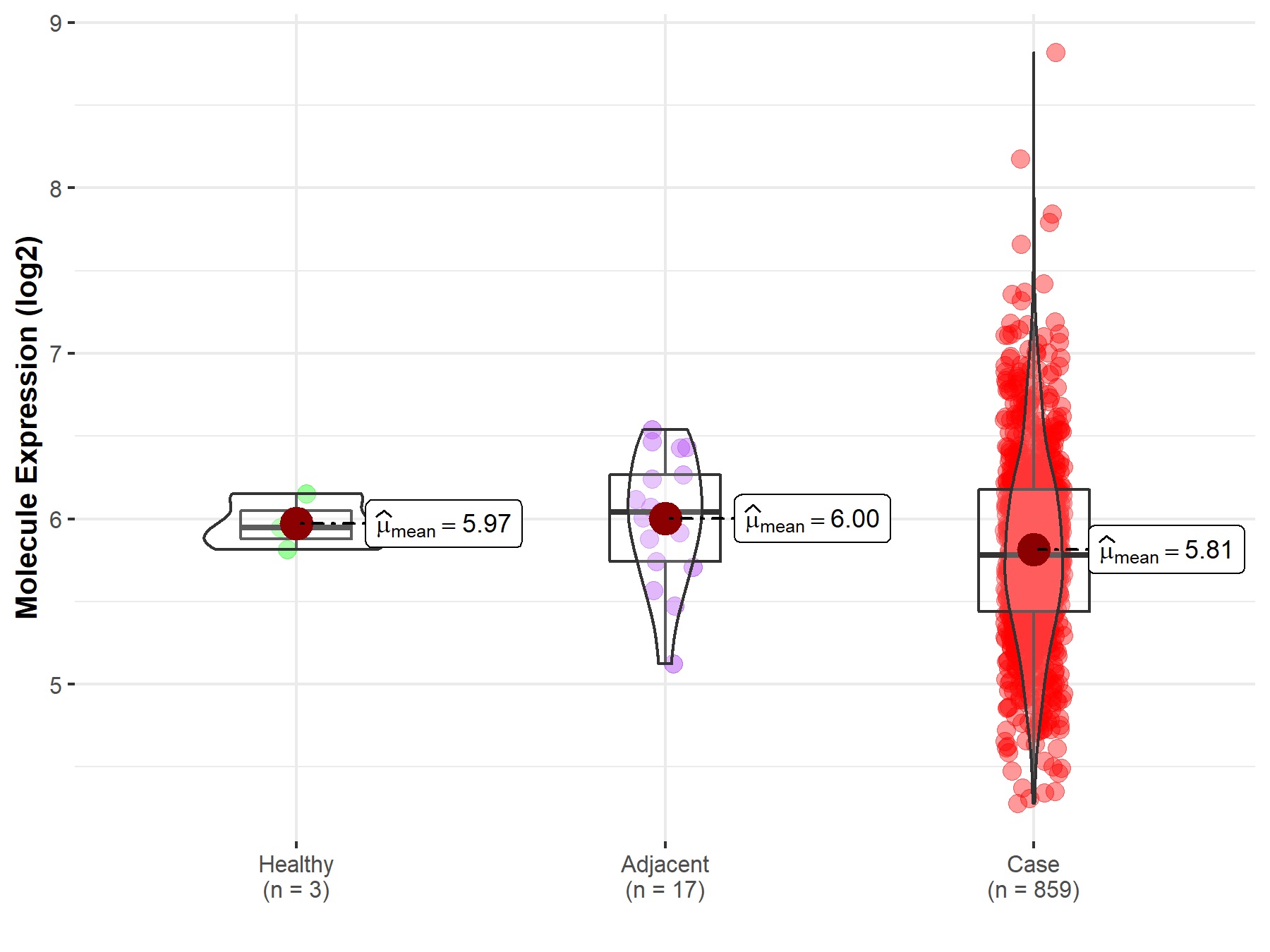
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Colon | |
| The Specified Disease | Colon cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.77E-56; Fold-change: 7.22E-01; Z-score: 1.93E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 9.73E-04; Fold-change: 2.68E-01; Z-score: 4.52E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
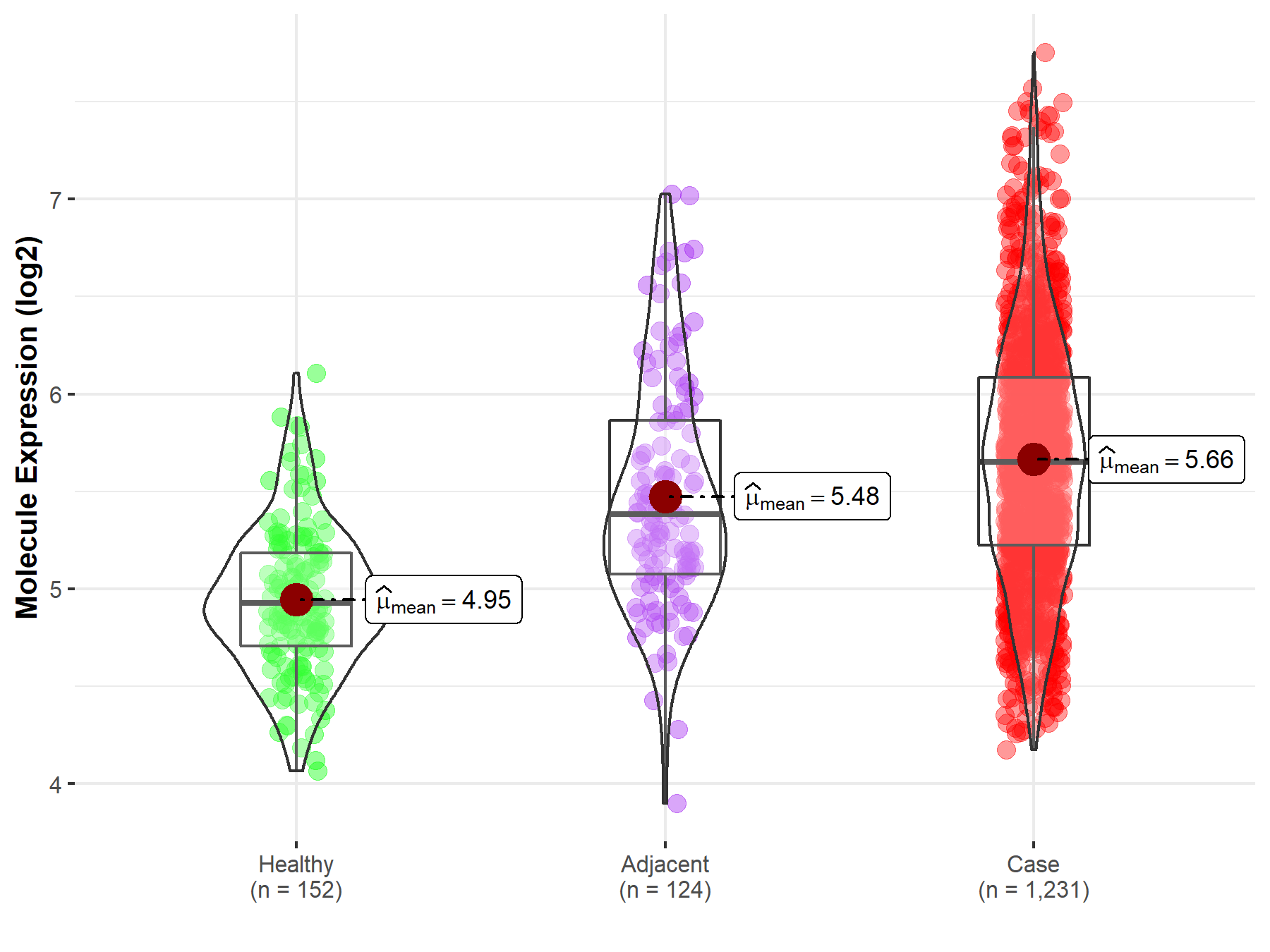
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Pancreas | |
| The Specified Disease | Pancreatic cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.38E-01; Fold-change: 2.78E-01; Z-score: 4.97E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 9.83E-02; Fold-change: -5.28E-02; Z-score: -1.13E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
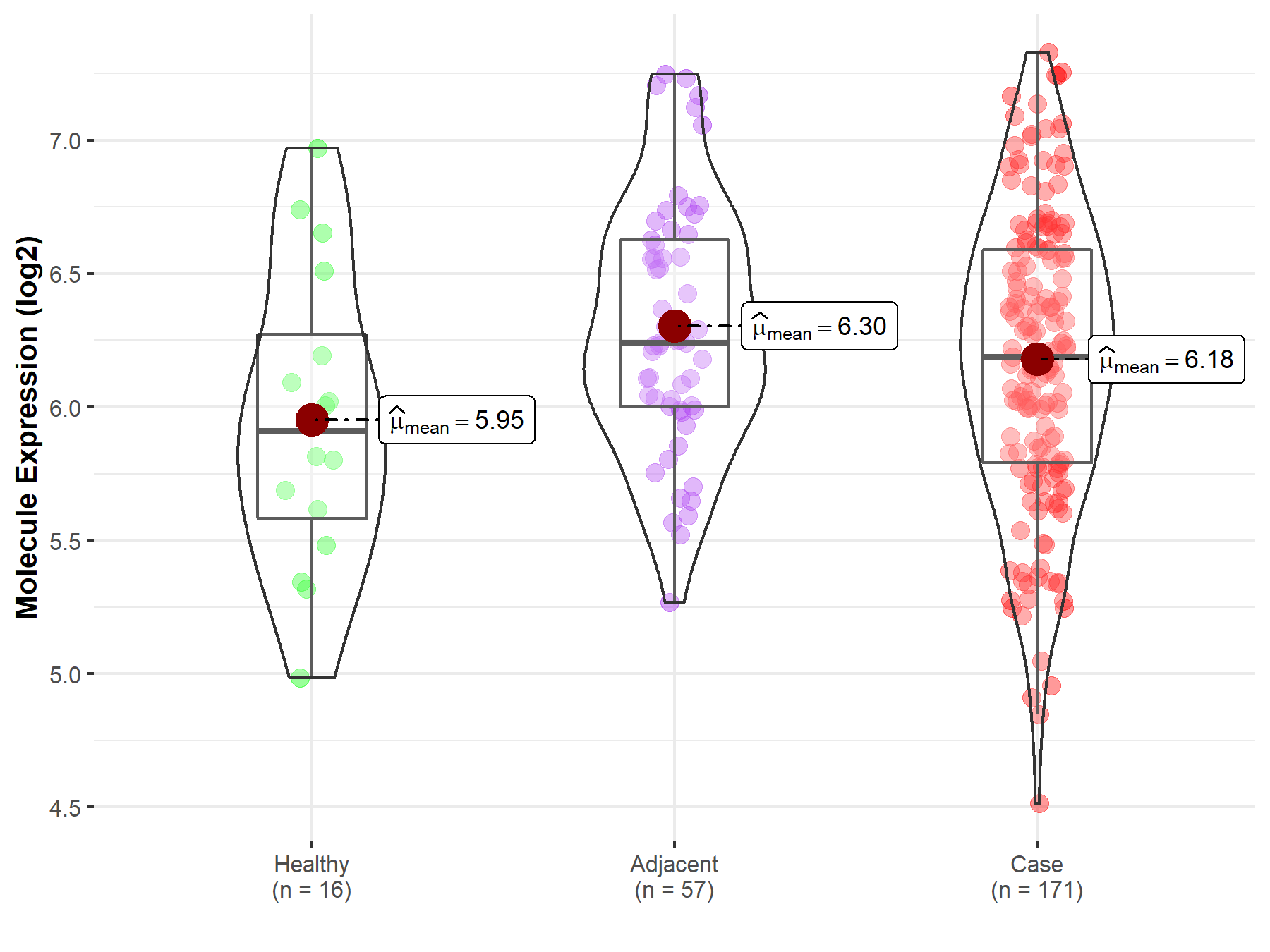
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Liver | |
| The Specified Disease | Liver cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.47E-07; Fold-change: 4.55E-01; Z-score: 8.68E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 3.11E-05; Fold-change: 2.15E-01; Z-score: 3.87E-01 | |
| The Expression Level of Disease Section Compare with the Other Disease Section | p-value: 5.15E-01; Fold-change: -2.35E-03; Z-score: -9.39E-03 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
Molecule expression in tissue other than the diseased tissue of patients
|
||
| Disease-specific Molecule Abundances |
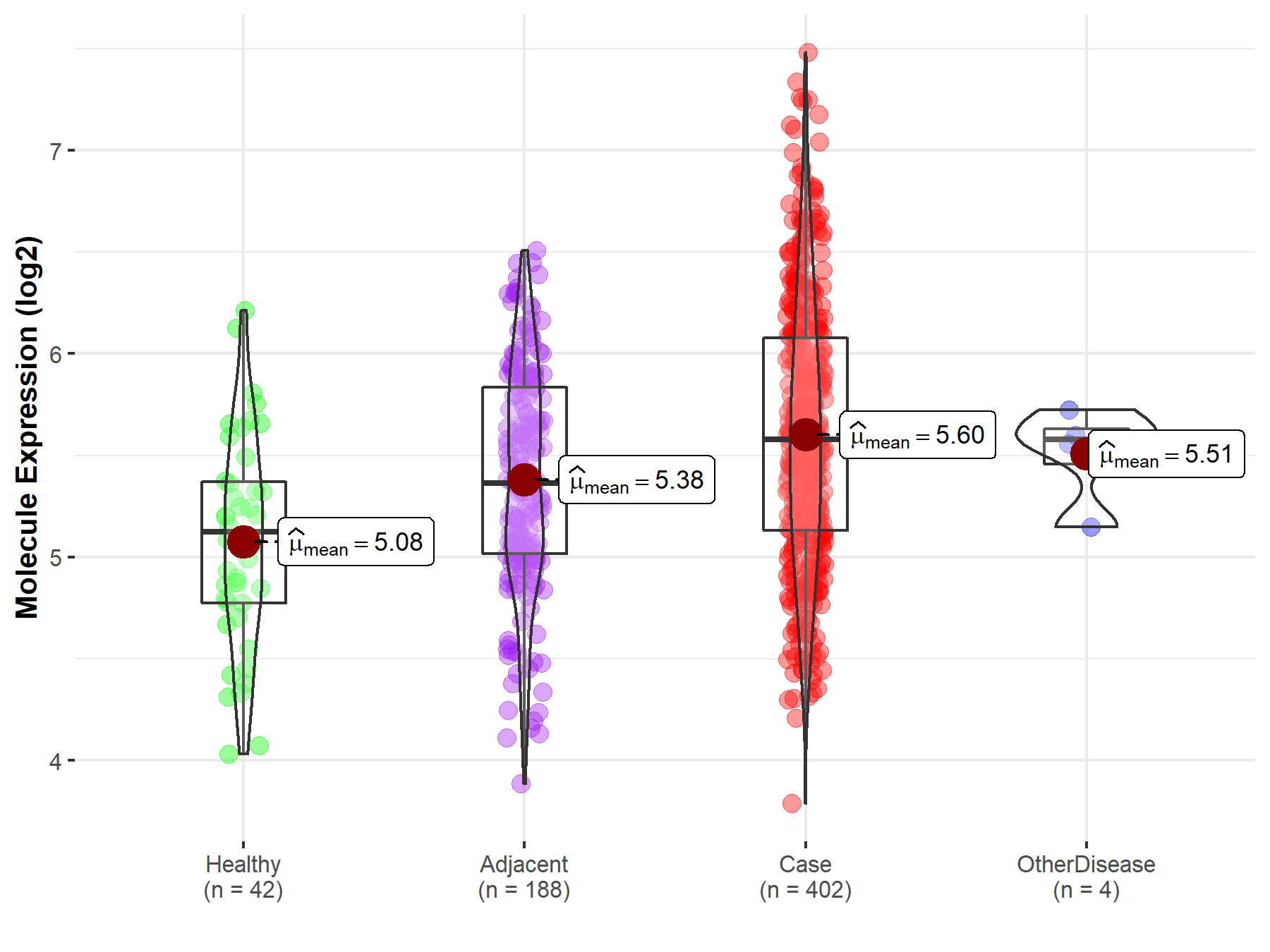
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Lung | |
| The Specified Disease | Lung cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.30E-20; Fold-change: 5.74E-01; Z-score: 9.81E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 3.37E-19; Fold-change: 6.19E-01; Z-score: 1.12E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Skin | |
| The Specified Disease | Melanoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.19E-02; Fold-change: 4.45E-01; Z-score: 7.27E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Ovary | |
| The Specified Disease | Ovarian cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.90E-02; Fold-change: 2.92E-01; Z-score: 5.18E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 6.19E-05; Fold-change: 5.48E-01; Z-score: 1.57E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
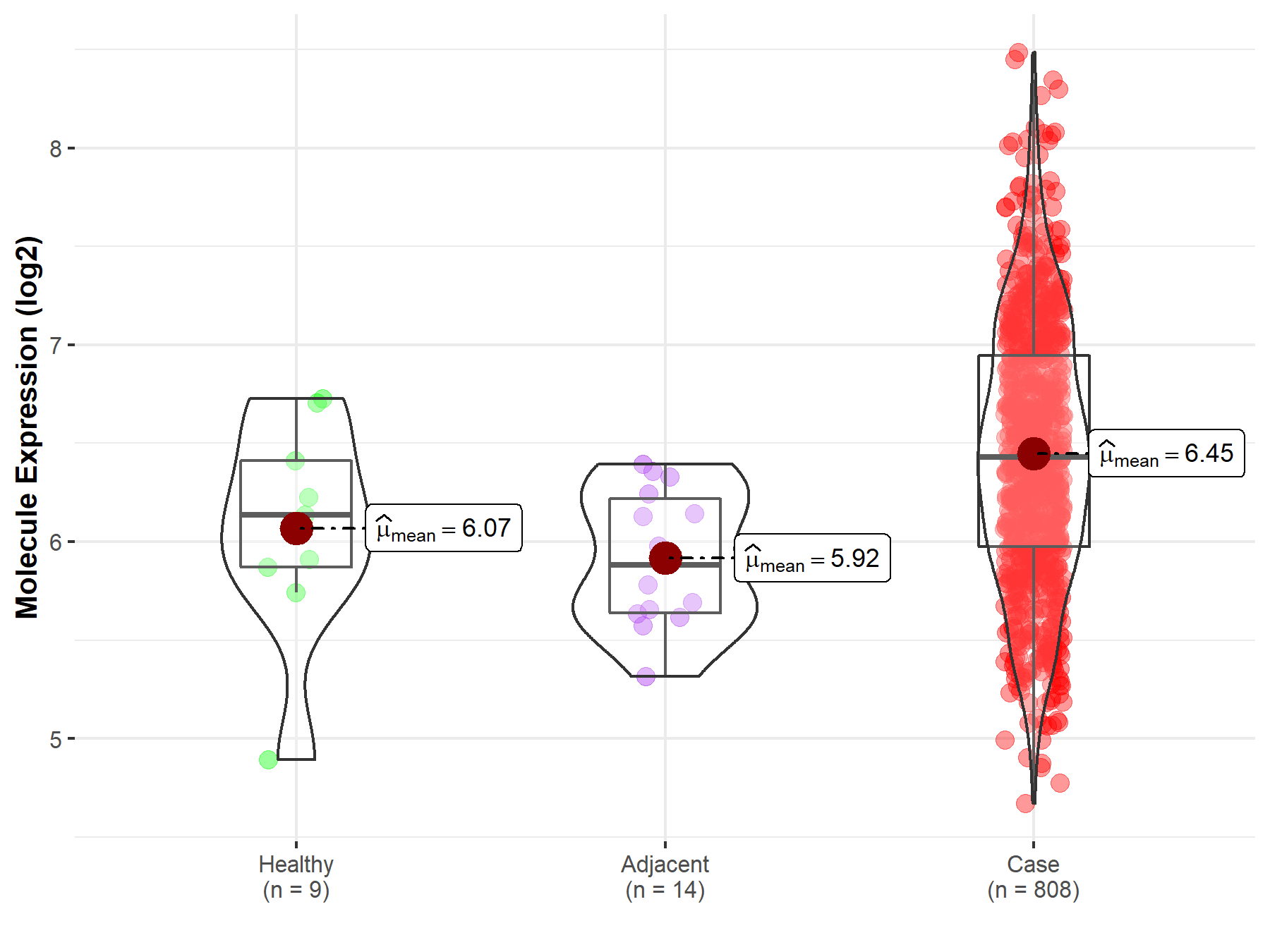
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Bladder tissue | |
| The Specified Disease | Bladder cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.48E-03; Fold-change: -6.98E-01; Z-score: -1.61E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
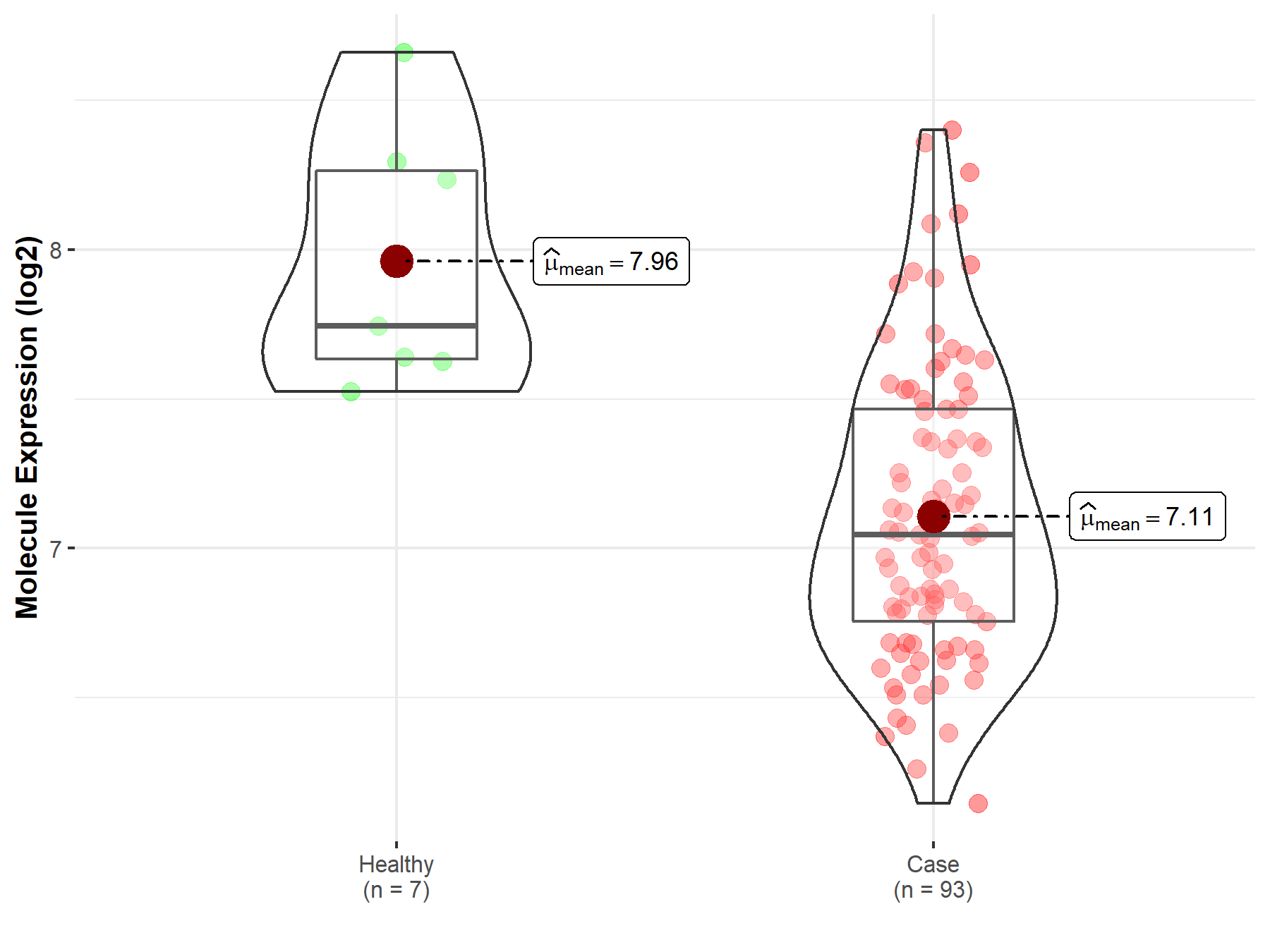
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Thyroid | |
| The Specified Disease | Thyroid cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.00E-01; Fold-change: 9.92E-02; Z-score: 2.05E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 9.41E-01; Fold-change: -2.90E-02; Z-score: -6.20E-02 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Head and neck tissue | |
| The Specified Disease | Head and neck cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.14E-02; Fold-change: -1.35E-01; Z-score: -2.67E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
