Drug Information
Drug (ID: DG00225) and It's Reported Resistant Information
| Name |
Ciprofloxacin XR
|
||||
|---|---|---|---|---|---|
| Synonyms |
Ciprofloxacin; 85721-33-1; Ciprofloxacine; Ciprobay; Ciproxan; Ciprofloxacina; Ciprofloxacinum; Ciprofloxacino; Cipro IV; Ciproxina; Ciprinol; Bernoflox; Ciprodar; Cifloxin; Septicide; Bacquinor; Ciproquinol; Cipromycin; Ciprocinol; Cipro XR; Superocin; Ciprowin; Ciprolon; Ciproflox; Ciprecu; BAY q 3939; Spitacin; Quintor; Quinolid; Proflaxin; Probiox; Ipiflox; Zumaflox; Ciproxine; Ciprolin; Roxytal; Italnik; Fimoflox; Corsacin; Citopcin; Ciprogis; Rancif; Ciriax; Ciplus; Baflox; Loxan; Cilab; Cycin; Cixan; Unex; GW1843; Ciprofloxacin Hydrochloride; Ciprofloxacin intratympanic - Otonomy
Click to Show/Hide
|
||||
| Indication |
In total 4 Indication(s)
|
||||
| Structure |
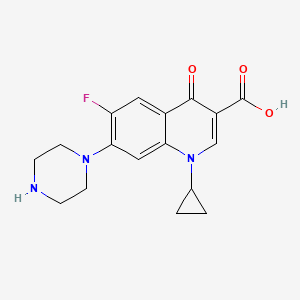
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Clinically Reported Resistance for This Drug
(24 diseases)
[3]
[7]
[8]
[9]
[12]
[17]
[18]
[19]
[20]
[21]
[22]
[23]
[24]
[25]
[11]
[26]
[27]
Disease(s) with Resistance Information Validated by in-vivo Model for This Drug
(3 diseases)
[28]
[29]
[30]
|
||||
| Target | Bacterial DNA gyrase (Bact gyrase) |
GYRA_STAAU
; GYRB_STAAU |
[1] | ||
| Bacterial Penicillin binding protein (Bact PBP) | NOUNIPROTAC | [1] | |||
| Candida Thymidylate synthase (Candi TMP1) | TYSY_CANAL | [2] | |||
| Staphylococcus Topoisomerase IV (Stap-coc parC) | PARC_STAAS | [2] | |||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C17H18FN3O3
|
||||
| IsoSMILES |
C1CC1N2C=C(C(=O)C3=CC(=C(C=C32)N4CCNCC4)F)C(=O)O
|
||||
| InChI |
1S/C17H18FN3O3/c18-13-7-11-14(8-15(13)20-5-3-19-4-6-20)21(10-1-2-10)9-12(16(11)22)17(23)24/h7-10,19H,1-6H2,(H,23,24)
|
||||
| InChIKey |
MYSWGUAQZAJSOK-UHFFFAOYSA-N
|
||||
| PubChem CID | |||||
| ChEBI ID | |||||
| TTD Drug ID | |||||
| VARIDT ID | |||||
| INTEDE ID | |||||
| DrugBank ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-01: Infectious/parasitic diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Erythromycin esterase (EREA2) | [8] | |||
| Resistant Disease | Vibrio cholerae infection [ICD-11: 1A00.0] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Vibrio cholerae PG153/1 | 666 | ||
| Experiment for Molecule Alteration |
PCR and DNA sequencing assay | |||
| Experiment for Drug Resistance |
Commercial antimicrobial discs assay | |||
| Mechanism Description | The expression of dfrA5, ereA2 lead to drug resistance. | |||
|
|
||||
| Key Molecule: Dihydrofolate reductase (DHFR) | [8] | |||
| Resistant Disease | Vibrio cholerae infection [ICD-11: 1A00.0] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Vibrio cholerae O62 strain AS438 | 666 | ||
| Vibrio cholerae PG224 | 666 | |||
| Vibrio cholerae PL1 | 666 | |||
| Vibrio cholerae PL61 | 666 | |||
| Vibrio cholerae PL78/6 | 666 | |||
| Vibrio cholerae PL141 | 666 | |||
| Experiment for Molecule Alteration |
PCR and DNA sequencing assay | |||
| Experiment for Drug Resistance |
Commercial antimicrobial discs assay | |||
| Mechanism Description | The expression of dfrA1 lead to drug resistance. | |||
| Key Molecule: Dihydrofolate reductase (DHFR) | [8] | |||
| Resistant Disease | Vibrio cholerae infection [ICD-11: 1A00.0] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Vibrio cholerae PG153/1 | 666 | ||
| Vibrio cholerae PG170 | 666 | |||
| Experiment for Molecule Alteration |
PCR and DNA sequencing assay | |||
| Experiment for Drug Resistance |
Commercial antimicrobial discs assay | |||
| Mechanism Description | The expression of dfrA15 lead to drug resistance. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: DNA gyrase subunit A (GYRA) | [31], [32] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.T83I |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Pseudomonas aeruginosa isolates | 287 | ||
| Pseudomonas aeruginosa ATCC10145 | 287 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Etest assay | |||
| Mechanism Description | The major mechanism of the resistance of this Pseudomonas aeruginosa to fluoroquinolones is the modification of type II topoisomerases (DNA gyrase and topoisomerase IV). | |||
| Key Molecule: DNA gyrase subunit A (GYRA) | [31], [32] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.H83R |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Pseudomonas aeruginosa isolates | 287 | ||
| Pseudomonas aeruginosa ATCC10145 | 287 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Etest assay | |||
| Mechanism Description | The major mechanism of the resistance of this Pseudomonas aeruginosa to fluoroquinolones is the modification of type II topoisomerases (DNA gyrase and topoisomerase IV). | |||
| Key Molecule: DNA gyrase subunit A (GYRA) | [33] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.S83L; p.S80L |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli ATCC 25922 | 1322345 | ||
| Pseudomonas aeruginosa ATCC 27853 | 287 | |||
| Experiment for Molecule Alteration |
ERIC-PCR | |||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | Mutations that occur in gyrA and parC genes were detected by DNA sequence analysis in 16 resistant strains representing each clone and subtype. | |||
| Key Molecule: DNA topoisomerase 4 subunit A (PARC) | [33] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.S83L; p.S80L |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli ATCC 25922 | 1322345 | ||
| Pseudomonas aeruginosa ATCC 27853 | 287 | |||
| Experiment for Molecule Alteration |
ERIC-PCR | |||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | Mutations that occur in gyrA and parC genes were detected by DNA sequence analysis in 16 resistant strains representing each clone and subtype. | |||
| Key Molecule: DNA topoisomerase 4 subunit B (PARE) | [34] | |||
| Resistant Disease | Morganella morganii infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.S463A |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Morganella morganii isolate | 582 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | The mutations in DNA gyrase (gyrA and gyrB) and topoisomerase IV (parC,parE) genes result in quinolone susceptibility. | |||
| Key Molecule: DNA topoisomerase 4 subunit B (PARE) | [34] | |||
| Resistant Disease | Morganella morganii infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.S464Y |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Morganella morganii isolate | 582 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | The mutations in DNA gyrase (gyrA and gyrB) and topoisomerase IV (parC,parE) genes result in quinolone susceptibility. | |||
| Key Molecule: DNA topoisomerase 4 subunit A (PARC) | [34] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.S80I |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Morganella morganii isolate | 582 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | The mutations in DNA gyrase (gyrA and gyrB) and topoisomerase IV (parC,parE) genes result in quinolone susceptibility. | |||
| Key Molecule: DNA gyrase subunit A (GYRA) | [4], [5], [6] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.S83L |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli strain kL16 | 1425342 | ||
| Escherichia coli strain N-112 | 562 | |||
| Escherichia coli strain N-118 | 562 | |||
| Escherichia coli strain N-119 | 562 | |||
| Escherichia coli strain N-51 | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Mechanism Description | Quinolones are considered to exert antibacterial activity by inhibiting DNA gyrase (EC 5.99.1.3), which catalyzes topological changes of DNA.DNA gyrase of Escherichia coli consists of subunits A and B, which are the products of the gyrA and gyrB genes, respectively. Mutations in either gene can cause quinolone resistance. | |||
| Key Molecule: DNA gyrase subunit A (GYRA) | [4], [5], [6] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.S83W |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli strain kL16 | 1425342 | ||
| Escherichia coli strain P-18 | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Mechanism Description | Quinolones are considered to exert antibacterial activity by inhibiting DNA gyrase (EC 5.99.1.3), which catalyzes topological changes of DNA.DNA gyrase of Escherichia coli consists of subunits A and B, which are the products of the gyrA and gyrB genes, respectively. Mutations in either gene can cause quinolone resistance. | |||
| Key Molecule: DNA gyrase subunit A (GYRA) | [4], [5], [6] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.D87N |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli strain kL16 | 1425342 | ||
| Escherichia coli strain N-113 | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Mechanism Description | Quinolones are considered to exert antibacterial activity by inhibiting DNA gyrase (EC 5.99.1.3), which catalyzes topological changes of DNA.DNA gyrase of Escherichia coli consists of subunits A and B, which are the products of the gyrA and gyrB genes, respectively. Mutations in either gene can cause quinolone resistance. | |||
| Key Molecule: DNA gyrase subunit A (GYRA) | [4], [5], [6] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.G81C |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli strain kL16 | 1425342 | ||
| Escherichia coli strain N-97 | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Mechanism Description | Quinolones are considered to exert antibacterial activity by inhibiting DNA gyrase (EC 5.99.1.3), which catalyzes topological changes of DNA.DNA gyrase of Escherichia coli consists of subunits A and B, which are the products of the gyrA and gyrB genes, respectively. Mutations in either gene can cause quinolone resistance. | |||
| Key Molecule: DNA gyrase subunit A (GYRA) | [4], [5], [6] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.A84P |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli strain kL16 | 1425342 | ||
| Escherichia coli strain P-5 | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Mechanism Description | Quinolones are considered to exert antibacterial activity by inhibiting DNA gyrase (EC 5.99.1.3), which catalyzes topological changes of DNA.DNA gyrase of Escherichia coli consists of subunits A and B, which are the products of the gyrA and gyrB genes, respectively. Mutations in either gene can cause quinolone resistance. | |||
| Key Molecule: DNA gyrase subunit A (GYRA) | [4], [5], [6] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.A67S |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli strain kL16 | 1425342 | ||
| Escherichia coli strain P-10 | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Mechanism Description | Quinolones are considered to exert antibacterial activity by inhibiting DNA gyrase (EC 5.99.1.3), which catalyzes topological changes of DNA.DNA gyrase of Escherichia coli consists of subunits A and B, which are the products of the gyrA and gyrB genes, respectively. Mutations in either gene can cause quinolone resistance. | |||
| Key Molecule: DNA gyrase subunit A (GYRA) | [4], [5], [6] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.Q106H |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli strain kL16 | 1425342 | ||
| Escherichia coli strain N-89 | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Mechanism Description | Quinolones are considered to exert antibacterial activity by inhibiting DNA gyrase (EC 5.99.1.3), which catalyzes topological changes of DNA.DNA gyrase of Escherichia coli consists of subunits A and B, which are the products of the gyrA and gyrB genes, respectively. Mutations in either gene can cause quinolone resistance. | |||
|
|
||||
| Key Molecule: Quinolone efflux pump (QEPA2) | [35] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.A99G+p.V134I |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli TOP10 | 83333 | ||
| Experiment for Molecule Alteration |
PCR amplification and sequence alignments assay | |||
| Experiment for Drug Resistance |
Disk diffusion assay | |||
| Mechanism Description | QepA confers decreased susceptibility to hydrophilic fluoroquinolones (e.g., norfloxacin, ciprofloxacin, and enrofloxacin) with a 32- to 64-fold increase of MICs. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: DNA topoisomerase 4 subunit B (PARE) | [30] | |||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | |||
| Molecule Alteration | Missense mutation | p.D476N |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli ECIS803 | 562 | ||
| Escherichia coli ATCC 43869 | 562 | |||
| Experiment for Molecule Alteration |
PCR; DNA sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Mutational substitutions in the quinolone target enzymes, namely DNA topoisomerase II (GyrA) and topoisomerase IV (ParC), are recognised to be the major mechanisms through which resistance develops. | |||
|
|
||||
| Key Molecule: Beta-lactamase (BLA) | [9] | |||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | |||
| Molecule Alteration | Expression | Acquired |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli JM109 | 562 | ||
| Experiment for Molecule Alteration |
PCR and molecular characterization assay | |||
| Experiment for Drug Resistance |
Disk diffusion method assay | |||
| Mechanism Description | CTX-M-55 is a novel ceftazidime-resistant CTX-M extended-spectrum Beta-lactamase, which reduced susceptibility. | |||
|
|
||||
| Key Molecule: DNA topoisomerase 4 subunit A (PARC) | [30] | |||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | |||
| Molecule Alteration | Missense mutation | p.S80l |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli ECIS803 | 562 | ||
| Escherichia coli ATCC 43869 | 562 | |||
| Experiment for Molecule Alteration |
PCR; DNA sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Mutational substitutions in the quinolone target enzymes, namely DNA topoisomerase II (GyrA) and topoisomerase IV (ParC), are recognised to be the major mechanisms through which resistance develops. | |||
| Key Molecule: DNA topoisomerase 4 subunit A (PARC) | [30] | |||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | |||
| Molecule Alteration | Missense mutation | p.E84G |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli ECIS803 | 562 | ||
| Escherichia coli ATCC 43869 | 562 | |||
| Experiment for Molecule Alteration |
PCR; DNA sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Mutational substitutions in the quinolone target enzymes, namely DNA topoisomerase II (GyrA) and topoisomerase IV (ParC), are recognised to be the major mechanisms through which resistance develops. | |||
|
|
||||
| Key Molecule: Quinolone resistance protein NorA (NORA) | [36] | |||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | |||
| Molecule Alteration | Expression | Acquired |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli HB101 | 634468 | ||
| Staphylococcus aureus strain SA113 | 1280 | |||
| Experiment for Molecule Alteration |
Dideoxy chain-termination method assay | |||
| Mechanism Description | The norA gene cloned from chromosomal DNA of quinolone-resistant Staphylococcus aureus Tk2566 conferred relatively high resistance to hydrophilic quinolones such as norfloxacin, enoxacin, ofloxacin, and ciprofloxacin, but only low or no resistance at all to hydrophobic ones such as nalidixic acid, oxolinic acid, and sparfloxacin in S. aureus and Escherichia coli. Escherichia coli strains containing one of the plasmids carrying the norA gene (pTUS1, pTUS180, pTUS829, and pTUS206) were 8 to 64 times more resistant to the hydrophilic quinolones than the parent quinolone-susceptible strain. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: DNA gyrase subunit A (GYRA) | [29] | |||
| Resistant Disease | Clostridium difficile infection [ICD-11: 1A04.0] | |||
| Molecule Alteration | Mutation | p.T82I |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| Mechanism Description | Mutations in the gyrA or gyrB gene within quinolone resistance-determining region lead to the reduction in fidelity or prevention of drug binding via the target conformation change. Although several amino acid substitutions have been noted in GyrA and/or GyrB, the most frequent amino acid change has been recognized at T82I in GyrA subunit. | |||
|
|
||||
| Key Molecule: Multidrug export protein MepA (cdeA) | [29] | |||
| Resistant Disease | Clostridium difficile infection [ICD-11: 1A04.0] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| Mechanism Description | In C. difficile, two secondary active transporters belonging to the MFS and MATE families have been reported to be associated with drug resistance. Heterologous expression of the clostridial Cme protein in the MFS subfamily promotes ERY resistance in Enterococcus faecalis. A sodium-dependent efflux pump of the MATE subfamily encoded by the cdeA gene of C. difficile attributes resistance to norfloxacin and ciprofloxacin when the gene was overexpressed in Escherichia coli. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: DNA gyrase subunit A (GYRA) | [11] | |||
| Resistant Disease | Typhoid fever [ICD-11: 1A07.0] | |||
| Molecule Alteration | Missense mutation | p.S83F |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Salmonella enterica subsp. enterica serovar Typhi isolates | 90370 | ||
| Experiment for Molecule Alteration |
PCR-RFLP | |||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | The targets of fluoroquinolones are the two enzymes, DNA gyrase and topoisomerase IV, whose subunits are encoded respectively by gyrA and gyrB and the parC and parE genes.The alteration caused by single point mutations within the QRDR of the DNA gyrase subunit gyrA gene leads to quinolone resistance. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: DNA gyrase subunit A (GYRA) | [10], [11] | |||
| Resistant Disease | Gastroenteritis [ICD-11: 1A40.0] | |||
| Molecule Alteration | Missense mutation | p.S97P |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Salmonella enteritidis isolates | 149539 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Etest assay | |||
| Mechanism Description | Quinolones target the bacterial DNA gyrase; this enzyme is a type II topoisomerase that is essential for bacterial DNA replication.This enzyme consists of 2A and 2B subunits encoded by gyrA and gyrB genes, respectively. | |||
| Key Molecule: DNA gyrase subunit A (GYRA) | [10], [11] | |||
| Resistant Disease | Gastroenteritis [ICD-11: 1A40.0] | |||
| Molecule Alteration | Missense mutation | p.S83F |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Salmonella enteritidis isolates | 149539 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Etest assay | |||
| Mechanism Description | Quinolones target the bacterial DNA gyrase; this enzyme is a type II topoisomerase that is essential for bacterial DNA replication.This enzyme consists of 2A and 2B subunits encoded by gyrA and gyrB genes, respectively. | |||
| Key Molecule: DNA gyrase subunit A (GYRA) | [10], [11] | |||
| Resistant Disease | Gastroenteritis [ICD-11: 1A40.0] | |||
| Molecule Alteration | Missense mutation | p.D87Y |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Salmonella enteritidis isolates | 149539 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Etest assay | |||
| Mechanism Description | Quinolones target the bacterial DNA gyrase; this enzyme is a type II topoisomerase that is essential for bacterial DNA replication.This enzyme consists of 2A and 2B subunits encoded by gyrA and gyrB genes, respectively. | |||
| Key Molecule: DNA gyrase subunit A (GYRA) | [10], [11] | |||
| Resistant Disease | Gastroenteritis [ICD-11: 1A40.0] | |||
| Molecule Alteration | Missense mutation | p.D87N |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Salmonella enteritidis isolates | 149539 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Etest assay | |||
| Mechanism Description | Quinolones target the bacterial DNA gyrase; this enzyme is a type II topoisomerase that is essential for bacterial DNA replication.This enzyme consists of 2A and 2B subunits encoded by gyrA and gyrB genes, respectively. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: DNA topoisomerase 4 subunit A (PARC) | [13], [14] | |||
| Resistant Disease | Gonococcal infection [ICD-11: 1A70.0] | |||
| Molecule Alteration | Missense mutation | p.E91G |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Neisseria gonorrhoeae isolates | 485 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Etest assay | |||
| Mechanism Description | Fluoroquinolones block DNA replication by inhibiting the enzymes DNA gyrase (topoisomerase II) and topoisomerase IV. DNA gyrase catalyzes the untwisting of DNA molecules during DNA replication, and consists of two type A subunits and two type B subunits encoded by gyrA and gyrB genes. Topoisomerase IV consists of two type C subunits and two type E subunits encoded by parC and parE genes.GyrA S91F, D95G/D95A and ParC E91G amino acid substitutions mediate high fluoroquinolone resistance in the analyzed kenyan GC. | |||
| Key Molecule: DNA gyrase subunit A (GYRA) | [13], [14] | |||
| Resistant Disease | Gonococcal infection [ICD-11: 1A70.0] | |||
| Molecule Alteration | Missense mutation | p.S91F+p.D95G/D95A |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Neisseria gonorrhoeae isolates | 485 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Etest assay | |||
| Mechanism Description | Fluoroquinolones block DNA replication by inhibiting the enzymes DNA gyrase (topoisomerase II) and topoisomerase IV. DNA gyrase catalyzes the untwisting of DNA molecules during DNA replication, and consists of two type A subunits and two type B subunits encoded by gyrA and gyrB genes. Topoisomerase IV consists of two type C subunits and two type E subunits encoded by parC and parE genes.GyrA S91F, D95G/D95A and ParC E91G amino acid substitutions mediate high fluoroquinolone resistance in the analyzed kenyan GC. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: DNA gyrase subunit A (GYRA) | [1], [2] | |||
| Resistant Disease | Ureaplasma parvum infection [ICD-11: 1A81.1] | |||
| Molecule Alteration | Missense mutation | p.S83L |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli k-12 JM109 | 83333 | ||
| Ureaplasma parvum serovar 3 isolates | 38504 | |||
| Ureaplasma parvum serovar 6 isolates | 95660 | |||
| Ureaplasma urealyticum isolates | 2130 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Quinolones are used for treating urogenital infections and interact in bacteria with the type II topoisomerases DNA gyrase and topoisomerase IV, both of which are composed of two A and two B subunits; these subunits are encoded by the gyrA and gyrB genes for DNA gyrase and parC and parE genes for topoisomerase IV.Out of 28 clinical Ureaplasma strains, we isolated 9 with high MICs of quinolones and found a single parC gene mutation, resulting in the change S83L. | |||
| Key Molecule: DNA gyrase subunit A (GYRA) | [1], [2] | |||
| Resistant Disease | Ureaplasma urealyticum infection [ICD-11: 1A81.2] | |||
| Molecule Alteration | Missense mutation | p.S83L |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli k-12 JM109 | 83333 | ||
| Ureaplasma parvum serovar 3 isolates | 38504 | |||
| Ureaplasma parvum serovar 6 isolates | 95660 | |||
| Ureaplasma urealyticum isolates | 2130 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Quinolones are used for treating urogenital infections and interact in bacteria with the type II topoisomerases DNA gyrase and topoisomerase IV, both of which are composed of two A and two B subunits; these subunits are encoded by the gyrA and gyrB genes for DNA gyrase and parC and parE genes for topoisomerase IV.Out of 28 clinical Ureaplasma strains, we isolated 9 with high MICs of quinolones and found a single parC gene mutation, resulting in the change S83L. | |||
| Key Molecule: DNA topoisomerase 4 subunit A (PARC) | [1], [2] | |||
| Resistant Disease | Nongonococcal urethritis [ICD-11: 1A81.3] | |||
| Molecule Alteration | Missense mutation | p.S83W |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli k-12 JM109 | 83333 | ||
| Ureaplasma parvum serovar 3 isolates | 38504 | |||
| Ureaplasma parvum serovar 6 isolates | 95660 | |||
| Ureaplasma urealyticum isolates | 2130 | |||
| Ureaplasma parvum isolates | 38504 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Quinolones are used for treating urogenital infections and interact in bacteria with the type II topoisomerases DNA gyrase and topoisomerase IV, both of which are composed of two A and two B subunits; these subunits are encoded by the gyrA and gyrB genes for DNA gyrase and parC and parE genes for topoisomerase IV.Out of 28 clinical Ureaplasma strains, we isolated 9 with high MICs of quinolones and found a single parC gene mutation, resulting in the change S83L. | |||
| Key Molecule: DNA topoisomerase 4 subunit A (PARC) | [1], [2] | |||
| Resistant Disease | Nongonococcal urethritis [ICD-11: 1A81.3] | |||
| Molecule Alteration | Missense mutation | p.S83P |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli k-12 JM109 | 83333 | ||
| Ureaplasma parvum serovar 3 isolates | 38504 | |||
| Ureaplasma parvum serovar 6 isolates | 95660 | |||
| Ureaplasma urealyticum isolates | 2130 | |||
| Ureaplasma parvum isolates | 38504 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Quinolones are used for treating urogenital infections and interact in bacteria with the type II topoisomerases DNA gyrase and topoisomerase IV, both of which are composed of two A and two B subunits; these subunits are encoded by the gyrA and gyrB genes for DNA gyrase and parC and parE genes for topoisomerase IV.Out of 28 clinical Ureaplasma strains, we isolated 9 with high MICs of quinolones and found a single parC gene mutation, resulting in the change S83L. | |||
| Key Molecule: DNA topoisomerase 4 subunit A (PARC) | [1], [2] | |||
| Resistant Disease | Nongonococcal urethritis [ICD-11: 1A81.3] | |||
| Molecule Alteration | Missense mutation | ParC p.S83L+GyrB p.P462S |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli k-12 JM109 | 83333 | ||
| Ureaplasma parvum serovar 3 isolates | 38504 | |||
| Ureaplasma parvum serovar 6 isolates | 95660 | |||
| Ureaplasma urealyticum isolates | 2130 | |||
| Ureaplasma parvum isolates | 38504 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Quinolones are used for treating urogenital infections and interact in bacteria with the type II topoisomerases DNA gyrase and topoisomerase IV, both of which are composed of two A and two B subunits; these subunits are encoded by the gyrA and gyrB genes for DNA gyrase and parC and parE genes for topoisomerase IV.Out of 28 clinical Ureaplasma strains, we isolated 9 with high MICs of quinolones and found a single parC gene mutation, resulting in the change S83L. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: DNA topoisomerase 4 subunit B (PARE) | [17] | |||
| Resistant Disease | Leprosy [ICD-11: 1B20.0] | |||
| Molecule Alteration | Missense mutation | p.D464N |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli BL21 (DE3) | 469008 | ||
| Escherichia coli Rosetta-gami 2 | 562 | |||
| Escherichia coli TOP-10 | 83333 | |||
| Mycobacterium leprae Thai-53 | 1769 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
DNA supercoiling assay; DNA cleavage assay | |||
| Mechanism Description | FQs are known to interact with both A and B subunits of DNA gyrase and inhibit supercoiling activity of this enzyme.The FQ-inhibited supercoiling assay and FQ-induced cleavage assay demonstrated the important roles of these amino acid substitutions in reduced sensitivity to FQ with marked influence by amino acid substitution, especially at position 502. | |||
| Key Molecule: DNA topoisomerase 4 subunit B (PARE) | [17] | |||
| Resistant Disease | Leprosy [ICD-11: 1B20.0] | |||
| Molecule Alteration | Missense mutation | p.N502D |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli BL21 (DE3) | 469008 | ||
| Escherichia coli Rosetta-gami 2 | 562 | |||
| Escherichia coli TOP-10 | 83333 | |||
| Mycobacterium leprae Thai-53 | 1769 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
DNA supercoiling assay; DNA cleavage assay | |||
| Mechanism Description | FQs are known to interact with both A and B subunits of DNA gyrase and inhibit supercoiling activity of this enzyme.The FQ-inhibited supercoiling assay and FQ-induced cleavage assay demonstrated the important roles of these amino acid substitutions in reduced sensitivity to FQ with marked influence by amino acid substitution, especially at position 502. | |||
| Key Molecule: DNA topoisomerase 4 subunit B (PARE) | [17] | |||
| Resistant Disease | Leprosy [ICD-11: 1B20.0] | |||
| Molecule Alteration | Missense mutation | p.E504V |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli BL21 (DE3) | 469008 | ||
| Escherichia coli Rosetta-gami 2 | 562 | |||
| Escherichia coli TOP-10 | 83333 | |||
| Mycobacterium leprae Thai-53 | 1769 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
DNA supercoiling assay; DNA cleavage assay | |||
| Mechanism Description | FQs are known to interact with both A and B subunits of DNA gyrase and inhibit supercoiling activity of this enzyme.The FQ-inhibited supercoiling assay and FQ-induced cleavage assay demonstrated the important roles of these amino acid substitutions in reduced sensitivity to FQ with marked influence by amino acid substitution, especially at position 502. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: OXA-23 carbapenemase (BLA OXA-23) | [19] | |||
| Resistant Disease | Cutaneous bacterial infection [ICD-11: 1B21.4] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Acinetobacter baumannii isolates | 470 | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Broth microdilution method assay; Agar dilution method assay | |||
| Mechanism Description | The isolate was resistant to antibiotics other than ampicillin-sulbactam and colistin, suggesting drug resistance due to carbapenemase production by OXA-23.carbapenem resistance in the isolated carbapenem-resistant A. baumannii strain was at least partially conferred by bla OXA-23-like carbapenemase. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Quinolone resistance protein NorB (NORB) | [24] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli | 668369 | ||
| Experiment for Molecule Alteration |
DNA microarray hybridization assay | |||
| Experiment for Drug Resistance |
Serial twofold agar dilutions assay | |||
| Mechanism Description | MgrA was an indirect regulator of norB expression. The mgrA norB double mutant was reproducibly twofold more susceptible to the tested quinolones than the mgrA mutant. | |||
| Key Molecule: Quinolone resistance protein NorA (NORA) | [36] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli HB101 | 634468 | ||
| Staphylococcus aureus strain SA113 | 1280 | |||
| Experiment for Molecule Alteration |
Dideoxy chain-termination method assay | |||
| Mechanism Description | The norA gene cloned from chromosomal DNA of quinolone-resistant Staphylococcus aureus Tk2566 conferred relatively high resistance to hydrophilic quinolones such as norfloxacin, enoxacin, ofloxacin, and ciprofloxacin, but only low or no resistance at all to hydrophobic ones such as nalidixic acid, oxolinic acid, and sparfloxacin in S. aureus and Escherichia coli. | |||
| Key Molecule: Quinolone resistance protein NorA (NORA) | [36] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Expression | Acquired |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli HB101 | 634468 | ||
| Staphylococcus aureus strain SA113 | 1280 | |||
| Experiment for Molecule Alteration |
Dideoxy chain-termination method assay | |||
| Mechanism Description | The norA gene cloned from chromosomal DNA of quinolone-resistant Staphylococcus aureus Tk2566 conferred relatively high resistance to hydrophilic quinolones such as norfloxacin, enoxacin, ofloxacin, and ciprofloxacin, but only low or no resistance at all to hydrophobic ones such as nalidixic acid, oxolinic acid, and sparfloxacin in S. aureus and Escherichia coli. S. aureus SA113 (pTUS20) harboring a plasmid carrying the staphylococcal norA gene was 16 to 64 times more resistant to relatively hydrophilic quinolones. | |||
|
|
||||
| Key Molecule: HTH-type transcriptional regulator MgrA (MGRA) | [24] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli | 668369 | ||
| Experiment for Molecule Alteration |
DNA microarray hybridization assay | |||
| Experiment for Drug Resistance |
Serial twofold agar dilutions assay | |||
| Mechanism Description | MgrA was an indirect regulator of norB expression. The mgrA norB double mutant was reproducibly twofold more susceptible to the tested quinolones than the mgrA mutant. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [25] | |||
| Resistant Disease | Staphylococcus infection [ICD-11: 1B7Y.3] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Pseudomonas aeruginosa isolates | 287 | ||
| Staphylococcus aureus isolates | 1280 | |||
| Klebsiella pneumoniae isolates | 573 | |||
| Acinetobacter isolates | 469 | |||
| Enterobacter cloacae isolates | 550 | |||
| Experiment for Drug Resistance |
Disk diffusion method assay | |||
| Mechanism Description | Up-regulation of P-glycoprotein led to ciprofloxacin resistance in the staphylococcus infection. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: DNA gyrase subunit A (GYRA) | [28] | |||
| Resistant Disease | Anthrax [ICD-11: 1B97.0] | |||
| Molecule Alteration | Mutation | p.S85+p.S85F+p.E89K+p.E89A |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli strain | 562 | ||
| Bacillus anthracis strain | 1392 | |||
| Experiment for Molecule Alteration |
DNA cleavage assay | |||
| Mechanism Description | The most common gyrase mutations in quinolone-resistant strains of B. anthracis are found at the conserved serine and glutamic acid residues (GyrAS85 and GyrAE89). In laboratory strains selected for resistance against ciprofloxacin and/or moxifloxacin (two widely prescribed quinolone antibacterials), approximately 80% of the isolates carried a GyrAS85L mutation (either alone or in combination with other gyrase/topoisomerase IV amino acid changes). The only other mutation reported to cause resistance without any other gyrase/topoisomerase IV changes was a GyrAE89K substitution. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: DNA gyrase subunit A (GYRA) | [7] | |||
| Resistant Disease | Bartonella bacilliformis infection [ICD-11: 1C11.0] | |||
| Molecule Alteration | Missense mutation | p.D95N |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Bartonella bacilliformis kC583 | 360095 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | The mutation of bartonella bacilliformis at asp-95 residue of gyrA QRDR resulted in the production of ciprofloxacin resistant strains. | |||
| Key Molecule: DNA gyrase subunit A (GYRA) | [7] | |||
| Resistant Disease | Bartonella bacilliformis infection [ICD-11: 1C11.0] | |||
| Molecule Alteration | Missense mutation | p.D90G |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Bartonella bacilliformis kC583 | 360095 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | The mutation of bartonella bacilliformis at asp-90 residue of gyrA QRDR resulted in the production of ciprofloxacin resistant strains. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: DNA topoisomerase 4 subunit B (PARE) | [15], [16] | |||
| Resistant Disease | HIV-infected patients with tuberculosis [ICD-11: 1C60.0] | |||
| Molecule Alteration | Missense mutation | p.N538D |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli | 668369 | ||
| Escherichia coli HB101 | 634468 | |||
| Mycobacterium smegmatis LR222 | 1772 | |||
| Mycobacterium tuberculosis MLB 262 | 1773 | |||
| Mycobacterium tuberculosis isolates | 1773 | |||
| Mycobacterium tuberculosis liquid | 1773 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay; disk diffusion test assay | |||
| Mechanism Description | DNA gyrase consists of two GyrA and two GyrB subunits encoded by gyrA and gyrB, respectively.Fluoroquinolone belong to the quinolone class of antibiotics which inhibit bacterial DNA gyrase and topoisomerase IV.Certain gyrA and gyrB mutations reported to confer cross-resistance to different FQ antibiotics based on clinical data have not yet been characterized in well-studied M. tuberculosis backgrounds. | |||
| Key Molecule: DNA topoisomerase 4 subunit B (PARE) | [15], [16] | |||
| Resistant Disease | HIV-infected patients with tuberculosis [ICD-11: 1C60.0] | |||
| Molecule Alteration | Missense mutation | p.E540V |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli | 668369 | ||
| Escherichia coli HB101 | 634468 | |||
| Mycobacterium smegmatis LR222 | 1772 | |||
| Mycobacterium tuberculosis MLB 262 | 1773 | |||
| Mycobacterium tuberculosis isolates | 1773 | |||
| Mycobacterium tuberculosis liquid | 1773 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay; disk diffusion test assay | |||
| Mechanism Description | DNA gyrase consists of two GyrA and two GyrB subunits encoded by gyrA and gyrB, respectively.Fluoroquinolone belong to the quinolone class of antibiotics which inhibit bacterial DNA gyrase and topoisomerase IV.Certain gyrA and gyrB mutations reported to confer cross-resistance to different FQ antibiotics based on clinical data have not yet been characterized in well-studied M. tuberculosis backgrounds. | |||
ICD-12: Respiratory system diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: DNA topoisomerase 4 subunit A (PARC) | [37] | |||
| Resistant Disease | Pneumocystis jirovecii infection [ICD-11: CA40.6] | |||
| Molecule Alteration | Missense mutation | p.D84H (GAT-CAT) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Streptococcus pneumoniae strain BM4203-BM4203-R | 1313 | ||
| Streptococcus pneumoniae strain BM4204-BM4204-R | 1313 | |||
| Experiment for Molecule Alteration |
Sequence analysis | |||
| Experiment for Drug Resistance |
Agar dilution assay | |||
| Mechanism Description | Mutations in parC were detected in the two resistant mutants obtained in vivo (BM4203-R andBM4204-R) as well as in two (BM4203-R1 and BM4203-R2) of the six mutants obtained in vitro. These mutations led to Ser-80-Tyr or Phe or to Asp-84-His substitutions(S. aureus coordinates) that are either identical or similar to those found in low-level-resistant parC mutations of S. aureus:Ser-80-Tyr or Phe and Glu-84-Lys or Leu. | |||
| Key Molecule: DNA topoisomerase 4 subunit A (PARC) | [37] | |||
| Resistant Disease | Pneumocystis jirovecii infection [ICD-11: CA40.6] | |||
| Molecule Alteration | Missense mutation | p.S80Y (TCT-TAT) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Streptococcus pneumoniae strain BM4203-BM4203-R | 1313 | ||
| Streptococcus pneumoniae strain BM4204-BM4204-R | 1313 | |||
| Experiment for Molecule Alteration |
Sequence analysis | |||
| Experiment for Drug Resistance |
Agar dilution assay | |||
| Mechanism Description | Mutations in parC were detected in the two resistant mutants obtained in vivo (BM4203-R andBM4204-R) as well as in two (BM4203-R1 and BM4203-R2) of the six mutants obtained in vitro. These mutations led to Ser-80-Tyr or Phe or to Asp-84-His substitutions(S. aureus coordinates) that are either identical or similar to those found in low-level-resistant parC mutations of S. aureus:Ser-80-Tyr or Phe and Glu-84-Lys or Leu. | |||
| Key Molecule: DNA topoisomerase 4 subunit A (PARC) | [37] | |||
| Resistant Disease | Pneumocystis jirovecii infection [ICD-11: CA40.6] | |||
| Molecule Alteration | Missense mutation | p.S80F (TCT-TTT) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Streptococcus pneumoniae strain BM4203-BM4203-R | 1313 | ||
| Streptococcus pneumoniae strain BM4204-BM4204-R | 1313 | |||
| Experiment for Molecule Alteration |
Sequence analysis | |||
| Experiment for Drug Resistance |
Agar dilution assay | |||
| Mechanism Description | Mutations in parC were detected in the two resistant mutants obtained in vivo (BM4203-R andBM4204-R) as well as in two (BM4203-R1 and BM4203-R2) of the six mutants obtained in vitro. These mutations led to Ser-80-Tyr or Phe or to Asp-84-His substitutions(S. aureus coordinates) that are either identical or similar to those found in low-level-resistant parC mutations of S. aureus:Ser-80-Tyr or Phe and Glu-84-Lys or Leu. | |||
| Key Molecule: DNA gyrase subunit A (GYRA) | [37] | |||
| Resistant Disease | Pneumocystis jirovecii infection [ICD-11: CA40.6] | |||
| Molecule Alteration | Missense mutation | p.S843F (TCC-TTC) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Streptococcus pneumoniae strain BM4203-BM4203-R | 1313 | ||
| Streptococcus pneumoniae strain BM4204-BM4204-R | 1313 | |||
| Experiment for Molecule Alteration |
Sequence analysis | |||
| Experiment for Drug Resistance |
Agar dilution assay | |||
| Mechanism Description | An additional mutant obtained in vitro, BM4205-R3, displayed a higher level of fluoroquinolone resistance and had a mutation in gyrA leading to a Ser-84-Phe change. | |||
|
|
||||
| Key Molecule: Beta-lactamase (BLA) | [9] | |||
| Resistant Disease | Klebsiella pneumoniae [ICD-11: CA40.0] | |||
| Molecule Alteration | Expression | Acquired |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Klebsiella pneumoniae isolates | 573 | ||
| Experiment for Molecule Alteration |
PCR and molecular characterization assay | |||
| Experiment for Drug Resistance |
Disk diffusion method assay | |||
| Mechanism Description | CTX-M-55 is a novel ceftazidime-resistant CTX-M extended-spectrum Beta-lactamase, which reduced susceptibility. | |||
|
|
||||
| Key Molecule: Multidrug efflux SMR transporter (ABES) | [38] | |||
| Resistant Disease | Acinetobacter baumannii infection [ICD-11: CA40.4] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli kAM32 | 562 | ||
| Experiment for Molecule Alteration |
Fluorometric efflux assay | |||
| Experiment for Drug Resistance |
Broth dilution assay | |||
| Mechanism Description | The abeS gene product conferred resistance to various antimicrobial compounds through an efflux mechanism. | |||
| Key Molecule: MATE family efflux transporter (ABEM) | [21] | |||
| Resistant Disease | Acinetobacter baumannii infection [ICD-11: CA40.4] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli kAM32 | 562 | ||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | AbeM was found to be an H+-coupled multidrug efflux pump and a unique member of the MATE family which lead to drug resistance. | |||
ICD-16: Genitourinary system diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Aminoglycoside (3'') (9) adenylyltransferase (AADA) | [27] | |||
| Resistant Disease | Urinary tract infection [ICD-11: GC08.1] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli serogroup O11 | 1095705 | ||
| Escherichia coli serogroup O17 | 1010800 | |||
| Escherichia coli serogroup O73 | 2170725 | |||
| Escherichia coli serogroup O77 | 562 | |||
| Experiment for Molecule Alteration |
PCR amplification and sequence alignments assay | |||
| Experiment for Drug Resistance |
Microdilution method assay | |||
| Mechanism Description | All the UTI outbreak CgA strains in this study contained the same class 1 integron dfrA17-aadA5 gene cassette arrangement with 100% sequence match, suggesting clonal spread of the bacterial strain itself. While aminoglycoside adenyltransferase A (aadA ) and dihydrofolate reductase A (dfrA ), encoding resistance to streptomycin and trimethoprim. | |||
| Key Molecule: Aminoglycoside (3'') (9) adenylyltransferase (AADA) | [27] | |||
| Resistant Disease | Urinary tract infection [ICD-11: GC08.1] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli serogroup O11 | 1095705 | ||
| Escherichia coli serogroup O17 | 1010800 | |||
| Escherichia coli serogroup O73 | 2170725 | |||
| Escherichia coli serogroup O77 | 562 | |||
| Experiment for Molecule Alteration |
PCR amplification and sequence alignments assay | |||
| Experiment for Drug Resistance |
Microdilution method assay | |||
| Mechanism Description | All the UTI outbreak CgA strains in this study contained the same class 1 integron dfrA17-aadA5 gene cassette arrangement with 100% sequence match, suggesting clonal spread of the bacterial strain itself. While aminoglycoside adenyltransferase A (aadA ) and dihydrofolate reductase A (dfrA ), encoding resistance to streptomycin and trimethoprim. | |||
|
|
||||
| Key Molecule: Dihydrofolate reductase (DHFR) | [27] | |||
| Resistant Disease | Urinary tract infection [ICD-11: GC08.1] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli serogroup O11 | 1095705 | ||
| Escherichia coli serogroup O17 | 1010800 | |||
| Escherichia coli serogroup O73 | 2170725 | |||
| Escherichia coli serogroup O77 | 562 | |||
| Experiment for Molecule Alteration |
PCR amplification and sequence alignments assay | |||
| Experiment for Drug Resistance |
Microdilution method assay | |||
| Mechanism Description | All the UTI outbreak CgA strains in this study contained the same class 1 integron dfrA17-aadA5 gene cassette arrangement with 100% sequence match, suggesting clonal spread of the bacterial strain itself. While aminoglycoside adenyltransferase A (aadA ) and dihydrofolate reductase A (dfrA ), encoding resistance to streptomycin and trimethoprim. | |||
| Key Molecule: Dihydrofolate reductase (DHFR) | [27] | |||
| Resistant Disease | Urinary tract infection [ICD-11: GC08.1] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli serogroup O11 | 1095705 | ||
| Escherichia coli serogroup O17 | 1010800 | |||
| Escherichia coli serogroup O73 | 2170725 | |||
| Escherichia coli serogroup O77 | 562 | |||
| Experiment for Molecule Alteration |
PCR amplification and sequence alignments assay | |||
| Experiment for Drug Resistance |
Microdilution method assay | |||
| Mechanism Description | All the UTI outbreak CgA strains in this study contained the same class 1 integron dfrA17-aadA5 gene cassette arrangement with 100% sequence match, suggesting clonal spread of the bacterial strain itself. While aminoglycoside adenyltransferase A (aadA ) and dihydrofolate reductase A (dfrA ), encoding resistance to streptomycin and trimethoprim. | |||
ICD-18: Pregnancy/Puerperium
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: DNA gyrase subunit A (GYRA) | [1], [2] | |||
| Resistant Disease | Ureaplasma urealyticum infection [ICD-11: 1A81.2] | |||
| Molecule Alteration | Missense mutation | p.S83L |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli k-12 JM109 | 83333 | ||
| Ureaplasma parvum serovar 3 isolates | 38504 | |||
| Ureaplasma parvum serovar 6 isolates | 95660 | |||
| Ureaplasma urealyticum isolates | 2130 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Quinolones are used for treating urogenital infections and interact in bacteria with the type II topoisomerases DNA gyrase and topoisomerase IV, both of which are composed of two A and two B subunits; these subunits are encoded by the gyrA and gyrB genes for DNA gyrase and parC and parE genes for topoisomerase IV.Out of 28 clinical Ureaplasma strains, we isolated 9 with high MICs of quinolones and found a single parC gene mutation, resulting in the change S83L. | |||
| Key Molecule: DNA gyrase subunit A (GYRA) | [1], [2] | |||
| Resistant Disease | Ureaplasma urealyticum infection [ICD-11: 1A81.2] | |||
| Molecule Alteration | Missense mutation | p.S83L |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli k-12 JM109 | 83333 | ||
| Ureaplasma parvum serovar 3 isolates | 38504 | |||
| Ureaplasma parvum serovar 6 isolates | 95660 | |||
| Ureaplasma urealyticum isolates | 2130 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Quinolones are used for treating urogenital infections and interact in bacteria with the type II topoisomerases DNA gyrase and topoisomerase IV, both of which are composed of two A and two B subunits; these subunits are encoded by the gyrA and gyrB genes for DNA gyrase and parC and parE genes for topoisomerase IV.Out of 28 clinical Ureaplasma strains, we isolated 9 with high MICs of quinolones and found a single parC gene mutation, resulting in the change S83L. | |||
| Key Molecule: DNA topoisomerase 4 subunit A (PARC) | [1], [2] | |||
| Resistant Disease | Miscarriage [ICD-11: JA00.0] | |||
| Molecule Alteration | Missense mutation | p.S83W |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli k-12 JM109 | 83333 | ||
| Ureaplasma parvum serovar 3 isolates | 38504 | |||
| Ureaplasma parvum serovar 6 isolates | 95660 | |||
| Ureaplasma urealyticum isolates | 2130 | |||
| Ureaplasma parvum isolates | 38504 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Quinolones are used for treating urogenital infections and interact in bacteria with the type II topoisomerases DNA gyrase and topoisomerase IV, both of which are composed of two A and two B subunits; these subunits are encoded by the gyrA and gyrB genes for DNA gyrase and parC and parE genes for topoisomerase IV.Out of 28 clinical Ureaplasma strains, we isolated 9 with high MICs of quinolones and found a single parC gene mutation, resulting in the change S83L. | |||
| Key Molecule: DNA topoisomerase 4 subunit A (PARC) | [1], [2] | |||
| Resistant Disease | Miscarriage [ICD-11: JA00.0] | |||
| Molecule Alteration | Missense mutation | p.S83P |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli k-12 JM109 | 83333 | ||
| Ureaplasma parvum serovar 3 isolates | 38504 | |||
| Ureaplasma parvum serovar 6 isolates | 95660 | |||
| Ureaplasma urealyticum isolates | 2130 | |||
| Ureaplasma parvum isolates | 38504 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Quinolones are used for treating urogenital infections and interact in bacteria with the type II topoisomerases DNA gyrase and topoisomerase IV, both of which are composed of two A and two B subunits; these subunits are encoded by the gyrA and gyrB genes for DNA gyrase and parC and parE genes for topoisomerase IV.Out of 28 clinical Ureaplasma strains, we isolated 9 with high MICs of quinolones and found a single parC gene mutation, resulting in the change S83L. | |||
| Key Molecule: DNA topoisomerase 4 subunit A (PARC) | [1], [2] | |||
| Resistant Disease | Miscarriage [ICD-11: JA00.0] | |||
| Molecule Alteration | Missense mutation | ParC p.S83L+GyrB p.P462S |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli k-12 JM109 | 83333 | ||
| Ureaplasma parvum serovar 3 isolates | 38504 | |||
| Ureaplasma parvum serovar 6 isolates | 95660 | |||
| Ureaplasma urealyticum isolates | 2130 | |||
| Ureaplasma parvum isolates | 38504 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Quinolones are used for treating urogenital infections and interact in bacteria with the type II topoisomerases DNA gyrase and topoisomerase IV, both of which are composed of two A and two B subunits; these subunits are encoded by the gyrA and gyrB genes for DNA gyrase and parC and parE genes for topoisomerase IV.Out of 28 clinical Ureaplasma strains, we isolated 9 with high MICs of quinolones and found a single parC gene mutation, resulting in the change S83L. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: DNA topoisomerase 4 subunit A (PARC) | [1], [2] | |||
| Resistant Disease | Preterm delivery with lung infection in neonates [ICD-11: JB00.0] | |||
| Molecule Alteration | Missense mutation | p.S83W |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli k-12 JM109 | 83333 | ||
| Ureaplasma parvum serovar 3 isolates | 38504 | |||
| Ureaplasma parvum serovar 6 isolates | 95660 | |||
| Ureaplasma urealyticum isolates | 2130 | |||
| Ureaplasma parvum isolates | 38504 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Quinolones are used for treating urogenital infections and interact in bacteria with the type II topoisomerases DNA gyrase and topoisomerase IV, both of which are composed of two A and two B subunits; these subunits are encoded by the gyrA and gyrB genes for DNA gyrase and parC and parE genes for topoisomerase IV.Out of 28 clinical Ureaplasma strains, we isolated 9 with high MICs of quinolones and found a single parC gene mutation, resulting in the change S83L. | |||
| Key Molecule: DNA topoisomerase 4 subunit A (PARC) | [1], [2] | |||
| Resistant Disease | Preterm delivery with lung infection in neonates [ICD-11: JB00.0] | |||
| Molecule Alteration | Missense mutation | p.S83P |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli k-12 JM109 | 83333 | ||
| Ureaplasma parvum serovar 3 isolates | 38504 | |||
| Ureaplasma parvum serovar 6 isolates | 95660 | |||
| Ureaplasma urealyticum isolates | 2130 | |||
| Ureaplasma parvum isolates | 38504 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Quinolones are used for treating urogenital infections and interact in bacteria with the type II topoisomerases DNA gyrase and topoisomerase IV, both of which are composed of two A and two B subunits; these subunits are encoded by the gyrA and gyrB genes for DNA gyrase and parC and parE genes for topoisomerase IV.Out of 28 clinical Ureaplasma strains, we isolated 9 with high MICs of quinolones and found a single parC gene mutation, resulting in the change S83L. | |||
| Key Molecule: DNA topoisomerase 4 subunit A (PARC) | [1], [2] | |||
| Resistant Disease | Preterm delivery with lung infection in neonates [ICD-11: JB00.0] | |||
| Molecule Alteration | Missense mutation | ParC p.S83L+GyrB p.P462S |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli k-12 JM109 | 83333 | ||
| Ureaplasma parvum serovar 3 isolates | 38504 | |||
| Ureaplasma parvum serovar 6 isolates | 95660 | |||
| Ureaplasma urealyticum isolates | 2130 | |||
| Ureaplasma parvum isolates | 38504 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Quinolones are used for treating urogenital infections and interact in bacteria with the type II topoisomerases DNA gyrase and topoisomerase IV, both of which are composed of two A and two B subunits; these subunits are encoded by the gyrA and gyrB genes for DNA gyrase and parC and parE genes for topoisomerase IV.Out of 28 clinical Ureaplasma strains, we isolated 9 with high MICs of quinolones and found a single parC gene mutation, resulting in the change S83L. | |||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
