Drug Information
Drug (ID: DG00297) and It's Reported Resistant Information
| Name |
Mitomycin
|
||||
|---|---|---|---|---|---|
| Synonyms |
Mitomycin C; mitomycin C; 1950/7/7; Mutamycin; Ametycine; Mitocin-C; Ametycin; Mitomycin-C; Mytozytrex; Mitomycinum; Mytomycin; Mitozytrex; Mitomycinum C; Mitocin C; Mitomycins; Mitamycin; MMC; Mitosol; Mitomycyna C; 7-Amino-9alpha-methoxymitosane; NSC-26980; Mitomycyna C [Polish]; Mito-C; Mit-C; Mitomycin (TN); Mitomycinum [INN-Latin]; Mitomycine [INN-French]; Mitomicina [INN-Spanish]; NCI-C04706; RCRA waste number U010; NSC26980; NSC 26980; Mitomycine; CCRIS 414; UNII-50SG953SK6; HSDB 3239; C15H18N4O5; EINECS 200-008-6; Mitomycin C,; Ametycin; Mitomicina; Muamycin; Mitomycin C from Streptomyces caespitosus; Mitomycin C (JP15); Mitomycin C, Streptomyces caespitosus; Muamycin (TN); Mitomycin (USP/INN); Mitomycin [USAN:INN:BAN]; Mitomycin C, Streptomyces caespitosus, Carrier-Free
Click to Show/Hide
|
||||
| Indication |
In total 3 Indication(s)
|
||||
| Structure |
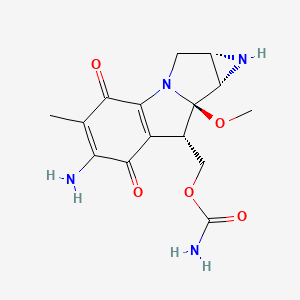
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Resistance Information Discovered by Cell Line Test for This Drug
(2 diseases)
[2]
[3]
|
||||
| Target | Human Deoxyribonucleic acid (hDNA) | NOUNIPROTAC | [1] | ||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C15H18N4O5
|
||||
| IsoSMILES |
CC1=C(C(=O)C2=C(C1=O)N3C[C@H]4[C@@H]([C@@]3([C@@H]2COC(=O)N)OC)N4)N
|
||||
| InChI |
1S/C15H18N4O5/c1-5-9(16)12(21)8-6(4-24-14(17)22)15(23-2)13-7(18-13)3-19(15)10(8)11(5)20/h6-7,13,18H,3-4,16H2,1-2H3,(H2,17,22)/t6-,7+,13+,15-/m1/s1
|
||||
| InChIKey |
NWIBSHFKIJFRCO-WUDYKRTCSA-N
|
||||
| PubChem CID | |||||
| ChEBI ID | |||||
| TTD Drug ID | |||||
| VARIDT ID | |||||
| DrugBank ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-02: Benign/in-situ/malignant neoplasm
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-34 | [4] | |||
| Sensitive Disease | Medulloblastoma [ICD-11: 2A00.10] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| p53 signaling pathway | Activation | hsa04115 | ||
| In Vitro Model | UW228 cells | Brain | Homo sapiens (Human) | CVCL_8585 |
| R262 cells | Bone marrow | Homo sapiens (Human) | CVCL_VU83 | |
| R300 cells | Bone marrow | Homo sapiens (Human) | CVCL_VU84 | |
| UW426 cells | Bone marrow | Homo sapiens (Human) | CVCL_DH82 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | The repression of MAGE-A by miR-34a results in increased expression of p53 thus lead to resistance. | |||
|
|
||||
| Key Molecule: Melanoma-associated antigen 12 (MAGEA12) | [4] | |||
| Sensitive Disease | Medulloblastoma [ICD-11: 2A00.10] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| p53 signaling pathway | Activation | hsa04115 | ||
| In Vitro Model | UW228 cells | Brain | Homo sapiens (Human) | CVCL_8585 |
| R262 cells | Bone marrow | Homo sapiens (Human) | CVCL_VU83 | |
| R300 cells | Bone marrow | Homo sapiens (Human) | CVCL_VU84 | |
| UW426 cells | Bone marrow | Homo sapiens (Human) | CVCL_DH82 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | The repression of MAGE-A by miR-34a results in increased expression of p53 thus lead to resistance. | |||
| Key Molecule: Melanoma-associated antigen 2 (MAGEA2) | [4] | |||
| Sensitive Disease | Medulloblastoma [ICD-11: 2A00.10] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| p53 signaling pathway | Activation | hsa04115 | ||
| In Vitro Model | UW228 cells | Brain | Homo sapiens (Human) | CVCL_8585 |
| R262 cells | Bone marrow | Homo sapiens (Human) | CVCL_VU83 | |
| R300 cells | Bone marrow | Homo sapiens (Human) | CVCL_VU84 | |
| UW426 cells | Bone marrow | Homo sapiens (Human) | CVCL_DH82 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | The repression of MAGE-A by miR-34a results in increased expression of p53 thus lead to resistance. | |||
| Key Molecule: Melanoma-associated antigen 3 (MAGEA3) | [4] | |||
| Sensitive Disease | Medulloblastoma [ICD-11: 2A00.10] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| p53 signaling pathway | Activation | hsa04115 | ||
| In Vitro Model | UW228 cells | Brain | Homo sapiens (Human) | CVCL_8585 |
| R262 cells | Bone marrow | Homo sapiens (Human) | CVCL_VU83 | |
| R300 cells | Bone marrow | Homo sapiens (Human) | CVCL_VU84 | |
| UW426 cells | Bone marrow | Homo sapiens (Human) | CVCL_DH82 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | The repression of MAGE-A by miR-34a results in increased expression of p53 thus lead to resistance. | |||
| Key Molecule: Melanoma-associated antigen 6 (MAGEA6) | [4] | |||
| Sensitive Disease | Medulloblastoma [ICD-11: 2A00.10] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| p53 signaling pathway | Activation | hsa04115 | ||
| In Vitro Model | UW228 cells | Brain | Homo sapiens (Human) | CVCL_8585 |
| R262 cells | Bone marrow | Homo sapiens (Human) | CVCL_VU83 | |
| R300 cells | Bone marrow | Homo sapiens (Human) | CVCL_VU84 | |
| UW426 cells | Bone marrow | Homo sapiens (Human) | CVCL_DH82 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | The repression of MAGE-A by miR-34a results in increased expression of p53 thus lead to resistance. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-223 | [5] | |||
| Sensitive Disease | Esophageal adenocarcinoma [ICD-11: 2B70.2] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | OE33 cellss | Esophagus | Homo sapiens (Human) | CVCL_0471 |
| HEEpiC cells | Esophagus | Homo sapiens (Human) | N.A. | |
| JHesoAD1 cells | Esophagus | Homo sapiens (Human) | CVCL_8098 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | The DNA damage repair protein poly(ADP-ribose) polymerase 1 (PARP1) is a bona fide target of miR-223, miR-223 up-regulation is also associated with reduced PARP1 transcripts, and an increased sensitivity to cis-diamminedichloroplatinum (II) (Cisplatin), Doxorubicin and Mitomycin C. | |||
|
|
||||
| Key Molecule: Poly[ADP-ribose] synthase 1 (PARP1) | [5] | |||
| Sensitive Disease | Esophageal adenocarcinoma [ICD-11: 2B70.2] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell migration | Activation | hsa04670 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | OE33 cellss | Esophagus | Homo sapiens (Human) | CVCL_0471 |
| HEEpiC cells | Esophagus | Homo sapiens (Human) | N.A. | |
| JHesoAD1 cells | Esophagus | Homo sapiens (Human) | CVCL_8098 | |
| Experiment for Molecule Alteration |
Luciferase reporter assay | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | The DNA damage repair protein poly(ADP-ribose) polymerase 1 (PARP1) is a bona fide target of miR-223, miR-223 up-regulation is also associated with reduced PARP1 transcripts, and an increased sensitivity to cis-diamminedichloroplatinum (II) (Cisplatin), Doxorubicin and Mitomycin C. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Hypoxia-inducible factor 2-alpha (EPAS1) | [2] | |||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | BGC-823 cells | Gastric | Homo sapiens (Human) | CVCL_3360 |
| BEL-7402 cells | Liver | Homo sapiens (Human) | CVCL_5492 | |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |
| Experiment for Molecule Alteration |
Western blotting assay | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | Over-expression of EPAS-1 increased the expression of PXR responsive genes, enhanced the proliferation of BGC-823 cells and boosted the resistance of BGC-823 cells against the cytotoxicity of chemotherapeutic drugs, e.g. Mitomycin C and Paclitaxel.EPAS-1 reduces BGC-823 cell apoptosis induced by Mitomycin C and Paclitaxel. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-495 | [1] | |||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| mTOR signaling pathway | Inhibition | hsa04150 | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | The miR-495 exerts promotive effects on GC chemosensitivity via inactivation of the mTOR signaling pathway by suppressing ERBB2. | |||
|
|
||||
| Key Molecule: Receptor tyrosine-protein kinase erbB-2 (ERBB2) | [1] | |||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| mTOR signaling pathway | Inhibition | hsa04150 | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | The miR-495 exerts promotive effects on GC chemosensitivity via inactivation of the mTOR signaling pathway by suppressing ERBB2. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-146a | [3] | |||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HCC Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The HCC Huh-7 cell line was treated with adramycin (ADM), cisplatin (DDP), carboplatin (CBP), mitomycin C (MMC) or vincristine (VCR) at increasing concentrations to develop drug-resistant sublines. Among these 51 upregulated and downregulated miRNAs, 12 miRNAs were upregulated and 13 miRNAs were downregulated in Huh-7/VCR. Upregulation of miR-27b, miR-181a, miR-146b-5p, miR-181d and miR-146a expression was verified using real-time RT-PCR in the parental and the five drug-resistant cell lines. | |||
| Key Molecule: hsa-miR-146b-5p | [3] | |||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HCC Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The HCC Huh-7 cell line was treated with adramycin (ADM), cisplatin (DDP), carboplatin (CBP), mitomycin C (MMC) or vincristine (VCR) at increasing concentrations to develop drug-resistant sublines. Among these 51 upregulated and downregulated miRNAs, 12 miRNAs were upregulated and 13 miRNAs were downregulated in Huh-7/VCR. Upregulation of miR-27b, miR-181a, miR-146b-5p, miR-181d and miR-146a expression was verified using real-time RT-PCR in the parental and the five drug-resistant cell lines. | |||
| Key Molecule: hsa-mir-181a | [3] | |||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HCC Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The HCC Huh-7 cell line was treated with adramycin (ADM), cisplatin (DDP), carboplatin (CBP), mitomycin C (MMC) or vincristine (VCR) at increasing concentrations to develop drug-resistant sublines. Among these 51 upregulated and downregulated miRNAs, 12 miRNAs were upregulated and 13 miRNAs were downregulated in Huh-7/VCR. Upregulation of miR-27b, miR-181a, miR-146b-5p, miR-181d and miR-146a expression was verified using real-time RT-PCR in the parental and the five drug-resistant cell lines. | |||
| Key Molecule: hsa-mir-181d | [3] | |||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HCC Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The HCC Huh-7 cell line was treated with adramycin (ADM), cisplatin (DDP), carboplatin (CBP), mitomycin C (MMC) or vincristine (VCR) at increasing concentrations to develop drug-resistant sublines. Among these 51 upregulated and downregulated miRNAs, 12 miRNAs were upregulated and 13 miRNAs were downregulated in Huh-7/VCR. Upregulation of miR-27b, miR-181a, miR-146b-5p, miR-181d and miR-146a expression was verified using real-time RT-PCR in the parental and the five drug-resistant cell lines. | |||
| Key Molecule: hsa-mir-27b | [3] | |||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HCC Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The HCC Huh-7 cell line was treated with adramycin (ADM), cisplatin (DDP), carboplatin (CBP), mitomycin C (MMC) or vincristine (VCR) at increasing concentrations to develop drug-resistant sublines. Among these 51 upregulated and downregulated miRNAs, 12 miRNAs were upregulated and 13 miRNAs were downregulated in Huh-7/VCR. Upregulation of miR-27b, miR-181a, miR-146b-5p, miR-181d and miR-146a expression was verified using real-time RT-PCR in the parental and the five drug-resistant cell lines. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-145 | [6] | |||
| Sensitive Disease | Cervical cancer [ICD-11: 2C77.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | p53 signaling pathway | Activation | hsa04115 | |
| In Vitro Model | C33A cells | Uterus | Homo sapiens (Human) | CVCL_1094 |
| Experiment for Molecule Alteration |
RT-PCR; Northern blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | p53 signalling pathway mediates the cytotoxic effects of chemotherapy, miR-145 augments p53-mediated cytotoxic effects. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-31 | [7] | |||
| Sensitive Disease | Bladder urothelial carcinoma [ICD-11: 2C94.2] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | AKT/ERK signaling pathway | Regulation | N.A. | |
| Cell apoptosis | Activation | hsa04210 | ||
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | 5637 cells | Bladder | Homo sapiens (Human) | CVCL_0126 |
| T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-31 expression brings about (+) sensitivity of UBC to MMC by suppressing ITGA5 and downstream pathways. | |||
|
|
||||
| Key Molecule: Integrin alpha-5 (ITGA5) | [7] | |||
| Sensitive Disease | Bladder urothelial carcinoma [ICD-11: 2C94.2] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | AKT/ERK signaling pathway | Regulation | N.A. | |
| Cell apoptosis | Activation | hsa04210 | ||
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | 5637 cells | Bladder | Homo sapiens (Human) | CVCL_0126 |
| T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-31 expression brings about (+) sensitivity of UBC to MMC by suppressing ITGA5 and downstream pathways. | |||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
