Drug Information
Drug (ID: DG00023) and It's Reported Resistant Information
| Name |
Daunorubicin
|
||||
|---|---|---|---|---|---|
| Synonyms |
Daunorubicin; Daunomycin; 20830-81-3; Rubidomycin; Cerubidine; Daunorubicine; Acetyladriamycin; Leukaemomycin C; Daunorubicinum; Daunarubicinum; Daunorrubicina; Daunamycin; Cerubidin; DaunoXome; Rubomycin C; (+)-Daunomycin; Daunoblastin; Anthracyline; Rubomycin; Daunorubicinum [INN-Latin]; RP 13057; Daunorubicin [INN:BAN]; RCRA waste no U059; FI6339; NSC-82151; DAUNORUBICIN HCL; DaunoXome (TN); UNII-ZS7284E0ZP; CCRIS 914; ZS7284E0ZP; CHEBI:41977; HSDB 5095; C27H29NO10; NCI-C04693; EINECS 244-069-7; Ondena; NSC 83142; Acetyladriamycin; Daunoblastine; Antibiotics from Streptomyces coeruleorubidus; DM1; FI 6339; Dauno-Rubidomycine; Daunorubicin (INN); Daunorubicin (liposomal); Daunorubicin, Hydrochloride; VS-103; (1S,3S)-3-acetyl-3,5,12-trihydroxy-10-(methyloxy)-6,11-dioxo-1,2,3,4,6,11-hexahydrotetracen-1-yl 3-amino-2,3,6-trideoxy-alpha-L-lyxo-hexopyranoside; (1S,3S)-3-acetyl-3,5,12-trihydroxy-10-methoxy-6,11-dioxo-1,2,3,4,6,11-hexahydrotetracen-1-yl 3-amino-2,3,6-trideoxy-a-L-lyxo-hexopyranoside; (7S,9R)-9-Acetyl-7-[(2S,4S,5S,6S)-4-amino-5-hydroxy-6-methyl-oxan-2-yl]oxy-6,9,11-trihydroxy-4-methoxy-8,10-dihydro-7H-tetracene-5,12-dione; (7S,9S)-9-acetyl-7-[(2R,4S,5S,6S)-4-amino-5-hydroxy-6-methyloxan-2-yl]oxy-6,9,11-trihydroxy-4-methoxy-8,10-dihydro-7H-tetracene-5,12-dione; (8S-cis)-8-Acetyl-10-((3-amino-2,3,6-trideoxy-alpha-L-lyxo-hexopyrannosyl)oxy)-7,8,9,10-tetrahydro-6,8,11-trihydroxy-1-methoxy-5,12-napthacenedione; (8S-cis)-8-Acetyl-10-[(3-amino-2,3,6-trideoxy-alpha-L-lyxo-hexopyranosyl)oxy]-7,8,9,10-tetrahydro-6,8,11-trihydroxy-1-methoxy-5,12-naphthacenedione; Anthracycline
Click to Show/Hide
|
||||
| Indication |
In total 3 Indication(s)
|
||||
| Structure |
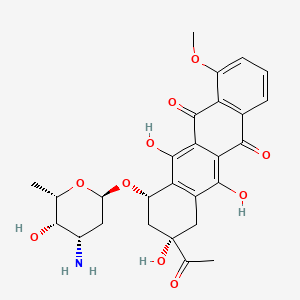
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Clinically Reported Resistance for This Drug
(3 diseases)
[2]
[3]
[6]
Disease(s) with Resistance Information Discovered by Cell Line Test for This Drug
(5 diseases)
[5]
[7]
[8]
[9]
[10]
|
||||
| Target | DNA replication (DNA repli) | NOUNIPROTAC | [1] | ||
| Human Deoxyribonucleic acid (hDNA) | NOUNIPROTAC | [1] | |||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C27H29NO10
|
||||
| IsoSMILES |
C[C@H]1[C@H]([C@H](C[C@@H](O1)O[C@H]2C[C@@](CC3=C2C(=C4C(=C3O)C(=O)C5=C(C4=O)C(=CC=C5)OC)O)(C(=O)C)O)N)O
|
||||
| InChI |
1S/C27H29NO10/c1-10-22(30)14(28)7-17(37-10)38-16-9-27(35,11(2)29)8-13-19(16)26(34)21-20(24(13)32)23(31)12-5-4-6-15(36-3)18(12)25(21)33/h4-6,10,14,16-17,22,30,32,34-35H,7-9,28H2,1-3H3/t10-,14-,16-,17-,22+,27-/m0/s1
|
||||
| InChIKey |
STQGQHZAVUOBTE-VGBVRHCVSA-N
|
||||
| PubChem CID | |||||
| ChEBI ID | |||||
| TTD Drug ID | |||||
| VARIDT ID | |||||
| INTEDE ID | |||||
| DrugBank ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-02: Benign/in-situ/malignant neoplasm
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [11] | |||
| Sensitive Disease | Renal cell carcinoma [ICD-11: 2C90.0] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Kidney cancer [ICD-11: 2C90] | |||
| The Specified Disease | Renal cell carcinoma | |||
| The Studied Tissue | Kidney | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.11E-46 Fold-change: -1.42E+00 Z-score: -1.91E+01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Flp-In-293/Mock cells | Kidney | Homo sapiens (Human) | CVCL_U421 |
| Flp-In-293/ABCB1 cells | Kidney | Homo sapiens (Human) | CVCL_U421 | |
| Experiment for Molecule Alteration |
ATPase assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Through calcein assays, we found that epimagnolin A inhibited the ABCB1-mediated export of calcein. This result suggests that epimagnolin A behaved as inhibitor or substrate for ABCB1. In ATPase assays, epimagnolin A stimulated ABCB1-dependent ATPase activity. This result indicates that epimagnolin A was recognised as a substrate by ABCB1, since ABCB1 utilises energy derived from ATP hydrolysis for substrate transport. Furthermore, in MTT assays we found that the cytotoxicity of daunorubicin, doxorubicin, vinblastine, and vincristine was enhanced by epimagnolin A in a manner comparable to verapamil, a typical substrate for ABCB1. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Eukaryotic translation initiation factor 5A-2 (EIF5A2) | [12] | |||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Acute myeloid leukemia [ICD-11: 2A60] | |||
| The Specified Disease | Acute myelocytic leukemia | |||
| The Studied Tissue | Bone marrow | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.74E-03 Fold-change: -4.62E-02 Z-score: -2.92E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | KG-1 cells | Bone marrow | Homo sapiens (Human) | CVCL_0374 |
| THP-1 cells | Blood | Homo sapiens (Human) | CVCL_0006 | |
| HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 | |
| Kasumi-1 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0589 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; EdU assay; Flow cytometry assay | |||
| Mechanism Description | miR-9 improved the anti-tumor effects of Dnr by inhibiting myeloid cell leukemia-1 (MCL-1) expression, which was dependent on downregulation of EIF5A2 expression. | |||
| Key Molecule: Induced myeloid leukemia cell differentiation protein Mcl-1 (MCL1) | [1] | |||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | KG-1 cells | Bone marrow | Homo sapiens (Human) | CVCL_0374 |
| THP-1 cells | Blood | Homo sapiens (Human) | CVCL_0006 | |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | MCL-1 participates in the regulation of DNR sensitivity mediated by miR-33b and overexpression of miR-33b enhances DNR sensitivity by downregulating MCL-1 in AML cells. | |||
| Key Molecule: Eukaryotic translation initiation factor 5A-2 (EIF5A2) | [1] | |||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | KG-1 cells | Bone marrow | Homo sapiens (Human) | CVCL_0374 |
| THP-1 cells | Blood | Homo sapiens (Human) | CVCL_0006 | |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | microRNA-33b regulates sensitivity to daunorubicin in acute myelocytic leukemia by regulating eukaryotic translation initiation factor 5A-2. | |||
|
|
||||
| Key Molecule: hsa-mir-33b | [1] | |||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | KG-1 cells | Bone marrow | Homo sapiens (Human) | CVCL_0374 |
| THP-1 cells | Blood | Homo sapiens (Human) | CVCL_0006 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | MCL-1 participates in the regulation of DNR sensitivity mediated by miR-33b and overexpression of miR-33b enhances DNR sensitivity by downregulating MCL-1 in AML cells. | |||
| Key Molecule: hsa-mir-9 | [12] | |||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | KG-1 cells | Bone marrow | Homo sapiens (Human) | CVCL_0374 |
| THP-1 cells | Blood | Homo sapiens (Human) | CVCL_0006 | |
| HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 | |
| Kasumi-1 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0589 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; EdU assay; Flow cytometry assay | |||
| Mechanism Description | miR-9 improved the anti-tumor effects of Dnr by inhibiting myeloid cell leukemia-1 (MCL-1) expression, which was dependent on downregulation of EIF5A2 expression. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Key Molecule: DNA (cytosine-5)-methyltransferase 3A (DNMT3A) | [3] | ||||||||||||
| Resistant Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Molecule Alteration | Missense mutation | p.R882H |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.40 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.44 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
A
A
E
E
630
|
K
K
R
R
K
K
P
P
I
I
R
R
V
V
L
L
S
S
L
L
640
|
F
F
D
D
G
G
I
I
A
A
T
T
G
G
L
L
L
L
V
V
650
|
L
L
K
K
D
D
L
L
G
G
I
I
Q
Q
V
V
D
D
R
R
660
|
Y
Y
I
I
A
A
S
S
E
E
V
V
C
C
E
E
D
D
S
S
670
|
I
I
T
T
V
V
G
G
M
M
V
V
R
R
H
H
Q
Q
G
G
680
|
K
K
I
I
M
M
Y
Y
V
V
G
G
D
D
V
V
R
R
S
S
690
|
V
V
T
T
Q
Q
K
K
H
H
I
I
Q
Q
E
E
W
W
G
G
700
|
P
P
F
F
D
D
L
L
V
V
I
I
G
G
G
G
S
S
P
P
710
|
C
C
N
N
D
D
L
L
S
S
I
I
V
V
N
N
P
P
A
A
720
|
R
R
K
K
G
G
L
L
Y
Y
E
E
G
G
T
T
G
G
R
R
730
|
L
L
F
F
F
F
E
E
F
F
Y
Y
R
R
L
L
L
L
H
H
740
|
D
D
A
A
R
R
P
P
K
K
E
E
G
G
D
D
D
D
R
R
750
|
P
P
F
F
F
F
W
W
L
L
F
F
E
E
N
N
V
V
V
V
760
|
A
A
M
M
G
G
V
V
S
S
D
D
K
K
R
R
D
D
I
I
770
|
S
S
R
R
F
F
L
L
E
E
S
S
N
N
P
P
V
V
M
M
780
|
I
I
D
D
A
A
K
K
E
E
V
V
S
S
A
A
A
A
H
H
790
|
R
R
A
A
R
R
Y
Y
F
F
W
W
G
G
N
N
L
L
P
P
800
|
G
G
M
M
N
N
R
R
P
P
L
L
A
A
S
S
T
T
V
V
810
|
N
N
D
D
K
K
L
L
E
E
L
L
Q
Q
E
E
C
C
L
L
820
|
E
E
H
H
G
G
R
R
I
I
A
A
K
K
F
F
S
S
K
K
830
|
V
V
R
R
T
T
I
I
T
T
T
T
R
R
S
S
N
N
S
S
840
|
I
I
K
K
Q
Q
G
G
K
K
D
D
Q
Q
H
H
F
F
P
P
850
|
V
V
F
F
M
M
N
N
E
E
K
K
E
E
D
D
I
I
L
L
860
|
W
W
C
C
T
T
E
E
M
M
E
E
R
R
V
V
F
F
G
G
870
|
F
F
P
P
V
V
H
H
Y
Y
T
T
D
D
V
V
S
S
N
N
880
|
M
M
S
S
R
H
L
L
A
A
R
R
Q
Q
R
R
L
L
L
L
890
|
G
G
R
R
S
S
W
W
S
S
V
V
P
P
V
V
I
I
R
R
900
|
H
H
L
L
F
F
A
A
P
P
L
L
K
K
E
E
Y
Y
F
F
910
|
A
A
C
C
V
V
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | ADAM9/EGFR signaling pathway | Inhibition | hsa01521 | ||||||||||
| AKT signaling pathway | Inhibition | hsa04151 | |||||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay | ||||||||||||
| Mechanism Description | DNMT3A mutations are most common in AML. DNMT3A mutant AML has been linked to anthracycline resistance and poor prognosis in some studies. Many of these mutations occur in genes with established roles in the regulation and maintenance of DNA methylation and/or chromatin modifications in hematopoietic stem/progenitor cells. | ||||||||||||
|
|
|||||||||||||
| Key Molecule: C-C motif chemokine 20 (CCL20) | [4] | ||||||||||||
| Resistant Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experiment for Molecule Alteration |
ELISA assay | ||||||||||||
| Mechanism Description | Our study has identified CCL20 as a pivotal factor in the promotion of chemoresistance in AML cells by M2 macrophages. The chemotherapeutic agent daunorubicin induces a marked increase in ROS and lipid peroxidation levels within AML cells. This is accompanied by the inhibition of the SLC7A11/GCL/GPX4 signaling axis, elevated levels of intracellular free iron, disrupted iron metabolism, and consequent mitochondrial damage, ultimately leading to ferroptosis. Notably, CCL20 enhances the ability of AML cells to maintain iron homeostasis by upregulating SLC7A11 protein activity, mitigating mitochondrial damage, and inhibiting ferroptosis, thereby contributing to chemotherapy resistance. Furthermore, in vivo experiments demonstrated that blocking CCL20 effectively restores the sensitivity of AML cells to daunorubicin chemotherapy. | ||||||||||||
|
|
|||||||||||||
| Key Molecule: Receptor-type tyrosine-protein kinase FLT3 (FLT3) | [5] | ||||||||||||
| Resistant Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Molecule Alteration | Mutation | . |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | MAPK-RSKs-YB-1 signaling pathway | Regulation | N.A. | ||||||||||
| In Vitro Model | MOLM-13 cells | Peripheral blood | Homo sapiens (Human) | CVCL_2119 | |||||||||
| MV-4-11 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0064 | ||||||||||
| Experiment for Drug Resistance |
Cell viability assay; Colony formation assay; Annexin-V/7-AAD double stain assay | ||||||||||||
| Mechanism Description | LJH-685 inhibited the proliferation and clone formation of AML cells, caused cell cycle arrest and induced the apoptosis of AML cells via inhibiting the RSK-YB-1 signaling pathway. MV4-11 and MOLM-13 cells carrying FLT3-ITD mutations were more sensitive to LJH-685 than that of other AML cell lines. Further studies suggested that LJH-685 combined with Daunorubicin or FF- 10101 synergistically inhibited the cell viability, promoted the apoptosis and caused cycle arrest of AML cells carrying FLT3-ITD mutations. | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: ATP-binding cassette sub-family B5 (ABCB5) | [13] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | CR1R12 cells | N.A. | Homo sapiens (Human) | N.A. |
| NIH-G185 cells | Ovary | Homo sapiens (Human) | CVCL_L991 | |
| Experiment for Drug Resistance |
propidium iodide staining assay | |||
| Mechanism Description | In a NIH-G185 cell line presenting an overexpressed amount of the human transporter P-gp, cholesterol caused dramatic inhibition of daunorubicin transport with an IC(50) of about 8 microM yet had no effect on the parent cell line nor rhodamine 123 transport. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: microRNA-18a-5p (miR-18a-5p) | [9] | |||
| Metabolic Type | Glucose metabolism | |||
| Resistant Disease | Chronic myeloid leukemia [ICD-11: 2A20.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | K562/ADM cells | Blood | Homo sapiens (Human) | CVCL_0004 |
| K563 cells | Blood | Homo sapiens (Human) | N.A. | |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | These results provided new evidence that miR-18a-5p may suppress the Warburg effect by targeting HIF-1alpha.Cells transfected with miR-18a-5p mimics were more sensitive to Adriamycin (AMD) compared with AMD group. | |||
| Key Molecule: Pyruvate kinase muscle isozyme 1 (PKM1) | [14] | |||
| Metabolic Type | Mitochondrial metabolism | |||
| Resistant Disease | Chronic myeloid leukemia [ICD-11: 2A20.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | K562/ADMR cells | Blood | Homo sapiens (Human) | CVCL_5950 |
| Experiment for Molecule Alteration |
Expression profiles | |||
| Experiment for Drug Resistance |
Cell viability assay | |||
| Mechanism Description | The overexpression of PKM1 resulted in resistance of the parental cells to 5-FU and oxaliplatin. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [15] | |||
| Sensitive Disease | Chronic myeloid leukemia [ICD-11: 2A20.0] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | NCI-H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 |
| K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 | |
| HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |
| K562-R cells | Pleural effusion | Homo sapiens (Human) | CVCL_5950 | |
| NCI-H460/VBL cells | Bone marrow | Homo sapiens (Human) | CVCL_0459 | |
| In Vivo Model | SCID beige mice | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | In ABCB1-overexpressing cell lines, HG-829 significantly enhanced cytotoxicity to daunorubicin, paclitaxel, vinblastine, vincristine, and etoposide. Coadministration of HG-829 fully restored in vivo antitumor activity of daunorubicin in mice without added toxicity. Functional assays showed that HG-829 is not a Pgp substrate or competitive inhibitor of Pgp-mediated drug efflux but rather acts as a noncompetitive modulator of P-glycoprotein transport function. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-125a | [16] | |||
| Resistant Disease | Leukemia [ICD-11: 2B33.6] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | THP-1 cells | Blood | Homo sapiens (Human) | CVCL_0006 |
| HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 | |
| HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 | |
| K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Luminescent cell viability assay | |||
| Mechanism Description | miR125a mediated daunorubicin resistance in leukemia cell lines through the decrease of GRk2 and Puma which were proved to be direct targets of miR125a. Overexpression of miR125a induced drug resistance in HL-60, k562, and THP-1cell lines through reducing apoptosis. | |||
| Key Molecule: hsa-mir-125b | [2] | |||
| Resistant Disease | Leukemia [ICD-11: 2B33.6] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | THP-1 cells | Blood | Homo sapiens (Human) | CVCL_0006 |
| Jurkat cells | Pleural effusion | Homo sapiens (Human) | CVCL_0065 | |
| K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 | |
| REH cells | Bone marrow | Homo sapiens (Human) | CVCL_1650 | |
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
Luminescent cell viability assay | |||
| Mechanism Description | miR-125b downregulated GRk2 and PUMA, which inhibited apoptosis and induced leukemia cell resistance to DNR. | |||
| Key Molecule: hsa-mir-21 | [7] | |||
| Resistant Disease | Leukemia [ICD-11: 2B33.6] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K/AKT signaling pathway | Activation | hsa04151 | |
| In Vitro Model | K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 |
| K562/DNR cells | Blood | Homo sapiens (Human) | CVCL_4T87 | |
| Experiment for Molecule Alteration |
RT-PCR; Northern blotting analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | DNR-induced drug resistance is associated with upregulation of miR-21 in the leukaemia cell line k562. miR-21 may regulate the survival of leukaemia cell lines by targeting PTEN expression and causing subsequent changes in the PI3k/Akt pathway. | |||
|
|
||||
| Key Molecule: Beta adrenoceptor kinase 1 (GRK2) | [16] | |||
| Resistant Disease | Leukemia [ICD-11: 2B33.6] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | THP-1 cells | Blood | Homo sapiens (Human) | CVCL_0006 |
| HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 | |
| HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 | |
| K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 | |
| Experiment for Molecule Alteration |
Luciferase reporter assay; Western blot analysis | |||
| Experiment for Drug Resistance |
Luminescent cell viability assay | |||
| Mechanism Description | miR125a mediated daunorubicin resistance in leukemia cell lines through the decrease of GRk2 and Puma which were proved to be direct targets of miR125a. Overexpression of miR125a induced drug resistance in HL-60, k562, and THP-1cell lines through reducing apoptosis. | |||
| Key Molecule: Bcl-2-binding component 3 (BBC3) | [16] | |||
| Resistant Disease | Leukemia [ICD-11: 2B33.6] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | THP-1 cells | Blood | Homo sapiens (Human) | CVCL_0006 |
| HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 | |
| HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 | |
| K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 | |
| Experiment for Molecule Alteration |
Luciferase reporter assay; Western blot analysis | |||
| Experiment for Drug Resistance |
Luminescent cell viability assay | |||
| Mechanism Description | miR125a mediated daunorubicin resistance in leukemia cell lines through the decrease of GRk2 and Puma which were proved to be direct targets of miR125a. Overexpression of miR125a induced drug resistance in HL-60, k562, and THP-1cell lines through reducing apoptosis. | |||
| Key Molecule: Beta adrenoceptor kinase 1 (GRK2) | [2] | |||
| Resistant Disease | Leukemia [ICD-11: 2B33.6] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | THP-1 cells | Blood | Homo sapiens (Human) | CVCL_0006 |
| Jurkat cells | Pleural effusion | Homo sapiens (Human) | CVCL_0065 | |
| K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 | |
| REH cells | Bone marrow | Homo sapiens (Human) | CVCL_1650 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Luminescent cell viability assay | |||
| Mechanism Description | miR-125b downregulated GRk2 and PUMA, which inhibited apoptosis and induced leukemia cell resistance to DNR. | |||
| Key Molecule: Bcl-2-binding component 3 (BBC3) | [2] | |||
| Resistant Disease | Leukemia [ICD-11: 2B33.6] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | THP-1 cells | Blood | Homo sapiens (Human) | CVCL_0006 |
| Jurkat cells | Pleural effusion | Homo sapiens (Human) | CVCL_0065 | |
| K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 | |
| REH cells | Bone marrow | Homo sapiens (Human) | CVCL_1650 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Luminescent cell viability assay | |||
| Mechanism Description | miR-125b downregulated GRk2 and PUMA, which inhibited apoptosis and induced leukemia cell resistance to DNR. | |||
| Key Molecule: Phosphatase and tensin homolog (PTEN) | [7] | |||
| Resistant Disease | Leukemia [ICD-11: 2B33.6] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K/AKT signaling pathway | Activation | hsa04151 | |
| In Vitro Model | K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 |
| K562/DNR cells | Blood | Homo sapiens (Human) | CVCL_4T87 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | DNR-induced drug resistance is associated with upregulation of miR-21 in the leukaemia cell line k562. miR-21 may regulate the survival of leukaemia cell lines by targeting PTEN expression and causing subsequent changes in the PI3k/Akt pathway. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: hsa-mir-210 | [17] | |||
| Sensitive Disease | Paediatric acute lymphocytic leukemia [ICD-11: 2B33.4] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | MLL/AF4+ RS4 cells | Blood | Homo sapiens (Human) | CVCL_0093 |
| TEL/AML1+ Reh cells | Blood | Homo sapiens (Human) | CVCL_ZV66 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
CellTiter 96 aqueous one solution cell proliferation assay | |||
| Mechanism Description | Functioning as a hypoxamir (i.e. a microRNA whose expression is upregulated by hypoxia), miR-210 targets many genes involved in a wide range of physiological processes, such as cell survival/proliferation, mitochondrial metabolism, protein modification/transport, DNA damage repair and angiogenesis. Increasing/decreasing miR-210 expression using agomir/antagomir could enhance or reduce the response of Reh cells and RS4;11 cells to daunorubicin/dexamethasone/L-asparaginase and daunorubicin/dexamethasone/vincristine, respectively. miR-210 may be a good prognostic factor and a useful predictor of drug sensitivity, and is a potential therapeutic target for pediatric ALL. | |||
| Key Molecule: hsa-mir-181a | [18] | |||
| Sensitive Disease | Leukemia [ICD-11: 2B33.6] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 |
| K562/A02 cells | Blood | Homo sapiens (Human) | CVCL_0368 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Anti-apoptotic BCL-2 contributes to the survival and chemoresistance of quiescent leukemia CD34+ cells, leukemia cells with decreased miR-181a expression and elevated BCL-2 protein expression were more resistant to DNR than the control cells. | |||
|
|
||||
| Key Molecule: Apoptosis regulator Bcl-2 (BCL2) | [18] | |||
| Sensitive Disease | Leukemia [ICD-11: 2B33.6] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 |
| K562/A02 cells | Blood | Homo sapiens (Human) | CVCL_0368 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Anti-apoptotic BCL-2 contributes to the survival and chemoresistance of quiescent leukemia CD34+ cells, leukemia cells with decreased miR-181a expression and elevated BCL-2 protein expression were more resistant to DNR than the control cells. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Nijmegen breakage syndrome protein 1 (NBS1) | [10] | |||
| Metabolic Type | Glucose metabolism | |||
| Resistant Disease | Gastric adenocarcinoma [ICD-11: 2B72.0] | |||
| Molecule Alteration | Lactylation | K388 |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | AGS cells | Gastric | Homo sapiens (Human) | CVCL_0139 |
| Experiment for Molecule Alteration |
LC-MS/MS analysis | |||
| Mechanism Description | Lactylation of NBS1 at lysine 388 (K388) is essential for MRE11-RAD50-NBS1 (MRN) complex formation and the accumulation of HR repair proteins at the sites of DNA double-strand breaks.It promotes DNA-damaging treatment resistance via HR repair. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: ATP-binding cassette sub-family G2 (ABCG2) | [8] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/AdrVp cells | Breast | Homo sapiens (Human) | CVCL_4Y46 | |
| Experiment for Molecule Alteration |
Northern blot analysis | |||
| Experiment for Drug Resistance |
Flow cytometric assay | |||
| Mechanism Description | The mRNA encodes a 663-aa member of the ATP-binding cassette superfamily of transporters that we term breast cancer resistance protein (BCRP). Enforced expression of the full-length BCRP cDNA in MCF-7 breast cancer cells confers resistance to mitoxantrone, doxorubicin, and daunorubicin, reduces daunorubicin accumulation and retention, and causes an ATP-dependent enhancement of the efflux of rhodamine 123 in the cloned transfected. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: ATP-binding cassette sub-family B5 (ABCB5) | [19] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7/DX1 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Sf9 cells | Ovary | Homo sapiens (Human) | CVCL_0549 | |
| HCMEC/D3 cells | Brain | Homo sapiens (Human) | CVCL_U985 | |
| Experiment for Drug Resistance |
Flow Cytometry assay | |||
| Mechanism Description | QT2C2Me2 (8) inhibited P-gp transport of R123, calcein-AM, doxorubicin, BODIPY-FL-verapamil, and [3H]-daunorubicin similarly to QT2C2 (1), with IC50 values in the low micromolar range. These IC50 values were 13- to 75-fold lower than those for the QT monomer. These results indicated that both dimers are effective P-gp inhibitor. | |||
ICD-12: Respiratory system diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: MATE family efflux transporter (ABEM) | [6] | |||
| Resistant Disease | Acinetobacter baumannii infection [ICD-11: CA40.4] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli kAM32 | 562 | ||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | AbeM was found to be an H+-coupled multidrug efflux pump and a unique member of the MATE family which lead to drug resistance. | |||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
