Molecule Information
General Information of the Molecule (ID: Mol00050)
| Name |
Cellular tumor antigen p53 (TP53)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Antigen NY-CO-13; Phosphoprotein p53; Tumor suppressor p53; P53
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
TP53
|
||||
| Gene ID | |||||
| Location |
chr17:7661779-7687538[-]
|
||||
| Sequence |
MEEPQSDPSVEPPLSQETFSDLWKLLPENNVLSPLPSQAMDDLMLSPDDIEQWFTEDPGP
DEAPRMPEAAPPVAPAPAAPTPAAPAPAPSWPLSSSVPSQKTYQGSYGFRLGFLHSGTAK SVTCTYSPALNKMFCQLAKTCPVQLWVDSTPPPGTRVRAMAIYKQSQHMTEVVRRCPHHE RCSDSDGLAPPQHLIRVEGNLRVEYLDDRNTFRHSVVVPYEPPEVGSDCTTIHYNYMCNS SCMGGMNRRPILTIITLEDSSGNLLGRNSFEVRVCACPGRDRRTEEENLRKKGEPHHELP PGSTKRALPNNTSSSPQPKKKPLDGEYFTLQIRGRERFEMFRELNEALELKDAQAGKEPG GSRAHSSHLKSKKGQSTSRHKKLMFKTEGPDSD Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Acts as a tumor suppressor in many tumor types; induces growth arrest or apoptosis depending on the physiological circumstances and cell type. Involved in cell cycle regulation as a trans-activator that acts to negatively regulate cell division by controlling a set of genes required for this process. One of the activated genes is an inhibitor of cyclin-dependent kinases. Apoptosis induction seems to be mediated either by stimulation of BAX and FAS antigen expression, or by repression of Bcl-2 expression. Its pro-apoptotic activity is activated via its interaction with PPP1R13B/ASPP1 or TP53BP2/ASPP2. However, this activity is inhibited when the interaction with PPP1R13B/ASPP1 or TP53BP2/ASPP2 is displaced by PPP1R13L/iASPP. In cooperation with mitochondrial PPIF is involved in activating oxidative stress-induced necrosis; the function is largely independent of transcription. Induces the transcription of long intergenic non-coding RNA p21 (lincRNA-p21) and lincRNA-Mkln1. LincRNA-p21 participates in TP53-dependent transcriptional repression leading to apoptosis and seems to have an effect on cell-cycle regulation. Implicated in Notch signaling cross-over. Prevents CDK7 kinase activity when associated to CAK complex in response to DNA damage, thus stopping cell cycle progression. Isoform 2 enhances the transactivation activity of isoform 1 from some but not all TP53-inducible promoters. Isoform 4 suppresses transactivation activity and impairs growth suppression mediated by isoform 1. Isoform 7 inhibits isoform 1-mediated apoptosis. Regulates the circadian clock by repressing CLOCK-ARNTL/BMAL1-mediated transcriptional activation of PER2.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
18 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hepatocellular carcinoma [ICD-11: 2C12.2] | [1] | |||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Sensitive Drug | IFN-alpha | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Liver cancer [ICD-11: 2C12] | |||
| The Specified Disease | Liver cancer | |||
| The Studied Tissue | Liver tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.64E-02 Fold-change: 5.45E-02 Z-score: 2.27E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| NF-kappaB p65/p53 signaling pathway | Regulation | N.A. | ||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| HCCLM3 cells | Liver | Homo sapiens (Human) | CVCL_6832 | |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| SMMC7721 cells | Uterus | Homo sapiens (Human) | CVCL_0534 | |
| PLC cells | Liver | Homo sapiens (Human) | CVCL_0485 | |
| MHCC97-H cells | Liver | Homo sapiens (Human) | CVCL_4972 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | LncRNA 00607 overexpression leads to decreased HCC cell proliferation in vitro and in vivo, enhanced apoptosis and chemotherapeutic drug sensitivity, inhibiting the p65 transcription by binding to the p65 promoter region, therefore contributing to increased p53 levels in HCC. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [2] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | Docetaxel | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Ovarian cancer [ICD-11: 2C73] | |||
| The Specified Disease | Ovarian cancer | |||
| The Studied Tissue | Ovarian tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.96E-01 Fold-change: 3.21E-02 Z-score: 7.12E-01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | 3AO cells | Ovary | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | LncRNA PVT1 boost the expression of p53 and TIMP 1 to enhance ovarian cancer cells chemosensitivity for carboplatin and docetaxel. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer bone metastasis [ICD-11: 2E03.1] | [9] | |||
| Resistant Disease | Breast cancer bone metastasis [ICD-11: 2E03.1] | |||
| Resistant Drug | Docetaxel | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Flow cytometric | |||
| Mechanism Description | Interleukin-6 (IL-6), a pro-inflammatory cytokine produced in the tumor microenvironment by stromal cells, fibroblasts, and cancer cells. Binding of IL-6 to its receptor IL-6R on the cell membrane activates Janus Kinase 2 (JAK2) kinases. Activated JAK2 mediates phosphorylation, dimerization, and nuclear translocation of Signal Transducer and Activator of Transcription 3 (STAT3). STAT3 signaling mediates the expression of various genes, including p53, Bcl-2, MRP1, and ABCG2. Bcl-2 and p53 are associated with regulation of apoptosis while overexpression of drug transporters MRP1, ABCG2 has been shown to mediate efflux of drugs from cancer cells, thus decreasing intracellular drug concentration leading to drug-resistance. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Glioma [ICD-11: 2A00.1] | [3] | |||
| Sensitive Disease | Glioma [ICD-11: 2A00.1] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Brain cancer [ICD-11: 2A00] | |||
| The Specified Disease | Brain cancer | |||
| The Studied Tissue | Nervous tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.42E-178 Fold-change: 2.91E-01 Z-score: 3.33E+01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | AKT signaling pathway | Inhibition | hsa04151 | |
| Cell apoptosis | Activation | hsa04210 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| U87-MG cells | Brain | Homo sapiens (Human) | CVCL_0022 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay; Flow cytometry assay | |||
| Mechanism Description | microRNA-127 silencing significantly affects cell growth and increases the sensitivity to adriamycin. microRNA-127 silencing arrests the cell cycle, potentiates adriamycin-induced apoptosis, and increases cellular Rh-123 uptake. microRNA-127 silencing down-regulates MDR1, MRP1, Runx2, Bcl-2, Survivin and ErbB4 expression while up-regulates p53 expression. microRNA-127 silencing inhibits AkT phosphorylation. | |||
| Disease Class: Hepatocellular carcinoma [ICD-11: 2C12.2] | [1] | |||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| NF-kappaB p65/p53 signaling pathway | Regulation | N.A. | ||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| HCCLM3 cells | Liver | Homo sapiens (Human) | CVCL_6832 | |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| SMMC7721 cells | Uterus | Homo sapiens (Human) | CVCL_0534 | |
| PLC cells | Liver | Homo sapiens (Human) | CVCL_0485 | |
| MHCC97-H cells | Liver | Homo sapiens (Human) | CVCL_4972 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | LncRNA 00607 overexpression leads to decreased HCC cell proliferation in vitro and in vivo, enhanced apoptosis and chemotherapeutic drug sensitivity, inhibiting the p65 transcription by binding to the p65 promoter region, therefore contributing to increased p53 levels in HCC. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Ewing sarcoma [ICD-11: 2B52.0] | [10] | |||
| Resistant Disease | Ewing sarcoma [ICD-11: 2B52.0] | |||
| Resistant Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| miR125b-p53/BAKT signaling pathway | Activation | hsa05206 | ||
| In Vitro Model | RD-ES cells | Bones | Homo sapiens (Human) | CVCL_2169 |
| Sk-ES cells | Bones | Homo sapiens (Human) | CVCL_0627 | |
| Sk-N-MC cells | Bones | Homo sapiens (Human) | CVCL_0530 | |
| TC-71 cells | Bones | Homo sapiens (Human) | CVCL_2213 | |
| VH-64 cells | Bones | Homo sapiens (Human) | CVCL_9672 | |
| WE-68 cells | Bones | Homo sapiens (Human) | CVCL_9717 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Celltiter-glo luminescent cell viability assay | |||
| Mechanism Description | miR-125b led to the development of chemoresistance by suppressing the expression of p53 and Bak, and repression of miR-125b sensitized EWS cells to apoptosis induced by treatment with various cytotoxic drugs. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lung cancer [ICD-11: 2C25.5] | [4] | |||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | |||
| The Specified Disease | Lung cancer | |||
| The Studied Tissue | Lung tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.24E-02 Fold-change: 2.19E-02 Z-score: 1.69E+00 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| Wnt/Beta-catenin signaling pathway | Inhibition | hsa04310 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Down-regulation of Meg3 enhances cisplatin resistance of lung cancer cells through activation of the WNT/beta-catenin signaling pathway.The present study detected that the expression levels of Meg3 were significantly lower in cisplatin-resistant A549/DDP lung cancer cells, compared with those in parental A549 cells. The results of the present study also demonstrated that the Meg3-mediated chemosensitivity enhancement was associated with the induction of cell-cycle arrest and increased apoptosis, through regulation of p53, beta-catenin and survivin, which is a target gene of the WNT/beta-catenin signaling pathway. | |||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [8] | |||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| p53/p66shc signaling pathway | Activation | hsa04115 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| A549/DDP cells | Lung | Homo sapiens (Human) | CVCL_0023 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometric analysis | |||
| Mechanism Description | Downregulated long non-coding RNA TRPM2-AS inhibits cisplatin resistance of non-small cell lung cancer cells via activation of p53- p66shc pathway. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Bladder cancer [ICD-11: 2C94.0] | [5] | |||
| Resistant Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Bladder cancer [ICD-11: 2C94] | |||
| The Specified Disease | Bladder cancer | |||
| The Studied Tissue | Bladder tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.97E-02 Fold-change: -4.78E-02 Z-score: -2.01E+00 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | 5637 cells | Bladder | Homo sapiens (Human) | CVCL_0126 |
| T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |
| SW780 cells | Bladder | Homo sapiens (Human) | CVCL_1728 | |
| Experiment for Molecule Alteration |
Western blot analysis; qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Upregulated HIF1A-AS2 hampers the p53 family proteins dependent apoptotic pathway to promote Cis resistance in bladder cancer. | |||
| Disease Class: Nasopharyngeal carcinoma [ICD-11: 2B6B.0] | [7] | |||
| Resistant Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B.0] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| p53 signaling pathway | Inhibition | hsa04115 | ||
| In Vitro Model | CNE1 cells | Throat | Homo sapiens (Human) | CVCL_6888 |
| CNE-2 cells | Nasopharynx | Homo sapiens (Human) | CVCL_6888 | |
| TW03 cells | Nasopharynx | Homo sapiens (Human) | CVCL_6010 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | In the TW03/DDP cells, the expression levels of miR 125a and miR 125b were upregulated, and this caused downregulation of p53. Ectopic expression of these miRNAs in the TW03 cell model sensitized TW03 to cisplatin by decreasing the protein expression levels of p53, whereas ectopic expression in the antisense oligos of these microRNAs demonstrated the opposite effect. In addition, the present demonstrated that the cisplatin induced expression of miR 125a and miR 125b inhibited cisplatin induced apoptosis in the TW03 cells by decreasing the protein expression levels of p53. Taken together, the present study revealed for the first time, to the best of our knowledge, that induction of the expression of miR 125a and miR 125b by treatment with cisplatin resulted in resistance to the cisplatin drug in the NPC cells. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Chronic lymphocytic leukemia [ICD-11: 2A82.0] | [6] | |||
| Resistant Disease | Chronic lymphocytic leukemia [ICD-11: 2A82.0] | |||
| Resistant Drug | Bendamustine hydrochloride | |||
| Molecule Alteration | Mutation | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | ||
| Experiment for Molecule Alteration |
Whole exome sequencing assay; Targeted deep sequencing assay; Sanger sequencing assay | |||
| Mechanism Description | Following exposure to chemoimmunotherapy, the resistant TP53 aberrant clones accumulate and dominate the tumour. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [2] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | Carboplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | 3AO cells | Ovary | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | LncRNA PVT1 boost the expression of p53 and TIMP 1 to enhance ovarian cancer cells chemosensitivity for carboplatin and docetaxel. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [11] | |||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Resistant Drug | Etoposide | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| Experiment for Molecule Alteration |
Western blot assay | |||
| Experiment for Drug Resistance |
Cell viability assay | |||
| Mechanism Description | M. arginini?and?M. salivarium, promoted the initiation of EMT and simultaneous suppression of the p53 tumor suppressor in A549 lung cancer cells. This led to an increase of cancer cell motility, resistance to the antitumor drug etoposide concomitantly with decreased autophagy. | |||
|
|
||||
| Disease Class: Ewing sarcoma [ICD-11: 2B52.0] | [10] | |||
| Resistant Disease | Ewing sarcoma [ICD-11: 2B52.0] | |||
| Resistant Drug | Etoposide | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| miR125b-p53/BAKT signaling pathway | Activation | hsa05206 | ||
| In Vitro Model | RD-ES cells | Bones | Homo sapiens (Human) | CVCL_2169 |
| Sk-ES cells | Bones | Homo sapiens (Human) | CVCL_0627 | |
| Sk-N-MC cells | Bones | Homo sapiens (Human) | CVCL_0530 | |
| TC-71 cells | Bones | Homo sapiens (Human) | CVCL_2213 | |
| VH-64 cells | Bones | Homo sapiens (Human) | CVCL_9672 | |
| WE-68 cells | Bones | Homo sapiens (Human) | CVCL_9717 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Celltiter-glo luminescent cell viability assay | |||
| Mechanism Description | miR-125b led to the development of chemoresistance by suppressing the expression of p53 and Bak, and repression of miR-125b sensitized EWS cells to apoptosis induced by treatment with various cytotoxic drugs. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Chronic lymphocytic leukemia [ICD-11: 2A82.0] | [12] | |||
| Resistant Disease | Chronic lymphocytic leukemia [ICD-11: 2A82.0] | |||
| Resistant Drug | Fludarabine | |||
| Molecule Alteration | Mutation | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | NF-kB signaling pathway | Inhibition | hsa04218 | |
| Experiment for Molecule Alteration |
Next-generation sequencing assay | |||
| Mechanism Description | Genes belonging to the DNA damage response and cell cycle control (TP53, ATM, POT1, BIRC3) happen to be more frequently mutated in uCLL cases. However, DNA-damaging chemotherapy results in the development of chemo-resistance in most of the cases, which has been initially attributed to the selection of driver mutations affecting genes of the DNA-damage response pathways, such as TP53 and ATM. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hepatocellular carcinoma [ICD-11: 2C12.2] | [1] | |||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Sensitive Drug | Fluorouracil | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| NF-kappaB p65/p53 signaling pathway | Regulation | N.A. | ||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| HCCLM3 cells | Liver | Homo sapiens (Human) | CVCL_6832 | |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| SMMC7721 cells | Uterus | Homo sapiens (Human) | CVCL_0534 | |
| PLC cells | Liver | Homo sapiens (Human) | CVCL_0485 | |
| MHCC97-H cells | Liver | Homo sapiens (Human) | CVCL_4972 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | LncRNA 00607 overexpression leads to decreased HCC cell proliferation in vitro and in vivo, enhanced apoptosis and chemotherapeutic drug sensitivity, inhibiting the p65 transcription by binding to the p65 promoter region, therefore contributing to increased p53 levels in HCC. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Lung cancer [ICD-11: 2C25.5] | [13] | ||||||||||||
| Resistant Disease | Lung cancer [ICD-11: 2C25.5] | ||||||||||||
| Resistant Drug | Gefitinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.Y163C |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.05 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.65 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
S
S
S
S
S
S
V
V
P
P
S
S
100
|
Q
Q
K
K
T
T
Y
Y
Q
Q
G
G
S
S
Y
Y
G
G
F
F
110
|
R
R
L
L
G
G
F
F
L
L
H
H
S
S
G
G
T
T
A
A
120
|
K
K
S
S
V
V
T
T
C
C
T
T
Y
Y
S
S
P
P
A
A
130
|
L
L
N
N
K
K
M
L
F
F
C
C
Q
Q
L
L
A
A
K
K
140
|
T
T
C
C
P
P
V
V
Q
Q
L
L
W
W
V
V
D
D
S
S
150
|
T
T
P
P
P
P
P
P
G
G
T
T
R
R
V
V
R
R
A
A
160
|
M
M
A
A
I
I
Y
C
K
K
Q
Q
S
S
Q
Q
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
R
C
C
P
P
H
H
H
H
180
|
E
E
R
R
C
C
S
S
D
D
S
S
D
D
G
G
L
L
A
A
190
|
P
P
P
P
Q
Q
H
H
L
L
I
I
R
R
V
V
E
E
G
G
200
|
N
N
L
L
R
R
V
A
E
E
Y
Y
L
L
D
D
D
D
R
R
210
|
N
N
T
T
F
F
R
R
H
H
S
S
V
V
V
V
V
V
P
P
220
|
Y
Y
E
E
P
P
P
P
E
E
V
V
G
G
S
S
D
D
C
C
230
|
T
T
T
T
I
I
H
H
Y
Y
N
N
Y
Y
M
M
C
C
N
Y
240
|
S
S
S
S
C
C
M
M
G
G
G
G
M
M
N
N
R
R
R
R
250
|
P
P
I
I
L
L
T
T
I
I
I
I
T
T
L
L
E
E
D
D
260
|
S
S
S
S
G
G
N
N
L
L
L
L
G
G
R
R
N
D
S
S
270
|
F
F
E
E
V
V
R
R
V
V
C
C
A
A
C
C
P
P
G
G
280
|
R
R
D
D
R
R
R
R
T
T
E
E
E
E
E
E
N
N
L
L
290
|
R
R
K
K
K
K
G
G
E
E
P
P
H
H
H
H
E
E
L
L
300
|
P
P
P
P
G
G
S
S
T
T
K
K
R
R
A
A
L
L
P
P
310
|
N
N
N
N
T
T
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | AXLK signaling pathway | Activation | hsa01521 | ||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Circulating-free DNA assay; Whole exome sequencing assay | ||||||||||||
| Mechanism Description | Quantification of allele fractions in plasma identified increased representation of mutant alleles in association with emergence of therapy resistance. | ||||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Rheumatoid arthritis [ICD-11: FA20.0] | [14] | |||
| Resistant Disease | Rheumatoid arthritis [ICD-11: FA20.0] | |||
| Resistant Drug | Leflunomide | |||
| Molecule Alteration | Mutation | p.P151S+p.R175H+p.G245C+p.R282W |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Mechanism Description | The wild-type p53 tumor suppressor (p53) is overexpressed in response to DNA damage and inflammation in RA fibroblast-like synoviocytes (FLS), which are highly specialized mesenchymal cells located in the internal lining of the synovium and are involved in the pathogenesis and progression of RA. In line with the effects of p53 gain-of-function mutation in tumor progression, mutation-mediated gain-of-function of p53 may contribute to the invasiveness and apoptosis-resistant feature of FLS in RA and the increased expression of cartilage degradative proteases, leading to degeneration of cartilage and bone. Gene knockout or gene transfer studies using a collagen-II-induced arthritis (CIA) model have established the crucial role of p53 in RA that provides a basis for additional research to fully characterize the clinical implications of p53 somatic mutations in drug-resistant RA. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Rheumatoid arthritis [ICD-11: FA20.0] | [14] | |||
| Resistant Disease | Rheumatoid arthritis [ICD-11: FA20.0] | |||
| Resistant Drug | Methotrexate | |||
| Molecule Alteration | Mutation | p.P151S+p.R175H+p.G245C+p.R282W |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Mechanism Description | The wild-type p53 tumor suppressor (p53) is overexpressed in response to DNA damage and inflammation in RA fibroblast-like synoviocytes (FLS), which are highly specialized mesenchymal cells located in the internal lining of the synovium and are involved in the pathogenesis and progression of RA. In line with the effects of p53 gain-of-function mutation in tumor progression, mutation-mediated gain-of-function of p53 may contribute to the invasiveness and apoptosis-resistant feature of FLS in RA and the increased expression of cartilage degradative proteases, leading to degeneration of cartilage and bone. Gene knockout or gene transfer studies using a collagen-II-induced arthritis (CIA) model have established the crucial role of p53 in RA that provides a basis for additional research to fully characterize the clinical implications of p53 somatic mutations in drug-resistant RA. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lung cancer [ICD-11: 2C25.5] | [15] | |||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Sensitive Drug | Methotrexate | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| P53/P21/EZH2/E-cad signaling pathway | Activation | hsa04115 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Transwell assay | |||
| Mechanism Description | Over expression of miR-200c reduced the resistance of A549/MTX cells to MTX with the mechanism of inducing apoptosis through the P53/P21 pathway. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lung cancer [ICD-11: 2C25.5] | [16] | |||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Sensitive Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| MEG3-P53 signaling pathway | Activation | hsa04115 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | The downregulation of MEG3 attenuated PTX-induced cytotoxicity, whereas upregulation of MEG3 induced cell death and increased P53 expression. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Chronic myelogenous leukemia [ICD-11: 2A20.3] | [17] | |||
| Resistant Disease | Chronic myelogenous leukemia [ICD-11: 2A20.3] | |||
| Resistant Drug | Ponatinib | |||
| Molecule Alteration | Missense mutation | p.V216M |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | JAKT2/STAT signaling pathway | Activation | hsa04030 | |
| RAF/KRAS/MEK signaling pathway | Activation | hsa04010 | ||
| In Vitro Model | HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 |
| U937 cells | Blood | Homo sapiens (Human) | CVCL_0007 | |
| K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 | |
| KCL-22 cells | Bone marrow | Homo sapiens (Human) | CVCL_2091 | |
| Sup-B15 cells | Bone marrow | Homo sapiens (Human) | CVCL_0103 | |
| HEL cells | Blood | Homo sapiens (Human) | CVCL_0001 | |
| HMC-1.2 cells | Blood | Homo sapiens (Human) | CVCL_H205 | |
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | ||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Sanger Sequencing assay | |||
| Mechanism Description | Po.tinib led to a decrease of ABL T315I positive transcripts from 47% before po.tinib treatment to 16% at the time of po.tinib resistance in this patient, suggesting that both TP53 and ABL mutations were present in the same clone and that the newly acquired TP53 mutation might have caused po.tinib resistance in this patient. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Chronic lymphocytic leukemia [ICD-11: 2A82.0] | [6] | |||
| Resistant Disease | Chronic lymphocytic leukemia [ICD-11: 2A82.0] | |||
| Resistant Drug | Rituximab | |||
| Molecule Alteration | Mutation | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | ||
| Experiment for Molecule Alteration |
Whole exome sequencing assay; Targeted deep sequencing assay; Sanger sequencing assay | |||
| Mechanism Description | Following exposure to chemoimmunotherapy, the resistant TP53 aberrant clones accumulate and dominate the tumour. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hereditary angioedema [ICD-11: 4B05.0] | [18] | |||
| Resistant Disease | Hereditary angioedema [ICD-11: 4B05.0] | |||
| Resistant Drug | Stanozolol | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vivo Model | Sprague Dawley male rats model | Rattus norvegicus | ||
| Experiment for Molecule Alteration |
Gene expression analysis | |||
| Mechanism Description | Stanozolol can increases levels of Bax, Bcl-2, P53, caspase 3 and Bax/Bcl-2 ratio. Resistance training, 50 and 100 mg/kg Tribulus terrestris and resistance training along with Tribulus terrestris can decrease the Bax, Bcl-2, P53, caspase 3 and Bax/Bcl-2 ratio in rats exposed to Stanozolol. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [19] | |||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Sensitive Drug | Venetoclax | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | H1048 shp53 cells | Lung | Homo sapiens (Human) | N.A. |
| H211 shp53 cells | Lung | Homo sapiens (Human) | N.A. | |
| Experiment for Drug Resistance |
Fluorescence-activated cell sorting assay; Cell viability assay | |||
| Mechanism Description | Down-Regulation of Onc-p53 Increases BIM Expression and Sensitizes to Venetoclax in SCLC-P Cells. Down-regulation of Onc-p53 increases the expression of a BH3-only pro-apoptotic BIM and sensitizes to venetoclax in SCLC-P cells | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Ewing sarcoma [ICD-11: 2B52.0] | [10] | |||
| Resistant Disease | Ewing sarcoma [ICD-11: 2B52.0] | |||
| Resistant Drug | Vincristine | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| miR125b-p53/BAKT signaling pathway | Activation | hsa05206 | ||
| In Vitro Model | RD-ES cells | Bones | Homo sapiens (Human) | CVCL_2169 |
| Sk-ES cells | Bones | Homo sapiens (Human) | CVCL_0627 | |
| Sk-N-MC cells | Bones | Homo sapiens (Human) | CVCL_0530 | |
| TC-71 cells | Bones | Homo sapiens (Human) | CVCL_2213 | |
| VH-64 cells | Bones | Homo sapiens (Human) | CVCL_9672 | |
| WE-68 cells | Bones | Homo sapiens (Human) | CVCL_9717 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Celltiter-glo luminescent cell viability assay | |||
| Mechanism Description | miR-125b led to the development of chemoresistance by suppressing the expression of p53 and Bak, and repression of miR-125b sensitized EWS cells to apoptosis induced by treatment with various cytotoxic drugs. | |||
Clinical Trial Drug(s)
7 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [20] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | Ganetespib | ||||||||||||
| Molecule Alteration | Missense mutation | p.R175H (c.524G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.37 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.38 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
H
C
C
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |||||||||
| H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | ||||||||||
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | ||||||||||
| ES2 cells | Ovary | Homo sapiens (Human) | CVCL_AX39 | ||||||||||
| DU145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 | ||||||||||
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | ||||||||||
| MDA-46 cells | N.A. | Homo sapiens (Human) | N.A. | ||||||||||
| HOC7 cells | Ascites | Homo sapiens (Human) | CVCL_5455 | ||||||||||
| EFO21 cells | Ascites | Homo sapiens (Human) | CVCL_0029 | ||||||||||
| COV434 cells | N.A. | Homo sapiens (Human) | CVCL_2010 | ||||||||||
| COLO704 cells | Ascites | Homo sapiens (Human) | CVCL_1994 | ||||||||||
| HOC7 cells | Ascites | Homo sapiens (Human) | CVCL_5455 | ||||||||||
| In Vivo Model | Athymic (nu/nu) male xenograft mouse model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; Quantitative PCR analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-blue cell viability assay | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [20] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | Ganetespib | ||||||||||||
| Molecule Alteration | Missense mutation | p.R280K (c.839G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.05 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.00 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
S
-
S
-
S
-
V
V
P
P
S
S
100
|
Q
Q
K
K
T
T
Y
Y
Q
Q
G
G
S
S
Y
Y
G
G
F
F
110
|
R
R
L
L
G
G
F
F
L
L
H
H
S
S
G
G
T
T
A
A
120
|
K
K
S
S
V
V
T
T
C
C
T
T
Y
Y
S
S
P
P
A
A
130
|
L
L
N
N
K
K
M
M
F
F
C
C
Q
Q
L
L
A
A
K
K
140
|
T
T
C
C
P
P
V
V
Q
Q
L
L
W
W
V
V
D
D
S
S
150
|
T
T
P
P
P
P
P
P
G
G
T
T
R
R
V
V
R
R
A
A
160
|
M
M
A
A
I
I
Y
Y
K
K
Q
Q
S
S
Q
Q
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
R
C
C
P
P
H
H
H
H
180
|
E
E
R
R
C
C
S
S
D
D
S
S
D
D
G
G
L
L
A
A
190
|
P
P
P
P
Q
Q
H
H
L
L
I
I
R
R
V
V
E
E
G
G
200
|
N
N
L
L
R
R
V
V
E
E
Y
Y
L
L
D
D
D
D
R
R
210
|
N
N
T
T
F
F
R
R
H
H
S
S
V
V
V
V
V
V
P
P
220
|
Y
Y
E
E
P
P
P
P
E
E
V
V
G
G
S
S
D
D
C
C
230
|
T
T
T
T
I
I
H
H
Y
Y
N
N
Y
Y
M
M
C
C
N
N
240
|
S
S
S
S
C
C
M
M
G
G
G
G
M
M
N
N
R
R
R
R
250
|
P
P
I
I
L
L
T
T
I
I
I
I
T
T
L
L
E
E
D
D
260
|
S
S
S
S
G
G
N
N
L
L
L
L
G
G
R
R
N
N
S
S
270
|
F
F
E
E
V
V
R
R
V
V
C
C
A
A
C
C
P
P
G
G
280
|
R
K
D
D
R
R
R
R
T
T
E
E
E
E
E
E
N
N
L
L
290
|
R
-
K
-
K
-
G
-
E
-
P
-
H
-
H
-
E
-
L
-
300
|
P
-
P
-
G
-
S
-
T
-
K
-
R
-
A
-
L
-
P
-
310
|
N
-
N
-
T
-
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |||||||||
| H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | ||||||||||
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | ||||||||||
| ES2 cells | Ovary | Homo sapiens (Human) | CVCL_AX39 | ||||||||||
| DU145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 | ||||||||||
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | ||||||||||
| MDA-46 cells | N.A. | Homo sapiens (Human) | N.A. | ||||||||||
| HOC7 cells | Ascites | Homo sapiens (Human) | CVCL_5455 | ||||||||||
| EFO21 cells | Ascites | Homo sapiens (Human) | CVCL_0029 | ||||||||||
| COV434 cells | N.A. | Homo sapiens (Human) | CVCL_2010 | ||||||||||
| COLO704 cells | Ascites | Homo sapiens (Human) | CVCL_1994 | ||||||||||
| HOC7 cells | Ascites | Homo sapiens (Human) | CVCL_5455 | ||||||||||
| In Vivo Model | Athymic (nu/nu) male xenograft mouse model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; Quantitative PCR analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-blue cell viability assay | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [20] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | Ganetespib | ||||||||||||
| Molecule Alteration | Missense mutation | p.L194F (c.580C>T) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |||||||||
| H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | ||||||||||
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | ||||||||||
| ES2 cells | Ovary | Homo sapiens (Human) | CVCL_AX39 | ||||||||||
| DU145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 | ||||||||||
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | ||||||||||
| MDA-46 cells | N.A. | Homo sapiens (Human) | N.A. | ||||||||||
| HOC7 cells | Ascites | Homo sapiens (Human) | CVCL_5455 | ||||||||||
| EFO21 cells | Ascites | Homo sapiens (Human) | CVCL_0029 | ||||||||||
| COV434 cells | N.A. | Homo sapiens (Human) | CVCL_2010 | ||||||||||
| COLO704 cells | Ascites | Homo sapiens (Human) | CVCL_1994 | ||||||||||
| HOC7 cells | Ascites | Homo sapiens (Human) | CVCL_5455 | ||||||||||
| In Vivo Model | Athymic (nu/nu) male xenograft mouse model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; Quantitative PCR analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-blue cell viability assay | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [20] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | Ganetespib | ||||||||||||
| Molecule Alteration | Missense mutation | p.R248Q (c.743G>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |||||||||
| H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | ||||||||||
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | ||||||||||
| ES2 cells | Ovary | Homo sapiens (Human) | CVCL_AX39 | ||||||||||
| DU145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 | ||||||||||
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | ||||||||||
| MDA-46 cells | N.A. | Homo sapiens (Human) | N.A. | ||||||||||
| HOC7 cells | Ascites | Homo sapiens (Human) | CVCL_5455 | ||||||||||
| EFO21 cells | Ascites | Homo sapiens (Human) | CVCL_0029 | ||||||||||
| COV434 cells | N.A. | Homo sapiens (Human) | CVCL_2010 | ||||||||||
| COLO704 cells | Ascites | Homo sapiens (Human) | CVCL_1994 | ||||||||||
| HOC7 cells | Ascites | Homo sapiens (Human) | CVCL_5455 | ||||||||||
| In Vivo Model | Athymic (nu/nu) male xenograft mouse model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; Quantitative PCR analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-blue cell viability assay | ||||||||||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [20] | ||||||||||||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | ||||||||||||
| Sensitive Drug | Ganetespib | ||||||||||||
| Molecule Alteration | Missense mutation | p.S241F (c.722C>T) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |||||||||
| H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | ||||||||||
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | ||||||||||
| ES2 cells | Ovary | Homo sapiens (Human) | CVCL_AX39 | ||||||||||
| DU145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 | ||||||||||
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | ||||||||||
| MDA-46 cells | N.A. | Homo sapiens (Human) | N.A. | ||||||||||
| HOC7 cells | Ascites | Homo sapiens (Human) | CVCL_5455 | ||||||||||
| EFO21 cells | Ascites | Homo sapiens (Human) | CVCL_0029 | ||||||||||
| COV434 cells | N.A. | Homo sapiens (Human) | CVCL_2010 | ||||||||||
| COLO704 cells | Ascites | Homo sapiens (Human) | CVCL_1994 | ||||||||||
| HOC7 cells | Ascites | Homo sapiens (Human) | CVCL_5455 | ||||||||||
| In Vivo Model | Athymic (nu/nu) male xenograft mouse model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; Quantitative PCR analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-blue cell viability assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [21] | |||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Sensitive Drug | Napabucasin | |||
| Molecule Alteration | Missense mutation | p.R248Q (c.743G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | JAKT2/STAT3 signaling pathway | Inhibition | hsa04030 | |
| In Vitro Model | HT29 Cells | Colon | Homo sapiens (Human) | CVCL_A8EZ |
| SW480 cells | Colon | Homo sapiens (Human) | CVCL_0546 | |
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |
| SW1116 cells | Colon | Homo sapiens (Human) | CVCL_0544 | |
| LS1034 cells | Colon | Homo sapiens (Human) | CVCL_1382 | |
| SW48 cells | Colon | Homo sapiens (Human) | CVCL_1724 | |
| Colo320 cells | Colon | Homo sapiens (Human) | CVCL_1989 | |
| SW837 cells | Colon | Homo sapiens (Human) | CVCL_1729 | |
| DLD-1 cells | Colon | Homo sapiens (Human) | CVCL_0248 | |
| SW1463 cells | Rectum | Homo sapiens (Human) | CVCL_1718 | |
| In Vivo Model | C57BL/6 mouse model | Mus musculus | ||
| Experiment for Molecule Alteration |
BCA protein assay; SDS-PAGE assay | |||
| Experiment for Drug Resistance |
Scratch assay; Transwell migration assay; Fluorescent in situ hybridization assay | |||
| Mechanism Description | The most common p53 mutant R248Q (mutp53) enhances Stat3 activation by binding to Stat3 and displacing SHP2 in colorectal cancer cells. Reduction of mutp53 genetically or by using the HSP90 inhibitor 17AAG reduces Stat3 signaling and the growth of mutp53-driven tumors. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [22] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Selumetinib | |||
| Molecule Alteration | Missense mutation | p.D259Y (c.775G>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 |
| Sk-Mel28 cells | Skin | Homo sapiens (Human) | CVCL_0526 | |
| A2058 cells | Skin | Homo sapiens (Human) | CVCL_1059 | |
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |
| WM2664 cells | Skin | Homo sapiens (Human) | CVCL_2765 | |
| SkMEL 30 cells | Skin | Homo sapiens (Human) | CVCL_0039 | |
| SkMEL 2 cells | Skin | Homo sapiens (Human) | CVCL_0069 | |
| SH4 cells | Skin | Mus musculus (Mouse) | CVCL_7702 | |
| MEXF-535 cells | Skin | Homo sapiens (Human) | N.A. | |
| MEXF-1792 cells | Skin | Homo sapiens (Human) | N.A. | |
| MEXF-1341 cells | Skin | Homo sapiens (Human) | N.A. | |
| M14 cells | Hypodermis | Homo sapiens (Human) | CVCL_1395 | |
| GAK cells | Lnguinal lymph node | Homo sapiens (Human) | CVCL_1225 | |
| Colo829 cells | Skin | Homo sapiens (Human) | CVCL_1137 | |
| In Vivo Model | Female NIH nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis; Crystallization assay; X-ray data collection and structure determination assay | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay; Enzymatic kinase assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [23] | |||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Sensitive Drug | APR-246 | |||
| Molecule Alteration | Missense mutation | p.S241F (c.722C>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| DMS456 cells | Lung | Homo sapiens (Human) | CVCL_0B92 | |
| In Vivo Model | Nude male NMRI mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The missense mutation p.S241F (c.722C>T) in gene TP53 cause the sensitivity of APR-246 by aberration of the drug's therapeutic target | |||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [23] | |||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Sensitive Drug | APR-246 | |||
| Molecule Alteration | Missense mutation | p.R273L (c.818G>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| DMS456 cells | Lung | Homo sapiens (Human) | CVCL_0B92 | |
| In Vivo Model | Nude male NMRI mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The missense mutation p.R273L (c.818G>T) in gene TP53 cause the sensitivity of APR-246 by aberration of the drug's therapeutic target | |||
|
|
||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [24] | |||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Sensitive Drug | APR-246 | |||
| Molecule Alteration | Missense mutation | p.V173M (c.517G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Blood | N.A. | ||
| Experiment for Molecule Alteration |
TP53 gene mutation status analysis | |||
| Experiment for Drug Resistance |
Pharmacokinetic Analysis | |||
| Mechanism Description | The missense mutation p.V173M (c.517G>A) in gene TP53 cause the sensitivity of APR-246 by unusual activation of pro-survival pathway | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [25] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Resistant Drug | Seliciclib | ||||||||||||
| Molecule Alteration | Missense mutation | p.Y220C (c.659A>G) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.05 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.24 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
S
S
S
S
S
S
V
V
P
P
S
S
100
|
Q
Q
K
K
T
T
Y
Y
Q
Q
G
G
S
S
Y
Y
G
G
F
F
110
|
R
R
L
L
G
G
F
F
L
L
H
H
S
S
G
G
T
T
A
A
120
|
K
K
S
S
V
V
T
T
C
C
T
T
Y
Y
S
S
P
P
A
A
130
|
L
L
N
N
K
K
M
L
F
F
C
C
Q
Q
L
L
A
A
K
K
140
|
T
T
C
C
P
P
V
V
Q
Q
L
L
W
W
V
V
D
D
S
S
150
|
T
T
P
P
P
P
P
P
G
G
T
T
R
R
V
V
R
R
A
A
160
|
M
M
A
A
I
I
Y
Y
K
K
Q
Q
S
S
Q
Q
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
R
C
C
P
P
H
H
H
H
180
|
E
E
R
R
C
C
S
S
D
D
S
S
D
D
G
G
L
L
A
A
190
|
P
P
P
P
Q
Q
H
H
L
L
I
I
R
R
V
V
E
E
G
G
200
|
N
N
L
L
R
R
V
A
E
E
Y
Y
L
L
D
D
D
D
R
R
210
|
N
N
T
T
F
F
R
R
H
H
S
S
V
V
V
V
V
V
P
P
220
|
Y
C
E
E
P
P
P
P
E
E
V
V
G
G
S
S
D
D
C
C
230
|
T
T
T
T
I
I
H
H
Y
Y
N
N
Y
Y
M
M
C
C
N
Y
240
|
S
S
S
S
C
C
M
M
G
G
G
G
M
M
N
N
R
R
R
R
250
|
P
P
I
I
L
L
T
T
I
I
I
I
T
T
L
L
E
E
D
D
260
|
S
S
S
S
G
G
N
N
L
L
L
L
G
G
R
R
N
D
S
S
270
|
F
F
E
E
V
V
R
R
V
V
C
C
A
A
C
C
P
P
G
G
280
|
R
R
D
D
R
R
R
R
T
T
E
E
E
E
E
E
N
N
L
L
290
|
R
R
K
K
K
K
G
G
E
E
P
P
H
H
H
H
E
E
L
L
300
|
P
P
P
P
G
G
S
S
T
T
K
K
R
R
A
A
L
L
P
P
310
|
N
N
N
N
T
T
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |||||||||
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | ||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Experiment for Drug Resistance |
Cell death detection ELISA assay | ||||||||||||
| Mechanism Description | The missense mutation p.Y220C (c.659A>G) in gene TP53 cause the resistance of Seliciclib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [25] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Resistant Drug | Seliciclib | ||||||||||||
| Molecule Alteration | Missense mutation | p.Y234C (c.701A>G) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.05 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.38 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
S
S
S
S
S
S
V
V
P
P
S
S
100
|
Q
Q
K
K
T
T
Y
Y
Q
Q
G
G
S
S
Y
Y
G
G
F
F
110
|
R
R
L
L
G
G
F
F
L
L
H
H
S
S
G
G
T
T
A
A
120
|
K
K
S
S
V
V
T
T
C
C
T
T
Y
Y
S
S
P
P
A
A
130
|
L
L
N
N
K
K
M
L
F
F
C
C
Q
Q
L
L
A
A
K
K
140
|
T
T
C
C
P
P
V
V
Q
Q
L
L
W
W
V
V
D
D
S
S
150
|
T
T
P
P
P
P
P
P
G
G
T
T
R
R
V
V
R
R
A
A
160
|
M
M
A
A
I
I
Y
Y
K
K
Q
Q
S
S
Q
Q
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
R
C
C
P
P
H
H
H
H
180
|
E
E
R
R
C
C
S
S
D
D
S
S
D
D
G
G
L
L
A
A
190
|
P
P
P
P
Q
Q
H
H
L
L
I
I
R
R
V
V
E
E
G
G
200
|
N
N
L
L
R
R
V
A
E
E
Y
Y
L
L
D
D
D
D
R
R
210
|
N
N
T
T
F
F
R
R
H
H
S
S
V
V
V
V
V
V
P
P
220
|
Y
Y
E
E
P
P
P
P
E
E
V
V
G
G
S
S
D
D
C
C
230
|
T
T
T
T
I
I
H
H
Y
C
N
N
Y
Y
M
M
C
C
N
Y
240
|
S
S
S
S
C
C
M
M
G
G
G
G
M
M
N
N
R
R
R
R
250
|
P
P
I
I
L
L
T
T
I
I
I
I
T
T
L
L
E
E
D
D
260
|
S
S
S
S
G
G
N
N
L
L
L
L
G
G
R
R
N
D
S
S
270
|
F
F
E
E
V
V
R
R
V
V
C
C
A
A
C
C
P
P
G
G
280
|
R
R
D
D
R
R
R
R
T
T
E
E
E
E
E
E
N
N
L
L
290
|
R
R
K
K
K
K
G
G
E
E
P
P
H
H
H
H
E
E
L
L
300
|
P
P
P
P
G
G
S
S
T
T
K
K
R
R
A
A
L
L
P
P
310
|
N
N
N
N
T
T
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |||||||||
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | ||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Experiment for Drug Resistance |
Cell death detection ELISA assay | ||||||||||||
| Mechanism Description | The missense mutation p.Y234C (c.701A>G) in gene TP53 cause the resistance of Seliciclib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [25] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Resistant Drug | Seliciclib | ||||||||||||
| Molecule Alteration | Missense mutation | p.R175H (c.524G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.37 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.38 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
H
C
C
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |||||||||
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | ||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Experiment for Drug Resistance |
Cell death detection ELISA assay | ||||||||||||
| Mechanism Description | The missense mutation p.R175H (c.524G>A) in gene TP53 cause the resistance of Seliciclib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [25] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Resistant Drug | Seliciclib | ||||||||||||
| Molecule Alteration | Missense mutation | p.P98A (c.292C>G) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |||||||||
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | ||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Experiment for Drug Resistance |
Cell death detection ELISA assay | ||||||||||||
| Mechanism Description | The missense mutation p.P98A (c.292C>G) in gene TP53 cause the resistance of Seliciclib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [25] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Resistant Drug | Seliciclib | ||||||||||||
| Molecule Alteration | Missense mutation | p.A159V (c.476C>T) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |||||||||
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | ||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Experiment for Drug Resistance |
Cell death detection ELISA assay | ||||||||||||
| Mechanism Description | The missense mutation p.A159V (c.476C>T) in gene TP53 cause the resistance of Seliciclib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [25] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Resistant Drug | Seliciclib | ||||||||||||
| Molecule Alteration | Missense mutation | p.S215G (c.643A>G) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |||||||||
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | ||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Experiment for Drug Resistance |
Cell death detection ELISA assay | ||||||||||||
| Mechanism Description | The missense mutation p.S215G (c.643A>G) in gene TP53 cause the resistance of Seliciclib by unusual activation of pro-survival pathway | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [21] | |||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Sensitive Drug | Tanespimycin | |||
| Molecule Alteration | Missense mutation | p.R248Q (c.743G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | JAKT2/STAT3 signaling pathway | Inhibition | hsa04030 | |
| In Vitro Model | HT29 Cells | Colon | Homo sapiens (Human) | CVCL_A8EZ |
| SW480 cells | Colon | Homo sapiens (Human) | CVCL_0546 | |
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |
| SW1116 cells | Colon | Homo sapiens (Human) | CVCL_0544 | |
| LS1034 cells | Colon | Homo sapiens (Human) | CVCL_1382 | |
| SW48 cells | Colon | Homo sapiens (Human) | CVCL_1724 | |
| Colo320 cells | Colon | Homo sapiens (Human) | CVCL_1989 | |
| SW837 cells | Colon | Homo sapiens (Human) | CVCL_1729 | |
| DLD-1 cells | Colon | Homo sapiens (Human) | CVCL_0248 | |
| SW1463 cells | Rectum | Homo sapiens (Human) | CVCL_1718 | |
| In Vivo Model | C57BL/6 mouse model | Mus musculus | ||
| Experiment for Molecule Alteration |
BCA protein assay; SDS-PAGE assay | |||
| Experiment for Drug Resistance |
Scratch assay; Transwell migration assay; Fluorescent in situ hybridization assay | |||
| Mechanism Description | The most common p53 mutant R248Q (mutp53) enhances Stat3 activation by binding to Stat3 and displacing SHP2 in colorectal cancer cells. Reduction of mutp53 genetically or by using the HSP90 inhibitor 17AAG reduces Stat3 signaling and the growth of mutp53-driven tumors. | |||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [20] | |||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Sensitive Drug | Tanespimycin | |||
| Molecule Alteration | Missense mutation | p.L194F (c.580C>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 |
| H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| ES2 cells | Ovary | Homo sapiens (Human) | CVCL_AX39 | |
| DU145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 | |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| MDA-46 cells | N.A. | Homo sapiens (Human) | N.A. | |
| HOC7 cells | Ascites | Homo sapiens (Human) | CVCL_5455 | |
| EFO21 cells | Ascites | Homo sapiens (Human) | CVCL_0029 | |
| COV434 cells | N.A. | Homo sapiens (Human) | CVCL_2010 | |
| COLO704 cells | Ascites | Homo sapiens (Human) | CVCL_1994 | |
| HOC7 cells | Ascites | Homo sapiens (Human) | CVCL_5455 | |
| In Vivo Model | Athymic (nu/nu) male xenograft mouse model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis; Quantitative PCR analysis | |||
| Experiment for Drug Resistance |
CellTiter-blue cell viability assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [22] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | LY3009120 | |||
| Molecule Alteration | Missense mutation | p.D259Y (c.775G>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 |
| Sk-Mel28 cells | Skin | Homo sapiens (Human) | CVCL_0526 | |
| A2058 cells | Skin | Homo sapiens (Human) | CVCL_1059 | |
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |
| WM2664 cells | Skin | Homo sapiens (Human) | CVCL_2765 | |
| SkMEL 30 cells | Skin | Homo sapiens (Human) | CVCL_0039 | |
| SkMEL 2 cells | Skin | Homo sapiens (Human) | CVCL_0069 | |
| SH4 cells | Skin | Mus musculus (Mouse) | CVCL_7702 | |
| MEXF-535 cells | Skin | Homo sapiens (Human) | N.A. | |
| MEXF-1792 cells | Skin | Homo sapiens (Human) | N.A. | |
| MEXF-1341 cells | Skin | Homo sapiens (Human) | N.A. | |
| M14 cells | Hypodermis | Homo sapiens (Human) | CVCL_1395 | |
| GAK cells | Lnguinal lymph node | Homo sapiens (Human) | CVCL_1225 | |
| Colo829 cells | Skin | Homo sapiens (Human) | CVCL_1137 | |
| In Vivo Model | Female NIH nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis; Crystallization assay; X-ray data collection and structure determination assay | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay; Enzymatic kinase assay | |||
Preclinical Drug(s)
10 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [26] | |||
| Resistant Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Resistant Drug | ALRN-6924 | |||
| Molecule Alteration | Missense mutation | p.R248Q (c.743G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [27] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | AMGMDS3 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R273H (c.818G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.05 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.98 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
S
S
S
S
S
S
V
V
P
P
S
S
100
|
Q
Q
K
K
T
T
Y
Y
Q
Q
G
G
S
S
Y
Y
G
G
F
F
110
|
R
R
L
L
G
G
F
F
L
L
H
H
S
S
G
G
T
T
A
A
120
|
K
K
S
S
V
V
T
T
C
C
T
T
Y
Y
S
S
P
P
A
A
130
|
L
L
N
N
K
K
M
L
F
F
C
C
Q
Q
L
L
A
A
K
K
140
|
T
T
C
C
P
P
V
V
Q
Q
L
L
W
W
V
V
D
D
S
S
150
|
T
T
P
P
P
P
P
P
G
G
T
T
R
R
V
V
R
R
A
A
160
|
M
M
A
A
I
I
Y
Y
K
K
Q
Q
S
S
Q
Q
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
R
C
C
P
P
H
H
H
H
180
|
E
E
R
R
C
C
S
S
D
D
S
S
D
D
G
G
L
L
A
A
190
|
P
P
P
P
Q
Q
H
H
L
L
I
I
R
R
V
V
E
E
G
G
200
|
N
N
L
L
R
R
V
A
E
E
Y
Y
L
L
D
D
D
D
R
R
210
|
N
N
T
T
F
F
R
R
H
H
S
S
V
V
V
V
V
V
P
P
220
|
Y
Y
E
E
P
P
P
P
E
E
V
V
G
G
S
S
D
D
C
C
230
|
T
T
T
T
I
I
H
H
Y
Y
N
N
Y
Y
M
M
C
C
N
Y
240
|
S
S
S
S
C
C
M
M
G
G
G
G
M
M
N
N
R
R
R
R
250
|
P
P
I
I
L
L
T
T
I
I
I
I
T
T
L
L
E
E
D
D
260
|
S
S
S
S
G
G
N
N
L
L
L
L
G
G
R
R
N
D
S
S
270
|
F
F
E
E
V
V
R
H
V
V
C
C
A
A
C
C
P
P
G
G
280
|
R
R
D
D
R
R
R
R
T
T
E
E
E
E
E
E
N
N
L
L
290
|
R
R
K
K
K
K
G
G
E
E
P
P
H
H
H
H
E
E
L
L
300
|
P
P
P
P
G
G
S
S
T
T
K
K
R
R
A
A
L
L
P
P
310
|
N
N
N
N
T
T
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| Capan-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0026 | ||||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MG-63 cells | Bone | Homo sapiens (Human) | CVCL_0426 | ||||||||||
| NCI-H82 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1591 | ||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Experiment for Drug Resistance |
Fluorescence microscopy assay | ||||||||||||
| Mechanism Description | The missense mutation p.R273H (c.818G>A) in gene TP53 cause the resistance of AMGMDS3 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [27] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | AMGMDS3 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V157F (c.469G>T) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.05 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.50 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
A
S
S
S
S
S
S
V
V
P
P
S
S
100
|
Q
Q
K
K
T
T
Y
Y
Q
Q
G
G
S
S
Y
Y
G
G
F
F
110
|
R
R
L
L
G
G
F
F
L
L
H
H
S
S
G
G
T
T
A
A
120
|
K
K
S
S
V
V
T
T
C
C
T
T
Y
Y
S
S
P
P
A
A
130
|
L
L
N
N
K
K
M
M
F
F
C
C
Q
Q
L
L
A
A
K
K
140
|
T
T
C
C
P
P
V
V
Q
Q
L
L
W
W
V
V
D
D
S
S
150
|
T
T
P
P
P
P
P
P
G
G
T
T
R
R
V
F
R
R
A
A
160
|
M
M
A
A
I
I
Y
Y
K
K
Q
Q
S
S
Q
Q
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
R
C
C
P
P
H
H
H
H
180
|
E
E
R
R
C
C
S
S
D
D
S
S
D
D
G
G
L
L
A
A
190
|
P
P
P
P
Q
Q
H
H
L
L
I
I
R
R
V
V
E
E
G
G
200
|
N
N
L
L
R
R
V
V
E
E
Y
Y
L
L
D
D
D
D
R
R
210
|
N
N
T
T
F
F
R
R
H
H
S
S
V
V
V
V
V
V
P
P
220
|
Y
Y
E
E
P
P
P
P
E
E
V
V
G
G
S
S
D
D
C
C
230
|
T
T
T
T
I
I
H
H
Y
Y
N
N
Y
Y
M
M
C
C
N
N
240
|
S
S
S
S
C
C
M
M
G
G
G
G
M
M
N
N
R
R
R
R
250
|
P
P
I
I
L
L
T
T
I
I
I
I
T
T
L
L
E
E
D
D
260
|
S
S
S
S
G
G
N
N
L
L
L
L
G
G
R
R
N
N
S
S
270
|
F
F
E
E
V
V
R
R
V
V
C
C
A
A
C
C
P
P
G
G
280
|
R
R
D
D
R
R
R
R
T
T
E
E
E
E
E
E
N
N
L
L
290
|
R
R
K
K
K
K
G
G
E
E
P
P
H
H
H
H
E
E
L
L
300
|
P
P
P
P
G
G
S
S
T
T
K
K
R
R
A
A
L
L
P
P
310
|
N
N
N
N
T
T
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| Capan-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0026 | ||||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MG-63 cells | Bone | Homo sapiens (Human) | CVCL_0426 | ||||||||||
| NCI-H82 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1591 | ||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Experiment for Drug Resistance |
Fluorescence microscopy assay | ||||||||||||
| Mechanism Description | The missense mutation p.V157F (c.469G>T) in gene TP53 cause the resistance of AMGMDS3 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [27] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | AMGMDS3 | ||||||||||||
| Molecule Alteration | Missense mutation | p.Y220C (c.659A>G) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.05 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.24 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
S
S
S
S
S
S
V
V
P
P
S
S
100
|
Q
Q
K
K
T
T
Y
Y
Q
Q
G
G
S
S
Y
Y
G
G
F
F
110
|
R
R
L
L
G
G
F
F
L
L
H
H
S
S
G
G
T
T
A
A
120
|
K
K
S
S
V
V
T
T
C
C
T
T
Y
Y
S
S
P
P
A
A
130
|
L
L
N
N
K
K
M
L
F
F
C
C
Q
Q
L
L
A
A
K
K
140
|
T
T
C
C
P
P
V
V
Q
Q
L
L
W
W
V
V
D
D
S
S
150
|
T
T
P
P
P
P
P
P
G
G
T
T
R
R
V
V
R
R
A
A
160
|
M
M
A
A
I
I
Y
Y
K
K
Q
Q
S
S
Q
Q
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
R
C
C
P
P
H
H
H
H
180
|
E
E
R
R
C
C
S
S
D
D
S
S
D
D
G
G
L
L
A
A
190
|
P
P
P
P
Q
Q
H
H
L
L
I
I
R
R
V
V
E
E
G
G
200
|
N
N
L
L
R
R
V
A
E
E
Y
Y
L
L
D
D
D
D
R
R
210
|
N
N
T
T
F
F
R
R
H
H
S
S
V
V
V
V
V
V
P
P
220
|
Y
C
E
E
P
P
P
P
E
E
V
V
G
G
S
S
D
D
C
C
230
|
T
T
T
T
I
I
H
H
Y
Y
N
N
Y
Y
M
M
C
C
N
Y
240
|
S
S
S
S
C
C
M
M
G
G
G
G
M
M
N
N
R
R
R
R
250
|
P
P
I
I
L
L
T
T
I
I
I
I
T
T
L
L
E
E
D
D
260
|
S
S
S
S
G
G
N
N
L
L
L
L
G
G
R
R
N
D
S
S
270
|
F
F
E
E
V
V
R
R
V
V
C
C
A
A
C
C
P
P
G
G
280
|
R
R
D
D
R
R
R
R
T
T
E
E
E
E
E
E
N
N
L
L
290
|
R
R
K
K
K
K
G
G
E
E
P
P
H
H
H
H
E
E
L
L
300
|
P
P
P
P
G
G
S
S
T
T
K
K
R
R
A
A
L
L
P
P
310
|
N
N
N
N
T
T
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| Capan-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0026 | ||||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MG-63 cells | Bone | Homo sapiens (Human) | CVCL_0426 | ||||||||||
| NCI-H82 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1591 | ||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Experiment for Drug Resistance |
Fluorescence microscopy assay | ||||||||||||
| Mechanism Description | The missense mutation p.Y220C (c.659A>G) in gene TP53 cause the resistance of AMGMDS3 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [27] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | AMGMDS3 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R249S (c.747G>C) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.92 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.90 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
P
-
L
-
S
S
S
S
S
S
V
V
P
P
S
S
100
|
Q
Q
K
K
T
T
Y
Y
Q
Q
G
G
S
S
Y
Y
G
G
F
F
110
|
R
R
L
L
G
G
F
F
L
L
H
H
S
S
G
G
T
T
A
A
120
|
K
K
S
S
V
V
T
T
C
C
T
T
Y
Y
S
S
P
P
A
A
130
|
L
L
N
N
K
K
M
L
F
F
C
C
Q
Q
L
L
A
A
K
K
140
|
T
T
C
C
P
P
V
V
Q
Q
L
L
W
W
V
V
D
D
S
S
150
|
T
T
P
P
P
P
P
P
G
G
T
T
R
R
V
V
R
R
A
A
160
|
M
M
A
A
I
I
Y
Y
K
K
Q
Q
S
S
Q
Q
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
R
C
C
P
P
H
H
H
H
180
|
E
E
R
R
C
C
S
S
D
D
S
S
D
D
G
G
L
L
A
A
190
|
P
P
P
P
Q
Q
H
H
L
L
I
I
R
R
V
V
E
E
G
G
200
|
N
N
L
L
R
R
V
A
E
E
Y
Y
L
L
D
D
D
D
R
R
210
|
N
N
T
T
F
F
R
R
H
H
S
S
V
V
V
V
V
V
P
P
220
|
Y
Y
E
E
P
P
P
P
E
E
V
V
G
G
S
S
D
D
C
C
230
|
T
T
T
T
I
I
H
H
Y
Y
N
N
Y
Y
M
M
C
C
N
Y
240
|
S
S
S
S
C
C
M
M
G
G
G
G
M
M
N
N
R
R
R
S
250
|
P
P
I
I
L
L
T
T
I
I
I
I
T
T
L
L
E
E
D
D
260
|
S
S
S
S
G
G
N
N
L
L
L
L
G
G
R
R
N
D
S
S
270
|
F
F
E
E
V
V
R
R
V
V
C
C
A
A
C
C
P
P
G
G
280
|
R
R
D
D
R
R
R
R
T
T
E
E
E
E
E
E
N
N
L
L
290
|
R
R
K
K
K
K
G
G
E
E
P
P
H
H
H
H
E
E
L
L
300
|
P
P
P
P
G
G
S
S
T
T
K
K
R
R
A
A
L
L
P
P
310
|
N
N
N
N
T
T
H
-
H
-
H
-
H
-
H
-
H
-
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| Capan-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0026 | ||||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MG-63 cells | Bone | Homo sapiens (Human) | CVCL_0426 | ||||||||||
| NCI-H82 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1591 | ||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Experiment for Drug Resistance |
Fluorescence microscopy assay | ||||||||||||
| Mechanism Description | The missense mutation p.R249S (c.747G>C) in gene TP53 cause the resistance of AMGMDS3 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [27] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | AMGMDS3 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R273C (c.817C>T) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.05 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.80 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
S
S
S
S
S
S
V
V
P
P
S
S
100
|
Q
Q
K
K
T
T
Y
Y
Q
Q
G
G
S
S
Y
Y
G
G
F
F
110
|
R
R
L
L
G
G
F
F
L
L
H
H
S
S
G
G
T
T
A
A
120
|
K
K
S
S
V
V
T
T
C
C
T
T
Y
Y
S
S
P
P
A
A
130
|
L
L
N
N
K
K
M
L
F
F
C
C
Q
Q
L
L
A
A
K
K
140
|
T
T
C
C
P
P
V
V
Q
Q
L
L
W
W
V
V
D
D
S
S
150
|
T
T
P
P
P
P
P
P
G
G
T
T
R
R
V
V
R
R
A
A
160
|
M
M
A
A
I
I
Y
Y
K
K
Q
Q
S
S
Q
Q
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
R
C
C
P
P
H
H
H
H
180
|
E
E
R
R
C
C
S
S
D
D
S
S
D
D
G
G
L
L
A
A
190
|
P
P
P
P
Q
Q
H
H
L
L
I
I
R
R
V
V
E
E
G
G
200
|
N
N
L
L
R
R
V
A
E
E
Y
Y
L
L
D
D
D
D
R
R
210
|
N
N
T
T
F
F
R
R
H
H
S
S
V
V
V
V
V
V
P
P
220
|
Y
Y
E
E
P
P
P
P
E
E
V
V
G
G
S
S
D
D
C
C
230
|
T
T
T
T
I
I
H
H
Y
Y
N
N
Y
Y
M
M
C
C
N
Y
240
|
S
S
S
S
C
C
M
M
G
G
G
G
M
M
N
N
R
R
R
R
250
|
P
P
I
I
L
L
T
T
I
I
I
I
T
T
L
L
E
E
D
D
260
|
S
S
S
S
G
G
N
N
L
L
L
L
G
G
R
R
N
D
S
S
270
|
F
F
E
E
V
V
R
C
V
V
C
C
A
A
C
C
P
P
G
G
280
|
R
R
D
D
R
R
R
R
T
T
E
E
E
E
E
E
N
N
L
L
290
|
R
R
K
K
K
K
G
G
E
E
P
P
H
H
H
H
E
E
L
L
300
|
P
P
P
P
G
G
S
S
T
T
K
K
R
R
A
A
L
L
P
P
310
|
N
N
N
N
T
T
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| Capan-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0026 | ||||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MG-63 cells | Bone | Homo sapiens (Human) | CVCL_0426 | ||||||||||
| NCI-H82 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1591 | ||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Experiment for Drug Resistance |
Fluorescence microscopy assay | ||||||||||||
| Mechanism Description | The missense mutation p.R273C (c.817C>T) in gene TP53 cause the resistance of AMGMDS3 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [27] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | AMGMDS3 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R175H (c.524G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.37 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.38 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
H
C
C
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| Capan-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0026 | ||||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MG-63 cells | Bone | Homo sapiens (Human) | CVCL_0426 | ||||||||||
| NCI-H82 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1591 | ||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Experiment for Drug Resistance |
Fluorescence microscopy assay | ||||||||||||
| Mechanism Description | The missense mutation p.R175H (c.524G>A) in gene TP53 cause the resistance of AMGMDS3 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [27] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | AMGMDS3 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R280K (c.839G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.05 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.00 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
S
-
S
-
S
-
V
V
P
P
S
S
100
|
Q
Q
K
K
T
T
Y
Y
Q
Q
G
G
S
S
Y
Y
G
G
F
F
110
|
R
R
L
L
G
G
F
F
L
L
H
H
S
S
G
G
T
T
A
A
120
|
K
K
S
S
V
V
T
T
C
C
T
T
Y
Y
S
S
P
P
A
A
130
|
L
L
N
N
K
K
M
M
F
F
C
C
Q
Q
L
L
A
A
K
K
140
|
T
T
C
C
P
P
V
V
Q
Q
L
L
W
W
V
V
D
D
S
S
150
|
T
T
P
P
P
P
P
P
G
G
T
T
R
R
V
V
R
R
A
A
160
|
M
M
A
A
I
I
Y
Y
K
K
Q
Q
S
S
Q
Q
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
R
C
C
P
P
H
H
H
H
180
|
E
E
R
R
C
C
S
S
D
D
S
S
D
D
G
G
L
L
A
A
190
|
P
P
P
P
Q
Q
H
H
L
L
I
I
R
R
V
V
E
E
G
G
200
|
N
N
L
L
R
R
V
V
E
E
Y
Y
L
L
D
D
D
D
R
R
210
|
N
N
T
T
F
F
R
R
H
H
S
S
V
V
V
V
V
V
P
P
220
|
Y
Y
E
E
P
P
P
P
E
E
V
V
G
G
S
S
D
D
C
C
230
|
T
T
T
T
I
I
H
H
Y
Y
N
N
Y
Y
M
M
C
C
N
N
240
|
S
S
S
S
C
C
M
M
G
G
G
G
M
M
N
N
R
R
R
R
250
|
P
P
I
I
L
L
T
T
I
I
I
I
T
T
L
L
E
E
D
D
260
|
S
S
S
S
G
G
N
N
L
L
L
L
G
G
R
R
N
N
S
S
270
|
F
F
E
E
V
V
R
R
V
V
C
C
A
A
C
C
P
P
G
G
280
|
R
K
D
D
R
R
R
R
T
T
E
E
E
E
E
E
N
N
L
L
290
|
R
-
K
-
K
-
G
-
E
-
P
-
H
-
H
-
E
-
L
-
300
|
P
-
P
-
G
-
S
-
T
-
K
-
R
-
A
-
L
-
P
-
310
|
N
-
N
-
T
-
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| Capan-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0026 | ||||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MG-63 cells | Bone | Homo sapiens (Human) | CVCL_0426 | ||||||||||
| NCI-H82 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1591 | ||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Experiment for Drug Resistance |
Fluorescence microscopy assay | ||||||||||||
| Mechanism Description | The missense mutation p.R280K (c.839G>A) in gene TP53 cause the resistance of AMGMDS3 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [27] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | AMGMDS3 | ||||||||||||
| Molecule Alteration | Missense mutation | p.G245S (c.733G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.05 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.15 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
S
S
S
S
S
S
V
V
P
P
S
S
100
|
Q
Q
K
K
T
T
Y
Y
Q
Q
G
G
S
S
Y
Y
G
G
F
F
110
|
R
R
L
L
G
G
F
F
L
L
H
H
S
S
G
G
T
T
A
A
120
|
K
K
S
S
V
V
T
T
C
C
T
T
Y
Y
S
S
P
P
A
A
130
|
L
L
N
N
K
K
M
M
F
F
C
C
Q
Q
L
L
A
A
K
K
140
|
T
T
C
C
P
P
V
V
Q
Q
L
L
W
W
V
V
D
D
S
S
150
|
T
T
P
P
P
P
P
P
G
G
T
T
R
R
V
V
R
R
A
A
160
|
M
M
A
A
I
I
Y
Y
K
K
Q
Q
S
S
Q
Q
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
R
C
C
P
P
H
H
H
H
180
|
E
E
R
R
C
C
S
S
D
D
S
S
D
D
G
G
L
L
A
A
190
|
P
P
P
P
Q
Q
H
H
L
L
I
I
R
R
V
V
E
E
G
G
200
|
N
N
L
L
R
R
V
V
E
E
Y
Y
L
L
D
D
D
D
R
R
210
|
N
N
T
T
F
F
R
R
H
H
S
S
V
V
V
V
V
V
P
P
220
|
Y
Y
E
E
P
P
P
P
E
E
V
V
G
G
S
S
D
D
C
C
230
|
T
T
T
T
I
I
H
H
Y
Y
N
N
Y
Y
M
M
C
C
N
N
240
|
S
S
S
S
C
C
M
M
G
G
G
S
M
M
N
N
R
R
R
R
250
|
P
P
I
I
L
L
T
T
I
I
I
I
T
T
L
L
E
E
D
D
260
|
S
S
S
S
G
G
N
N
L
L
L
L
G
G
R
R
N
N
S
S
270
|
F
F
E
E
V
V
R
R
V
V
C
C
A
A
C
C
P
P
G
G
280
|
R
R
D
D
R
R
R
R
T
T
E
E
E
E
E
E
N
N
L
L
290
|
R
R
K
K
K
K
G
G
E
-
P
-
H
-
H
-
E
-
L
-
300
|
P
-
P
-
G
-
S
-
T
-
K
-
R
-
A
-
L
-
P
-
310
|
N
-
N
-
T
-
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| Capan-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0026 | ||||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MG-63 cells | Bone | Homo sapiens (Human) | CVCL_0426 | ||||||||||
| NCI-H82 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1591 | ||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Experiment for Drug Resistance |
Fluorescence microscopy assay | ||||||||||||
| Mechanism Description | The missense mutation p.G245S (c.733G>A) in gene TP53 cause the resistance of AMGMDS3 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [27] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | AMGMDS3 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R158H (c.473G>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| Capan-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0026 | ||||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MG-63 cells | Bone | Homo sapiens (Human) | CVCL_0426 | ||||||||||
| NCI-H82 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1591 | ||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Experiment for Drug Resistance |
Fluorescence microscopy assay | ||||||||||||
| Mechanism Description | The missense mutation p.R158H (c.473G>A) in gene TP53 cause the resistance of AMGMDS3 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [27] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | AMGMDS3 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R158L (c.473G>T) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| Capan-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0026 | ||||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MG-63 cells | Bone | Homo sapiens (Human) | CVCL_0426 | ||||||||||
| NCI-H82 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1591 | ||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Experiment for Drug Resistance |
Fluorescence microscopy assay | ||||||||||||
| Mechanism Description | The missense mutation p.R158L (c.473G>T) in gene TP53 cause the resistance of AMGMDS3 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [27] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | AMGMDS3 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R248W (c.742C>T) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| Capan-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0026 | ||||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MG-63 cells | Bone | Homo sapiens (Human) | CVCL_0426 | ||||||||||
| NCI-H82 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1591 | ||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Experiment for Drug Resistance |
Fluorescence microscopy assay | ||||||||||||
| Mechanism Description | The missense mutation p.R248W (c.742C>T) in gene TP53 cause the resistance of AMGMDS3 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [27] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | AMGMDS3 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R248Q (c.743G>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| Capan-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0026 | ||||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MG-63 cells | Bone | Homo sapiens (Human) | CVCL_0426 | ||||||||||
| NCI-H82 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1591 | ||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Experiment for Drug Resistance |
Fluorescence microscopy assay | ||||||||||||
| Mechanism Description | The missense mutation p.R248Q (c.743G>A) in gene TP53 cause the resistance of AMGMDS3 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [27] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | AMGMDS3 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R280T (c.839G>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| Capan-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0026 | ||||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MG-63 cells | Bone | Homo sapiens (Human) | CVCL_0426 | ||||||||||
| NCI-H82 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1591 | ||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Experiment for Drug Resistance |
Fluorescence microscopy assay | ||||||||||||
| Mechanism Description | The missense mutation p.R280T (c.839G>C) in gene TP53 cause the resistance of AMGMDS3 by unusual activation of pro-survival pathway | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [28] | ||||||||||||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | ||||||||||||
| Sensitive Drug | APR-246/Cisplatin | ||||||||||||
| Molecule Alteration | Missense mutation | p.R280K (c.839G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.05 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.00 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
S
-
S
-
S
-
V
V
P
P
S
S
100
|
Q
Q
K
K
T
T
Y
Y
Q
Q
G
G
S
S
Y
Y
G
G
F
F
110
|
R
R
L
L
G
G
F
F
L
L
H
H
S
S
G
G
T
T
A
A
120
|
K
K
S
S
V
V
T
T
C
C
T
T
Y
Y
S
S
P
P
A
A
130
|
L
L
N
N
K
K
M
M
F
F
C
C
Q
Q
L
L
A
A
K
K
140
|
T
T
C
C
P
P
V
V
Q
Q
L
L
W
W
V
V
D
D
S
S
150
|
T
T
P
P
P
P
P
P
G
G
T
T
R
R
V
V
R
R
A
A
160
|
M
M
A
A
I
I
Y
Y
K
K
Q
Q
S
S
Q
Q
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
R
C
C
P
P
H
H
H
H
180
|
E
E
R
R
C
C
S
S
D
D
S
S
D
D
G
G
L
L
A
A
190
|
P
P
P
P
Q
Q
H
H
L
L
I
I
R
R
V
V
E
E
G
G
200
|
N
N
L
L
R
R
V
V
E
E
Y
Y
L
L
D
D
D
D
R
R
210
|
N
N
T
T
F
F
R
R
H
H
S
S
V
V
V
V
V
V
P
P
220
|
Y
Y
E
E
P
P
P
P
E
E
V
V
G
G
S
S
D
D
C
C
230
|
T
T
T
T
I
I
H
H
Y
Y
N
N
Y
Y
M
M
C
C
N
N
240
|
S
S
S
S
C
C
M
M
G
G
G
G
M
M
N
N
R
R
R
R
250
|
P
P
I
I
L
L
T
T
I
I
I
I
T
T
L
L
E
E
D
D
260
|
S
S
S
S
G
G
N
N
L
L
L
L
G
G
R
R
N
N
S
S
270
|
F
F
E
E
V
V
R
R
V
V
C
C
A
A
C
C
P
P
G
G
280
|
R
K
D
D
R
R
R
R
T
T
E
E
E
E
E
E
N
N
L
L
290
|
R
-
K
-
K
-
G
-
E
-
P
-
H
-
H
-
E
-
L
-
300
|
P
-
P
-
G
-
S
-
T
-
K
-
R
-
A
-
L
-
P
-
310
|
N
-
N
-
T
-
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Ascitic fluid | N.A. | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
FMCA assay | ||||||||||||
| Disease Class: Peritoneal cancer [ICD-11: 2C51.0] | [28] | ||||||||||||
| Sensitive Disease | Peritoneal cancer [ICD-11: 2C51.0] | ||||||||||||
| Sensitive Drug | APR-246/Cisplatin | ||||||||||||
| Molecule Alteration | Missense mutation | p.C135Y (c.404G>A) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Ascitic fluid | N.A. | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
FMCA assay | ||||||||||||
| Disease Class: Fallopian tube cancer [ICD-11: 2C74.0] | [28] | ||||||||||||
| Sensitive Disease | Fallopian tube cancer [ICD-11: 2C74.0] | ||||||||||||
| Sensitive Drug | APR-246/Cisplatin | ||||||||||||
| Molecule Alteration | Missense mutation | p.C238F (c.713G>T) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Ascitic fluid | N.A. | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
FMCA assay | ||||||||||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [28] | ||||||||||||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | ||||||||||||
| Sensitive Drug | APR-246/Cisplatin | ||||||||||||
| Molecule Alteration | Missense mutation | p.L111Q (c.332T>A) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Ascitic fluid | N.A. | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
FMCA assay | ||||||||||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [28] | ||||||||||||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | ||||||||||||
| Sensitive Drug | APR-246/Cisplatin | ||||||||||||
| Molecule Alteration | Missense mutation | p.P151H (c.452C>A) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Ascitic fluid | N.A. | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
FMCA assay | ||||||||||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [28] | ||||||||||||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | ||||||||||||
| Sensitive Drug | APR-246/Cisplatin | ||||||||||||
| Molecule Alteration | Missense mutation | p.Y163H (c.487T>C) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Ascitic fluid | N.A. | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
FMCA assay | ||||||||||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [28] | ||||||||||||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | ||||||||||||
| Sensitive Drug | APR-246/Cisplatin | ||||||||||||
| Molecule Alteration | Missense mutation | p.P278R (c.833C>G) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Ascitic fluid | N.A. | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
FMCA assay | ||||||||||||
|
|
|||||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [29] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Sensitive Drug | APR-246/Cisplatin | ||||||||||||
| Molecule Alteration | Missense mutation | p.Y163C (c.488A>G) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.05 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.65 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
S
S
S
S
S
S
V
V
P
P
S
S
100
|
Q
Q
K
K
T
T
Y
Y
Q
Q
G
G
S
S
Y
Y
G
G
F
F
110
|
R
R
L
L
G
G
F
F
L
L
H
H
S
S
G
G
T
T
A
A
120
|
K
K
S
S
V
V
T
T
C
C
T
T
Y
Y
S
S
P
P
A
A
130
|
L
L
N
N
K
K
M
L
F
F
C
C
Q
Q
L
L
A
A
K
K
140
|
T
T
C
C
P
P
V
V
Q
Q
L
L
W
W
V
V
D
D
S
S
150
|
T
T
P
P
P
P
P
P
G
G
T
T
R
R
V
V
R
R
A
A
160
|
M
M
A
A
I
I
Y
C
K
K
Q
Q
S
S
Q
Q
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
R
C
C
P
P
H
H
H
H
180
|
E
E
R
R
C
C
S
S
D
D
S
S
D
D
G
G
L
L
A
A
190
|
P
P
P
P
Q
Q
H
H
L
L
I
I
R
R
V
V
E
E
G
G
200
|
N
N
L
L
R
R
V
A
E
E
Y
Y
L
L
D
D
D
D
R
R
210
|
N
N
T
T
F
F
R
R
H
H
S
S
V
V
V
V
V
V
P
P
220
|
Y
Y
E
E
P
P
P
P
E
E
V
V
G
G
S
S
D
D
C
C
230
|
T
T
T
T
I
I
H
H
Y
Y
N
N
Y
Y
M
M
C
C
N
Y
240
|
S
S
S
S
C
C
M
M
G
G
G
G
M
M
N
N
R
R
R
R
250
|
P
P
I
I
L
L
T
T
I
I
I
I
T
T
L
L
E
E
D
D
260
|
S
S
S
S
G
G
N
N
L
L
L
L
G
G
R
R
N
D
S
S
270
|
F
F
E
E
V
V
R
R
V
V
C
C
A
A
C
C
P
P
G
G
280
|
R
R
D
D
R
R
R
R
T
T
E
E
E
E
E
E
N
N
L
L
290
|
R
R
K
K
K
K
G
G
E
E
P
P
H
H
H
H
E
E
L
L
300
|
P
P
P
P
G
G
S
S
T
T
K
K
R
R
A
A
L
L
P
P
310
|
N
N
N
N
T
T
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A2780-CP20 cells | Ovary | Homo sapiens (Human) | CVCL_A5PS | |||||||||
| In Vivo Model | Female CD-1 Nu/Nu mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
FMCA assay; WST-1 assay; Cell Titer-Glo assay; MTS assay | ||||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [29] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Sensitive Drug | APR-246/Cisplatin | ||||||||||||
| Molecule Alteration | Missense mutation | p.R273H (c.818G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.05 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.98 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
S
S
S
S
S
S
V
V
P
P
S
S
100
|
Q
Q
K
K
T
T
Y
Y
Q
Q
G
G
S
S
Y
Y
G
G
F
F
110
|
R
R
L
L
G
G
F
F
L
L
H
H
S
S
G
G
T
T
A
A
120
|
K
K
S
S
V
V
T
T
C
C
T
T
Y
Y
S
S
P
P
A
A
130
|
L
L
N
N
K
K
M
L
F
F
C
C
Q
Q
L
L
A
A
K
K
140
|
T
T
C
C
P
P
V
V
Q
Q
L
L
W
W
V
V
D
D
S
S
150
|
T
T
P
P
P
P
P
P
G
G
T
T
R
R
V
V
R
R
A
A
160
|
M
M
A
A
I
I
Y
Y
K
K
Q
Q
S
S
Q
Q
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
R
C
C
P
P
H
H
H
H
180
|
E
E
R
R
C
C
S
S
D
D
S
S
D
D
G
G
L
L
A
A
190
|
P
P
P
P
Q
Q
H
H
L
L
I
I
R
R
V
V
E
E
G
G
200
|
N
N
L
L
R
R
V
A
E
E
Y
Y
L
L
D
D
D
D
R
R
210
|
N
N
T
T
F
F
R
R
H
H
S
S
V
V
V
V
V
V
P
P
220
|
Y
Y
E
E
P
P
P
P
E
E
V
V
G
G
S
S
D
D
C
C
230
|
T
T
T
T
I
I
H
H
Y
Y
N
N
Y
Y
M
M
C
C
N
Y
240
|
S
S
S
S
C
C
M
M
G
G
G
G
M
M
N
N
R
R
R
R
250
|
P
P
I
I
L
L
T
T
I
I
I
I
T
T
L
L
E
E
D
D
260
|
S
S
S
S
G
G
N
N
L
L
L
L
G
G
R
R
N
D
S
S
270
|
F
F
E
E
V
V
R
H
V
V
C
C
A
A
C
C
P
P
G
G
280
|
R
R
D
D
R
R
R
R
T
T
E
E
E
E
E
E
N
N
L
L
290
|
R
R
K
K
K
K
G
G
E
E
P
P
H
H
H
H
E
E
L
L
300
|
P
P
P
P
G
G
S
S
T
T
K
K
R
R
A
A
L
L
P
P
310
|
N
N
N
N
T
T
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A2780-CP20 cells | Ovary | Homo sapiens (Human) | CVCL_A5PS | |||||||||
| In Vivo Model | Female CD-1 Nu/Nu mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
FMCA assay; WST-1 assay; Cell Titer-Glo assay; MTS assay | ||||||||||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [29] | ||||||||||||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | ||||||||||||
| Sensitive Drug | APR-246/Cisplatin | ||||||||||||
| Molecule Alteration | Missense mutation | p.R248Q (c.743G>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A2780-CP20 cells | Ovary | Homo sapiens (Human) | CVCL_A5PS | |||||||||
| In Vivo Model | Female CD-1 Nu/Nu mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
FMCA assay; WST-1 assay; Cell Titer-Glo assay; MTS assay | ||||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [29] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Sensitive Drug | APR-246/Cisplatin | ||||||||||||
| Molecule Alteration | Missense mutation | p.R248W (c.742C>T) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A2780-CP20 cells | Ovary | Homo sapiens (Human) | CVCL_A5PS | |||||||||
| In Vivo Model | Female CD-1 Nu/Nu mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
FMCA assay; WST-1 assay; Cell Titer-Glo assay; MTS assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Cholangiocarcinoma [ICD-11: 2C12.0] | [30] | ||||||||||||
| Sensitive Disease | Cholangiocarcinoma [ICD-11: 2C12.0] | ||||||||||||
| Sensitive Drug | CP-31398 | ||||||||||||
| Molecule Alteration | Missense mutation | p.Y220C (c.659A>G) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.05 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.24 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
S
S
S
S
S
S
V
V
P
P
S
S
100
|
Q
Q
K
K
T
T
Y
Y
Q
Q
G
G
S
S
Y
Y
G
G
F
F
110
|
R
R
L
L
G
G
F
F
L
L
H
H
S
S
G
G
T
T
A
A
120
|
K
K
S
S
V
V
T
T
C
C
T
T
Y
Y
S
S
P
P
A
A
130
|
L
L
N
N
K
K
M
L
F
F
C
C
Q
Q
L
L
A
A
K
K
140
|
T
T
C
C
P
P
V
V
Q
Q
L
L
W
W
V
V
D
D
S
S
150
|
T
T
P
P
P
P
P
P
G
G
T
T
R
R
V
V
R
R
A
A
160
|
M
M
A
A
I
I
Y
Y
K
K
Q
Q
S
S
Q
Q
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
R
C
C
P
P
H
H
H
H
180
|
E
E
R
R
C
C
S
S
D
D
S
S
D
D
G
G
L
L
A
A
190
|
P
P
P
P
Q
Q
H
H
L
L
I
I
R
R
V
V
E
E
G
G
200
|
N
N
L
L
R
R
V
A
E
E
Y
Y
L
L
D
D
D
D
R
R
210
|
N
N
T
T
F
F
R
R
H
H
S
S
V
V
V
V
V
V
P
P
220
|
Y
C
E
E
P
P
P
P
E
E
V
V
G
G
S
S
D
D
C
C
230
|
T
T
T
T
I
I
H
H
Y
Y
N
N
Y
Y
M
M
C
C
N
Y
240
|
S
S
S
S
C
C
M
M
G
G
G
G
M
M
N
N
R
R
R
R
250
|
P
P
I
I
L
L
T
T
I
I
I
I
T
T
L
L
E
E
D
D
260
|
S
S
S
S
G
G
N
N
L
L
L
L
G
G
R
R
N
D
S
S
270
|
F
F
E
E
V
V
R
R
V
V
C
C
A
A
C
C
P
P
G
G
280
|
R
R
D
D
R
R
R
R
T
T
E
E
E
E
E
E
N
N
L
L
290
|
R
R
K
K
K
K
G
G
E
E
P
P
H
H
H
H
E
E
L
L
300
|
P
P
P
P
G
G
S
S
T
T
K
K
R
R
A
A
L
L
P
P
310
|
N
N
N
N
T
T
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Disease Class: Cholangiocarcinoma [ICD-11: 2C12.0] | [30] | ||||||||||||
| Sensitive Disease | Cholangiocarcinoma [ICD-11: 2C12.0] | ||||||||||||
| Sensitive Drug | CP-31398 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R249S (c.747G>C) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.92 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.90 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
P
-
L
-
S
S
S
S
S
S
V
V
P
P
S
S
100
|
Q
Q
K
K
T
T
Y
Y
Q
Q
G
G
S
S
Y
Y
G
G
F
F
110
|
R
R
L
L
G
G
F
F
L
L
H
H
S
S
G
G
T
T
A
A
120
|
K
K
S
S
V
V
T
T
C
C
T
T
Y
Y
S
S
P
P
A
A
130
|
L
L
N
N
K
K
M
L
F
F
C
C
Q
Q
L
L
A
A
K
K
140
|
T
T
C
C
P
P
V
V
Q
Q
L
L
W
W
V
V
D
D
S
S
150
|
T
T
P
P
P
P
P
P
G
G
T
T
R
R
V
V
R
R
A
A
160
|
M
M
A
A
I
I
Y
Y
K
K
Q
Q
S
S
Q
Q
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
R
C
C
P
P
H
H
H
H
180
|
E
E
R
R
C
C
S
S
D
D
S
S
D
D
G
G
L
L
A
A
190
|
P
P
P
P
Q
Q
H
H
L
L
I
I
R
R
V
V
E
E
G
G
200
|
N
N
L
L
R
R
V
A
E
E
Y
Y
L
L
D
D
D
D
R
R
210
|
N
N
T
T
F
F
R
R
H
H
S
S
V
V
V
V
V
V
P
P
220
|
Y
Y
E
E
P
P
P
P
E
E
V
V
G
G
S
S
D
D
C
C
230
|
T
T
T
T
I
I
H
H
Y
Y
N
N
Y
Y
M
M
C
C
N
Y
240
|
S
S
S
S
C
C
M
M
G
G
G
G
M
M
N
N
R
R
R
S
250
|
P
P
I
I
L
L
T
T
I
I
I
I
T
T
L
L
E
E
D
D
260
|
S
S
S
S
G
G
N
N
L
L
L
L
G
G
R
R
N
D
S
S
270
|
F
F
E
E
V
V
R
R
V
V
C
C
A
A
C
C
P
P
G
G
280
|
R
R
D
D
R
R
R
R
T
T
E
E
E
E
E
E
N
N
L
L
290
|
R
R
K
K
K
K
G
G
E
E
P
P
H
H
H
H
E
E
L
L
300
|
P
P
P
P
G
G
S
S
T
T
K
K
R
R
A
A
L
L
P
P
310
|
N
N
N
N
T
T
H
-
H
-
H
-
H
-
H
-
H
-
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |||||||||
| Experiment for Molecule Alteration |
qRT-PCR | ||||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [31] | ||||||||||||
| Resistant Disease | Ovarian cancer [ICD-11: 2C73.0] | ||||||||||||
| Resistant Drug | DS-7423 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R175H (c.524G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.37 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.38 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
H
C
C
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | ES-2 cells | Ovary | Homo sapiens (Human) | CVCL_3509 | |||||||||
| TOV-21 cells | Ovary | Homo sapiens (Human) | CVCL_3613 | ||||||||||
| SKOV5 cells | Ovary | Homo sapiens (Human) | N.A. | ||||||||||
| RMG-I cells | Ascites | Homo sapiens (Human) | CVCL_1662 | ||||||||||
| OVTOKO cells | Spleen | Homo sapiens (Human) | CVCL_3117 | ||||||||||
| OVSAHO cells | Abdomen | Homo sapiens (Human) | CVCL_3114 | ||||||||||
| OVMANA cells | Ovary | Homo sapiens (Human) | CVCL_3111 | ||||||||||
| OVKATE cells | Ovary | Homo sapiens (Human) | CVCL_3110 | ||||||||||
| OVISE cells | Pelvi | Homo sapiens (Human) | CVCL_3116 | ||||||||||
| OV1063 cells | Ascites | Homo sapiens (Human) | CVCL_4366 | ||||||||||
| SKOV5 cells | Ovary | Homo sapiens (Human) | N.A. | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase assay | ||||||||||||
| Experiment for Drug Resistance |
CCK-8 assay | ||||||||||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [31] | ||||||||||||
| Resistant Disease | Ovarian cancer [ICD-11: 2C73.0] | ||||||||||||
| Resistant Drug | DS-7423 | ||||||||||||
| Molecule Alteration | Missense mutation | p.S241F (c.722C>T) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | ES-2 cells | Ovary | Homo sapiens (Human) | CVCL_3509 | |||||||||
| TOV-21 cells | Ovary | Homo sapiens (Human) | CVCL_3613 | ||||||||||
| SKOV6 cells | Uterus | Homo sapiens (Human) | CVCL_A457 | ||||||||||
| RMG-I cells | Ascites | Homo sapiens (Human) | CVCL_1662 | ||||||||||
| OVTOKO cells | Spleen | Homo sapiens (Human) | CVCL_3117 | ||||||||||
| OVSAHO cells | Abdomen | Homo sapiens (Human) | CVCL_3114 | ||||||||||
| OVMANA cells | Ovary | Homo sapiens (Human) | CVCL_3111 | ||||||||||
| OVKATE cells | Ovary | Homo sapiens (Human) | CVCL_3110 | ||||||||||
| OVISE cells | Pelvi | Homo sapiens (Human) | CVCL_3116 | ||||||||||
| OV1063 cells | Ascites | Homo sapiens (Human) | CVCL_4366 | ||||||||||
| SKOV6 cells | Uterus | Homo sapiens (Human) | CVCL_A457 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase assay | ||||||||||||
| Experiment for Drug Resistance |
CCK-8 assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [20] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | HSP90 inhibitor | ||||||||||||
| Molecule Alteration | Missense mutation | p.R175H (c.524G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.37 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.38 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
H
C
C
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |||||||||
| H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | ||||||||||
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | ||||||||||
| ES2 cells | Ovary | Homo sapiens (Human) | CVCL_AX39 | ||||||||||
| DU145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 | ||||||||||
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | ||||||||||
| MDA-46 cells | N.A. | Homo sapiens (Human) | N.A. | ||||||||||
| HOC7 cells | Ascites | Homo sapiens (Human) | CVCL_5455 | ||||||||||
| EFO21 cells | Ascites | Homo sapiens (Human) | CVCL_0029 | ||||||||||
| COV434 cells | N.A. | Homo sapiens (Human) | CVCL_2010 | ||||||||||
| COLO704 cells | Ascites | Homo sapiens (Human) | CVCL_1994 | ||||||||||
| HOC7 cells | Ascites | Homo sapiens (Human) | CVCL_5455 | ||||||||||
| In Vivo Model | Athymic (nu/nu) male xenograft mouse model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; Quantitative PCR analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-blue cell viability assay | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [20] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | HSP90 inhibitor | ||||||||||||
| Molecule Alteration | Missense mutation | p.R248Q (c.743G>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |||||||||
| H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | ||||||||||
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | ||||||||||
| ES2 cells | Ovary | Homo sapiens (Human) | CVCL_AX39 | ||||||||||
| DU145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 | ||||||||||
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | ||||||||||
| MDA-46 cells | N.A. | Homo sapiens (Human) | N.A. | ||||||||||
| HOC7 cells | Ascites | Homo sapiens (Human) | CVCL_5455 | ||||||||||
| EFO21 cells | Ascites | Homo sapiens (Human) | CVCL_0029 | ||||||||||
| COV434 cells | N.A. | Homo sapiens (Human) | CVCL_2010 | ||||||||||
| COLO704 cells | Ascites | Homo sapiens (Human) | CVCL_1994 | ||||||||||
| HOC7 cells | Ascites | Homo sapiens (Human) | CVCL_5455 | ||||||||||
| In Vivo Model | Athymic (nu/nu) male xenograft mouse model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; Quantitative PCR analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-blue cell viability assay | ||||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [32] | |||
| Resistant Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Resistant Drug | NSC319726 | |||
| Molecule Alteration | Missense mutation | p.R273W (c.817_819delCGTinsTGG) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | (MEF) (10)3 cells | N.A. | Mus musculus (Mouse) | N.A. |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | The missense mutation p.R273W (c.817_819delCGTinsTGG) in gene TP53 cause the resistance of NSC319726 by aberration of the drug's therapeutic target | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [32] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | NSC319726 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R175H (c.524G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.37 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.38 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
H
C
C
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | (MEF) (10)3 cells | N.A. | Mus musculus (Mouse) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTS assay | ||||||||||||
| Mechanism Description | The missense mutation p.R175H (c.524G>A) in gene TP53 cause the sensitivity of NSC319726 by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [32] | ||||||||||||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | ||||||||||||
| Sensitive Drug | NSC319726 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R175H (c.524G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.37 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.38 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
H
C
C
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | (MEF) (10)3 cells | N.A. | Mus musculus (Mouse) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTS assay | ||||||||||||
| Mechanism Description | The missense mutation p.R175H (c.524G>A) in gene TP53 cause the sensitivity of NSC319726 by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [32] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Sensitive Drug | NSC319726 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R175L (c.524G>T) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | (MEF) (10)3 cells | N.A. | Mus musculus (Mouse) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTS assay | ||||||||||||
| Mechanism Description | The missense mutation p.R175L (c.524G>T) in gene TP53 cause the sensitivity of NSC319726 by aberration of the drug's therapeutic target | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [33] | ||||||||||||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | ||||||||||||
| Sensitive Drug | NSC59984 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R175H (c.524G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.37 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.38 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
H
C
C
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | SW480 cells | Colon | Homo sapiens (Human) | CVCL_0546 | |||||||||
| HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | ||||||||||
| DLD-1 cells | Colon | Homo sapiens (Human) | CVCL_0248 | ||||||||||
| RXF393 cells | Kidney | Homo sapiens (Human) | CVCL_1673 | ||||||||||
| MRC5 cells | Fetal lung | Homo sapiens (Human) | CVCL_0440 | ||||||||||
| Wi38 cells | Fetal lung | Homo sapiens (Human) | CVCL_0579 | ||||||||||
| Hop92 cells | Lung | Homo sapiens (Human) | CVCL_1286 | ||||||||||
| In Vivo Model | Female CRL nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Semi-quantitative RT-PCR; Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay; Colony formation assay | ||||||||||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [33] | ||||||||||||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | ||||||||||||
| Sensitive Drug | NSC59984 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R175L (c.524G>T) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | SW480 cells | Colon | Homo sapiens (Human) | CVCL_0546 | |||||||||
| HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | ||||||||||
| DLD-1 cells | Colon | Homo sapiens (Human) | CVCL_0248 | ||||||||||
| RXF393 cells | Kidney | Homo sapiens (Human) | CVCL_1673 | ||||||||||
| MRC5 cells | Fetal lung | Homo sapiens (Human) | CVCL_0440 | ||||||||||
| Wi38 cells | Fetal lung | Homo sapiens (Human) | CVCL_0579 | ||||||||||
| Hop92 cells | Lung | Homo sapiens (Human) | CVCL_1286 | ||||||||||
| In Vivo Model | Female CRL nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Semi-quantitative RT-PCR; Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay; Colony formation assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [34] | ||||||||||||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | ||||||||||||
| Sensitive Drug | ReACp53 | ||||||||||||
| Molecule Alteration | Missense mutation | p.Y234C (c.701A>G) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.05 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.38 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
S
S
S
S
S
S
V
V
P
P
S
S
100
|
Q
Q
K
K
T
T
Y
Y
Q
Q
G
G
S
S
Y
Y
G
G
F
F
110
|
R
R
L
L
G
G
F
F
L
L
H
H
S
S
G
G
T
T
A
A
120
|
K
K
S
S
V
V
T
T
C
C
T
T
Y
Y
S
S
P
P
A
A
130
|
L
L
N
N
K
K
M
L
F
F
C
C
Q
Q
L
L
A
A
K
K
140
|
T
T
C
C
P
P
V
V
Q
Q
L
L
W
W
V
V
D
D
S
S
150
|
T
T
P
P
P
P
P
P
G
G
T
T
R
R
V
V
R
R
A
A
160
|
M
M
A
A
I
I
Y
Y
K
K
Q
Q
S
S
Q
Q
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
R
C
C
P
P
H
H
H
H
180
|
E
E
R
R
C
C
S
S
D
D
S
S
D
D
G
G
L
L
A
A
190
|
P
P
P
P
Q
Q
H
H
L
L
I
I
R
R
V
V
E
E
G
G
200
|
N
N
L
L
R
R
V
A
E
E
Y
Y
L
L
D
D
D
D
R
R
210
|
N
N
T
T
F
F
R
R
H
H
S
S
V
V
V
V
V
V
P
P
220
|
Y
Y
E
E
P
P
P
P
E
E
V
V
G
G
S
S
D
D
C
C
230
|
T
T
T
T
I
I
H
H
Y
C
N
N
Y
Y
M
M
C
C
N
Y
240
|
S
S
S
S
C
C
M
M
G
G
G
G
M
M
N
N
R
R
R
R
250
|
P
P
I
I
L
L
T
T
I
I
I
I
T
T
L
L
E
E
D
D
260
|
S
S
S
S
G
G
N
N
L
L
L
L
G
G
R
R
N
D
S
S
270
|
F
F
E
E
V
V
R
R
V
V
C
C
A
A
C
C
P
P
G
G
280
|
R
R
D
D
R
R
R
R
T
T
E
E
E
E
E
E
N
N
L
L
290
|
R
R
K
K
K
K
G
G
E
E
P
P
H
H
H
H
E
E
L
L
300
|
P
P
P
P
G
G
S
S
T
T
K
K
R
R
A
A
L
L
P
P
310
|
N
N
N
N
T
T
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | S1 GODL cells | N.A. | Homo sapiens (Human) | N.A. | |||||||||
| In Vivo Model | Immunocompromised NSG mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
In vitro 3D organoid assay | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [34] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | ReACp53 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R175H (c.524G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.37 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.38 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
H
C
C
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | S1 GODL cells | N.A. | Homo sapiens (Human) | N.A. | |||||||||
| In Vivo Model | Immunocompromised NSG mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
In vitro 3D organoid assay | ||||||||||||
| Disease Class: Head and neck cancer [ICD-11: 2D42.0] | [34] | ||||||||||||
| Sensitive Disease | Head and neck cancer [ICD-11: 2D42.0] | ||||||||||||
| Sensitive Drug | ReACp53 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R175H (c.524G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.37 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.38 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
H
C
C
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | S1 GODL cells | N.A. | Homo sapiens (Human) | N.A. | |||||||||
| In Vivo Model | Immunocompromised NSG mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
In vitro 3D organoid assay | ||||||||||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [34] | ||||||||||||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | ||||||||||||
| Sensitive Drug | ReACp53 | ||||||||||||
| Molecule Alteration | Missense mutation | p.Y327L (c.979_980delTAinsCT) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | S1 GODL cells | N.A. | Homo sapiens (Human) | N.A. | |||||||||
| In Vivo Model | Immunocompromised NSG mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
In vitro 3D organoid assay | ||||||||||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [34] | ||||||||||||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | ||||||||||||
| Sensitive Drug | ReACp53 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R248Q (c.743G>A) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | S1 GODL cells | N.A. | Homo sapiens (Human) | N.A. | |||||||||
| In Vivo Model | Immunocompromised NSG mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
In vitro 3D organoid assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: ER negative breast cancer [ICD-11: 2C60.7] | [35] | ||||||||||||
| Sensitive Disease | ER negative breast cancer [ICD-11: 2C60.7] | ||||||||||||
| Sensitive Drug | YW3-56 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R280K (c.839G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.05 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.00 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
S
-
S
-
S
-
V
V
P
P
S
S
100
|
Q
Q
K
K
T
T
Y
Y
Q
Q
G
G
S
S
Y
Y
G
G
F
F
110
|
R
R
L
L
G
G
F
F
L
L
H
H
S
S
G
G
T
T
A
A
120
|
K
K
S
S
V
V
T
T
C
C
T
T
Y
Y
S
S
P
P
A
A
130
|
L
L
N
N
K
K
M
M
F
F
C
C
Q
Q
L
L
A
A
K
K
140
|
T
T
C
C
P
P
V
V
Q
Q
L
L
W
W
V
V
D
D
S
S
150
|
T
T
P
P
P
P
P
P
G
G
T
T
R
R
V
V
R
R
A
A
160
|
M
M
A
A
I
I
Y
Y
K
K
Q
Q
S
S
Q
Q
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
R
C
C
P
P
H
H
H
H
180
|
E
E
R
R
C
C
S
S
D
D
S
S
D
D
G
G
L
L
A
A
190
|
P
P
P
P
Q
Q
H
H
L
L
I
I
R
R
V
V
E
E
G
G
200
|
N
N
L
L
R
R
V
V
E
E
Y
Y
L
L
D
D
D
D
R
R
210
|
N
N
T
T
F
F
R
R
H
H
S
S
V
V
V
V
V
V
P
P
220
|
Y
Y
E
E
P
P
P
P
E
E
V
V
G
G
S
S
D
D
C
C
230
|
T
T
T
T
I
I
H
H
Y
Y
N
N
Y
Y
M
M
C
C
N
N
240
|
S
S
S
S
C
C
M
M
G
G
G
G
M
M
N
N
R
R
R
R
250
|
P
P
I
I
L
L
T
T
I
I
I
I
T
T
L
L
E
E
D
D
260
|
S
S
S
S
G
G
N
N
L
L
L
L
G
G
R
R
N
N
S
S
270
|
F
F
E
E
V
V
R
R
V
V
C
C
A
A
C
C
P
P
G
G
280
|
R
K
D
D
R
R
R
R
T
T
E
E
E
E
E
E
N
N
L
L
290
|
R
-
K
-
K
-
G
-
E
-
P
-
H
-
H
-
E
-
L
-
300
|
P
-
P
-
G
-
S
-
T
-
K
-
R
-
A
-
L
-
P
-
310
|
N
-
N
-
T
-
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |||||||||
| 1833TR cells | N.A. | N.A. | N.A. | ||||||||||
| In Vivo Model | Female xenograft nude mouse model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
Investigative Drug(s)
3 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Gastric adenocarcinoma [ICD-11: 2B72.0] | [36] | ||||||||||||
| Sensitive Disease | Gastric adenocarcinoma [ICD-11: 2B72.0] | ||||||||||||
| Sensitive Drug | EAP Protocol | ||||||||||||
| Molecule Alteration | Missense mutation | p.R175H (c.524G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.37 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.38 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
H
H
M
M
170
|
T
T
E
E
V
V
V
V
R
R
R
H
C
C
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Stomach | N.A. | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Mechanism Description | The missense mutation p.R175H (c.524G>A) in gene TP53 cause the sensitivity of EAP Protocol by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Gastric adenocarcinoma [ICD-11: 2B72.0] | [36] | ||||||||||||
| Sensitive Disease | Gastric adenocarcinoma [ICD-11: 2B72.0] | ||||||||||||
| Sensitive Drug | EAP Protocol | ||||||||||||
| Molecule Alteration | Missense mutation | p.R282L (c.845G>T) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Stomach | N.A. | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Mechanism Description | The missense mutation p.R282L (c.845G>T) in gene TP53 cause the sensitivity of EAP Protocol by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Gastric adenocarcinoma [ICD-11: 2B72.0] | [36] | ||||||||||||
| Sensitive Disease | Gastric adenocarcinoma [ICD-11: 2B72.0] | ||||||||||||
| Sensitive Drug | EAP Protocol | ||||||||||||
| Molecule Alteration | Missense mutation | p.R213P (c.638G>C) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Stomach | N.A. | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Mechanism Description | The missense mutation p.R213P (c.638G>C) in gene TP53 cause the sensitivity of EAP Protocol by unusual activation of pro-survival pathway | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [37] | |||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Sensitive Drug | PKI-587 | |||
| Molecule Alteration | Missense mutation | p.R158G (c.472C>G) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Chronic lymphocytic leukemia [ICD-11: 2A82.0] | [38] | |||
| Resistant Disease | Chronic lymphocytic leukemia [ICD-11: 2A82.0] | |||
| Resistant Drug | Purine analogues | |||
| Molecule Alteration | Mutation | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Bone marrow | Blood | Homo sapiens (Human) | N.A. |
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | ||
| Experiment for Molecule Alteration |
Sanger sequencing assay; Next-generation sequencing assay | |||
| Experiment for Drug Resistance |
karyotyping assay | |||
| Mechanism Description | TP53 abnormalities lead to resistance to purine a.logues and are found in over 40% of patients with refractory chronic lymphocytic leukemia (CLL). | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Nervous tissue | |
| The Specified Disease | Brain cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.42E-178; Fold-change: 1.18E+00; Z-score: 2.70E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.93E-02; Fold-change: 2.69E-01; Z-score: 2.34E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| The Studied Tissue | White matter | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.95E-02; Fold-change: 3.97E-01; Z-score: 7.90E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
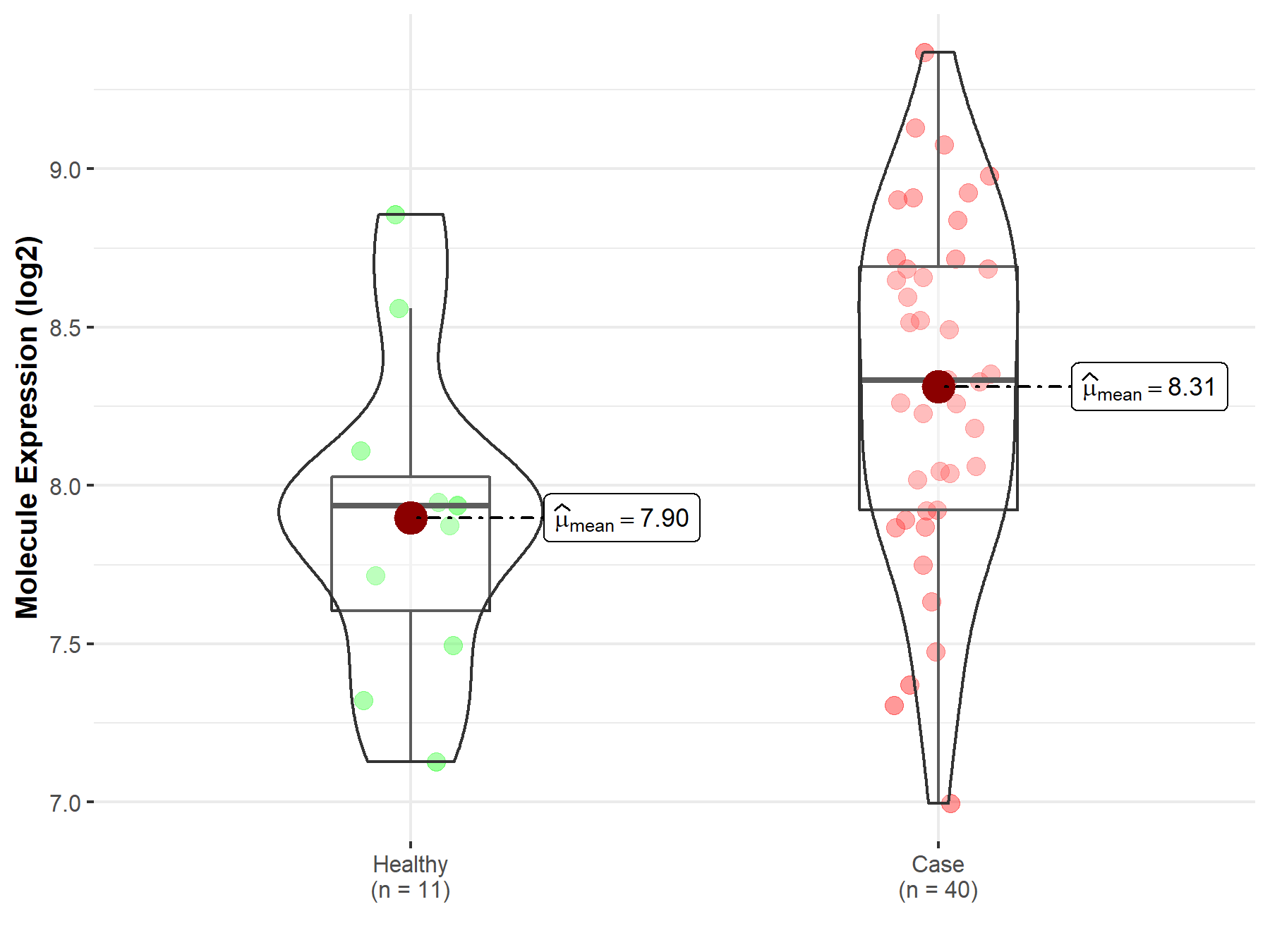
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Neuroectodermal tumor | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.74E-01; Fold-change: -1.33E-01; Z-score: -6.77E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
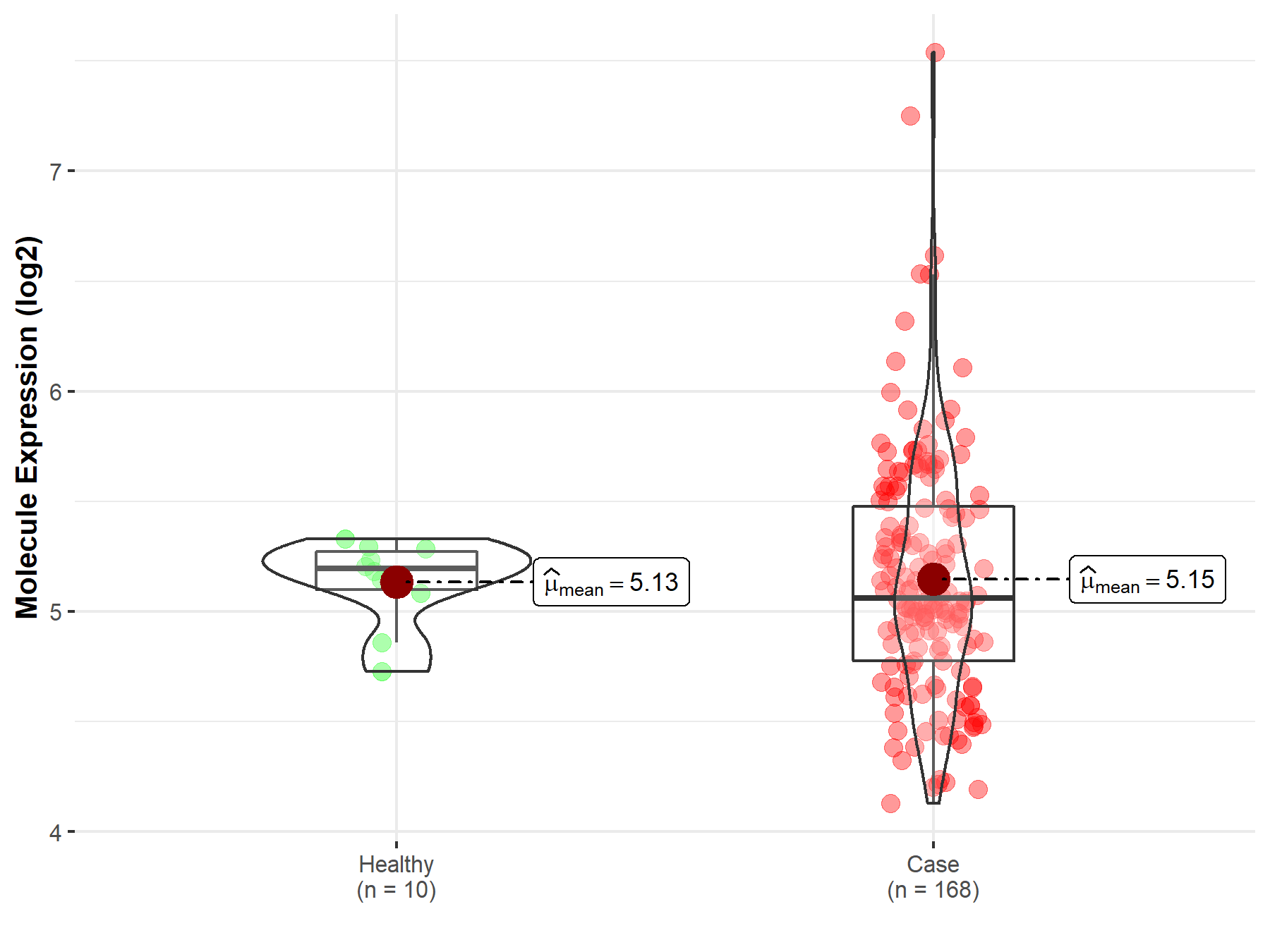
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Whole blood | |
| The Specified Disease | Myelofibrosis | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.02E-05; Fold-change: -6.91E-01; Z-score: -3.13E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Whole blood | |
| The Specified Disease | Polycythemia vera | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.56E-24; Fold-change: -7.55E-01; Z-score: -3.26E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Bone marrow | |
| The Specified Disease | Acute myeloid leukemia | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.90E-07; Fold-change: 2.95E-01; Z-score: 5.95E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Gastric tissue | |
| The Specified Disease | Gastric cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.43E-01; Fold-change: 2.14E-01; Z-score: 5.31E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.31E-04; Fold-change: 1.18E+00; Z-score: 1.51E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
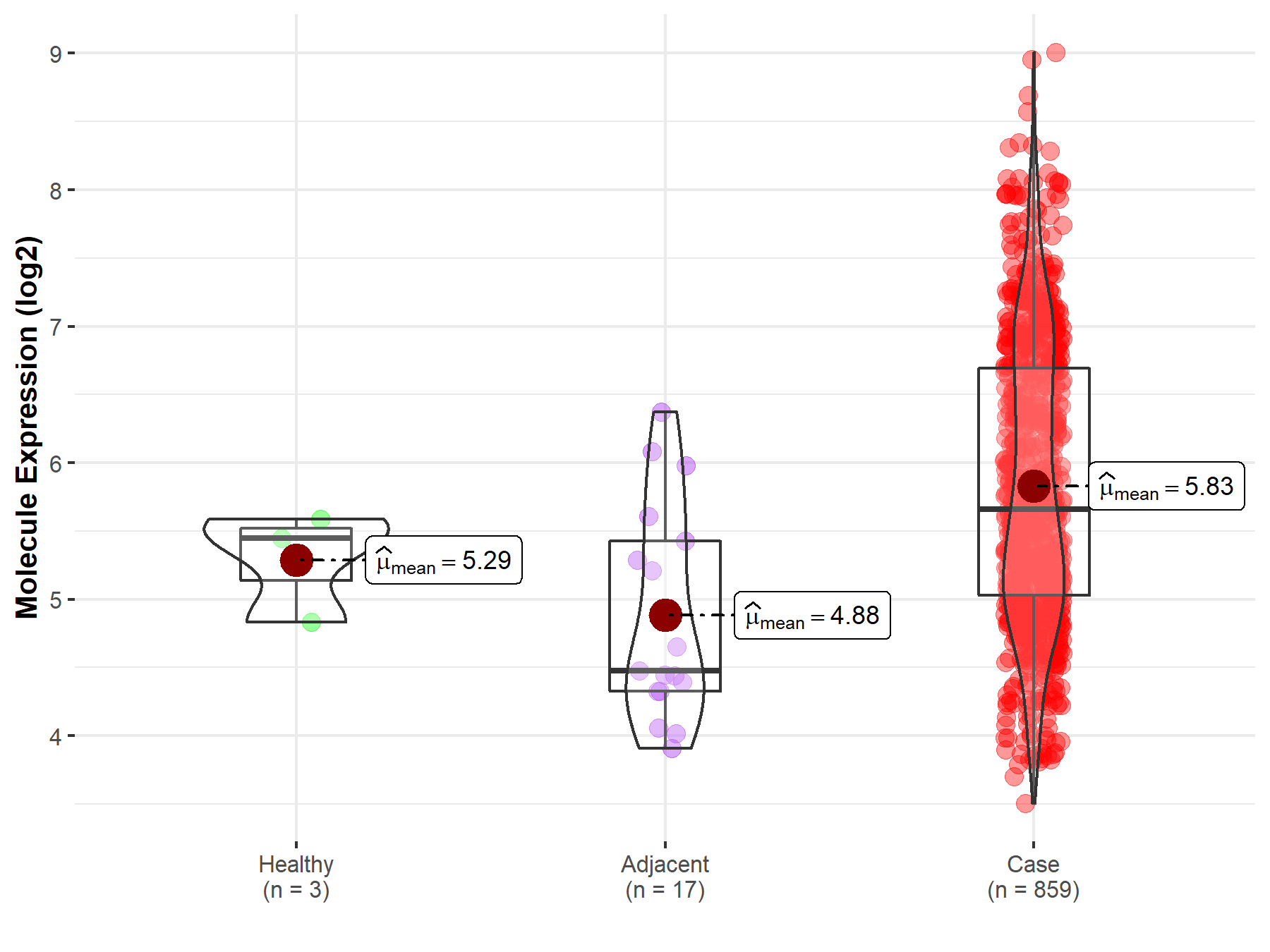
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Liver | |
| The Specified Disease | Liver cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.64E-02; Fold-change: 1.46E-01; Z-score: 2.81E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 3.27E-17; Fold-change: 7.65E-01; Z-score: 1.14E+00 | |
| The Expression Level of Disease Section Compare with the Other Disease Section | p-value: 7.04E-01; Fold-change: 2.46E-01; Z-score: 3.20E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
Molecule expression in tissue other than the diseased tissue of patients
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Lung | |
| The Specified Disease | Lung cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.24E-02; Fold-change: -1.28E-02; Z-score: -2.15E-02 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.01E-01; Fold-change: -3.27E-01; Z-score: -4.78E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
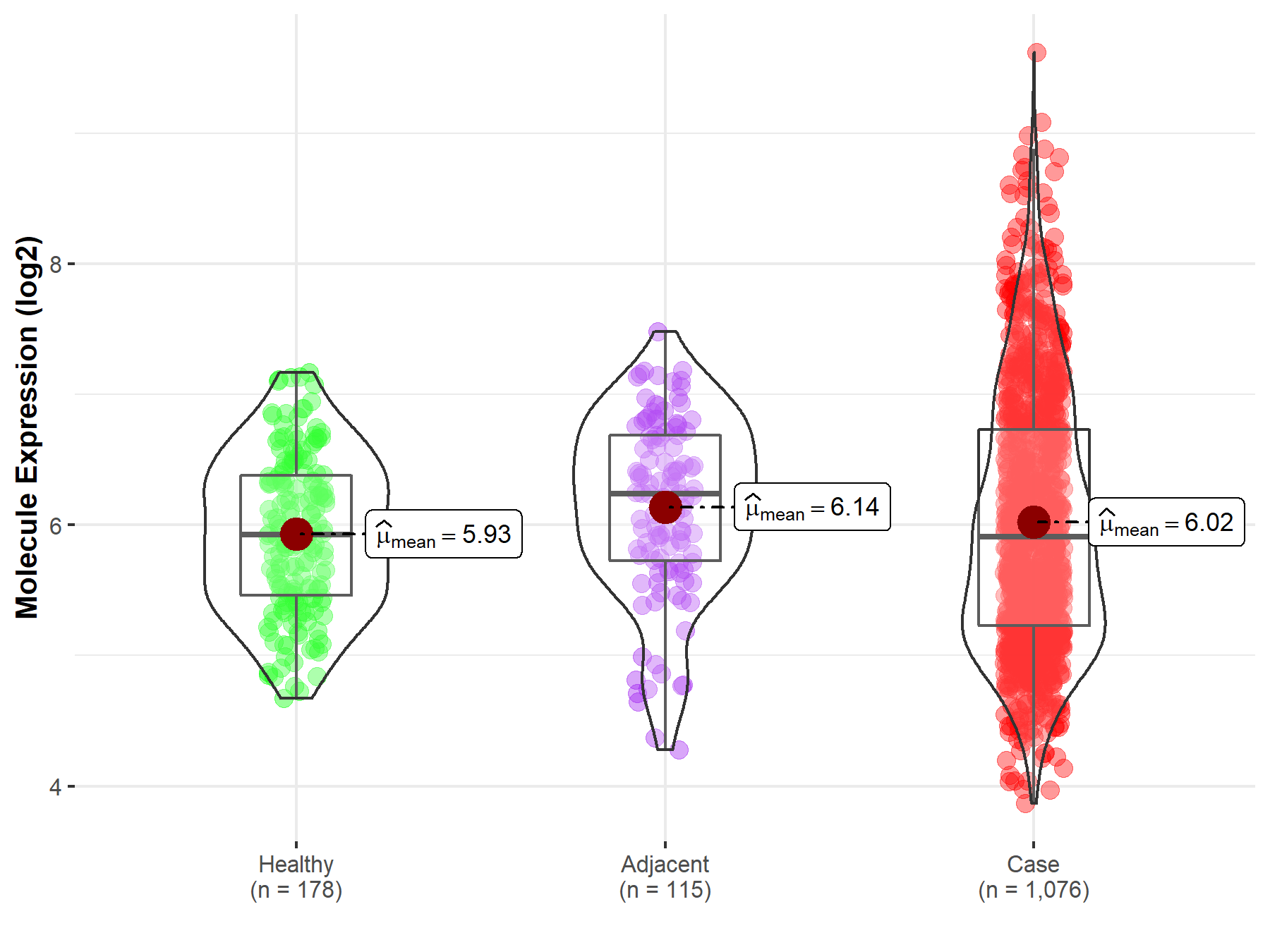
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Skin | |
| The Specified Disease | Melanoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.47E-01; Fold-change: 1.46E-01; Z-score: 2.20E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.63E-01; Fold-change: 1.62E-02; Z-score: 1.76E-02 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 7.38E-02; Fold-change: -1.63E-01; Z-score: -1.55E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
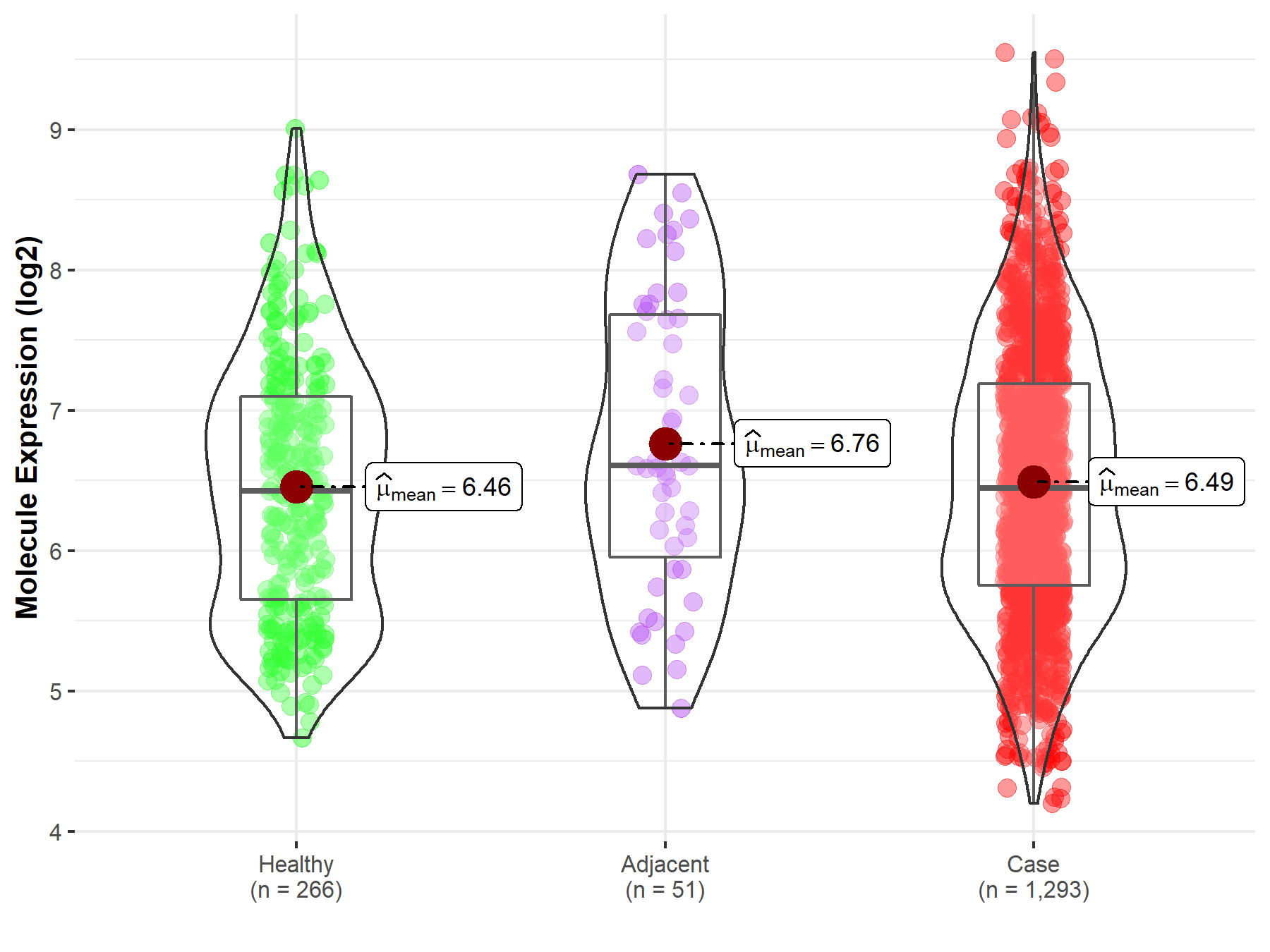
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Ovary | |
| The Specified Disease | Ovarian cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.96E-01; Fold-change: 7.27E-02; Z-score: 1.17E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 3.50E-04; Fold-change: 1.14E+00; Z-score: 1.72E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
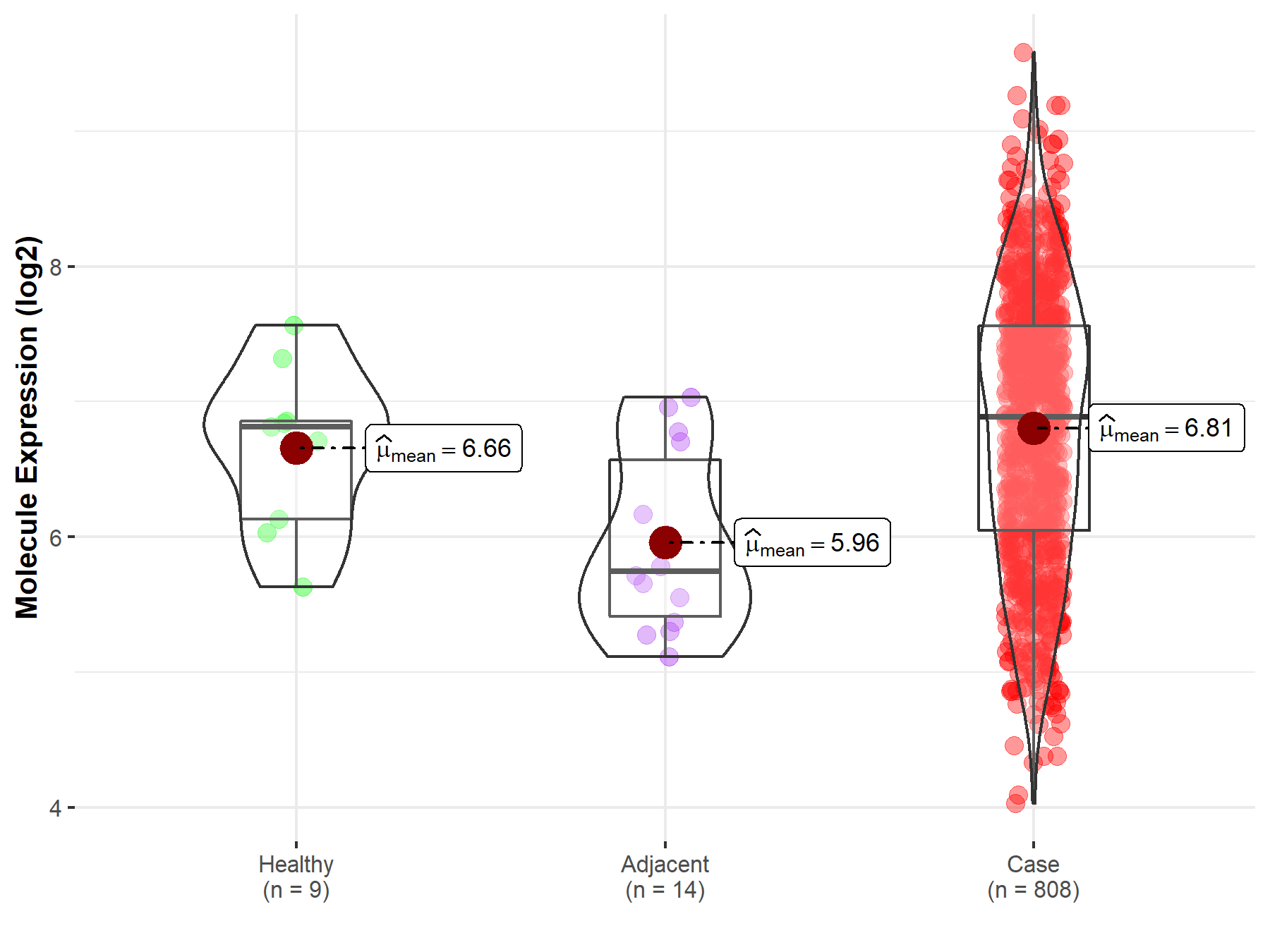
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Bladder tissue | |
| The Specified Disease | Bladder cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.97E-02; Fold-change: -1.24E-01; Z-score: -4.64E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
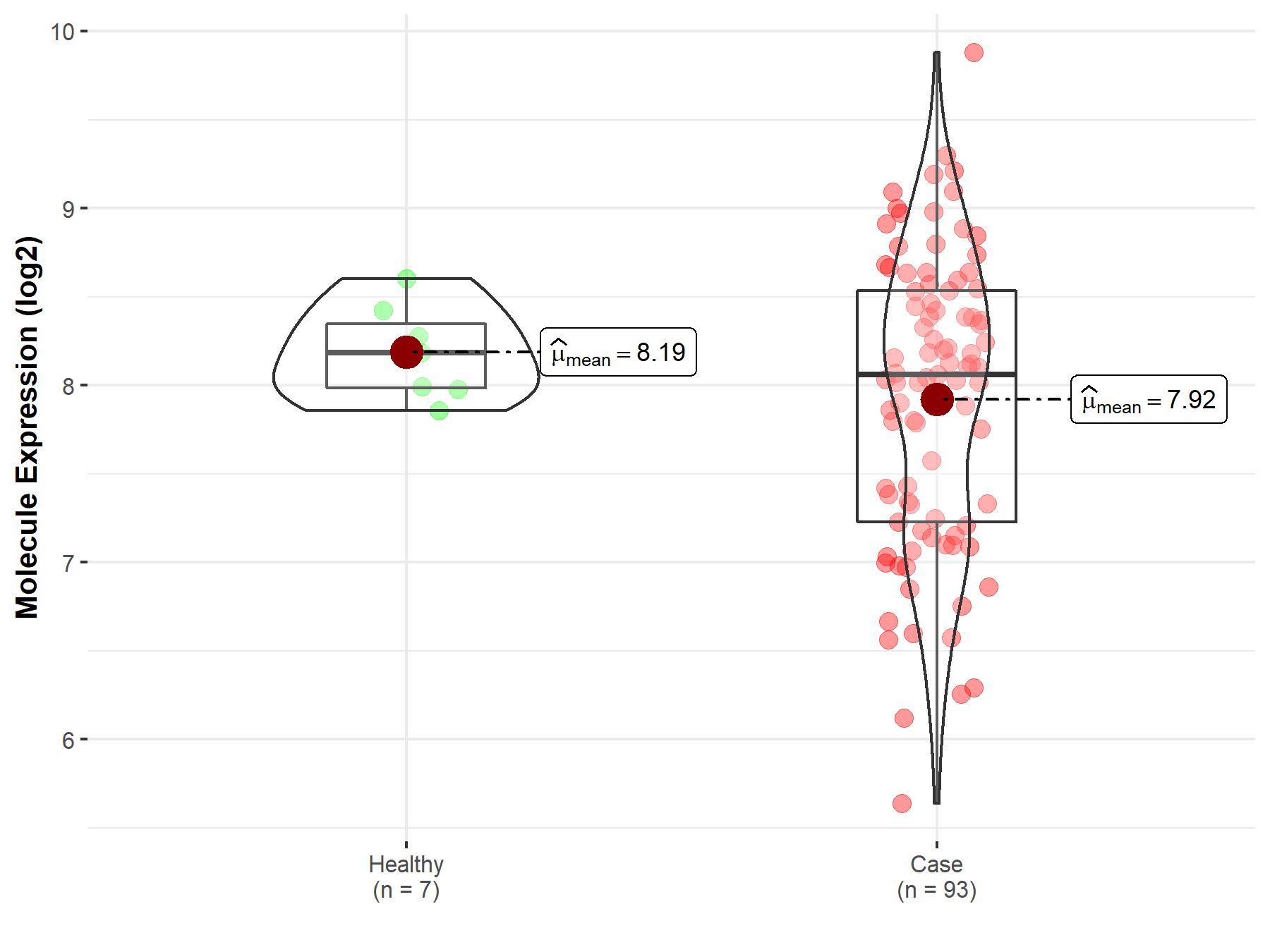
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Head and neck tissue | |
| The Specified Disease | Head and neck cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.22E-01; Fold-change: 2.20E-01; Z-score: 4.71E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
ICD Disease Classification 15

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Peripheral blood | |
| The Specified Disease | Rheumatoid arthritis | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.42E-04; Fold-change: -5.49E-02; Z-score: -1.83E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
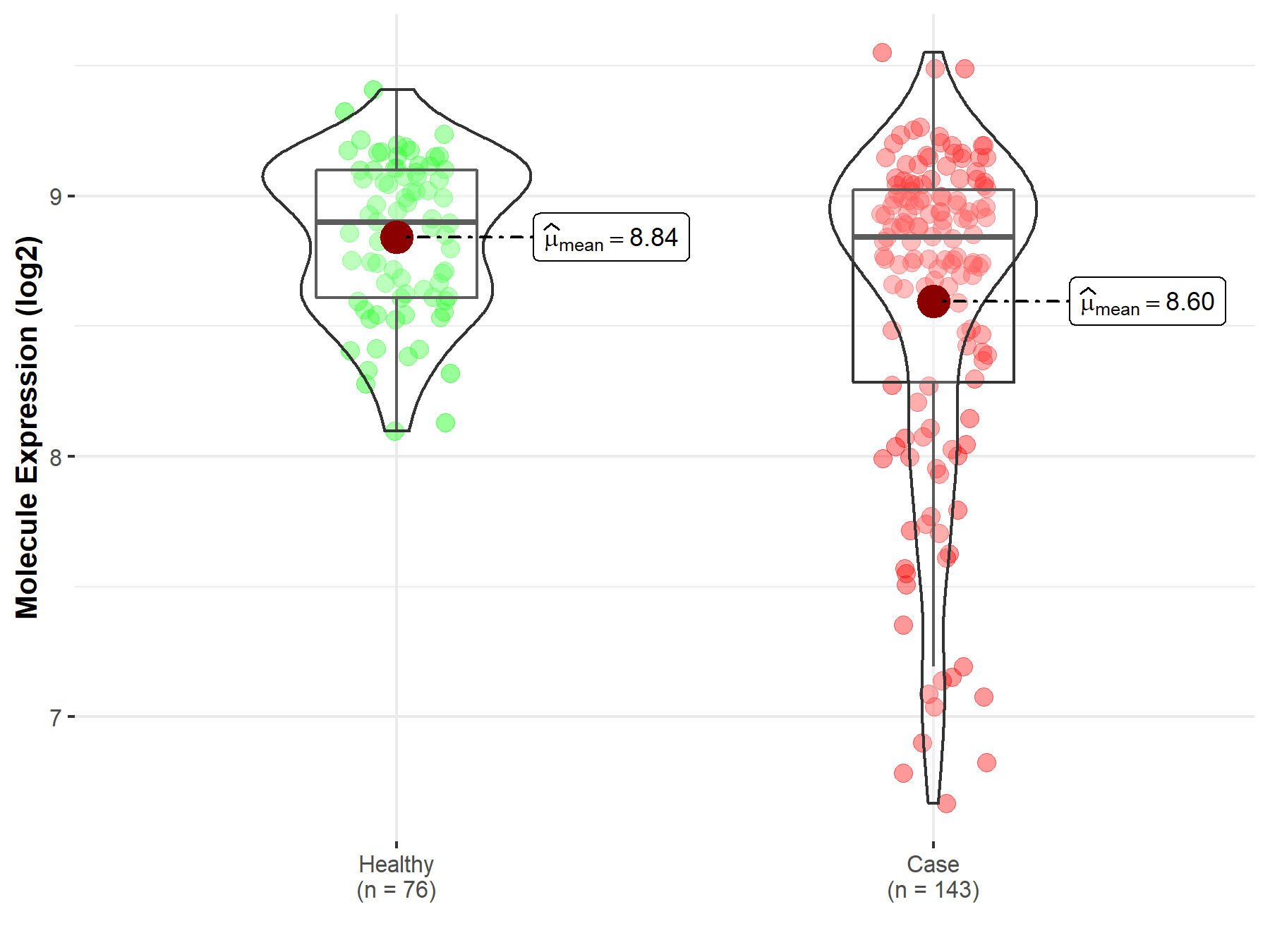
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Synovial tissue | |
| The Specified Disease | Rheumatoid arthritis | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.88E-11; Fold-change: 2.48E+00; Z-score: 1.06E+01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
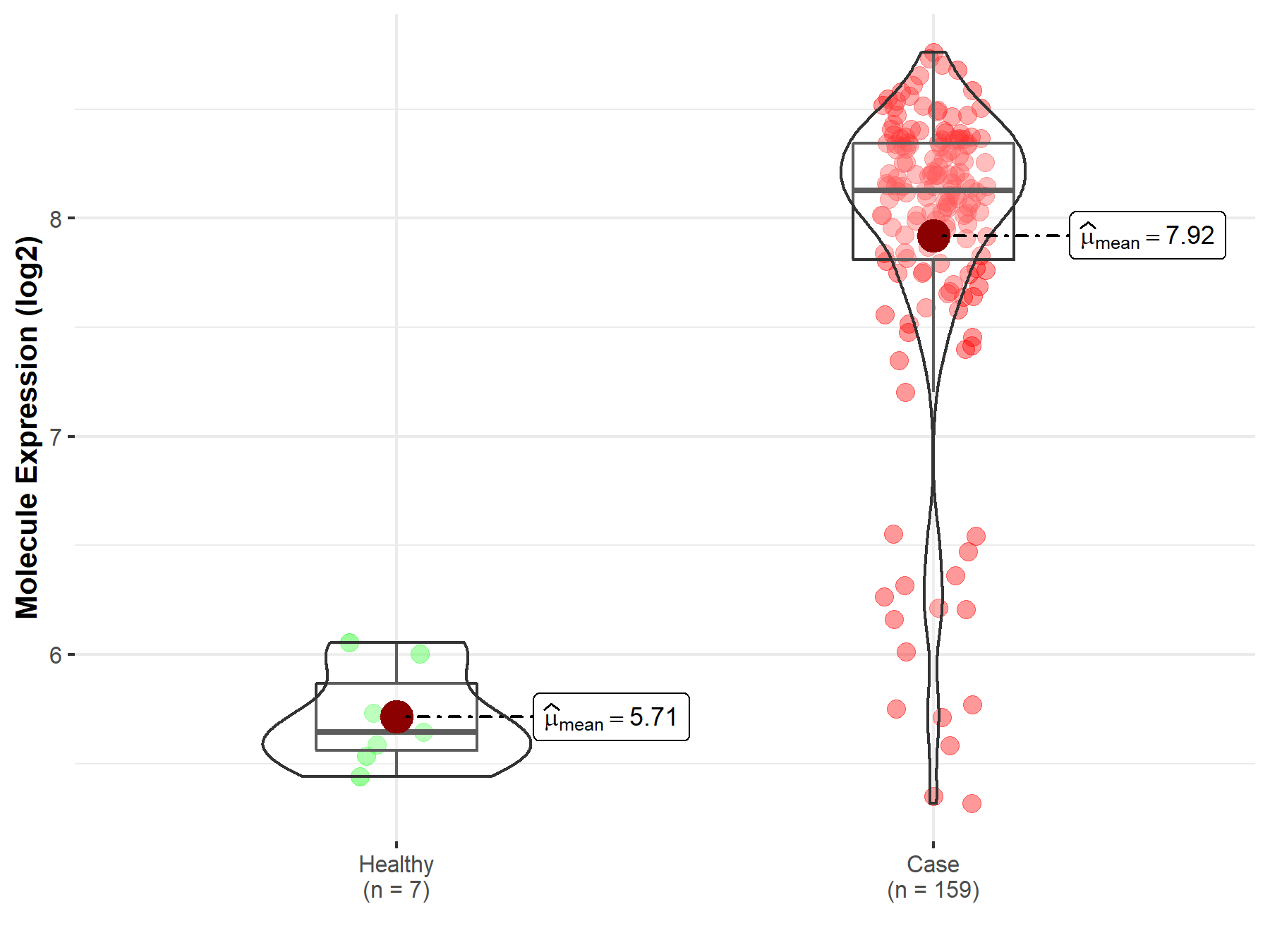
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals


|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
