Molecule Information
General Information of the Molecule (ID: Mol00562)
| Name |
PI3-kinase alpha (PIK3CA)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
PI3-kinase subunit alpha; PI3K-alpha; PI3Kalpha; PtdIns-3-kinase subunit alpha; Phosphatidylinositol 4;5-bisphosphate 3-kinase 110 kDa catalytic subunit alpha; PtdIns-3-kinase subunit p110-alpha; p110alpha; Phosphoinositide 3-kinase alpha; Phosphoinositide-3-kinase catalytic alpha polypeptide; Serine/threonine protein kinase PIK3CA
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
PIK3CA
|
||||
| Gene ID | |||||
| Location |
chr3:179148114-179240093[+]
|
||||
| Sequence |
MPPRPSSGELWGIHLMPPRILVECLLPNGMIVTLECLREATLITIKHELFKEARKYPLHQ
LLQDESSYIFVSVTQEAEREEFFDETRRLCDLRLFQPFLKVIEPVGNREEKILNREIGFA IGMPVCEFDMVKDPEVQDFRRNILNVCKEAVDLRDLNSPHSRAMYVYPPNVESSPELPKH IYNKLDKGQIIVVIWVIVSPNNDKQKYTLKINHDCVPEQVIAEAIRKKTRSMLLSSEQLK LCVLEYQGKYILKVCGCDEYFLEKYPLSQYKYIRSCIMLGRMPNLMLMAKESLYSQLPMD CFTMPSYSRRISTATPYMNGETSTKSLWVINSALRIKILCATYVNVNIRDIDKIYVRTGI YHGGEPLCDNVNTQRVPCSNPRWNEWLNYDIYIPDLPRAARLCLSICSVKGRKGAKEEHC PLAWGNINLFDYTDTLVSGKMALNLWPVPHGLEDLLNPIGVTGSNPNKETPCLELEFDWF SSVVKFPDMSVIEEHANWSVSREAGFSYSHAGLSNRLARDNELRENDKEQLKAISTRDPL SEITEQEKDFLWSHRHYCVTIPEILPKLLLSVKWNSRDEVAQMYCLVKDWPPIKPEQAME LLDCNYPDPMVRGFAVRCLEKYLTDDKLSQYLIQLVQVLKYEQYLDNLLVRFLLKKALTN QRIGHFFFWHLKSEMHNKTVSQRFGLLLESYCRACGMYLKHLNRQVEAMEKLINLTDILK QEKKDETQKVQMKFLVEQMRRPDFMDALQGFLSPLNPAHQLGNLRLEECRIMSSAKRPLW LNWENPDIMSELLFQNNEIIFKNGDDLRQDMLTLQIIRIMENIWQNQGLDLRMLPYGCLS IGDCVGLIEVVRNSHTIMQIQCKGGLKGALQFNSHTLHQWLKDKNKGEIYDAAIDLFTRS CAGYCVATFILGIGDRHNSNIMVKDDGQLFHIDFGHFLDHKKKKFGYKRERVPFVLTQDF LIVISKGAQECTKTREFERFQEMCYKAYLAIRQHANLFINLFSMMLGSGMPELQSFDDIA YIRKTLALDKTEQEALEYFMKQMNDAHHGGWTTKMDWIFHTIKQHALN Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Phosphoinositide-3-kinase (PI3K) phosphorylates phosphatidylinositol (PI) and its phosphorylated derivatives at position 3 of the inositol ring to produce 3-phosphoinositides. Uses ATP and PtdIns(4,5)P2 (phosphatidylinositol 4,5-bisphosphate) to generate phosphatidylinositol 3,4,5-trisphosphate (PIP3). PIP3 plays a key role by recruiting PH domain-containing proteins to the membrane, including AKT1 and PDPK1, activating signaling cascades involved in cell growth, survival, proliferation, motility and morphology. Participates in cellular signaling in response to various growth factors. Involved in the activation of AKT1 upon stimulation by receptor tyrosine kinases ligands such as EGF, insulin, IGF1, VEGFA and PDGF. Involved in signaling via insulin-receptor substrate (IRS) proteins. Essential in endothelial cell migration during vascular development through VEGFA signaling, possibly by regulating RhoA activity. Required for lymphatic vasculature development, possibly by binding to RAS and by activation by EGF and FGF2, but not by PDGF. Regulates invadopodia formation through the PDPK1-AKT1 pathway. Participates in cardiomyogenesis in embryonic stem cells through a AKT1 pathway. Participates in vasculogenesis in embryonic stem cells through PDK1 and protein kinase C pathway. In addition to its lipid kinase activity, it displays a serine-protein kinase activity that results in the autophosphorylation of the p85alpha regulatory subunit as well as phosphorylation of other proteins such as 4EBP1, H-Ras, the IL-3 beta c receptor and possibly others. Plays a role in the positive regulation of phagocytosis and pinocytosis.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Clinical Trial Drug(s)
19 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [1] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Saracatinib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Breast cancer [ICD-11: 2C60] | |||
| The Specified Disease | Breast cancer | |||
| The Studied Tissue | Breast tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.59E-38 Fold-change: -1.23E-01 Z-score: -1.46E+01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell viability | Inhibition | hsa05200 | ||
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| BT474 cells | Breast | Homo sapiens (Human) | CVCL_0179 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Transwell assay; Flow cytometry assay | |||
| Mechanism Description | miR-19b-3p increases saracatinib sensitivity by inhibiting the PI3k/Akt pathway and miR-19b-3p directly bound to the 3'-UTR of PIk3CA and inhibited PIk3CA expression. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [15] | |||
| Resistant Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Resistant Drug | Buparlisib | |||
| Molecule Alteration | Missense mutation | p.N345I (c.1034A>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | PI3K signaling pathway | Inhibition | hsa04151 | |
| In Vitro Model | HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 |
| Experiment for Molecule Alteration |
Ion Torrent sequencing assay | |||
| Experiment for Drug Resistance |
IC50 assay; Proliferation assay | |||
| Mechanism Description | Deregulation of the phosphoinositide 3-kinase (PI3K) pathway contributes to the development and progression of tumors. These PIK3CA and PIK3R1 impactful mutations exhibit a mutually exclusive pattern, leading to oncogenesis and hyperactivity of PI3K pathway. The PIK3CA impactful mutations are tightly associated with hormone receptor positivity. | |||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [15] | |||
| Resistant Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Resistant Drug | Buparlisib | |||
| Molecule Alteration | Missense mutation | p.E39K (c.115G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | PI3K signaling pathway | Inhibition | hsa04151 | |
| In Vitro Model | HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 |
| Experiment for Molecule Alteration |
Ion Torrent sequencing assay | |||
| Experiment for Drug Resistance |
IC50 assay; Proliferation assay | |||
| Mechanism Description | Deregulation of the phosphoinositide 3-kinase (PI3K) pathway contributes to the development and progression of tumors. These PIK3CA and PIK3R1 impactful mutations exhibit a mutually exclusive pattern, leading to oncogenesis and hyperactivity of PI3K pathway. The PIK3CA impactful mutations are tightly associated with hormone receptor positivity. | |||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [15] | |||
| Resistant Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Resistant Drug | Buparlisib | |||
| Molecule Alteration | Missense mutation | p.E453K (c.1357G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | PI3K signaling pathway | Inhibition | hsa04151 | |
| In Vitro Model | HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 |
| Experiment for Molecule Alteration |
Ion Torrent sequencing assay | |||
| Experiment for Drug Resistance |
IC50 assay; Proliferation assay | |||
| Mechanism Description | Deregulation of the phosphoinositide 3-kinase (PI3K) pathway contributes to the development and progression of tumors. These PIK3CA and PIK3R1 impactful mutations exhibit a mutually exclusive pattern, leading to oncogenesis and hyperactivity of PI3K pathway. The PIK3CA impactful mutations are tightly associated with hormone receptor positivity. | |||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [15] | |||
| Resistant Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Resistant Drug | Buparlisib | |||
| Molecule Alteration | Missense mutation | p.E542K (c.1624G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | PI3K signaling pathway | Inhibition | hsa04151 | |
| In Vitro Model | HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 |
| Experiment for Molecule Alteration |
Ion Torrent sequencing assay | |||
| Experiment for Drug Resistance |
IC50 assay; Proliferation assay | |||
| Mechanism Description | Deregulation of the phosphoinositide 3-kinase (PI3K) pathway contributes to the development and progression of tumors. These PIK3CA and PIK3R1 impactful mutations exhibit a mutually exclusive pattern, leading to oncogenesis and hyperactivity of PI3K pathway. The PIK3CA impactful mutations are tightly associated with hormone receptor positivity. | |||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [15] | |||
| Resistant Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Resistant Drug | Buparlisib | |||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | PI3K signaling pathway | Inhibition | hsa04151 | |
| In Vitro Model | HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 |
| Experiment for Molecule Alteration |
Ion Torrent sequencing assay | |||
| Experiment for Drug Resistance |
IC50 assay; Proliferation assay | |||
| Mechanism Description | Deregulation of the phosphoinositide 3-kinase (PI3K) pathway contributes to the development and progression of tumors. These PIK3CA and PIK3R1 impactful mutations exhibit a mutually exclusive pattern, leading to oncogenesis and hyperactivity of PI3K pathway. The PIK3CA impactful mutations are tightly associated with hormone receptor positivity. | |||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [15] | |||
| Resistant Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Resistant Drug | Buparlisib | |||
| Molecule Alteration | Missense mutation | p.G1049R (c.3145G>C) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | PI3K signaling pathway | Inhibition | hsa04151 | |
| In Vitro Model | HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 |
| Experiment for Molecule Alteration |
Ion Torrent sequencing assay | |||
| Experiment for Drug Resistance |
IC50 assay; Proliferation assay | |||
| Mechanism Description | Deregulation of the phosphoinositide 3-kinase (PI3K) pathway contributes to the development and progression of tumors. These PIK3CA and PIK3R1 impactful mutations exhibit a mutually exclusive pattern, leading to oncogenesis and hyperactivity of PI3K pathway. The PIK3CA impactful mutations are tightly associated with hormone receptor positivity. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [16] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | Buparlisib | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |||||||||
| BT474 cells | Breast | Homo sapiens (Human) | CVCL_0179 | ||||||||||
| MDA-MB-361 cells | Breast | Homo sapiens (Human) | CVCL_0620 | ||||||||||
| In Vivo Model | Athymic mouse PDX model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
qRT-PCR; Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Promega assay | ||||||||||||
| Mechanism Description | Brain metastases evade phosphatidylinositide 3-kinase (PI3K) inhibition despite drug accumulation in the brain lesions. In comparison to extracranial disease, increased HER3 expression and phosphorylation in brain lesions. HER3 blockade overcame the resistance of HER2-amplified and/or PIK3CA-mutant breast cancer brain metastases to PI3K inhibitors, resulting in marked tumor growth delay and improvement in mouse survival. | ||||||||||||
|
|
|||||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [17] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Sensitive Drug | Buparlisib | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | SQCLC SKMES-1 cells | Pleura | Homo sapiens (Human) | CVCL_0630 | |||||||||
| HCC2450 cells | Lung | Homo sapiens (Human) | CVCL_5133 | ||||||||||
| H596 cells | Lung | Homo sapiens (Human) | CVCL_1571 | ||||||||||
| In Vivo Model | Balb/c-Nude female xenograft mouse model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [17] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Sensitive Drug | Buparlisib | ||||||||||||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | SQCLC SKMES-1 cells | Pleura | Homo sapiens (Human) | CVCL_0630 | |||||||||
| HCC2450 cells | Lung | Homo sapiens (Human) | CVCL_5133 | ||||||||||
| H596 cells | Lung | Homo sapiens (Human) | CVCL_1571 | ||||||||||
| In Vivo Model | Balb/c-Nude female xenograft mouse model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Gastric adenocarcinoma [ICD-11: 2B72.0] | [18] | ||||||||||||
| Sensitive Disease | Gastric adenocarcinoma [ICD-11: 2B72.0] | ||||||||||||
| Sensitive Drug | Capivasertib | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Stomach | N.A. | |||||||||||
| In Vivo Model | GC xenograft (PDGCX) mouse model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTS assay | ||||||||||||
| Disease Class: Gastrointestinal system cancer [ICD-11: 2C11.Y] | [18] | ||||||||||||
| Sensitive Disease | Gastrointestinal system cancer [ICD-11: 2C11.Y] | ||||||||||||
| Sensitive Drug | Capivasertib | ||||||||||||
| Molecule Alteration | Missense mutation | p.E542K (c.1624G>A) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Stomach | N.A. | |||||||||||
| In Vivo Model | GC xenograft (PDGCX) mouse model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTS assay | ||||||||||||
| Disease Class: Gastrointestinal system cancer [ICD-11: 2C11.Y] | [18] | ||||||||||||
| Sensitive Disease | Gastrointestinal system cancer [ICD-11: 2C11.Y] | ||||||||||||
| Sensitive Drug | Capivasertib | ||||||||||||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Stomach | N.A. | |||||||||||
| In Vivo Model | GC xenograft (PDGCX) mouse model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTS assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Head and neck squamous cell carcinoma [ICD-11: 2D42.1] | [19] | ||||||||||||
| Sensitive Disease | Head and neck squamous cell carcinoma [ICD-11: 2D42.1] | ||||||||||||
| Sensitive Drug | Rigosertib | ||||||||||||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HNSCC cells | Neck | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
DNA sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Sulforhodamine B colorimetric assay | ||||||||||||
| Mechanism Description | The missense mutation p.E545K (c.1633G>A) in gene PIK3CA cause the sensitivity of Rigosertib by aberration of the drug's therapeutic target | ||||||||||||
|
|
|||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [20] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | Rigosertib | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | RAS/RAF/MEK/ERK signaling pathway | Inhibition | hsa01521 | ||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 | ||||||||||
| A431 cells | Skin | Homo sapiens (Human) | CVCL_0037 | ||||||||||
| HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 | ||||||||||
| MIA PaCa-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0428 | ||||||||||
| WM1617 cells | Lymph node | Homo sapiens (Human) | CVCL_6791 | ||||||||||
| In Vivo Model | Female nu/nu mouse PDX model | Mus musculus | |||||||||||
| Mechanism Description | Rigosertib, a styryl-benzyl sulfone, acts as a RAS-mimetic and interacts with the RBDs of RAF kinases, resulting in their inability to bind to RAS, disruption of RAF activation, and inhibition of the RAS-RAF-MEK pathway. We also find that rigosertib binds to the RBDs of Ral-GDS and PI3Ks. | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [20] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | Rigosertib | ||||||||||||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | RAS/RAF/MEK/ERK signaling pathway | Inhibition | hsa01521 | ||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 | ||||||||||
| A431 cells | Skin | Homo sapiens (Human) | CVCL_0037 | ||||||||||
| HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 | ||||||||||
| MIA PaCa-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0428 | ||||||||||
| WM1617 cells | Lymph node | Homo sapiens (Human) | CVCL_6791 | ||||||||||
| In Vivo Model | Female nu/nu mouse PDX model | Mus musculus | |||||||||||
| Mechanism Description | Rigosertib, a styryl-benzyl sulfone, acts as a RAS-mimetic and interacts with the RBDs of RAF kinases, resulting in their inability to bind to RAS, disruption of RAF activation, and inhibition of the RAS-RAF-MEK pathway. We also find that rigosertib binds to the RBDs of Ral-GDS and PI3Ks. | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Head and neck squamous cell carcinoma [ICD-11: 2D42.1] | [21] | ||||||||||||
| Sensitive Disease | Head and neck squamous cell carcinoma [ICD-11: 2D42.1] | ||||||||||||
| Sensitive Drug | Taselisib | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | FaDu cells | Pharynx | Homo sapiens (Human) | CVCL_1218 | |||||||||
| SCC25 cells | Oral | Homo sapiens (Human) | CVCL_1682 | ||||||||||
| SCC4 cells | Tongue | Homo sapiens (Human) | CVCL_1684 | ||||||||||
| SCC9 cells | Tongue | Homo sapiens (Human) | CVCL_1685 | ||||||||||
| SCC15 cells | Tongue | Homo sapiens (Human) | CVCL_1681 | ||||||||||
| UPCI-SCC-90 cells | Tongue | Homo sapiens (Human) | CVCL_1899 | ||||||||||
| UPCI-SCC-154 cells | Tongue | Homo sapiens (Human) | CVCL_2230 | ||||||||||
| UM-SCC-47 cells | Tongue | Homo sapiens (Human) | CVCL_7759 | ||||||||||
| UM-SCC-104 cells | Cervical lymph node | Homo sapiens (Human) | CVCL_7712 | ||||||||||
| UD-SCC-2 cells | Neck | Homo sapiens (Human) | CVCL_E325 | ||||||||||
| SNU46 cells | Larynx | Homo sapiens (Human) | CVCL_5063 | ||||||||||
| SNU cells | Stomach | Homo sapiens (Human) | CVCL_0099 | ||||||||||
| HSC-4 cells | Cervical lymph node | Homo sapiens (Human) | CVCL_1289 | ||||||||||
| HSC-3 cells | Cervical lymph node | Homo sapiens (Human) | CVCL_1288 | ||||||||||
| HSC-2 cells | Cervical lymph node | Homo sapiens (Human) | CVCL_1287 | ||||||||||
| Detroit 562 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1171 | ||||||||||
| Cal-33 cells | Tongue | Homo sapiens (Human) | CVCL_1108 | ||||||||||
| BICR-31 cells | Tongue | Homo sapiens (Human) | CVCL_2312 | ||||||||||
| BICR-22 cells | Lymph node | Homo sapiens (Human) | CVCL_2310 | ||||||||||
| BICR-18 cells | Lymph Node | Homo sapiens (Human) | CVCL_2309 | ||||||||||
| BICR-16 cells | Tongue | Homo sapiens (Human) | CVCL_2308 | ||||||||||
| 93-VU-147T cells | Oral cavity | Homo sapiens (Human) | CVCL_L895 | ||||||||||
| In Vivo Model | Nu/Nu mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Crystal violet staining assay | ||||||||||||
|
|
|||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [22] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | Taselisib | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | PI3K signaling pathway | Inhibition | hsa04151 | ||||||||||
| In Vitro Model | KPL-4 cells | Breast | Homo sapiens (Human) | CVCL_5310 | |||||||||
| In Vivo Model | SCID beige mouse PDX model | Mus musculus | |||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [22] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Sensitive Drug | Taselisib | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047X (c.3139_3141) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | PI3K signaling pathway | Inhibition | hsa04151 | ||||||||||
| In Vitro Model | KPL-4 cells | Breast | Homo sapiens (Human) | CVCL_5310 | |||||||||
| In Vivo Model | SCID beige mouse PDX model | Mus musculus | |||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Malignant pleural mesothelioma [ICD-11: 2C26.0] | [23] | |||
| Sensitive Disease | Malignant pleural mesothelioma [ICD-11: 2C26.0] | |||
| Sensitive Drug | Apitolisib | |||
| Molecule Alteration | Missense mutation | p.R88Q (c.263G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | . | N.A. | ||
| Experiment for Molecule Alteration |
DxS allele-specific PCR; qRT-PCR assays; Sanger sequencing assay | |||
| Experiment for Drug Resistance |
CTCAE assay | |||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [23] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Apitolisib | |||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Experiment for Molecule Alteration |
DxS allele-specific PCR; qRT-PCR assays; Sanger sequencing assay | |||
| Experiment for Drug Resistance |
CTCAE assay | |||
| Disease Class: Head and neck squamous cell carcinoma [ICD-11: 2D42.1] | [23] | |||
| Sensitive Disease | Head and neck squamous cell carcinoma [ICD-11: 2D42.1] | |||
| Sensitive Drug | Apitolisib | |||
| Molecule Alteration | Missense mutation | p.E542K (c.1624G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Experiment for Molecule Alteration |
DxS allele-specific PCR; qRT-PCR assays; Sanger sequencing assay | |||
| Experiment for Drug Resistance |
CTCAE assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Acute lymphocytic leukemia [ICD-11: 2B33.0] | [24] | |||
| Sensitive Disease | Acute lymphocytic leukemia [ICD-11: 2B33.0] | |||
| Sensitive Drug | LY-3023414 | |||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K signaling pathway | Inhibition | hsa04151 | |
| In Vitro Model | A2780 cells | Ovary | Homo sapiens (Human) | CVCL_0134 |
| THP-1 cells | Blood | Homo sapiens (Human) | CVCL_0006 | |
| NCI-H446 cells | Lung | Homo sapiens (Human) | CVCL_1562 | |
| AsPC-1 cells | Pancreas | Homo sapiens (Human) | CVCL_0152 | |
| SJSA-1 cells | Bone | Homo sapiens (Human) | CVCL_1697 | |
| NCI-H1650 cells | Lung | Homo sapiens (Human) | CVCL_1483 | |
| MSTO-211H cells | Lung | Homo sapiens (Human) | CVCL_1430 | |
| 786-O cells | Kidney | Homo sapiens (Human) | CVCL_1051 | |
| NCI-H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | |
| NCI-H520 cells | Lung | Homo sapiens (Human) | CVCL_1566 | |
| NCI-H1734 cells | Lung | Homo sapiens (Human) | CVCL_1491 | |
| NCI-H1395 cells | Lung | Homo sapiens (Human) | CVCL_1467 | |
| NCI-H226 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1544 | |
| NCI-H1993 cells | Lymph node | Homo sapiens (Human) | CVCL_1512 | |
| NCI-H1703 cells | Lung | Homo sapiens (Human) | CVCL_1490 | |
| NCI-H1299 cells | Lymph node | Homo sapiens (Human) | CVCL_0060 | |
| NCI- H460 cells | Pleural effusion | Homo sapiens (Human) | CVCL_0459 | |
| ACC-MESO-4 cells | Pleural epithelium | Homo sapiens (Human) | CVCL_5114 | |
| ACC-MESO-1 cells | Pleural epithelium | Homo sapiens (Human) | CVCL_5113 | |
| Experiment for Drug Resistance |
CellTitre-Glo assay; Caspase-Glo 3/7 assay | |||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [24] | |||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Sensitive Drug | LY-3023414 | |||
| Molecule Alteration | Missense mutation | p.G118D (c.353G>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K signaling pathway | Inhibition | hsa04151 | |
| In Vitro Model | A2780 cells | Ovary | Homo sapiens (Human) | CVCL_0134 |
| THP-1 cells | Blood | Homo sapiens (Human) | CVCL_0006 | |
| NCI-H446 cells | Lung | Homo sapiens (Human) | CVCL_1562 | |
| AsPC-1 cells | Pancreas | Homo sapiens (Human) | CVCL_0152 | |
| SJSA-1 cells | Bone | Homo sapiens (Human) | CVCL_1697 | |
| NCI-H1650 cells | Lung | Homo sapiens (Human) | CVCL_1483 | |
| MSTO-211H cells | Lung | Homo sapiens (Human) | CVCL_1430 | |
| 786-O cells | Kidney | Homo sapiens (Human) | CVCL_1051 | |
| NCI-H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | |
| NCI-H520 cells | Lung | Homo sapiens (Human) | CVCL_1566 | |
| NCI-H1734 cells | Lung | Homo sapiens (Human) | CVCL_1491 | |
| NCI-H1395 cells | Lung | Homo sapiens (Human) | CVCL_1467 | |
| NCI-H226 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1544 | |
| NCI-H1993 cells | Lymph node | Homo sapiens (Human) | CVCL_1512 | |
| NCI-H1703 cells | Lung | Homo sapiens (Human) | CVCL_1490 | |
| NCI-H1299 cells | Lymph node | Homo sapiens (Human) | CVCL_0060 | |
| NCI- H460 cells | Pleural effusion | Homo sapiens (Human) | CVCL_0459 | |
| ACC-MESO-4 cells | Pleural epithelium | Homo sapiens (Human) | CVCL_5114 | |
| ACC-MESO-1 cells | Pleural epithelium | Homo sapiens (Human) | CVCL_5113 | |
| Experiment for Drug Resistance |
CellTiter-Glo assay; Caspase-Glo 3/7 assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [25] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | Miransertib | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | DU-145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 | |||||||||
| LNCaP cells | Prostate | Homo sapiens (Human) | CVCL_0395 | ||||||||||
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | ||||||||||
| Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 | ||||||||||
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | ||||||||||
| NCI-H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | ||||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | ||||||||||
| HCC70 cells | Breast | Homo sapiens (Human) | CVCL_1270 | ||||||||||
| A2058 cells | Skin | Homo sapiens (Human) | CVCL_1059 | ||||||||||
| Caco2 cells | Colon | Homo sapiens (Human) | CVCL_0025 | ||||||||||
| MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 | ||||||||||
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | ||||||||||
| NCI-60 cells | N.A. | Homo sapiens (Human) | N.A. | ||||||||||
| KU-19 cells | Blood | Bos taurus (Bovine) | CVCL_VN09 | ||||||||||
| EVSA-T cells | Ascites | Homo sapiens (Human) | CVCL_1207 | ||||||||||
| CAL-120 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1104 | ||||||||||
| BT-549 cells | Breast | Homo sapiens (Human) | CVCL_1092 | ||||||||||
| BT-474 cells | Breast | Homo sapiens (Human) | CVCL_0179 | ||||||||||
| BT-20 cells | Mammary gland | Homo sapiens (Human) | CVCL_0178 | ||||||||||
| B16F10 cells | Skin | Mus musculus (Mouse) | CVCL_0159 | ||||||||||
| In Vivo Model | Female NMRI (nu/nu) mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Mechanism Description | The missense mutation p.H1047R (c.3140A>G) in gene PIK3CA cause the sensitivity of Miransertib by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [25] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | Miransertib | ||||||||||||
| Molecule Alteration | Missense mutation | p.E542K (c.1624G>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | DU-145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 | |||||||||
| LNCaP cells | Prostate | Homo sapiens (Human) | CVCL_0395 | ||||||||||
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | ||||||||||
| Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 | ||||||||||
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | ||||||||||
| NCI-H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | ||||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | ||||||||||
| HCC70 cells | Breast | Homo sapiens (Human) | CVCL_1270 | ||||||||||
| A2058 cells | Skin | Homo sapiens (Human) | CVCL_1059 | ||||||||||
| Caco2 cells | Colon | Homo sapiens (Human) | CVCL_0025 | ||||||||||
| MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 | ||||||||||
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | ||||||||||
| NCI-60 cells | N.A. | Homo sapiens (Human) | N.A. | ||||||||||
| KU-19 cells | Blood | Bos taurus (Bovine) | CVCL_VN09 | ||||||||||
| EVSA-T cells | Ascites | Homo sapiens (Human) | CVCL_1207 | ||||||||||
| CAL-120 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1104 | ||||||||||
| BT-549 cells | Breast | Homo sapiens (Human) | CVCL_1092 | ||||||||||
| BT-474 cells | Breast | Homo sapiens (Human) | CVCL_0179 | ||||||||||
| BT-20 cells | Mammary gland | Homo sapiens (Human) | CVCL_0178 | ||||||||||
| B16F10 cells | Skin | Mus musculus (Mouse) | CVCL_0159 | ||||||||||
| In Vivo Model | Female NMRI (nu/nu) mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Mechanism Description | The missense mutation p.E542K (c.1624G>A) in gene PIK3CA cause the sensitivity of Miransertib by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [25] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | Miransertib | ||||||||||||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | DU-145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 | |||||||||
| LNCaP cells | Prostate | Homo sapiens (Human) | CVCL_0395 | ||||||||||
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | ||||||||||
| Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 | ||||||||||
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | ||||||||||
| NCI-H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | ||||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | ||||||||||
| HCC70 cells | Breast | Homo sapiens (Human) | CVCL_1270 | ||||||||||
| A2058 cells | Skin | Homo sapiens (Human) | CVCL_1059 | ||||||||||
| Caco2 cells | Colon | Homo sapiens (Human) | CVCL_0025 | ||||||||||
| MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 | ||||||||||
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | ||||||||||
| NCI-60 cells | N.A. | Homo sapiens (Human) | N.A. | ||||||||||
| KU-19 cells | Blood | Bos taurus (Bovine) | CVCL_VN09 | ||||||||||
| EVSA-T cells | Ascites | Homo sapiens (Human) | CVCL_1207 | ||||||||||
| CAL-120 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1104 | ||||||||||
| BT-549 cells | Breast | Homo sapiens (Human) | CVCL_1092 | ||||||||||
| BT-474 cells | Breast | Homo sapiens (Human) | CVCL_0179 | ||||||||||
| BT-20 cells | Mammary gland | Homo sapiens (Human) | CVCL_0178 | ||||||||||
| B16F10 cells | Skin | Mus musculus (Mouse) | CVCL_0159 | ||||||||||
| In Vivo Model | Female NMRI (nu/nu) mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Mechanism Description | The missense mutation p.E545K (c.1633G>A) in gene PIK3CA cause the sensitivity of Miransertib by aberration of the drug's therapeutic target | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [26] | |||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Sensitive Drug | MK-2206 | |||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Immunoblotting analysis | |||
| Experiment for Drug Resistance |
Soft-agar colony formation assay | |||
| Mechanism Description | The missense mutation p.E545K (c.1633G>A) in gene PIK3CA cause the sensitivity of MK-2206 by aberration of the drug's therapeutic target | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [27] | ||||||||||||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | ||||||||||||
| Sensitive Drug | PF-04691502 | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | 293-MSR cells | Fetal kidney | Homo sapiens (Human) | CVCL_KS18 | |||||||||
| In Vivo Model | Female nu/nu mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Caspase-Glo 3/7 assay | ||||||||||||
| Mechanism Description | The missense mutation p.H1047R (c.3140A>G) in gene PIK3CA cause the sensitivity of PF-04691502 by aberration of the drug's therapeutic target | ||||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Bladder cancer [ICD-11: 2C94.0] | [28] | |||
| Resistant Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Resistant Drug | Pictilisib | |||
| Molecule Alteration | Missense mutation | p.D549Y (c.1645G>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | MEK/ERK signaling pathway | Activation | hsa04011 | |
| In Vitro Model | 5637 cells | Bladder | Homo sapiens (Human) | CVCL_0126 |
| J82 cells | Bladder | Homo sapiens (Human) | CVCL_0359 | |
| RT4 cells | Bladder | Homo sapiens (Human) | CVCL_0036 | |
| T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |
| TCCSuP cells | Bladder | Homo sapiens (Human) | CVCL_1738 | |
| In Vivo Model | NSG mouse PDX model | Mus musculus | ||
| Experiment for Drug Resistance |
MTS assay; FACS assay | |||
| Mechanism Description | Pictilisib activated the compensatory MEK/ERK pathways that likely contributed to pictilisib resistance, which was reversed by co-treatment with the RAF inhibitor sorafenib. RNA-sequencing of tumors resistant to treatment suggested that LSP1 down-regulation correlated with drug resistance. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [26] | |||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Sensitive Drug | Pictilisib | |||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| In Vivo Model | Female NCR nu/nu mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
PCR DNA sequencing assay | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay; IC50 assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [29] | |||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Sensitive Drug | Voxtalisib | |||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| U87-MG cells | Brain | Homo sapiens (Human) | CVCL_0022 | |
| LS174T cells | Colon | Homo sapiens (Human) | CVCL_1384 | |
| MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 | |
| Ramos cells | Ascites | Homo sapiens (Human) | CVCL_0597 | |
| OVCAR-3 cells | Ascites | Homo sapiens (Human) | CVCL_0465 | |
| In Vivo Model | Athymic nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
ViaLight HS assay; Apo-ONE Homogeneous Caspase-3/7 assay; Soft agar assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [30] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | CH-5132799 | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | KPL-4 cells | Breast | Homo sapiens (Human) | CVCL_5310 | |||||||||
| IGROV1 cells | Ovary | Homo sapiens (Human) | CVCL_1304 | ||||||||||
| GXF97 cells | N.A. | N.A. | N.A. | ||||||||||
| In Vivo Model | Female BALB-nu/nu mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
CCK-8 assay | ||||||||||||
| Mechanism Description | The missense mutation p.H1047R (c.3140A>G) in gene PIK3CA cause the sensitivity of CH-5132799 by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [31] | ||||||||||||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | ||||||||||||
| Sensitive Drug | CH-5132799 | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
ARRAY system assay | ||||||||||||
| Experiment for Drug Resistance |
Presto blue assay | ||||||||||||
| Disease Class: Prostate cancer [ICD-11: 2C82.0] | [30] | ||||||||||||
| Sensitive Disease | Prostate cancer [ICD-11: 2C82.0] | ||||||||||||
| Sensitive Drug | CH-5132799 | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q546R (c.1637A>G) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | KPL-4 cells | Breast | Homo sapiens (Human) | CVCL_5310 | |||||||||
| IGROV1 cells | Ovary | Homo sapiens (Human) | CVCL_1304 | ||||||||||
| GXF97 cells | N.A. | N.A. | N.A. | ||||||||||
| In Vivo Model | Female BALB-nu/nu mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
CCK-8 assay | ||||||||||||
| Mechanism Description | The missense mutation p.Q546R (c.1637A>G) in gene PIK3CA cause the sensitivity of CH-5132799 by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [30] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | CH-5132799 | ||||||||||||
| Molecule Alteration | Missense mutation | p.E542K (c.1624G>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | KPL-4 cells | Breast | Homo sapiens (Human) | CVCL_5310 | |||||||||
| IGROV1 cells | Ovary | Homo sapiens (Human) | CVCL_1304 | ||||||||||
| GXF97 cells | N.A. | N.A. | N.A. | ||||||||||
| In Vivo Model | Female BALB-nu/nu mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
CCK-8 assay | ||||||||||||
| Mechanism Description | The missense mutation p.E542K (c.1624G>A) in gene PIK3CA cause the sensitivity of CH-5132799 by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Endometrial adenocarcinoma [ICD-11: 2C76.0] | [30] | ||||||||||||
| Sensitive Disease | Endometrial adenocarcinoma [ICD-11: 2C76.0] | ||||||||||||
| Sensitive Drug | CH-5132799 | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047Y (c.3139C>T) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | KPL-4 cells | Breast | Homo sapiens (Human) | CVCL_5310 | |||||||||
| IGROV1 cells | Ovary | Homo sapiens (Human) | CVCL_1304 | ||||||||||
| GXF97 cells | N.A. | N.A. | N.A. | ||||||||||
| In Vivo Model | Female BALB-nu/nu mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
CCK-8 assay | ||||||||||||
| Mechanism Description | The missense mutation p.H1047Y (c.3139C>T) in gene PIK3CA cause the sensitivity of CH-5132799 by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [30] | ||||||||||||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | ||||||||||||
| Sensitive Drug | CH-5132799 | ||||||||||||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | KPL-4 cells | Breast | Homo sapiens (Human) | CVCL_5310 | |||||||||
| IGROV1 cells | Ovary | Homo sapiens (Human) | CVCL_1304 | ||||||||||
| GXF97 cells | N.A. | N.A. | N.A. | ||||||||||
| In Vivo Model | Female BALB-nu/nu mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
CCK-8 assay | ||||||||||||
| Mechanism Description | The missense mutation p.E545K (c.1633G>A) in gene PIK3CA cause the sensitivity of CH-5132799 by aberration of the drug's therapeutic target | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [32] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | BAY1125976 | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | LNCaP cells | Prostate | Homo sapiens (Human) | CVCL_0395 | |||||||||
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | ||||||||||
| Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 | ||||||||||
| NCI-H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | ||||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | ||||||||||
| HCC70 cells | Breast | Homo sapiens (Human) | CVCL_1270 | ||||||||||
| A2058 cells | Skin | Homo sapiens (Human) | CVCL_1059 | ||||||||||
| Caco2 cells | Colon | Homo sapiens (Human) | CVCL_0025 | ||||||||||
| MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 | ||||||||||
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | ||||||||||
| DU-145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 | ||||||||||
| NCI-60 cells | N.A. | Homo sapiens (Human) | N.A. | ||||||||||
| EVSA-T cells | Ascites | Homo sapiens (Human) | CVCL_1207 | ||||||||||
| KU-19 cells | Blood | Bos taurus (Bovine) | CVCL_VN09 | ||||||||||
| T47D cells | Pleural effusion | Homo sapiens (Human) | CVCL_0553 | ||||||||||
| CAL-120 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1104 | ||||||||||
| BT-549 cells | Breast | Homo sapiens (Human) | CVCL_1092 | ||||||||||
| BT-474 cells | Breast | Homo sapiens (Human) | CVCL_0179 | ||||||||||
| BT-20 cells | Mammary gland | Homo sapiens (Human) | CVCL_0178 | ||||||||||
| B16F10 cells | Skin | Mus musculus (Mouse) | CVCL_0159 | ||||||||||
| In Vivo Model | Female NMRI (nu/nu) mouse xenograft model | Mus musculus | |||||||||||
| Mechanism Description | The missense mutation p.H1047R (c.3140A>G) in gene PIK3CA cause the sensitivity of BAY1125976 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [32] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | BAY1125976 | ||||||||||||
| Molecule Alteration | Missense mutation | p.K111N (c.333G>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | LNCaP cells | Prostate | Homo sapiens (Human) | CVCL_0395 | |||||||||
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | ||||||||||
| Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 | ||||||||||
| NCI-H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | ||||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | ||||||||||
| HCC70 cells | Breast | Homo sapiens (Human) | CVCL_1270 | ||||||||||
| A2058 cells | Skin | Homo sapiens (Human) | CVCL_1059 | ||||||||||
| Caco2 cells | Colon | Homo sapiens (Human) | CVCL_0025 | ||||||||||
| MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 | ||||||||||
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | ||||||||||
| DU-145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 | ||||||||||
| NCI-60 cells | N.A. | Homo sapiens (Human) | N.A. | ||||||||||
| EVSA-T cells | Ascites | Homo sapiens (Human) | CVCL_1207 | ||||||||||
| KU-19 cells | Blood | Bos taurus (Bovine) | CVCL_VN09 | ||||||||||
| T47D cells | Pleural effusion | Homo sapiens (Human) | CVCL_0553 | ||||||||||
| CAL-120 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1104 | ||||||||||
| BT-549 cells | Breast | Homo sapiens (Human) | CVCL_1092 | ||||||||||
| BT-474 cells | Breast | Homo sapiens (Human) | CVCL_0179 | ||||||||||
| BT-20 cells | Mammary gland | Homo sapiens (Human) | CVCL_0178 | ||||||||||
| B16F10 cells | Skin | Mus musculus (Mouse) | CVCL_0159 | ||||||||||
| In Vivo Model | Female NMRI (nu/nu) mouse xenograft model | Mus musculus | |||||||||||
| Mechanism Description | The missense mutation p.K111N (c.333G>C) in gene PIK3CA cause the sensitivity of BAY1125976 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [32] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | BAY1125976 | ||||||||||||
| Molecule Alteration | Missense mutation | p.P539R (c.1616C>G) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | LNCaP cells | Prostate | Homo sapiens (Human) | CVCL_0395 | |||||||||
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | ||||||||||
| Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 | ||||||||||
| NCI-H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | ||||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | ||||||||||
| HCC70 cells | Breast | Homo sapiens (Human) | CVCL_1270 | ||||||||||
| A2058 cells | Skin | Homo sapiens (Human) | CVCL_1059 | ||||||||||
| Caco2 cells | Colon | Homo sapiens (Human) | CVCL_0025 | ||||||||||
| MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 | ||||||||||
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | ||||||||||
| DU-145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 | ||||||||||
| NCI-60 cells | N.A. | Homo sapiens (Human) | N.A. | ||||||||||
| EVSA-T cells | Ascites | Homo sapiens (Human) | CVCL_1207 | ||||||||||
| KU-19 cells | Blood | Bos taurus (Bovine) | CVCL_VN09 | ||||||||||
| T47D cells | Pleural effusion | Homo sapiens (Human) | CVCL_0553 | ||||||||||
| CAL-120 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1104 | ||||||||||
| BT-549 cells | Breast | Homo sapiens (Human) | CVCL_1092 | ||||||||||
| BT-474 cells | Breast | Homo sapiens (Human) | CVCL_0179 | ||||||||||
| BT-20 cells | Mammary gland | Homo sapiens (Human) | CVCL_0178 | ||||||||||
| B16F10 cells | Skin | Mus musculus (Mouse) | CVCL_0159 | ||||||||||
| In Vivo Model | Female NMRI (nu/nu) mouse xenograft model | Mus musculus | |||||||||||
| Mechanism Description | The missense mutation p.P539R (c.1616C>G) in gene PIK3CA cause the sensitivity of BAY1125976 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [32] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | BAY1125976 | ||||||||||||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | LNCaP cells | Prostate | Homo sapiens (Human) | CVCL_0395 | |||||||||
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | ||||||||||
| Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 | ||||||||||
| NCI-H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | ||||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | ||||||||||
| HCC70 cells | Breast | Homo sapiens (Human) | CVCL_1270 | ||||||||||
| A2058 cells | Skin | Homo sapiens (Human) | CVCL_1059 | ||||||||||
| Caco2 cells | Colon | Homo sapiens (Human) | CVCL_0025 | ||||||||||
| MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 | ||||||||||
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | ||||||||||
| DU-145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 | ||||||||||
| NCI-60 cells | N.A. | Homo sapiens (Human) | N.A. | ||||||||||
| EVSA-T cells | Ascites | Homo sapiens (Human) | CVCL_1207 | ||||||||||
| KU-19 cells | Blood | Bos taurus (Bovine) | CVCL_VN09 | ||||||||||
| T47D cells | Pleural effusion | Homo sapiens (Human) | CVCL_0553 | ||||||||||
| CAL-120 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1104 | ||||||||||
| BT-549 cells | Breast | Homo sapiens (Human) | CVCL_1092 | ||||||||||
| BT-474 cells | Breast | Homo sapiens (Human) | CVCL_0179 | ||||||||||
| BT-20 cells | Mammary gland | Homo sapiens (Human) | CVCL_0178 | ||||||||||
| B16F10 cells | Skin | Mus musculus (Mouse) | CVCL_0159 | ||||||||||
| In Vivo Model | Female NMRI (nu/nu) mouse xenograft model | Mus musculus | |||||||||||
| Mechanism Description | The missense mutation p.E545K (c.1633G>A) in gene PIK3CA cause the sensitivity of BAY1125976 by unusual activation of pro-survival pathway | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Oesophagus adenocarcinoma [ICD-11: 2B70.0] | [33] | |||
| Sensitive Disease | Oesophagus adenocarcinoma [ICD-11: 2B70.0] | |||
| Sensitive Drug | LY-294002 | |||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | TE cells | Eye | Oreochromis mossambicus (Mozambique tilapia) | CVCL_YD05 |
| KYSE cells | N.A. | N.A. | N.A. | |
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The missense mutation p.E545K (c.1633G>A) in gene PIK3CA cause the sensitivity of LY-294002 by aberration of the drug's therapeutic target | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colon cancer [ICD-11: 2B90.1] | [34] | |||
| Sensitive Disease | Colon cancer [ICD-11: 2B90.1] | |||
| Sensitive Drug | LY2780301 | |||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | . | N.A. | ||
| In Vivo Model | Female athymic nude-Foxn1 nu mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Pharmacokinetic studies; Pharmacokinetic studies; Antitumor activity | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [35] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | M2698 | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | PI3K signaling pathway | Inhibition | hsa04151 | ||||||||||
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 | |||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | ||||||||||
| JIMT-1 cells | Breast | Homo sapiens (Human) | CVCL_2077 | ||||||||||
| HCC1419 cells | Breast | Homo sapiens (Human) | CVCL_1251 | ||||||||||
| In Vivo Model | Nude mouse PDX model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
SRB staining assay | ||||||||||||
| Mechanism Description | M2698 blocked p70S6K to provide potent PAM pathway inhibition while simultaneously targeting Akt to overcome the compensatory feedback loop. | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Thyroid gland cancer [ICD-11: 2D10.0] | [36] | ||||||||||||
| Sensitive Disease | Thyroid gland cancer [ICD-11: 2D10.0] | ||||||||||||
| Sensitive Drug | Perifosine | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | SW1736 cells | Thyroid | Homo sapiens (Human) | CVCL_3883 | |||||||||
| C643 cells | Thyroid gland | Homo sapiens (Human) | CVCL_5969 | ||||||||||
| HTH7 cells | Thyroid gland | Homo sapiens (Human) | CVCL_6289 | ||||||||||
| SW1736 cells | Thyroid | Homo sapiens (Human) | CVCL_3883 | ||||||||||
| HTH7 cells | Thyroid gland | Homo sapiens (Human) | CVCL_6289 | ||||||||||
| Hth74 cells | Thyroid gland | Homo sapiens (Human) | CVCL_6288 | ||||||||||
| Hth74 cells | Thyroid gland | Homo sapiens (Human) | CVCL_6288 | ||||||||||
| In Vivo Model | Nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Mechanism Description | The missense mutation p.H1047R (c.3140A>G) in gene PIK3CA cause the sensitivity of Perifosine by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Thyroid gland cancer [ICD-11: 2D10.0] | [36] | ||||||||||||
| Sensitive Disease | Thyroid gland cancer [ICD-11: 2D10.0] | ||||||||||||
| Sensitive Drug | Perifosine | ||||||||||||
| Molecule Alteration | Missense mutation | p.E542K (c.1624G>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | SW1736 cells | Thyroid | Homo sapiens (Human) | CVCL_3883 | |||||||||
| C643 cells | Thyroid gland | Homo sapiens (Human) | CVCL_5969 | ||||||||||
| HTH7 cells | Thyroid gland | Homo sapiens (Human) | CVCL_6289 | ||||||||||
| SW1736 cells | Thyroid | Homo sapiens (Human) | CVCL_3883 | ||||||||||
| HTH7 cells | Thyroid gland | Homo sapiens (Human) | CVCL_6289 | ||||||||||
| Hth74 cells | Thyroid gland | Homo sapiens (Human) | CVCL_6288 | ||||||||||
| Hth74 cells | Thyroid gland | Homo sapiens (Human) | CVCL_6288 | ||||||||||
| In Vivo Model | Nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Mechanism Description | The missense mutation p.E542K (c.1624G>A) in gene PIK3CA cause the sensitivity of Perifosine by unusual activation of pro-survival pathway | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colon cancer [ICD-11: 2B90.1] | [37] | |||
| Sensitive Disease | Colon cancer [ICD-11: 2B90.1] | |||
| Sensitive Drug | VS-5584 | |||
| Molecule Alteration | Missense mutation | p.E542K (c.1624G>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SNU869 cells | Biliary tract | Homo sapiens (Human) | CVCL_5101 |
| SNU478 cells | Biliary tract | Homo sapiens (Human) | CVCL_5065 | |
| SNU308 cells | Biliary tract | Homo sapiens (Human) | CVCL_5048 | |
| SNU245 cells | Biliary tract | Homo sapiens (Human) | CVCL_5038 | |
| SNU1196 cells | Biliary tract | Homo sapiens (Human) | CVCL_5015 | |
| SNU1079 cells | Biliary tract | Homo sapiens (Human) | CVCL_5008 | |
| SET-2 cells | Peripheral blood | Homo sapiens (Human) | CVCL_2187 | |
| In Vivo Model | Athymic BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay | |||
| Mechanism Description | The missense mutation p.E542K (c.1624G>A) in gene PIK3CA cause the sensitivity of VS-5584 by aberration of the drug's therapeutic target | |||
Approved Drug(s)
10 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Tongue squamous cell carcinoma [ICD-11: 2B62.1] | [3] | |||
| Sensitive Disease | Tongue squamous cell carcinoma [ICD-11: 2B62.1] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| PIk3CA signaling pathway | Regulation | N.A. | ||
| In Vitro Model | Tca8113 cells | Tongue | Homo sapiens (Human) | CVCL_6851 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-203 expression is much lower in the clinical tongue cancer samples. In the surviving Tca8113 cells after cisplatin treatment, miR-203 expression was much lower. Delivery of miR-203 sensitized the Tca8113 cells to cisplatin induced cell death through downregulation of PIk3CA. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [4] | |||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Resistant Drug | Erlotinib | |||
| Molecule Alteration | Mutation | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Experiment for Molecule Alteration |
Low throughput experiment assay | |||
| Experiment for Drug Resistance |
Progression-free survival assay | |||
| Mechanism Description | Acquired resistance can occur through failure of drug delivery to the target, as in isolated central nervous system (CNS) progression, or by selection of biological variants during TkI exposure. | |||
| Disease Class: EGFR-mutant non-small cell lung cancer [ICD-11: 2C25.7] | [5] | |||
| Resistant Disease | EGFR-mutant non-small cell lung cancer [ICD-11: 2C25.7] | |||
| Resistant Drug | Erlotinib | |||
| Molecule Alteration | Mutation | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Experiment for Molecule Alteration |
Next generation sequencing assay | |||
| Experiment for Drug Resistance |
Multivariate analysis of overall or disease-free survival assay | |||
| Mechanism Description | Quantification of allele fractions in plasma identified increased representation of mutantalleles in association with emergence of therapy resistance. These included an activating mutation in PIk3CA. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: EGFR-mutant non-small cell lung cancer [ICD-11: 2C25.7] | [6] | |||
| Resistant Disease | EGFR-mutant non-small cell lung cancer [ICD-11: 2C25.7] | |||
| Resistant Drug | Gefitinib | |||
| Molecule Alteration | Mutation | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | ||
| Experiment for Molecule Alteration |
MGB SNP detection kit assay; Mutation Detection assay | |||
| Experiment for Drug Resistance |
Digital PCR assay | |||
| Mechanism Description | Resistance mechanisms to EGFR-TkI therapy in EGFR-mutated NSCLC include secondary EGFR T790M mutation, c-Met amplification, PIk3CA mutation, and transformation to small-cell lung cancer. | |||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [4] | |||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Resistant Drug | Gefitinib | |||
| Molecule Alteration | Mutation | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Experiment for Molecule Alteration |
Low throughput experiment assay | |||
| Experiment for Drug Resistance |
Progression-free survival assay | |||
| Mechanism Description | Acquired resistance can occur through failure of drug delivery to the target, as in isolated central nervous system (CNS) progression, or by selection of biological variants during TkI exposure. | |||
| Disease Class: EGFR-mutant non-small cell lung cancer [ICD-11: 2C25.7] | [5] | |||
| Resistant Disease | EGFR-mutant non-small cell lung cancer [ICD-11: 2C25.7] | |||
| Resistant Drug | Gefitinib | |||
| Molecule Alteration | Mutation | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Experiment for Molecule Alteration |
Next generation sequencing assay | |||
| Experiment for Drug Resistance |
Multivariate analysis of overall or disease-free survival assay | |||
| Mechanism Description | Quantification of allele fractions in plasma identified increased representation of mutantalleles in association with emergence of therapy resistance. These included an activating mutation in PIk3CA. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Esophageal squamous cell carcinoma [ICD-11: 2B70.3] | [7] | |||
| Sensitive Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.3] | |||
| Sensitive Drug | Gefitinib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| PI3K/AKT/survivin signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | TE-1 cells | Esophagus | Homo sapiens (Human) | CVCL_1759 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | Exogenous expression of miR 1 inhibited growth, arrested cell cycle in the G1 phase and increased apoptosis in ESCC cells, whereas it decreased PIk3CA protein expression levels. Furthermore, overexpression of miR 1 increased the sensitivity of ESCC cells to the anticancer drug, gefitinib. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Merkel cell carcinoma [ICD-11: 2C34.0] | [8] | |||
| Sensitive Disease | Merkel cell carcinoma [ICD-11: 2C34.0] | |||
| Sensitive Drug | Idelalisib | |||
| Molecule Alteration | Missense mutation | p.P471L (c.1412C>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Skin | N.A. | ||
| Experiment for Molecule Alteration |
Real-time PCR | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [9] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Lapatinib | |||
| Molecule Alteration | Mutation | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | HER2-amplified breast cancer cells | Breast | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Multi-region sequencing assay; Single-cell sequencing assay | |||
| Mechanism Description | Similarly, PTEN loss or PIk3CA mutation was found to lower the clinical benefit of lapatinib in HER2-amplified metastatic breast cancer and to be responsible for lapatinib resistance in breast cancer cell lines. Tumor-promoting mutations seem to be involved in three major biological processes: cell survival, sensitive to mutations in EGFR, HER2, PIk3CA, BRAF, PTEN, MYC and others; cell fate, influenced by mutations in APC, NOTCH, AR, GATA2, kLF4 and genomic stability, altered by mutations in TP53, ATM, BRCA1, BRCA2 and others. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Esophageal squamous cell carcinoma [ICD-11: 2B70.3] | [10] | |||
| Resistant Disease | Esophageal squamous cell carcinoma [ICD-11: 2B70.3] | |||
| Resistant Drug | Nimotuzumab | |||
| Molecule Alteration | Mutation | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | PI3K/AKT/mTOR signaling pathway | Activation | hsa04151 | |
| Mechanism Description | NGS examination of this patient demonstrated that PIK3CA mutation and a RICTOR amplification might participate in primary and acquired resistance to nimotuzumab, respectively, via the PI3K/AKT/mTOR signaling pathway. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [11] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Paclitaxel | |||
| Molecule Alteration | Missense mutation | p.E545K |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Experiment for Molecule Alteration |
Circulating-free DNA assay; Whole exome sequencing assay | |||
| Mechanism Description | Quantification of allele fractions in plasma identified increased representation of mutant alleles in association with emergence of therapy resistance. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [12] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.N345K (c.1035T>G) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| In Vivo Model | Female nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Mechanism Description | The missense mutation p.N345K (c.1035T>G) in gene PIK3CA cause the sensitivity of Trametinib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [12] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.E542K (c.1624G>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| In Vivo Model | Female nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Mechanism Description | The missense mutation p.E542K (c.1624G>A) in gene PIK3CA cause the sensitivity of Trametinib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [12] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| In Vivo Model | Female nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Mechanism Description | The missense mutation p.E545K (c.1633G>A) in gene PIK3CA cause the sensitivity of Trametinib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [12] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q546K (c.1636C>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| In Vivo Model | Female nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Mechanism Description | The missense mutation p.Q546K (c.1636C>A) in gene PIK3CA cause the sensitivity of Trametinib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [12] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| In Vivo Model | Female nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Mechanism Description | The missense mutation p.H1047R (c.3140A>G) in gene PIK3CA cause the sensitivity of Trametinib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [12] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| In Vivo Model | Female nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Mechanism Description | The missense mutation p.H1047R (c.3140A>G) in gene PIK3CA cause the sensitivity of Trametinib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [12] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047L (c.3140A>T) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.77 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.96 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
A
A
H
L
H
H
G
G
1050
|
G
G
-
W
T
T
T
T
K
K
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| In Vivo Model | Female nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Mechanism Description | The missense mutation p.H1047L (c.3140A>T) in gene PIK3CA cause the sensitivity of Trametinib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [12] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.G1049R (c.3145G>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| In Vivo Model | Female nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Mechanism Description | The missense mutation p.G1049R (c.3145G>C) in gene PIK3CA cause the sensitivity of Trametinib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [12] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.N345K (c.1035T>G) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| In Vivo Model | Female nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Mechanism Description | The missense mutation p.N345K (c.1035T>G) in gene PIK3CA cause the sensitivity of Trametinib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [12] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| In Vivo Model | Female nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Mechanism Description | The missense mutation p.E545K (c.1633G>A) in gene PIK3CA cause the sensitivity of Trametinib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [12] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q546K (c.1636C>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| In Vivo Model | Female nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Mechanism Description | The missense mutation p.Q546K (c.1636C>A) in gene PIK3CA cause the sensitivity of Trametinib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [12] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.G1049R (c.3145G>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| In Vivo Model | Female nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Mechanism Description | The missense mutation p.G1049R (c.3145G>C) in gene PIK3CA cause the sensitivity of Trametinib by unusual activation of pro-survival pathway | ||||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [13] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Trastuzumab | |||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vivo Model | Female athymic nude xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Immunoblotting analysis | |||
| Experiment for Drug Resistance |
WST-1 assay | |||
| Mechanism Description | The missense mutation p.E545K (c.1633G>A) in gene PIK3CA cause the resistance of Trastuzumab by unusual activation of pro-survival pathway | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [14] | |||
| Resistant Disease | Melanoma [ICD-11: 2C30.0] | |||
| Resistant Drug | Vemurafenib | |||
| Molecule Alteration | Missense mutation | p.E545K |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Experiment for Molecule Alteration |
Next-generation sequencing assay | |||
| Experiment for Drug Resistance |
Computerized tomography assay | |||
| Mechanism Description | In patient #11, sequential biopsies showed three mutations that were not detected in the pretreatment biopsy, including an activating mutation in PIk3CA E545k readily explaining the resistance. | |||
Discontinued Drug(s)
2 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [38], [39] | |||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Resistant Drug | Rociletinib | |||
| Molecule Alteration | Missense mutation | p.E545K |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Experiment for Molecule Alteration |
Circulating tumour DNA analysis; Next-generation sequencing assay | |||
| Experiment for Drug Resistance |
Progression-free survival assay | |||
| Mechanism Description | Similarly,resistance to the third-generation inhibitor rociletinib may not only be mediated by EGFR (L798I, C797S) mutations, but also by alterations of MET, PIk3CA, ERRB2, and kRAS, and by the negative selection of T790M-mutant subclones. | |||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [38], [39] | |||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Resistant Drug | Rociletinib | |||
| Molecule Alteration | Missense mutation | p.E542K |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Experiment for Molecule Alteration |
Circulating tumour DNA analysis; Next-generation sequencing assay | |||
| Experiment for Drug Resistance |
Progression-free survival assay | |||
| Mechanism Description | Similarly,resistance to the third-generation inhibitor rociletinib may not only be mediated by EGFR (L798I, C797S) mutations, but also by alterations of MET, PIk3CA, ERRB2, and kRAS, and by the negative selection of T790M-mutant subclones. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: EGFR-mutant non-small cell lung cancer [ICD-11: 2C25.7] | [6] | |||
| Resistant Disease | EGFR-mutant non-small cell lung cancer [ICD-11: 2C25.7] | |||
| Resistant Drug | S-1 | |||
| Molecule Alteration | Mutation | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | ||
| Experiment for Molecule Alteration |
MGB SNP detection kit assay; Mutation Detection assay | |||
| Experiment for Drug Resistance |
Digital PCR assay | |||
| Mechanism Description | Resistance mechanisms to EGFR-TkI therapy in EGFR-mutated NSCLC include secondary EGFR T790M mutation, c-Met amplification, PIk3CA mutation, and transformation to small-cell lung cancer. | |||
Preclinical Drug(s)
20 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [40] | |||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Resistant Drug | A66 | |||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Skin | N.A. | ||
| In Vivo Model | Mouse xenograft model | Mus musculus | ||
| Mechanism Description | The missense mutation p.E545K (c.1633G>A) in gene PIK3CA cause the resistance of A66 by aberration of the drug's therapeutic target | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [40] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | A66 | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | Skin | N.A. | |||||||||||
| In Vivo Model | Mouse xenograft model | Mus musculus | |||||||||||
| Mechanism Description | The missense mutation p.H1047R (c.3140A>G) in gene PIK3CA cause the sensitivity of A66 by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [40] | ||||||||||||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | ||||||||||||
| Sensitive Drug | A66 | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | Skin | N.A. | |||||||||||
| In Vivo Model | Mouse xenograft model | Mus musculus | |||||||||||
| Mechanism Description | The missense mutation p.H1047R (c.3140A>G) in gene PIK3CA cause the sensitivity of A66 by aberration of the drug's therapeutic target | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [25] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | ARQ 751 | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | DU-145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 | |||||||||
| LNCaP cells | Prostate | Homo sapiens (Human) | CVCL_0395 | ||||||||||
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | ||||||||||
| Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 | ||||||||||
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | ||||||||||
| NCI-H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | ||||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | ||||||||||
| HCC70 cells | Breast | Homo sapiens (Human) | CVCL_1270 | ||||||||||
| A2058 cells | Skin | Homo sapiens (Human) | CVCL_1059 | ||||||||||
| Caco2 cells | Colon | Homo sapiens (Human) | CVCL_0025 | ||||||||||
| MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 | ||||||||||
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | ||||||||||
| NCI-60 cells | N.A. | Homo sapiens (Human) | N.A. | ||||||||||
| KU-19 cells | Blood | Bos taurus (Bovine) | CVCL_VN09 | ||||||||||
| EVSA-T cells | Ascites | Homo sapiens (Human) | CVCL_1207 | ||||||||||
| CAL-120 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1104 | ||||||||||
| BT-549 cells | Breast | Homo sapiens (Human) | CVCL_1092 | ||||||||||
| BT-474 cells | Breast | Homo sapiens (Human) | CVCL_0179 | ||||||||||
| BT-20 cells | Mammary gland | Homo sapiens (Human) | CVCL_0178 | ||||||||||
| B16F10 cells | Skin | Mus musculus (Mouse) | CVCL_0159 | ||||||||||
| In Vivo Model | Female NMRI (nu/nu) mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Mechanism Description | The missense mutation p.H1047R (c.3140A>G) in gene PIK3CA cause the sensitivity of ARQ 751 by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [25] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | ARQ 751 | ||||||||||||
| Molecule Alteration | Missense mutation | p.K111N (c.333G>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | DU-145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 | |||||||||
| LNCaP cells | Prostate | Homo sapiens (Human) | CVCL_0395 | ||||||||||
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | ||||||||||
| Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 | ||||||||||
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | ||||||||||
| NCI-H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | ||||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | ||||||||||
| HCC70 cells | Breast | Homo sapiens (Human) | CVCL_1270 | ||||||||||
| A2058 cells | Skin | Homo sapiens (Human) | CVCL_1059 | ||||||||||
| Caco2 cells | Colon | Homo sapiens (Human) | CVCL_0025 | ||||||||||
| MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 | ||||||||||
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | ||||||||||
| NCI-60 cells | N.A. | Homo sapiens (Human) | N.A. | ||||||||||
| KU-19 cells | Blood | Bos taurus (Bovine) | CVCL_VN09 | ||||||||||
| EVSA-T cells | Ascites | Homo sapiens (Human) | CVCL_1207 | ||||||||||
| CAL-120 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1104 | ||||||||||
| BT-549 cells | Breast | Homo sapiens (Human) | CVCL_1092 | ||||||||||
| BT-474 cells | Breast | Homo sapiens (Human) | CVCL_0179 | ||||||||||
| BT-20 cells | Mammary gland | Homo sapiens (Human) | CVCL_0178 | ||||||||||
| B16F10 cells | Skin | Mus musculus (Mouse) | CVCL_0159 | ||||||||||
| In Vivo Model | Female NMRI (nu/nu) mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Mechanism Description | The missense mutation p.K111N (c.333G>C) in gene PIK3CA cause the sensitivity of ARQ 751 by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [25] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | ARQ 751 | ||||||||||||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | DU-145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 | |||||||||
| LNCaP cells | Prostate | Homo sapiens (Human) | CVCL_0395 | ||||||||||
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | ||||||||||
| Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 | ||||||||||
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | ||||||||||
| NCI-H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | ||||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | ||||||||||
| HCC70 cells | Breast | Homo sapiens (Human) | CVCL_1270 | ||||||||||
| A2058 cells | Skin | Homo sapiens (Human) | CVCL_1059 | ||||||||||
| Caco2 cells | Colon | Homo sapiens (Human) | CVCL_0025 | ||||||||||
| MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 | ||||||||||
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | ||||||||||
| NCI-60 cells | N.A. | Homo sapiens (Human) | N.A. | ||||||||||
| KU-19 cells | Blood | Bos taurus (Bovine) | CVCL_VN09 | ||||||||||
| EVSA-T cells | Ascites | Homo sapiens (Human) | CVCL_1207 | ||||||||||
| CAL-120 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1104 | ||||||||||
| BT-549 cells | Breast | Homo sapiens (Human) | CVCL_1092 | ||||||||||
| BT-474 cells | Breast | Homo sapiens (Human) | CVCL_0179 | ||||||||||
| BT-20 cells | Mammary gland | Homo sapiens (Human) | CVCL_0178 | ||||||||||
| B16F10 cells | Skin | Mus musculus (Mouse) | CVCL_0159 | ||||||||||
| In Vivo Model | Female NMRI (nu/nu) mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Mechanism Description | The missense mutation p.E545K (c.1633G>A) in gene PIK3CA cause the sensitivity of ARQ 751 by aberration of the drug's therapeutic target | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: ER positive breast cancer [ICD-11: 2C60.6] | [41] | |||
| Sensitive Disease | ER positive breast cancer [ICD-11: 2C60.6] | |||
| Sensitive Drug | AZD-8835 | |||
| Molecule Alteration | Missense mutation | p.K111N (c.333G>C) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K signaling pathway | Inhibition | hsa04151 | |
| In Vivo Model | Female Swiss athymic nude mouse PDX model | Mus musculus | ||
| Disease Class: ER positive breast cancer [ICD-11: 2C60.6] | [41] | |||
| Sensitive Disease | ER positive breast cancer [ICD-11: 2C60.6] | |||
| Sensitive Drug | AZD-8835 | |||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K signaling pathway | Inhibition | hsa04151 | |
| In Vivo Model | Female Swiss athymic nude mouse PDX model | Mus musculus | ||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [42] | ||||||||||||
| Sensitive Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Sensitive Drug | AZD5363/Trastuzumab | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTS assay | ||||||||||||
| Mechanism Description | The missense mutation p.H1047R (c.3140A>G) in gene PIK3CA cause the sensitivity of AZD5363 + Trastuzumab by unusual activation of pro-survival pathway | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Bladder cancer [ICD-11: 2C94.0] | [28] | ||||||||||||
| Sensitive Disease | Bladder cancer [ICD-11: 2C94.0] | ||||||||||||
| Sensitive Drug | Cisplatin/Pictilisib | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | MEK/ERK signaling pathway | Inhibition | hsa04011 | ||||||||||
| In Vitro Model | 5637 cells | Bladder | Homo sapiens (Human) | CVCL_0126 | |||||||||
| J82 cells | Bladder | Homo sapiens (Human) | CVCL_0359 | ||||||||||
| RT4 cells | Bladder | Homo sapiens (Human) | CVCL_0036 | ||||||||||
| T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | ||||||||||
| TCCSuP cells | Bladder | Homo sapiens (Human) | CVCL_1738 | ||||||||||
| In Vivo Model | NSG mouse PDX model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
MTS assay; FACS assay | ||||||||||||
| Mechanism Description | Pictilisib activated the compensatory MEK/ERK pathways that likely contributed to pictilisib resistance, which was reversed by co-treatment with the RAF inhibitor sorafenib. RNA-sequencing of tumors resistant to treatment suggested that LSP1 down-regulation correlated with drug resistance. | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [43] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | CYH33 | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | PI3K signaling pathway | Inhibition | hsa04151 | ||||||||||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 | |||||||||
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | ||||||||||
| AU565 cells | Breast | Homo sapiens (Human) | CVCL_1074 | ||||||||||
| In Vivo Model | MMTV-Cre mouse PDX model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; Immunohistochemistry analysis | ||||||||||||
| Experiment for Drug Resistance |
Sulforhodamine B assay; FACS assay | ||||||||||||
| Mechanism Description | Sensitivity to CYH33 has been further revealed to be associated with induction of G1 phase arrest and simultaneous inhibition of Akt and ERK. Sensitivity of patient-derived xenograft to CYH33 was also positively correlated with decrease in phosphorylated ERK. Taken together, CYH33 is a promising PI3Kalpha inhibitor for breast cancer treatment and decrease in ERK phosphorylation may indicate its efficacy, which provides useful clues for rational design of the ongoing clinical trials. | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [43] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | CYH33 | ||||||||||||
| Molecule Alteration | Missense mutation | p.P539R (c.1616C>G) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | PI3K signaling pathway | Inhibition | hsa04151 | ||||||||||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 | |||||||||
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | ||||||||||
| AU565 cells | Breast | Homo sapiens (Human) | CVCL_1074 | ||||||||||
| In Vivo Model | MMTV-Cre mouse PDX model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; Immunohistochemistry analysis | ||||||||||||
| Experiment for Drug Resistance |
Sulforhodamine B assay; FACS assay | ||||||||||||
| Mechanism Description | Sensitivity to CYH33 has been further revealed to be associated with induction of G1 phase arrest and simultaneous inhibition of Akt and ERK. Sensitivity of patient-derived xenograft to CYH33 was also positively correlated with decrease in phosphorylated ERK. Taken together, CYH33 is a promising PI3Kalpha inhibitor for breast cancer treatment and decrease in ERK phosphorylation may indicate its efficacy, which provides useful clues for rational design of the ongoing clinical trials. | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [43] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | CYH33 | ||||||||||||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | PI3K signaling pathway | Inhibition | hsa04151 | ||||||||||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 | |||||||||
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | ||||||||||
| AU565 cells | Breast | Homo sapiens (Human) | CVCL_1074 | ||||||||||
| In Vivo Model | MMTV-Cre mouse PDX model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; Immunohistochemistry analysis | ||||||||||||
| Experiment for Drug Resistance |
Sulforhodamine B assay; FACS assay | ||||||||||||
| Mechanism Description | Sensitivity to CYH33 has been further revealed to be associated with induction of G1 phase arrest and simultaneous inhibition of Akt and ERK. Sensitivity of patient-derived xenograft to CYH33 was also positively correlated with decrease in phosphorylated ERK. Taken together, CYH33 is a promising PI3Kalpha inhibitor for breast cancer treatment and decrease in ERK phosphorylation may indicate its efficacy, which provides useful clues for rational design of the ongoing clinical trials. | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [44] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | DHM25 | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 | |||||||||
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | ||||||||||
| BT474 cells | Breast | Homo sapiens (Human) | CVCL_0179 | ||||||||||
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | ||||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | ||||||||||
| BT549 cells | Breast | Homo sapiens (Human) | CVCL_1092 | ||||||||||
| T47-D cells | Pleural effusion | Homo sapiens (Human) | CVCL_0553 | ||||||||||
| In Vivo Model | NOD/SCID/gamac null mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [44] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | DHM25 | ||||||||||||
| Molecule Alteration | Missense mutation | p.K111N (c.333G>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 | |||||||||
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | ||||||||||
| BT474 cells | Breast | Homo sapiens (Human) | CVCL_0179 | ||||||||||
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | ||||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | ||||||||||
| BT549 cells | Breast | Homo sapiens (Human) | CVCL_1092 | ||||||||||
| T47-D cells | Pleural effusion | Homo sapiens (Human) | CVCL_0553 | ||||||||||
| In Vivo Model | NOD/SCID/gamac null mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [44] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | DHM25 | ||||||||||||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 | |||||||||
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | ||||||||||
| BT474 cells | Breast | Homo sapiens (Human) | CVCL_0179 | ||||||||||
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | ||||||||||
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | ||||||||||
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | ||||||||||
| BT549 cells | Breast | Homo sapiens (Human) | CVCL_1092 | ||||||||||
| T47-D cells | Pleural effusion | Homo sapiens (Human) | CVCL_0553 | ||||||||||
| In Vivo Model | NOD/SCID/gamac null mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [45] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | DS-7423 | |||
| Molecule Alteration | Missense mutation | p.C420R (c.1258T>C) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SkOV3 cells | Ovary | Homo sapiens (Human) | CVCL_0532 |
| ES-2 cells | Ovary | Homo sapiens (Human) | CVCL_3509 | |
| SkOV3 cells | Ovary | Homo sapiens (Human) | CVCL_0532 | |
| TOV-21 cells | Ovary | Homo sapiens (Human) | CVCL_3613 | |
| RMG-I cells | Ascites | Homo sapiens (Human) | CVCL_1662 | |
| OVTOKO cells | Spleen | Homo sapiens (Human) | CVCL_3117 | |
| OVSAHO cells | Abdomen | Homo sapiens (Human) | CVCL_3114 | |
| OVMANA cells | Ovary | Homo sapiens (Human) | CVCL_3111 | |
| OVKATE cells | Ovary | Homo sapiens (Human) | CVCL_3110 | |
| OVISE cells | Pelvi | Homo sapiens (Human) | CVCL_3116 | |
| OV1063 cells | Ascites | Homo sapiens (Human) | CVCL_4366 | |
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase assay | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [45] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | DS-7423 | |||
| Molecule Alteration | Missense mutation | p.E545V (c.1634A>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | ES-2 cells | Ovary | Homo sapiens (Human) | CVCL_3509 |
| TOV-21 cells | Ovary | Homo sapiens (Human) | CVCL_3613 | |
| SKOV4 cells | Uterus | Homo sapiens (Human) | CVCL_X008 | |
| RMG-I cells | Ascites | Homo sapiens (Human) | CVCL_1662 | |
| OVTOKO cells | Spleen | Homo sapiens (Human) | CVCL_3117 | |
| OVSAHO cells | Abdomen | Homo sapiens (Human) | CVCL_3114 | |
| OVMANA cells | Ovary | Homo sapiens (Human) | CVCL_3111 | |
| OVKATE cells | Ovary | Homo sapiens (Human) | CVCL_3110 | |
| OVISE cells | Pelvi | Homo sapiens (Human) | CVCL_3116 | |
| OV1063 cells | Ascites | Homo sapiens (Human) | CVCL_4366 | |
| SKOV4 cells | Uterus | Homo sapiens (Human) | CVCL_X008 | |
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase assay | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [12] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | MK2206 | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| In Vivo Model | Female nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Mechanism Description | The missense mutation p.H1047R (c.3140A>G) in gene PIK3CA cause the sensitivity of MK2206 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [12] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | MK2206 | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047L (c.3140A>T) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.77 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.96 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
A
A
H
L
H
H
G
G
1050
|
G
G
-
W
T
T
T
T
K
K
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| In Vivo Model | Female nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Mechanism Description | The missense mutation p.H1047L (c.3140A>T) in gene PIK3CA cause the sensitivity of MK2206 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [12] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | MK2206 | ||||||||||||
| Molecule Alteration | Missense mutation | p.N345K (c.1035T>G) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| In Vivo Model | Female nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Mechanism Description | The missense mutation p.N345K (c.1035T>G) in gene PIK3CA cause the sensitivity of MK2206 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [12] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | MK2206 | ||||||||||||
| Molecule Alteration | Missense mutation | p.E542K (c.1624G>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| In Vivo Model | Female nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Mechanism Description | The missense mutation p.E542K (c.1624G>A) in gene PIK3CA cause the sensitivity of MK2206 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [12] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | MK2206 | ||||||||||||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| In Vivo Model | Female nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Mechanism Description | The missense mutation p.E545K (c.1633G>A) in gene PIK3CA cause the sensitivity of MK2206 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [12] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | MK2206 | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q546K (c.1636C>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| In Vivo Model | Female nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Mechanism Description | The missense mutation p.Q546K (c.1636C>A) in gene PIK3CA cause the sensitivity of MK2206 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [12] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | MK2206 | ||||||||||||
| Molecule Alteration | Missense mutation | p.G1049R (c.3145G>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| In Vivo Model | Female nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Mechanism Description | The missense mutation p.G1049R (c.3145G>C) in gene PIK3CA cause the sensitivity of MK2206 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Thyroid gland cancer [ICD-11: 2D10.0] | [46] | ||||||||||||
| Sensitive Disease | Thyroid gland cancer [ICD-11: 2D10.0] | ||||||||||||
| Sensitive Drug | MK2206 | ||||||||||||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | SW1736 cells | Thyroid | Homo sapiens (Human) | CVCL_3883 | |||||||||
| C643 cells | Thyroid gland | Homo sapiens (Human) | CVCL_5969 | ||||||||||
| HTH7 cells | Thyroid gland | Homo sapiens (Human) | CVCL_6289 | ||||||||||
| Hth74 cells | Thyroid gland | Homo sapiens (Human) | CVCL_6288 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Mechanism Description | The missense mutation p.E545K (c.1633G>A) in gene PIK3CA cause the sensitivity of MK2206 by unusual activation of pro-survival pathway | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Thyroid gland cancer [ICD-11: 2D10.0] | [46] | ||||||||||||
| Sensitive Disease | Thyroid gland cancer [ICD-11: 2D10.0] | ||||||||||||
| Sensitive Drug | MK2206/Temsirolimus | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | SW1736 cells | Thyroid | Homo sapiens (Human) | CVCL_3883 | |||||||||
| C643 cells | Thyroid gland | Homo sapiens (Human) | CVCL_5969 | ||||||||||
| HTH7 cells | Thyroid gland | Homo sapiens (Human) | CVCL_6289 | ||||||||||
| Hth74 cells | Thyroid gland | Homo sapiens (Human) | CVCL_6288 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Mechanism Description | The missense mutation p.H1047R (c.3140A>G) in gene PIK3CA cause the sensitivity of MK2206 + Temsirolimus by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Thyroid gland cancer [ICD-11: 2D10.0] | [46] | ||||||||||||
| Sensitive Disease | Thyroid gland cancer [ICD-11: 2D10.0] | ||||||||||||
| Sensitive Drug | MK2206/Temsirolimus | ||||||||||||
| Molecule Alteration | Missense mutation | p.E542K (c.1624G>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | SW1736 cells | Thyroid | Homo sapiens (Human) | CVCL_3883 | |||||||||
| C643 cells | Thyroid gland | Homo sapiens (Human) | CVCL_5969 | ||||||||||
| HTH7 cells | Thyroid gland | Homo sapiens (Human) | CVCL_6289 | ||||||||||
| Hth74 cells | Thyroid gland | Homo sapiens (Human) | CVCL_6288 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
MTT assay | ||||||||||||
| Mechanism Description | The missense mutation p.E542K (c.1624G>A) in gene PIK3CA cause the sensitivity of MK2206 + Temsirolimus by unusual activation of pro-survival pathway | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [47] | |||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Sensitive Drug | PD-0325901/Pictilisib | |||
| Molecule Alteration | Missense mutation | p.E542K (c.1624G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | HT29 Cells | Colon | Homo sapiens (Human) | CVCL_A8EZ |
| HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |
| In Vivo Model | Female athymic CD1 mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Tumor volume measurement assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [48] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Sensitive Drug | PI103 | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [48] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Sensitive Drug | PI103 | ||||||||||||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Bladder cancer [ICD-11: 2C94.0] | [28] | |||
| Sensitive Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Sensitive Drug | Pictilisib/Sorafenib | |||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | MEK/ERK signaling pathway | Inhibition | hsa04011 | |
| In Vitro Model | 5637 cells | Bladder | Homo sapiens (Human) | CVCL_0126 |
| J82 cells | Bladder | Homo sapiens (Human) | CVCL_0359 | |
| RT4 cells | Bladder | Homo sapiens (Human) | CVCL_0036 | |
| T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |
| TCCSuP cells | Bladder | Homo sapiens (Human) | CVCL_1738 | |
| In Vivo Model | NSG mouse PDX model | Mus musculus | ||
| Experiment for Drug Resistance |
MTS assay; FACS assay | |||
| Mechanism Description | Pictilisib activated the compensatory MEK/ERK pathways that likely contributed to pictilisib resistance, which was reversed by co-treatment with the RAF inhibitor sorafenib. RNA-sequencing of tumors resistant to treatment suggested that LSP1 down-regulation correlated with drug resistance. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [49] | |||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Sensitive Drug | Pilaralisib | |||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| U87-MG cells | Brain | Homo sapiens (Human) | CVCL_0022 | |
| LS174T cells | Colon | Homo sapiens (Human) | CVCL_1384 | |
| MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 | |
| Ramos cells | Ascites | Homo sapiens (Human) | CVCL_0597 | |
| OVCAR-3 cells | Ascites | Homo sapiens (Human) | CVCL_0465 | |
| In Vivo Model | Athymic nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
ViaLight HS assay; Apo-ONE Homogeneous Caspase-3/7 assay; Soft agar assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [50] | |||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Sensitive Drug | PKI-402 | |||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HT29 Cells | Colon | Homo sapiens (Human) | CVCL_A8EZ |
| DLD1 cells | Colon | Homo sapiens (Human) | CVCL_0248 | |
| PC3 cells | Prostate | Homo sapiens (Human) | CVCL_0035 | |
| HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| BT474 cells | Breast | Homo sapiens (Human) | CVCL_0179 | |
| NCI-H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| H157 cells | Lung | Homo sapiens (Human) | CVCL_2458 | |
| U87MG cells | Brain | Homo sapiens (Human) | CVCL_GP63 | |
| A498 cells | Kidney | Homo sapiens (Human) | CVCL_1056 | |
| NCI-H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | |
| KB cells | Gastric | Homo sapiens (Human) | CVCL_0372 | |
| MDA-MB-361 cells | Breast | Homo sapiens (Human) | CVCL_0620 | |
| MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 | |
| MIA PaCa-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0428 | |
| NCI-H2170 cells | Lung | Homo sapiens (Human) | CVCL_1535 | |
| NCI- H1650 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1483 | |
| 786-0 cells | Kidney | Homo sapiens (Human) | CVCL_1051 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Mechanism Description | The missense mutation p.E545K (c.1633G>A) in gene PIK3CA cause the sensitivity of PKI-402 by aberration of the drug's therapeutic target | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Cervical cancer [ICD-11: 2C77.0] | [51] | |||
| Sensitive Disease | Cervical cancer [ICD-11: 2C77.0] | |||
| Sensitive Drug | PW12 | |||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Immunoblotting analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | The missense mutation p.E545K (c.1633G>A) in gene PIK3CA cause the sensitivity of PW12 by aberration of the drug's therapeutic target | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Head and neck squamous cell carcinoma [ICD-11: 2D42.1] | [52] | ||||||||||||
| Sensitive Disease | Head and neck squamous cell carcinoma [ICD-11: 2D42.1] | ||||||||||||
| Sensitive Drug | SHR-A1307 | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | SW480 cells | Colon | Homo sapiens (Human) | CVCL_0546 | |||||||||
| SW620 cells | Colon | Homo sapiens (Human) | CVCL_0547 | ||||||||||
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | ||||||||||
| H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | ||||||||||
| HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | ||||||||||
| LOVO cells | Colon | Homo sapiens (Human) | CVCL_0399 | ||||||||||
| HCC827 cells | Lung | Homo sapiens (Human) | CVCL_2063 | ||||||||||
| MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | ||||||||||
| HT-29 cells | Colon | Homo sapiens (Human) | CVCL_0320 | ||||||||||
| A431 cells | Skin | Homo sapiens (Human) | CVCL_0037 | ||||||||||
| DiFi cells | Colon | Homo sapiens (Human) | CVCL_6895 | ||||||||||
| Detroit562 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1171 | ||||||||||
| Colo-205 cells | Ascites | Homo sapiens (Human) | CVCL_0218 | ||||||||||
| In Vivo Model | Female BALB/c nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay; IC50 assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Head and neck squamous cell carcinoma [ICD-11: 2D42.1] | [53] | ||||||||||||
| Sensitive Disease | Head and neck squamous cell carcinoma [ICD-11: 2D42.1] | ||||||||||||
| Sensitive Drug | Sirolimus/Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | PI3K/mTOR signaling pathway | Inhibition | hsa04151 | ||||||||||
| In Vitro Model | CAL27 cells | Oral | Homo sapiens (Human) | CVCL_1107 | |||||||||
| UM-SCC-17B cells | Cervical lymph node | Homo sapiens (Human) | CVCL_7725 | ||||||||||
| Detroit 562 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1171 | ||||||||||
| In Vivo Model | Athymic nude mouse tumor xenografts model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Experiment for Drug Resistance |
Alamar blue cell viability reagent assay | ||||||||||||
| Mechanism Description | mTOR and MEK inhibition display a synergistic growth inhibitory activity in HNSCC cells genetically engineered to express activating KRAS and PIK3CA mutations. Antitumoral activity of the rapamycin and trametinib combination therapy increase in genetically engineered HNSCC cells expressing activating RAS or PIK3CA mutations | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [54] | |||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Sensitive Drug | WYE-125132 | |||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 |
| LNCaP cells | Prostate | Homo sapiens (Human) | CVCL_0395 | |
| HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |
| Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 | |
| H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | |
| MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| H157 cells | Lung | Homo sapiens (Human) | CVCL_2458 | |
| BT549 cells | Breast | Homo sapiens (Human) | CVCL_1092 | |
| HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |
| 786-O cells | Kidney | Homo sapiens (Human) | CVCL_1051 | |
| U87MG cells | Brain | Homo sapiens (Human) | CVCL_GP63 | |
| A498 cells | Kidney | Homo sapiens (Human) | CVCL_1056 | |
| MDA-MB-361 cells | Breast | Homo sapiens (Human) | CVCL_0620 | |
| Rat1 cells | Whole embryo | Rattus norvegicus (Rat) | CVCL_0492 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Mechanism Description | The missense mutation p.E545K (c.1633G>A) in gene PIK3CA cause the sensitivity of WYE-125132 by unusual activation of pro-survival pathway | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: FGFR-tacc positive glioblastoma [ICD-11: 2A00.01] | [55] | |||
| Sensitive Disease | FGFR-tacc positive glioblastoma [ICD-11: 2A00.01] | |||
| Sensitive Drug | YM-024 | |||
| Molecule Alteration | Missense mutation | p.H1047Y (c.3139C>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | SK-MG-26 cell | N.A. | Homo sapiens (Human) | CVCL_D701 |
| SK-MG-17 cells | N.A. | Homo sapiens (Human) | CVCL_8574 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter96 AQueous assay; Soft-agar colony formation assay | |||
Investigative Drug(s)
7 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: HER2 negative breast cancer [ICD-11: 2C60.11] | [56] | ||||||||||||
| Sensitive Disease | HER2 negative breast cancer [ICD-11: 2C60.11] | ||||||||||||
| Sensitive Drug | Alpelisib/Fulvestrant | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: HER2 negative breast cancer [ICD-11: 2C60.11] | [56] | ||||||||||||
| Sensitive Disease | HER2 negative breast cancer [ICD-11: 2C60.11] | ||||||||||||
| Sensitive Drug | Alpelisib/Fulvestrant | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047L (c.3140A>T) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.77 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.96 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
A
A
H
L
H
H
G
G
1050
|
G
G
-
W
T
T
T
T
K
K
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: HER2 negative breast cancer [ICD-11: 2C60.11] | [56] | ||||||||||||
| Sensitive Disease | HER2 negative breast cancer [ICD-11: 2C60.11] | ||||||||||||
| Sensitive Drug | Alpelisib/Fulvestrant | ||||||||||||
| Molecule Alteration | Missense mutation | p.C420R (c.1258T>C) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: HER2 negative breast cancer [ICD-11: 2C60.11] | [56] | ||||||||||||
| Sensitive Disease | HER2 negative breast cancer [ICD-11: 2C60.11] | ||||||||||||
| Sensitive Drug | Alpelisib/Fulvestrant | ||||||||||||
| Molecule Alteration | Missense mutation | p.E542K (c.1624G>A) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: HER2 negative breast cancer [ICD-11: 2C60.11] | [56] | ||||||||||||
| Sensitive Disease | HER2 negative breast cancer [ICD-11: 2C60.11] | ||||||||||||
| Sensitive Drug | Alpelisib/Fulvestrant | ||||||||||||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: HER2 negative breast cancer [ICD-11: 2C60.11] | [56] | ||||||||||||
| Sensitive Disease | HER2 negative breast cancer [ICD-11: 2C60.11] | ||||||||||||
| Sensitive Drug | Alpelisib/Fulvestrant | ||||||||||||
| Molecule Alteration | Missense mutation | p.E545A (c.1634A>C) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: HER2 negative breast cancer [ICD-11: 2C60.11] | [56] | ||||||||||||
| Sensitive Disease | HER2 negative breast cancer [ICD-11: 2C60.11] | ||||||||||||
| Sensitive Drug | Alpelisib/Fulvestrant | ||||||||||||
| Molecule Alteration | Missense mutation | p.E545G (c.1634A>G) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: HER2 negative breast cancer [ICD-11: 2C60.11] | [56] | ||||||||||||
| Sensitive Disease | HER2 negative breast cancer [ICD-11: 2C60.11] | ||||||||||||
| Sensitive Drug | Alpelisib/Fulvestrant | ||||||||||||
| Molecule Alteration | Missense mutation | p.E545D (c.1635G>C) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: HER2 negative breast cancer [ICD-11: 2C60.11] | [56] | ||||||||||||
| Sensitive Disease | HER2 negative breast cancer [ICD-11: 2C60.11] | ||||||||||||
| Sensitive Drug | Alpelisib/Fulvestrant | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q546E (c.1636C>G) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: HER2 negative breast cancer [ICD-11: 2C60.11] | [56] | ||||||||||||
| Sensitive Disease | HER2 negative breast cancer [ICD-11: 2C60.11] | ||||||||||||
| Sensitive Drug | Alpelisib/Fulvestrant | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q546R (c.1637A>G) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: HER2 negative breast cancer [ICD-11: 2C60.11] | [56] | ||||||||||||
| Sensitive Disease | HER2 negative breast cancer [ICD-11: 2C60.11] | ||||||||||||
| Sensitive Drug | Alpelisib/Fulvestrant | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047Y (c.3139C>T) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [57] | |||
| Resistant Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Resistant Drug | Bevacizumab/Temsirolimus | |||
| Molecule Alteration | Missense mutation | p.H1047X (c.3139_3141) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Mechanism Description | The missense mutation p.H1047X (c.3139_3141) in gene PIK3CA cause the resistance of Bevacizumab + Temsirolimus by unusual activation of pro-survival pathway | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [58] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | Buparlisib/Paclitaxel | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Cervical cancer [ICD-11: 2C77.0] | [59] | |||
| Sensitive Disease | Cervical cancer [ICD-11: 2C77.0] | |||
| Sensitive Drug | Gemcitabine/LY2780301 | |||
| Molecule Alteration | Missense mutation | p.E542K (c.1624G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | PI3K signaling pathway | Inhibition | hsa04151 | |
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [60], [61] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | HER2 inhibitors | |||
| Molecule Alteration | Mutation | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | AKT signaling pathway | Activation | hsa04151 | |
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | ||
| Experiment for Molecule Alteration |
Qubit HS dsDNA assay; Illumina sequencing assay; Sanger sequencing assay | |||
| Mechanism Description | PIk3CA mutations may have independent driver properties in a HER2+ context and have been implicated in resistance to anti-HER2 therapies. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [62] | |||
| Resistant Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Resistant Drug | PI3K pathway inhibitors | |||
| Molecule Alteration | Copy number gain | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | KPL-4 cells | Breast | Homo sapiens (Human) | CVCL_5310 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter Glo luminescence assay; PathScan RTK signaling antibody array assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [63] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | PKI-587 | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HT29 Cells | Colon | Homo sapiens (Human) | CVCL_A8EZ | |||||||||
| DLD1 cells | Colon | Homo sapiens (Human) | CVCL_0248 | ||||||||||
| H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | ||||||||||
| PC3 cells | Prostate | Homo sapiens (Human) | CVCL_0035 | ||||||||||
| HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | ||||||||||
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | ||||||||||
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | ||||||||||
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | ||||||||||
| BT474 cells | Breast | Homo sapiens (Human) | CVCL_0179 | ||||||||||
| U87 cells | Brain | Homo sapiens (Human) | CVCL_0022 | ||||||||||
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | ||||||||||
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | ||||||||||
| H157 cells | Lung | Homo sapiens (Human) | CVCL_2458 | ||||||||||
| H2170 cells | Lung | Homo sapiens (Human) | CVCL_1535 | ||||||||||
| MDA-MB-361 cells | Breast | Homo sapiens (Human) | CVCL_0620 | ||||||||||
| H1650 cells | Pleural effusion | Homo sapiens (Human) | CVCL_4V01 | ||||||||||
| MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 | ||||||||||
| MIA PaCa-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0428 | ||||||||||
| HTB44 cells | Kidney | Homo sapiens (Human) | N.A. | ||||||||||
| H1666 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1485 | ||||||||||
| 786-0 cells | Kidney | Homo sapiens (Human) | CVCL_1051 | ||||||||||
| In Vivo Model | Female nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
IC50 assay | ||||||||||||
| Disease Class: Head and neck squamous cell carcinoma [ICD-11: 2D42.1] | [64] | ||||||||||||
| Sensitive Disease | Head and neck squamous cell carcinoma [ICD-11: 2D42.1] | ||||||||||||
| Sensitive Drug | PKI-587 | ||||||||||||
| Molecule Alteration | Missense mutation | p.H1047R (c.3140A>G) |
|||||||||||
| Wild Type Structure | Method: Electron microscopy | Resolution: 2.41 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.61 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
-
S
-
Y
-
Y
-
H
-
H
-
H
-
-20
|
H
-
H
-
H
-
D
-
Y
-
D
-
I
-
P
-
T
-
T
-
-10
|
E
-
N
-
L
-
Y
-
F
-
Q
-
G
G
A
A
M
M
G
G
0
|
S
S
M
M
P
P
P
P
R
R
P
P
S
S
S
S
G
G
E
E
10
|
L
L
W
W
G
G
I
I
H
H
L
L
M
M
P
P
P
P
R
R
20
|
I
I
L
L
V
V
E
E
C
C
L
L
L
L
P
P
N
N
G
G
30
|
M
M
I
I
V
V
T
T
L
L
E
E
C
C
L
L
R
R
E
E
40
|
A
A
T
T
L
L
I
I
T
T
I
I
K
K
H
H
E
E
L
L
50
|
F
F
K
K
E
E
A
A
R
R
K
K
Y
Y
P
P
L
L
H
H
60
|
Q
Q
L
L
L
L
Q
Q
D
D
E
E
S
S
S
S
Y
Y
I
I
70
|
F
F
V
V
S
S
V
V
T
T
Q
Q
E
E
A
A
E
E
R
R
80
|
E
E
E
E
F
F
F
F
D
D
E
E
T
T
R
R
R
R
L
L
90
|
C
C
D
D
L
L
R
R
L
L
F
F
Q
Q
P
P
F
F
L
L
100
|
K
K
V
V
I
I
E
E
P
P
V
V
G
G
N
N
R
R
E
E
110
|
E
E
K
K
I
I
L
L
N
N
R
R
E
E
I
I
G
G
F
F
120
|
A
A
I
I
G
G
M
M
P
P
V
V
C
C
E
E
F
F
D
D
130
|
M
M
V
V
K
K
D
D
P
P
E
E
V
V
Q
Q
D
D
F
F
140
|
R
R
R
R
N
N
I
I
L
L
N
N
V
V
C
C
K
K
E
E
150
|
A
A
V
V
D
D
L
L
R
R
D
D
L
L
N
N
S
S
P
P
160
|
H
H
S
S
R
R
A
A
M
M
Y
Y
V
V
Y
Y
P
P
P
P
170
|
N
N
V
V
E
E
S
S
S
S
P
P
E
E
L
L
P
P
K
K
180
|
H
H
I
I
Y
Y
N
N
K
K
L
L
D
D
K
K
G
G
Q
Q
190
|
I
I
I
I
V
V
V
V
I
I
W
W
V
V
I
I
V
V
S
S
200
|
P
P
N
N
N
N
D
D
K
K
Q
Q
K
K
Y
Y
T
T
L
L
210
|
K
K
I
I
N
N
H
H
D
D
C
C
V
V
P
P
E
E
Q
Q
220
|
V
V
I
I
A
A
E
E
A
A
I
I
R
R
K
K
K
K
T
T
230
|
R
R
S
S
M
M
L
L
L
L
S
S
S
S
E
E
Q
Q
L
L
240
|
K
K
L
L
C
C
V
V
L
L
E
E
Y
Y
Q
Q
G
G
K
K
250
|
Y
Y
I
I
L
L
K
K
V
V
C
C
G
G
C
C
D
D
E
E
260
|
Y
Y
F
F
L
L
E
E
K
K
Y
Y
P
P
L
L
S
S
Q
Q
270
|
Y
Y
K
K
Y
Y
I
I
R
R
S
S
C
C
I
I
M
M
L
L
280
|
G
G
R
R
M
M
P
P
N
N
L
L
M
M
L
L
M
M
A
A
290
|
K
K
E
E
S
S
L
L
Y
Y
S
S
Q
Q
L
L
P
P
M
M
300
|
D
D
C
C
F
F
T
T
M
M
P
P
S
S
Y
Y
S
S
R
R
310
|
R
R
I
I
S
S
T
T
A
A
T
T
P
P
Y
Y
M
M
N
N
320
|
G
G
E
E
T
T
S
S
T
T
K
K
S
S
L
L
W
W
V
V
330
|
I
I
N
N
S
S
A
A
L
L
R
R
I
I
K
K
I
I
L
L
340
|
C
C
A
A
T
T
Y
Y
V
V
N
N
V
V
N
N
I
I
R
R
350
|
D
D
I
I
D
D
K
K
I
I
Y
Y
V
V
R
R
T
T
G
G
360
|
I
I
Y
Y
H
H
G
G
G
G
E
E
P
P
L
L
C
C
D
D
370
|
N
N
V
V
N
N
T
T
Q
Q
R
R
V
V
P
P
C
C
S
S
380
|
N
N
P
P
R
R
W
W
N
N
E
E
W
W
L
L
N
N
Y
Y
390
|
D
D
I
I
Y
Y
I
I
P
P
D
D
L
L
P
P
R
R
A
A
400
|
A
A
R
R
L
L
C
C
L
L
S
S
I
I
C
C
S
S
V
V
410
|
K
K
G
G
R
R
K
K
G
G
A
A
K
K
E
E
E
E
H
H
420
|
C
C
P
P
L
L
A
A
W
W
G
G
N
N
I
I
N
N
L
L
430
|
F
F
D
D
Y
Y
T
T
D
D
T
T
L
L
V
V
S
S
G
G
440
|
K
K
M
M
A
A
L
L
N
N
L
L
W
W
P
P
V
V
P
P
450
|
H
H
G
G
L
L
E
E
D
D
L
L
L
L
N
N
P
P
I
I
460
|
G
G
V
V
T
T
G
G
S
S
N
N
P
P
N
N
K
K
E
E
470
|
T
T
P
P
C
C
L
L
E
E
L
L
E
E
F
F
D
D
W
W
480
|
F
F
S
S
S
S
V
V
V
V
K
K
F
F
P
P
D
D
M
M
490
|
S
S
V
V
I
I
E
E
E
E
H
H
A
A
N
N
W
W
S
S
500
|
V
V
S
S
R
R
E
E
A
A
G
G
F
F
S
S
Y
Y
S
S
510
|
H
H
A
A
G
G
L
L
S
S
N
N
R
R
L
L
A
A
R
R
520
|
D
D
N
N
E
E
L
L
R
R
E
E
N
N
D
D
K
K
E
E
530
|
Q
Q
L
L
K
K
A
A
I
I
S
S
T
T
R
R
D
D
P
P
540
|
L
L
S
S
E
E
I
I
T
T
E
E
Q
Q
E
E
K
K
D
D
550
|
F
F
L
L
W
W
S
S
H
H
R
R
H
H
Y
Y
C
C
V
V
560
|
T
T
I
I
P
P
E
E
I
I
L
L
P
P
K
K
L
L
L
L
570
|
L
L
S
S
V
V
K
K
W
W
N
N
S
S
R
R
D
D
E
E
580
|
V
V
A
A
Q
Q
M
M
Y
Y
C
C
L
L
V
V
K
K
D
D
590
|
W
W
P
P
P
P
I
I
K
K
P
P
E
E
Q
Q
A
A
M
M
600
|
E
E
L
L
L
L
D
D
C
C
N
N
Y
Y
P
P
D
D
P
P
610
|
M
M
V
V
R
R
G
G
F
F
A
A
V
V
R
R
C
C
L
L
620
|
E
E
K
K
Y
Y
L
L
T
T
D
D
D
D
K
K
L
L
S
S
630
|
Q
Q
Y
Y
L
L
I
I
Q
Q
L
L
V
V
Q
Q
V
V
L
L
640
|
K
K
Y
Y
E
E
Q
Q
Y
Y
L
L
D
D
N
N
L
L
L
L
650
|
V
V
R
R
F
F
L
L
L
L
K
K
K
K
A
A
L
L
T
T
660
|
N
N
Q
Q
R
R
I
I
G
G
H
H
F
F
F
F
F
F
W
W
670
|
H
H
L
L
K
K
S
S
E
E
M
M
H
H
N
N
K
K
T
T
680
|
V
V
S
S
Q
Q
R
R
F
F
G
G
L
L
L
L
L
L
E
E
690
|
S
S
Y
Y
C
C
R
R
A
A
C
C
G
G
M
M
Y
Y
L
L
700
|
K
K
H
H
L
L
N
N
R
R
Q
Q
V
V
E
E
A
A
M
M
710
|
E
E
K
K
L
L
I
I
N
N
L
L
T
T
D
D
I
I
L
L
720
|
K
K
Q
Q
E
E
K
K
K
K
D
D
E
E
T
T
Q
Q
K
K
730
|
V
V
Q
Q
M
M
K
K
F
F
L
L
V
V
E
E
Q
Q
M
M
740
|
R
R
R
R
P
P
D
D
F
F
M
M
D
D
A
A
L
L
Q
Q
750
|
G
G
F
F
L
L
S
S
P
P
L
L
N
N
P
P
A
A
H
H
760
|
Q
Q
L
L
G
G
N
N
L
L
R
R
L
L
E
E
E
E
C
C
770
|
R
R
I
I
M
M
S
S
S
S
A
A
K
K
R
R
P
P
L
L
780
|
W
W
L
L
N
N
W
W
E
E
N
N
P
P
D
D
I
I
M
M
790
|
S
S
E
E
L
L
L
L
F
F
Q
Q
N
N
N
N
E
E
I
I
800
|
I
I
F
F
K
K
N
N
G
G
D
D
D
D
L
L
R
R
Q
Q
810
|
D
D
M
M
L
L
T
T
L
L
Q
Q
I
I
I
I
R
R
I
I
820
|
M
M
E
E
N
N
I
I
W
W
Q
Q
N
N
Q
Q
G
G
L
L
830
|
D
D
L
L
R
R
M
M
L
L
P
P
Y
Y
G
G
C
C
L
L
840
|
S
S
I
I
G
G
D
D
C
C
V
V
G
G
L
L
I
I
E
E
850
|
V
V
V
V
R
R
N
N
S
S
H
H
T
T
I
I
M
M
Q
Q
860
|
I
I
Q
Q
C
C
K
K
G
G
G
G
L
L
K
K
G
G
A
A
870
|
L
L
Q
Q
F
F
N
N
S
S
H
H
T
T
L
L
H
H
Q
Q
880
|
W
W
L
L
K
K
D
D
K
K
N
N
K
K
G
G
E
E
I
I
890
|
Y
Y
D
D
A
A
A
A
I
I
D
D
L
L
F
F
T
T
R
R
900
|
S
S
C
C
A
A
G
G
Y
Y
C
C
V
V
A
A
T
T
F
F
910
|
I
I
L
L
G
G
I
I
G
G
D
D
R
R
H
H
N
N
S
S
920
|
N
N
I
I
M
M
V
V
K
K
D
D
D
D
G
G
Q
Q
L
L
930
|
F
F
H
H
I
I
D
D
F
F
G
G
H
H
F
F
L
L
D
D
940
|
H
H
K
K
K
K
K
K
K
K
F
F
G
G
Y
Y
K
K
R
R
950
|
E
E
R
R
V
V
P
P
F
F
V
V
L
L
T
T
Q
Q
D
D
960
|
F
F
L
L
I
I
V
V
I
I
S
S
K
K
G
G
A
A
Q
Q
970
|
E
E
C
C
T
T
K
K
T
T
R
R
E
E
F
F
E
E
R
R
980
|
F
F
Q
Q
E
E
M
M
C
C
Y
Y
K
K
A
A
Y
Y
L
L
990
|
A
A
I
I
R
R
Q
Q
H
H
A
A
N
N
L
L
F
F
I
I
1000
|
N
N
L
L
F
F
S
S
M
M
M
M
L
L
G
G
S
S
G
G
1010
|
M
M
P
P
E
E
L
L
Q
Q
S
S
F
F
D
D
D
D
I
I
1020
|
A
A
Y
Y
I
I
R
R
K
K
T
T
L
L
A
A
L
L
D
D
1030
|
K
K
T
T
E
E
Q
Q
E
E
A
A
L
L
E
E
Y
Y
F
F
1040
|
M
M
K
K
Q
Q
M
M
N
N
D
D
A
A
H
R
H
H
G
G
1050
|
G
G
W
W
T
T
T
T
K
K
M
M
D
D
W
W
I
I
F
F
1060
|
H
H
T
T
I
I
K
K
Q
Q
H
H
A
A
L
L
N
N
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | UMSCC cells | Oral cavity | Homo sapiens (Human) | CVCL_7707 | |||||||||
| In Vivo Model | SCID/NCr-Balb/c mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; qPCR | ||||||||||||
| Experiment for Drug Resistance |
XTT assay | ||||||||||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [63] | ||||||||||||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | ||||||||||||
| Sensitive Drug | PKI-587 | ||||||||||||
| Molecule Alteration | Missense mutation | p.E545K (c.1633G>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HT29 Cells | Colon | Homo sapiens (Human) | CVCL_A8EZ | |||||||||
| DLD1 cells | Colon | Homo sapiens (Human) | CVCL_0248 | ||||||||||
| H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | ||||||||||
| PC3 cells | Prostate | Homo sapiens (Human) | CVCL_0035 | ||||||||||
| HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | ||||||||||
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | ||||||||||
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | ||||||||||
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | ||||||||||
| BT474 cells | Breast | Homo sapiens (Human) | CVCL_0179 | ||||||||||
| U87 cells | Brain | Homo sapiens (Human) | CVCL_0022 | ||||||||||
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | ||||||||||
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | ||||||||||
| H157 cells | Lung | Homo sapiens (Human) | CVCL_2458 | ||||||||||
| H2170 cells | Lung | Homo sapiens (Human) | CVCL_1535 | ||||||||||
| MDA-MB-361 cells | Breast | Homo sapiens (Human) | CVCL_0620 | ||||||||||
| H1650 cells | Pleural effusion | Homo sapiens (Human) | CVCL_4V01 | ||||||||||
| MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 | ||||||||||
| MIA PaCa-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0428 | ||||||||||
| HTB44 cells | Kidney | Homo sapiens (Human) | N.A. | ||||||||||
| H1666 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1485 | ||||||||||
| 786-0 cells | Kidney | Homo sapiens (Human) | CVCL_1051 | ||||||||||
| In Vivo Model | Female nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
IC50 assay | ||||||||||||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Nervous tissue | |
| The Specified Disease | Brain cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.99E-16; Fold-change: 2.52E-01; Z-score: 4.65E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.60E-01; Fold-change: 5.84E-01; Z-score: 1.44E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
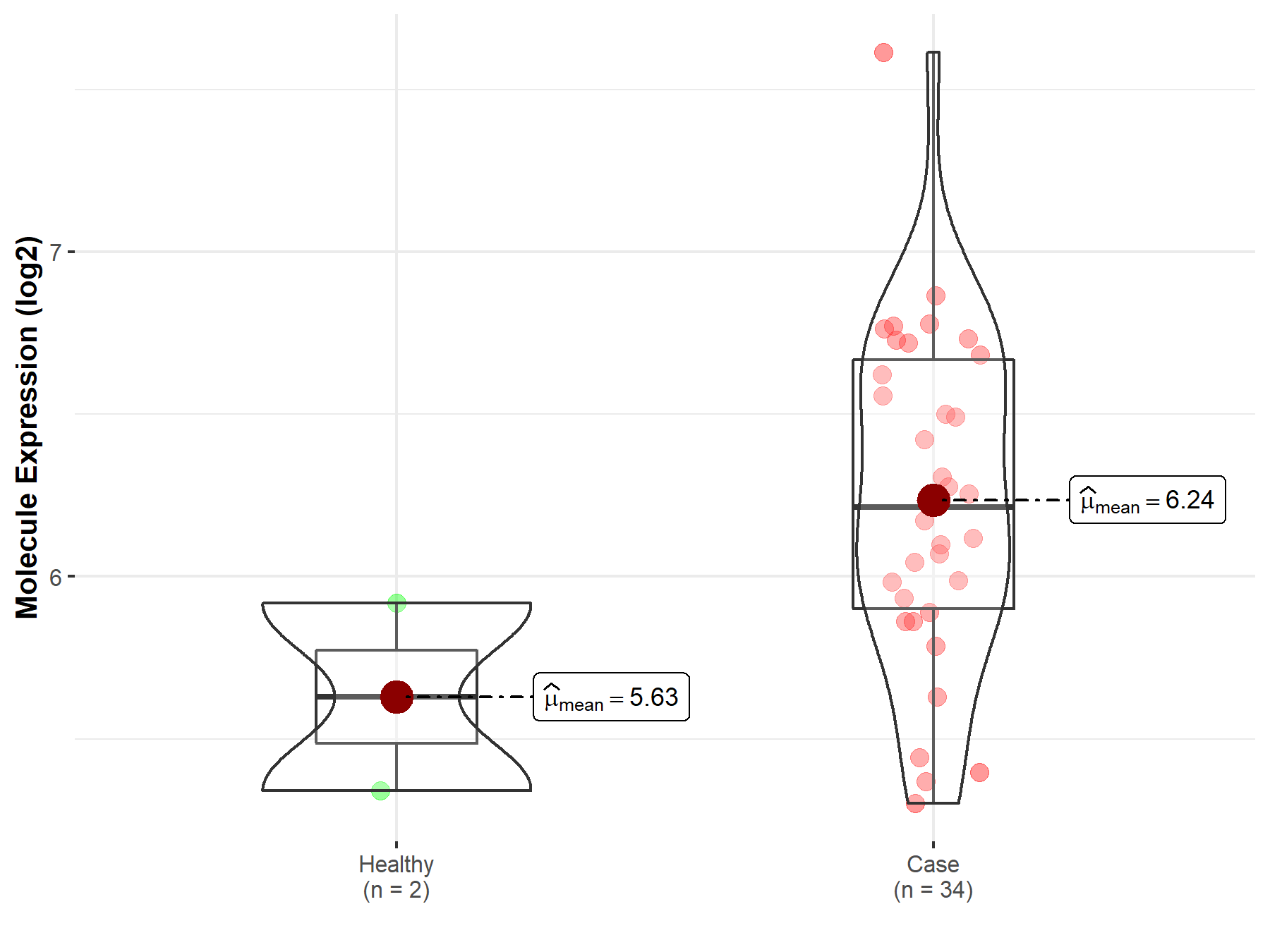
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | White matter | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.07E-05; Fold-change: 9.05E-01; Z-score: 1.91E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Neuroectodermal tumor | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.15E-08; Fold-change: 1.75E+00; Z-score: 4.45E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
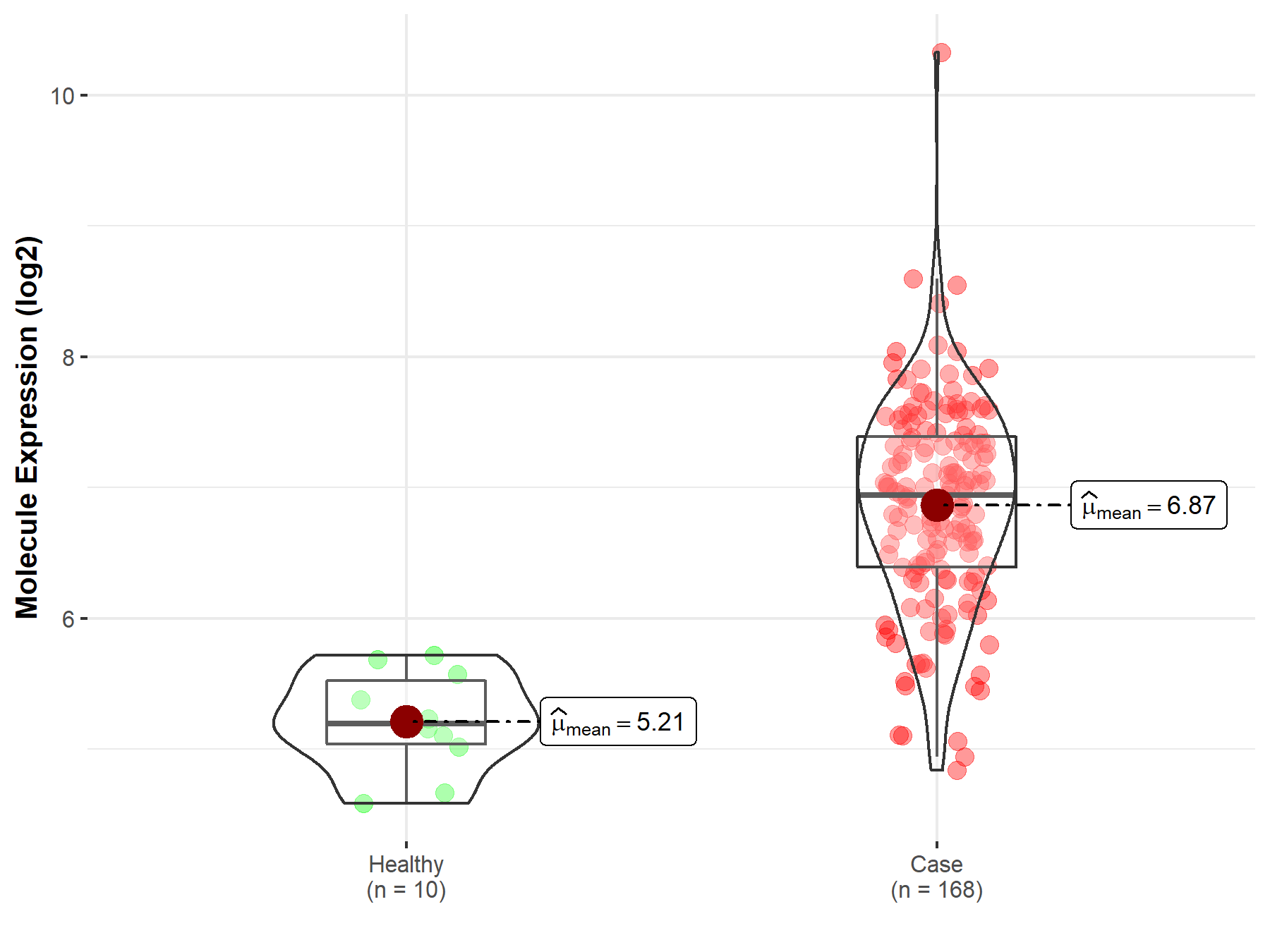
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Esophagus | |
| The Specified Disease | Esophageal cancer | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.86E-01; Fold-change: -1.85E-01; Z-score: -5.12E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
|
||
| Disease-specific Molecule Abundances |
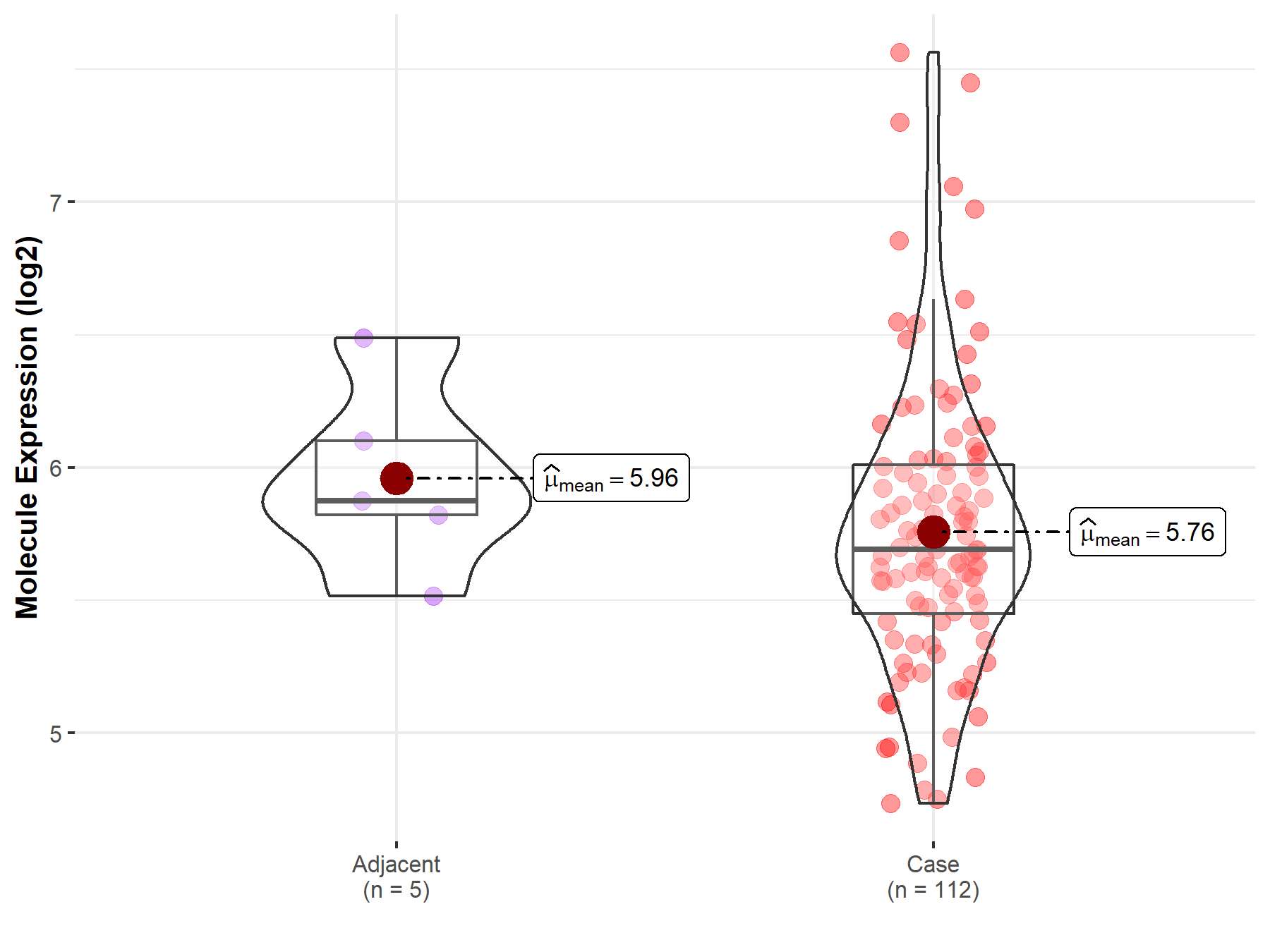
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Gastric tissue | |
| The Specified Disease | Gastric cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.13E-01; Fold-change: 1.71E-01; Z-score: 2.92E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 5.18E-01; Fold-change: 3.73E-02; Z-score: 1.07E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
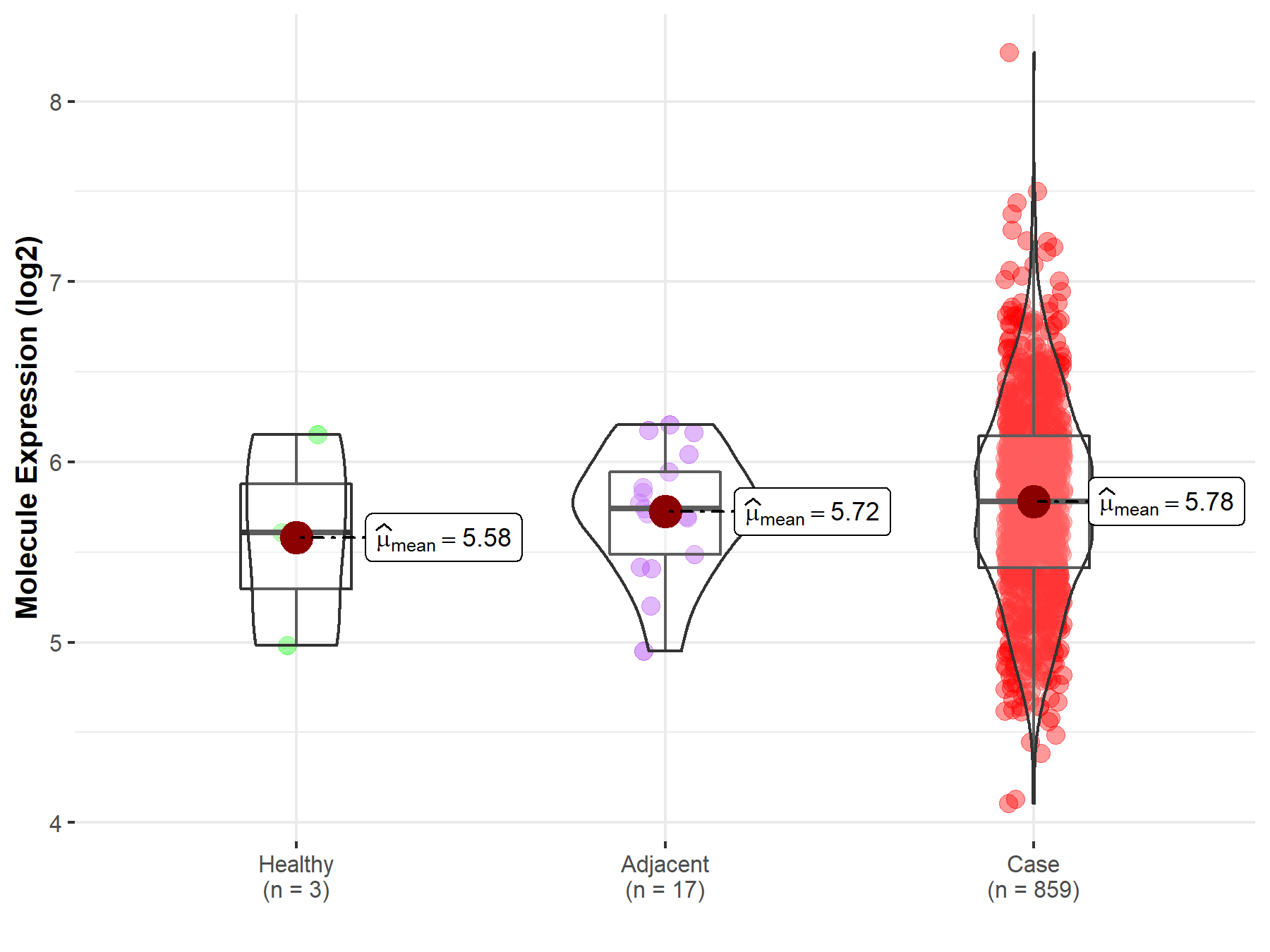
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Colon | |
| The Specified Disease | Colon cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.96E-12; Fold-change: 1.58E-01; Z-score: 5.08E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.93E-01; Fold-change: 6.10E-03; Z-score: 1.55E-02 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
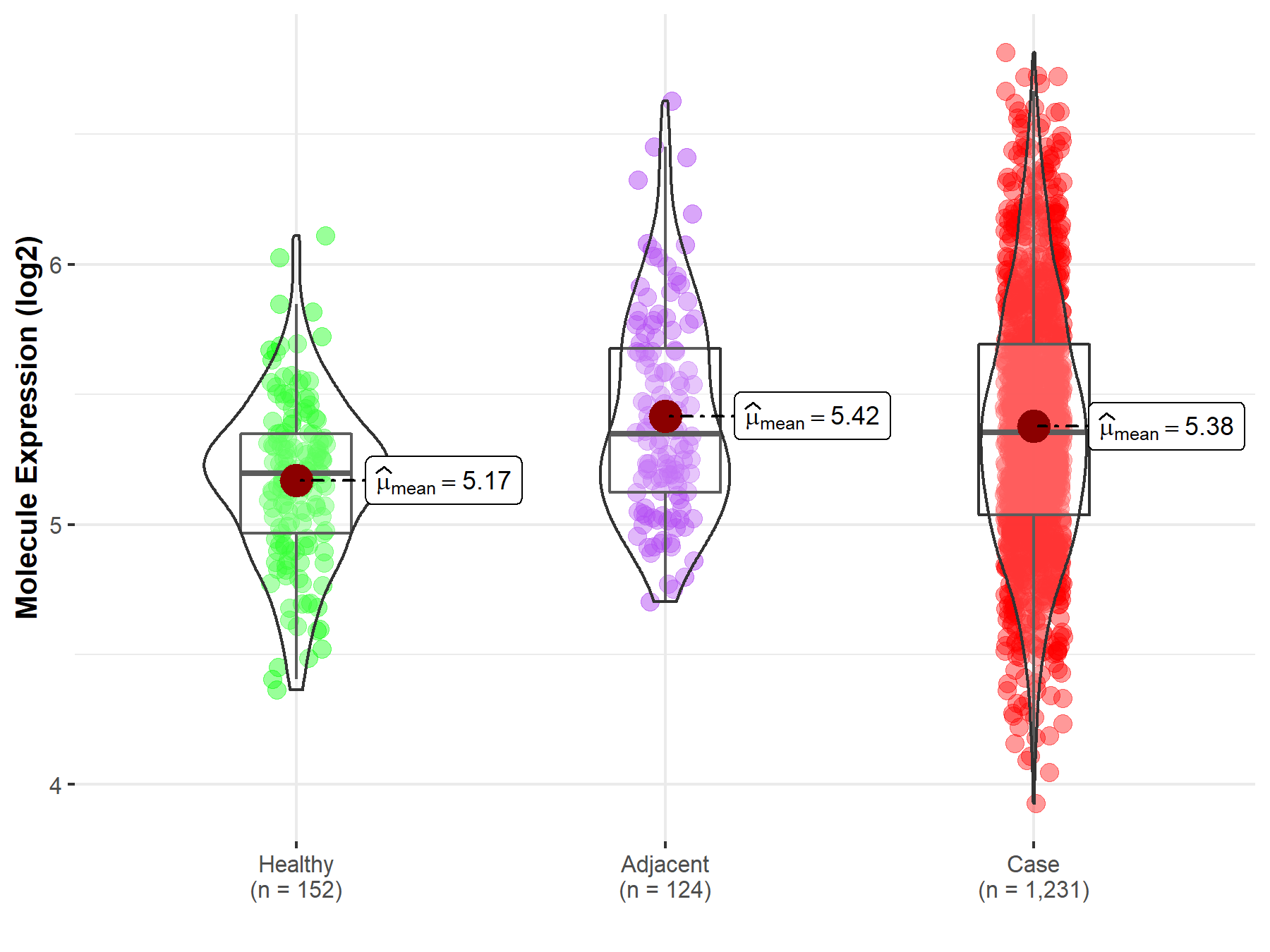
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Lung | |
| The Specified Disease | Lung cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.17E-13; Fold-change: -2.41E-01; Z-score: -7.91E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 6.76E-11; Fold-change: -3.01E-01; Z-score: -7.03E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Skin | |
| The Specified Disease | Melanoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.70E-01; Fold-change: -1.51E-01; Z-score: -3.12E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
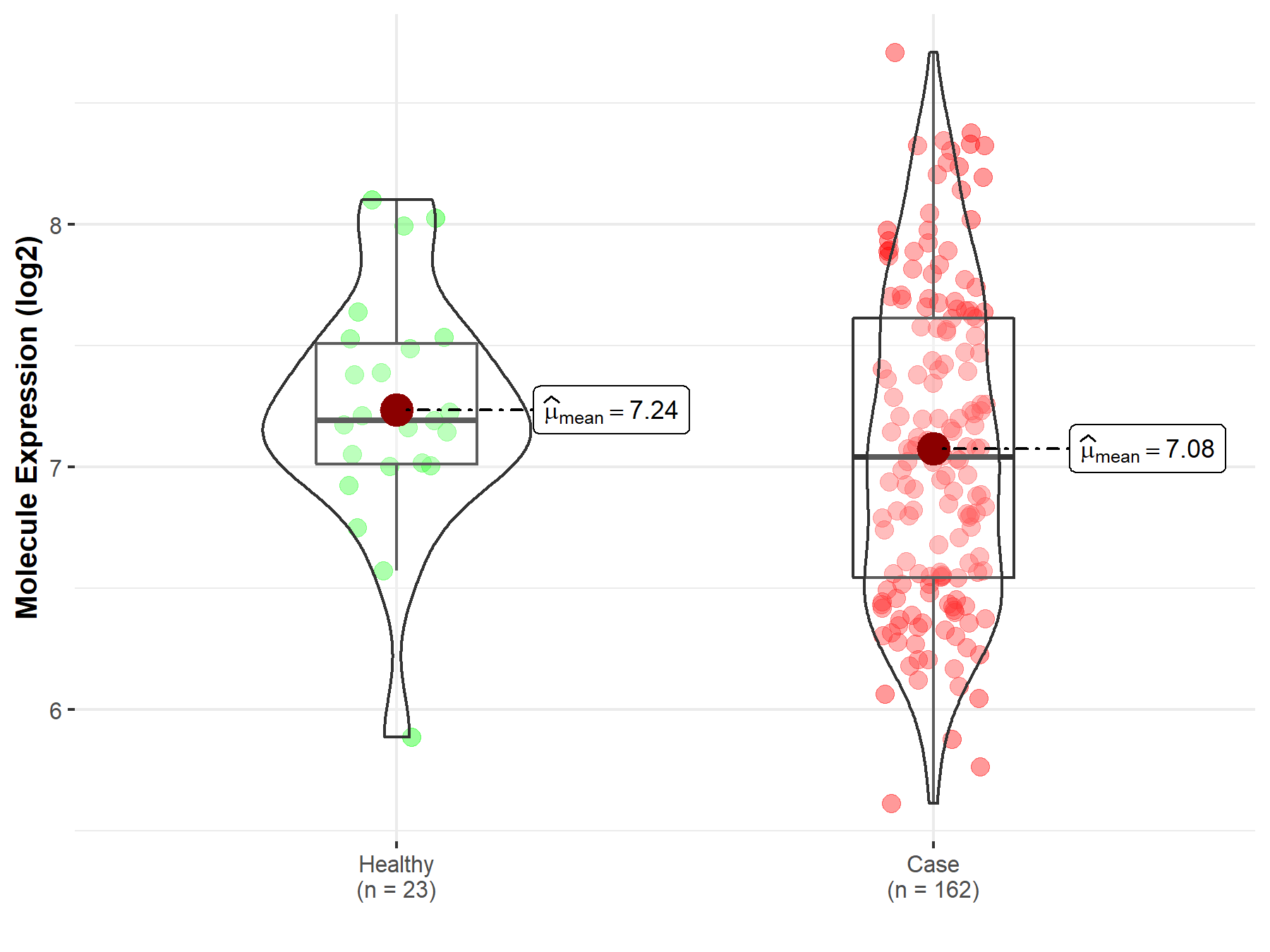
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.59E-38; Fold-change: -6.47E-01; Z-score: -1.11E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 4.29E-03; Fold-change: -4.33E-01; Z-score: -7.01E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Ovary | |
| The Specified Disease | Ovarian cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.39E-01; Fold-change: -1.90E-02; Z-score: -3.32E-02 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 8.18E-04; Fold-change: 5.53E-01; Z-score: 1.13E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
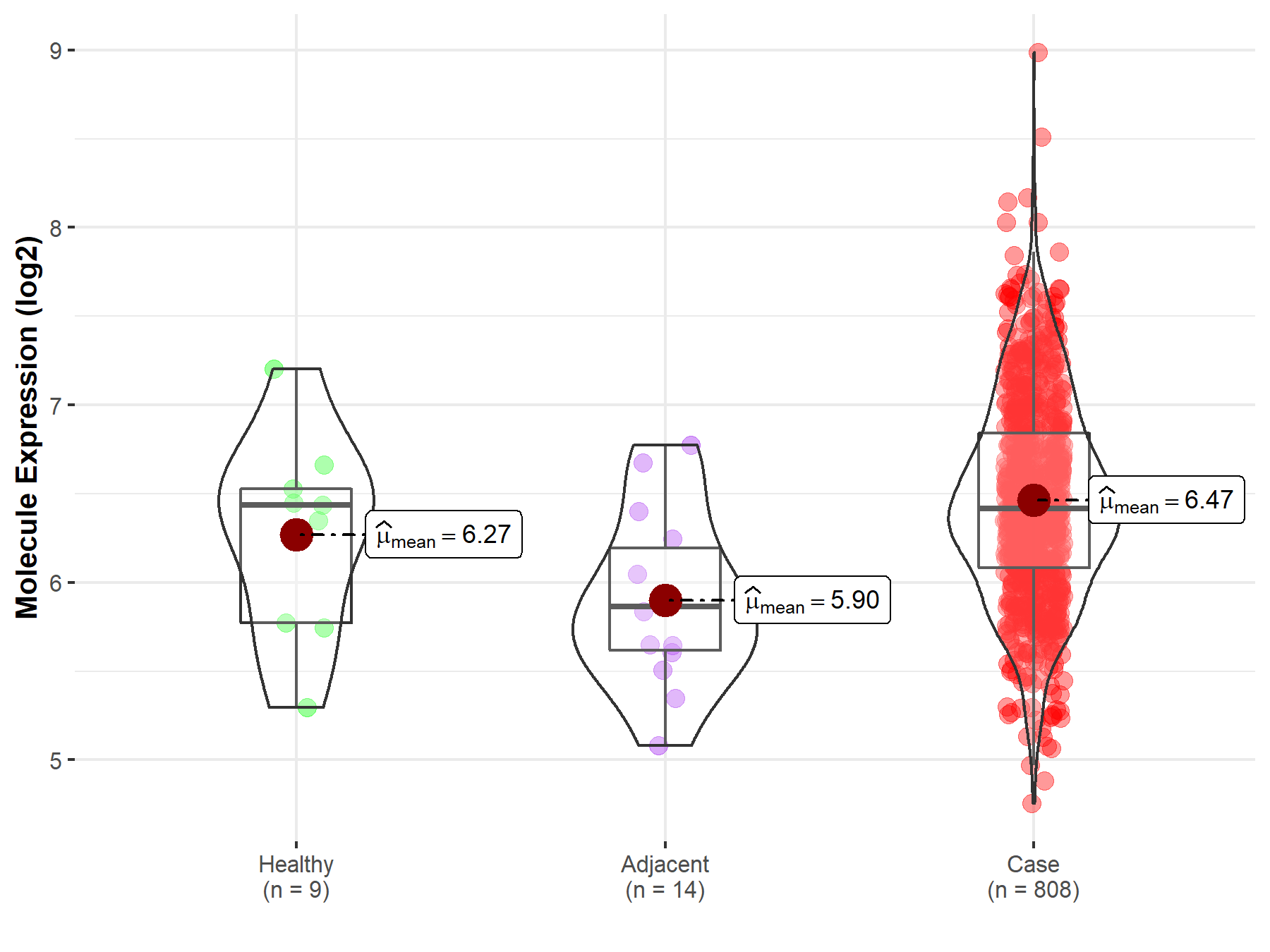
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Cervix uteri | |
| The Specified Disease | Cervical cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.32E-05; Fold-change: 4.20E-01; Z-score: 1.10E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
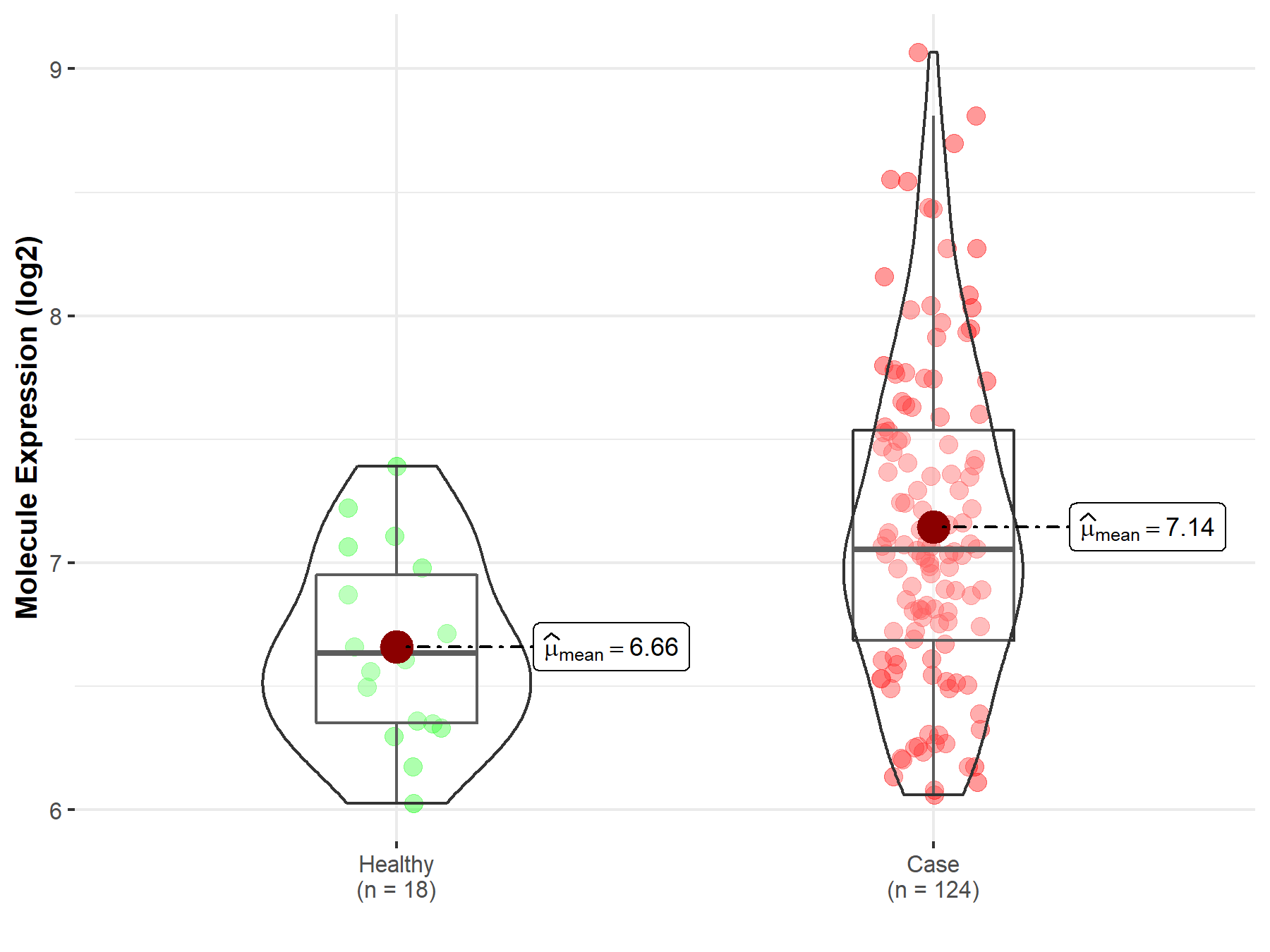
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Prostate | |
| The Specified Disease | Prostate cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.43E-02; Fold-change: 9.14E-01; Z-score: 9.00E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
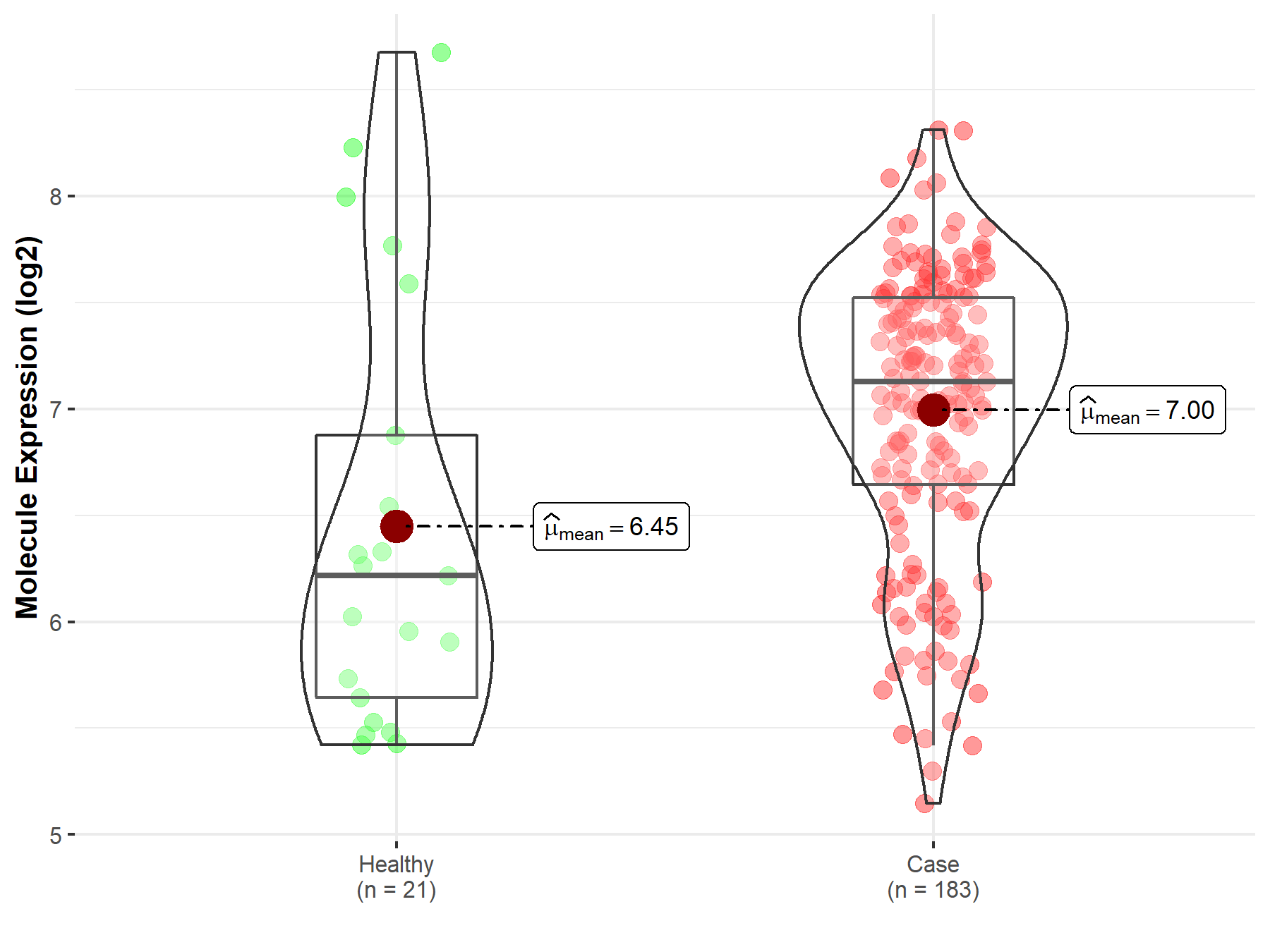
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Bladder tissue | |
| The Specified Disease | Bladder cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.42E-12; Fold-change: -1.06E+00; Z-score: -7.76E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
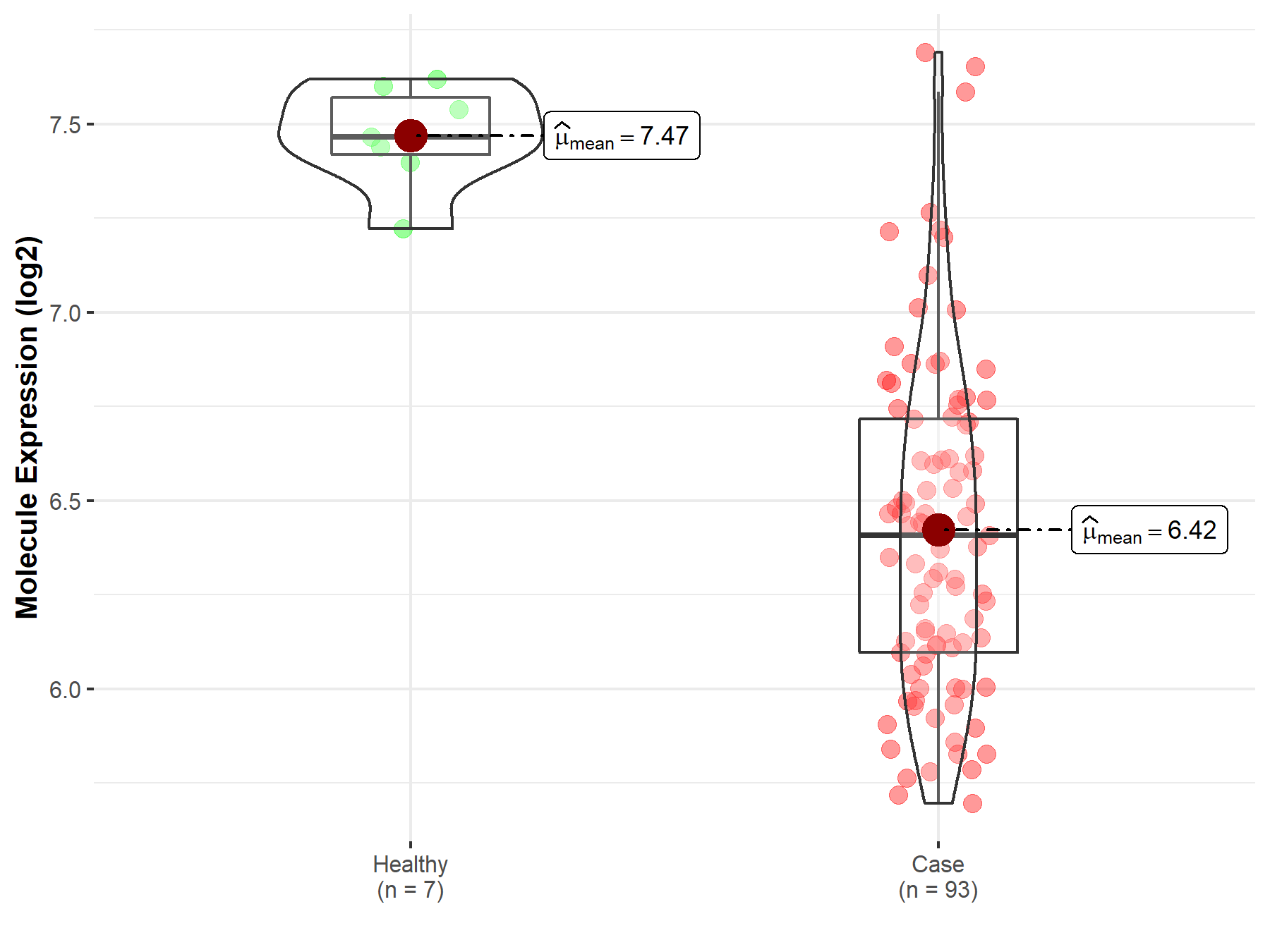
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Thyroid | |
| The Specified Disease | Thyroid cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.40E-01; Fold-change: 3.75E-02; Z-score: 1.20E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 7.23E-03; Fold-change: 1.42E-01; Z-score: 4.14E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
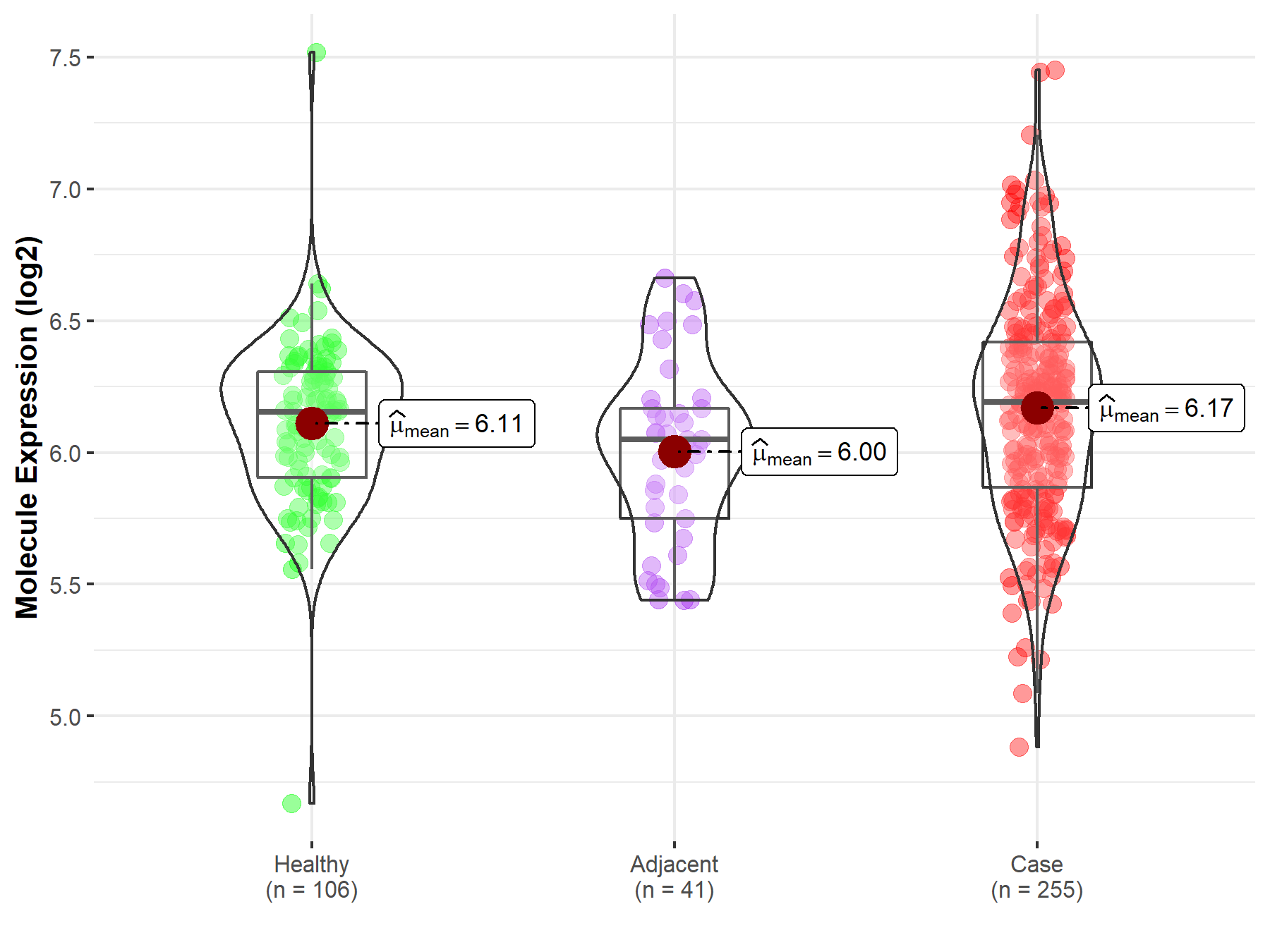
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

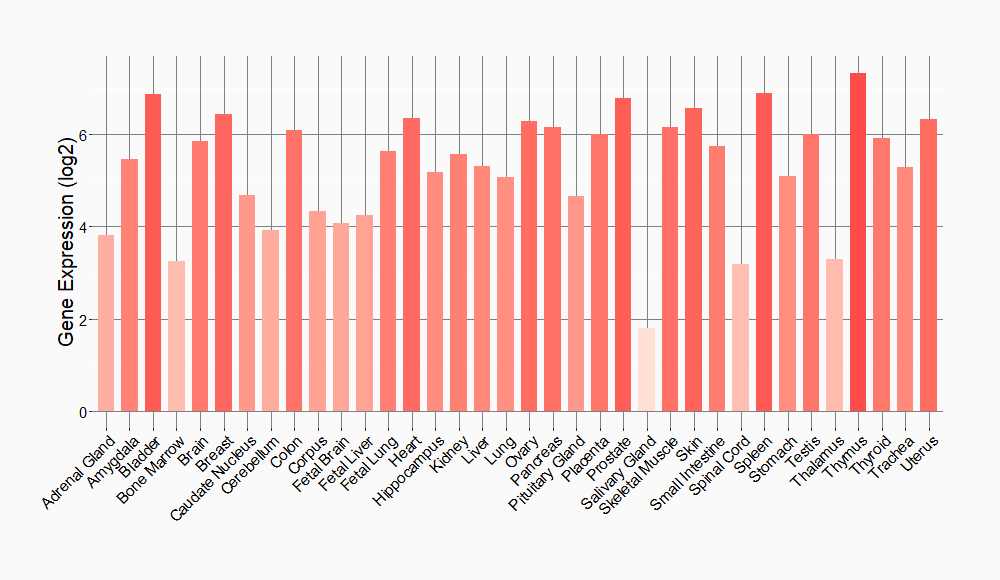
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
