Drug Information
Drug (ID: DG00237) and It's Reported Resistant Information
| Name |
Lincomycin
|
||||
|---|---|---|---|---|---|
| Synonyms |
Cillimycin; Epilincomycin; Jiemycin; LCM; Lincocin; Lincocine; Lincolcina; Lincolnensin; Lincomicina; Lincomix; Lincomycine; Lincomycinum; Lincomyocin; Lincorex; Mycivin; Lincomycin A; Lincomycine [French]; CBMicro_021584; Lincomix 20; Pura Ject 100; Lincocin (TN); Lincomicina [INN-Spanish]; Lincomycine [INN-French]; Lincomycinum [INN-Latin]; U 10,149A; Lincomycin (USAN/INN); Lincomycin [USAN:INN:BAN]; Lincomycin, (2S-cis)-Isomer; Methyl 6,8-dideoxy-6-[(1-methyl-4-propylprolyl)amino]-1-thiooctopyranoside
Click to Show/Hide
|
||||
| Indication |
In total 1 Indication(s)
|
||||
| Structure |
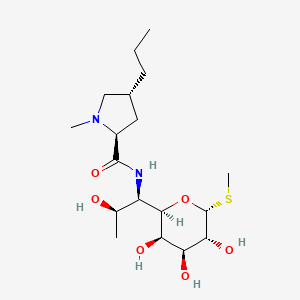
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Clinically Reported Resistance for This Drug
(10 diseases)
[6]
[7]
[8]
[7]
[7]
[7]
[7]
[7]
[7]
[7]
Disease(s) with Resistance Information Validated by in-vivo Model for This Drug
(1 diseases)
[9]
|
||||
| Target | Bacterial 50S ribosomal RNA (Bact 50S rRNA) | NOUNIPROTAC | [1] | ||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C18H34N2O6S
|
||||
| IsoSMILES |
CCC[C@@H]1C[C@H](N(C1)C)C(=O)N[C@@H]([C@@H]2[C@@H]([C@@H]([C@H]([C@H](O2)SC)O)O)O)[C@@H](C)O
|
||||
| InChI |
1S/C18H34N2O6S/c1-5-6-10-7-11(20(3)8-10)17(25)19-12(9(2)21)16-14(23)13(22)15(24)18(26-16)27-4/h9-16,18,21-24H,5-8H2,1-4H3,(H,19,25)/t9-,10-,11+,12-,13+,14-,15-,16-,18-/m1/s1
|
||||
| InChIKey |
OJMMVQQUTAEWLP-KIDUDLJLSA-N
|
||||
| PubChem CID | |||||
| ChEBI ID | |||||
| TTD Drug ID | |||||
| DrugBank ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-01: Infectious/parasitic diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: rRNA adenine N-6-methyltransferase ermE (ERME) | [1], [2], [3] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli AS19 | 562 | ||
| Escherichia coli AS19-RrmA- | 562 | |||
| Escherichia coli DH10B | 316385 | |||
| Escherichia coli JC7623 | 562 | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | Methylation of specific nucleotides in rRNA is one of the means by which bacteria achieve resistance to macrolides-lincosamides-streptogramin B (MLSB) and ketolide antibiotics.ErmE dimethylation confers high resistance to all the MLSB and ketolide drugs. | |||
| Key Molecule: 23S ribosomal RNA methyltransferase Erm36 (ERM36) | [10] | |||
| Resistant Disease | Micrococcus luteus infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Micrococcus luteus MAW843 | 1270 | ||
| Experiment for Molecule Alteration |
Sequence analysis | |||
| Experiment for Drug Resistance |
Agar diffusion test assay | |||
| Mechanism Description | Erm(36) was most related (about 52-54% identity) to erythromycin-resistance proteins found in high-G+C Gram-positive bacteria and lead to drug resistance. | |||
| Key Molecule: erm(X)cj (Unclear) | [8] | |||
| Resistant Disease | Corynebacterium jeikeium infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Frameshift mutation | Codon 216 frame shift |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Corynebacterium glutamicum ATCC 13032 | 196627 | ||
| Staphylococcus aureus ATCC 29213 | 1280 | |||
| Corynebacterium diphtheriae isolate | 1717 | |||
| Corynebacterium glutamicum kO8 | 1718 | |||
| Corynebacterium jeikeium isolates | 38289 | |||
| Escherichia coli ATCC 25923 | 562 | |||
| Escherichia coli strain XL1-Blue MRF9 | 562 | |||
| Experiment for Molecule Alteration |
Southern blotting assay | |||
| Experiment for Drug Resistance |
Disk diffusion methods assay; agar dilution methods assay | |||
| Mechanism Description | Abundant amplificationproducts of slightly less than 400 bp were generated from DNAisolated from the 17 MLSb-resistant strains, whereas no am-plification products were generated with the DNA isolatedfrom the three susceptible strains. The DNA sequences of the amplification products showed 95% identity to the erm(X) gene isolated from a C. xerosis strain,erm(X)cx or ermCX. Thus, MLSb resistance in C. jeikeiumis associated with the presence of an allele, erm(X)cj, of the class Xermgenes. The first 215 amino acids of the predicted polypeptides for strains CJ12 and CJ21 are 93.5 and 98.6% identical to Erm(X)cx, the Erm protein from C. xerosi. The major difference between the two Erm(X)cj polypeptides and the Erm(X)cx polypeptide is a frame shift within codon 216. This results in the Erm(X)cj polypeptides being 31 amino acids longer than Erm(X)cx. | |||
|
|
||||
| Key Molecule: Lincosamide nucleotidyltransferase (LNUG) | [9] | |||
| Resistant Disease | Enterococcus faecalis infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Enterococcus faecalis | 1351 | ||
| Experiment for Molecule Alteration |
PCR; DNA sequence assay | |||
| Mechanism Description | A novel resistance gene, designated lnu(G), which encodes a putative lincosamide nucleotidyltransferase, was found in E. faecalis E531. | |||
|
|
||||
| Key Molecule: ABC superfamily ATP binding cassette transporter (ABCCT) | [11], [12] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli TOP10 | 83333 | ||
| Staphylococcus aureus RN4220 | 1280 | |||
| Staphylococcus saprophyticus ATCC 15305 | 342451 | |||
| Staphylococcus sciuri ATCC 29059 | 1296 | |||
| Staphylococcus sciuri ATCC 29062 | 1296 | |||
| Staphylococcus sciuri ATCC 700058 | 1296 | |||
| Staphylococcus sciuri ATCC 700061 | 1296 | |||
| Staphylococcus sciuri BL2 | 1296 | |||
| Staphylococcus sciuri SS226 | 1296 | |||
| Staphylococcus sciuri SVv1 | 1296 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Disk diffusion test assay; E-strip test assay | |||
| Mechanism Description | Efflux-mediated resistance to MLS antibiotics in staphylococci relies on the ATPase activity of a very special kind of ATP-binding cassette (ABC) protein.By whole-genome sequencing of strain ATCC 29059, we identified a candidate gene that encodes an ATP-binding cassette protein similar to the Lsa and VmlR resistance determinants. Isolation and reverse transcription-quantitative PCR (qRT-PCR) expression studies confirmed that Sal(A) can confer a moderate resistance to lincosamides (8 times the MIC of lincomycin) and a high-level resistance to streptogramins A. The chromosomal location of sal(A) between two housekeeping genes of the staphylococcal core genome supports the gene's ancient origins and thus innate resistance to these antimicrobials within S. sciuri subspecies. | |||
| Key Molecule: ABC transporter ATP-binding protein (ABCP) | [13], [14] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.T450I |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli TOP10 | 83333 | ||
| Enterococcus faecium HM1070 | 1352 | |||
| Enterococcus faecium UCN80 | 1352 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Mechanism Description | ABC systems constitute one of the largest families of proteins, with most of them being involved in import and export, often called ABC transporters.Several of these class 2 ABC systems have been involved in MLS resistance, such as Msr-, Vga-, or Lsa-like proteins.The observed profile of cross-resistance to lincosamides, streptogramins A, and pleuromutilins conferred by Eat(A)v was similar to those conferred by other Lsa-like proteins. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: ABC protein lsaC (lsaC-Unclear) | [7] | |||
| Resistant Disease | Streptococcus agalactiae infection [ICD-11: 1B21.2] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli TOP10 | 83333 | ||
| Staphylococcus aureus ATCC 29213 | 1280 | |||
| Streptococcus agalactiae UCN70 | 1311 | |||
| Streptococcus agalactiae isolates | 1311 | |||
| Streptococcus agalactiae BM132 | 1319 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Expression of this novel gene, named lsa(C), in S. agalactiae BM132 after cloning led to an increase in MICs of lincomycin (0.06 to 4 ug/ml), clindamycin (0.03 to 2 ug/ml), dalfopristin (2 to >32 ug/ml), and tiamulin (0.12 to 32 ug/ml), whereas no change in MICs of erythromycin (0.06 ug/ml), azithromycin (0.03 ug/ml), spiramycin (0.25 ug/ml), telithromycin (0.03 ug/ml), and quinupristin (8 ug/ml) was observed. The phenotype was renamed the LS(A)P phenotype on the basis of cross-resistance to lincosamides, streptogramins A, and pleuromutilins. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: ABC protein lsaC (lsaC-Unclear) | [7] | |||
| Resistant Disease | Streptococcus agalactiae infection [ICD-11: 1B21.2] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli TOP10 | 83333 | ||
| Staphylococcus aureus ATCC 29213 | 1280 | |||
| Streptococcus agalactiae UCN70 | 1311 | |||
| Streptococcus agalactiae isolates | 1311 | |||
| Streptococcus agalactiae BM132 | 1319 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Expression of this novel gene, named lsa(C), in S. agalactiae BM132 after cloning led to an increase in MICs of lincomycin (0.06 to 4 ug/ml), clindamycin (0.03 to 2 ug/ml), dalfopristin (2 to >32 ug/ml), and tiamulin (0.12 to 32 ug/ml), whereas no change in MICs of erythromycin (0.06 ug/ml), azithromycin (0.03 ug/ml), spiramycin (0.25 ug/ml), telithromycin (0.03 ug/ml), and quinupristin (8 ug/ml) was observed. The phenotype was renamed the LS(A)P phenotype on the basis of cross-resistance to lincosamides, streptogramins A, and pleuromutilins. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: 23S ribosomal RNA methyltransferase Erm34 (ERM34) | [6] | |||
| Resistant Disease | Bacillus clausii infection [ICD-11: 1C4Y.1] | |||
| Molecule Alteration | Methylation | Ribosomal methylation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Bacillus clausii ATCC 21536 | 79880 | ||
| Experiment for Molecule Alteration |
Cloning experiments and gene seqencing assay | |||
| Experiment for Drug Resistance |
Agar dilution assay | |||
| Mechanism Description | This pattern of resistance generally due to the presence of an erm gene encoding a ribosomal methylase. | |||
ICD-11: Circulatory system diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: ABC protein lsaC (lsaC-Unclear) | [7] | |||
| Resistant Disease | Streptococcus agalactiae infection [ICD-11: 1B21.2] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli TOP10 | 83333 | ||
| Staphylococcus aureus ATCC 29213 | 1280 | |||
| Streptococcus agalactiae UCN70 | 1311 | |||
| Streptococcus agalactiae isolates | 1311 | |||
| Streptococcus agalactiae BM132 | 1319 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Expression of this novel gene, named lsa(C), in S. agalactiae BM132 after cloning led to an increase in MICs of lincomycin (0.06 to 4 ug/ml), clindamycin (0.03 to 2 ug/ml), dalfopristin (2 to >32 ug/ml), and tiamulin (0.12 to 32 ug/ml), whereas no change in MICs of erythromycin (0.06 ug/ml), azithromycin (0.03 ug/ml), spiramycin (0.25 ug/ml), telithromycin (0.03 ug/ml), and quinupristin (8 ug/ml) was observed. The phenotype was renamed the LS(A)P phenotype on the basis of cross-resistance to lincosamides, streptogramins A, and pleuromutilins. | |||
ICD-12: Respiratory system diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: ABC protein lsaC (lsaC-Unclear) | [7] | |||
| Resistant Disease | Streptococcus agalactiae infection [ICD-11: 1B21.2] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli TOP10 | 83333 | ||
| Staphylococcus aureus ATCC 29213 | 1280 | |||
| Streptococcus agalactiae UCN70 | 1311 | |||
| Streptococcus agalactiae isolates | 1311 | |||
| Streptococcus agalactiae BM132 | 1319 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Expression of this novel gene, named lsa(C), in S. agalactiae BM132 after cloning led to an increase in MICs of lincomycin (0.06 to 4 ug/ml), clindamycin (0.03 to 2 ug/ml), dalfopristin (2 to >32 ug/ml), and tiamulin (0.12 to 32 ug/ml), whereas no change in MICs of erythromycin (0.06 ug/ml), azithromycin (0.03 ug/ml), spiramycin (0.25 ug/ml), telithromycin (0.03 ug/ml), and quinupristin (8 ug/ml) was observed. The phenotype was renamed the LS(A)P phenotype on the basis of cross-resistance to lincosamides, streptogramins A, and pleuromutilins. | |||
ICD-15: Musculoskeletal/connective-tissue diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: ABC protein lsaC (lsaC-Unclear) | [7] | |||
| Resistant Disease | Streptococcus agalactiae infection [ICD-11: 1B21.2] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli TOP10 | 83333 | ||
| Staphylococcus aureus ATCC 29213 | 1280 | |||
| Streptococcus agalactiae UCN70 | 1311 | |||
| Streptococcus agalactiae isolates | 1311 | |||
| Streptococcus agalactiae BM132 | 1319 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Expression of this novel gene, named lsa(C), in S. agalactiae BM132 after cloning led to an increase in MICs of lincomycin (0.06 to 4 ug/ml), clindamycin (0.03 to 2 ug/ml), dalfopristin (2 to >32 ug/ml), and tiamulin (0.12 to 32 ug/ml), whereas no change in MICs of erythromycin (0.06 ug/ml), azithromycin (0.03 ug/ml), spiramycin (0.25 ug/ml), telithromycin (0.03 ug/ml), and quinupristin (8 ug/ml) was observed. The phenotype was renamed the LS(A)P phenotype on the basis of cross-resistance to lincosamides, streptogramins A, and pleuromutilins. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: ABC protein lsaC (lsaC-Unclear) | [7] | |||
| Resistant Disease | Streptococcus agalactiae inection [ICD-11: FB84.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli TOP10 | 83333 | ||
| Staphylococcus aureus ATCC 29213 | 1280 | |||
| Streptococcus agalactiae UCN70 | 1311 | |||
| Streptococcus agalactiae isolates | 1311 | |||
| Streptococcus agalactiae BM132 | 1319 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Expression of this novel gene, named lsa(C), in S. agalactiae BM132 after cloning led to an increase in MICs of lincomycin (0.06 to 4 ug/ml), clindamycin (0.03 to 2 ug/ml), dalfopristin (2 to >32 ug/ml), and tiamulin (0.12 to 32 ug/ml), whereas no change in MICs of erythromycin (0.06 ug/ml), azithromycin (0.03 ug/ml), spiramycin (0.25 ug/ml), telithromycin (0.03 ug/ml), and quinupristin (8 ug/ml) was observed. The phenotype was renamed the LS(A)P phenotype on the basis of cross-resistance to lincosamides, streptogramins A, and pleuromutilins. | |||
ICD-16: Genitourinary system diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: ABC protein lsaC (lsaC-Unclear) | [7] | |||
| Resistant Disease | Klebsiella pneumoniae [ICD-11: CA40.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli TOP10 | 83333 | ||
| Staphylococcus aureus ATCC 29213 | 1280 | |||
| Streptococcus agalactiae UCN70 | 1311 | |||
| Streptococcus agalactiae isolates | 1311 | |||
| Streptococcus agalactiae BM132 | 1319 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Expression of this novel gene, named lsa(C), in S. agalactiae BM132 after cloning led to an increase in MICs of lincomycin (0.06 to 4 ug/ml), clindamycin (0.03 to 2 ug/ml), dalfopristin (2 to >32 ug/ml), and tiamulin (0.12 to 32 ug/ml), whereas no change in MICs of erythromycin (0.06 ug/ml), azithromycin (0.03 ug/ml), spiramycin (0.25 ug/ml), telithromycin (0.03 ug/ml), and quinupristin (8 ug/ml) was observed. The phenotype was renamed the LS(A)P phenotype on the basis of cross-resistance to lincosamides, streptogramins A, and pleuromutilins. | |||
ICD-21: Symptoms/clinical signs/unclassified clinical findings
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: ABC protein lsaC (lsaC-Unclear) | [7] | |||
| Resistant Disease | Streptococcus agalactiae infection [ICD-11: 1B21.2] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli TOP10 | 83333 | ||
| Staphylococcus aureus ATCC 29213 | 1280 | |||
| Streptococcus agalactiae UCN70 | 1311 | |||
| Streptococcus agalactiae isolates | 1311 | |||
| Streptococcus agalactiae BM132 | 1319 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Expression of this novel gene, named lsa(C), in S. agalactiae BM132 after cloning led to an increase in MICs of lincomycin (0.06 to 4 ug/ml), clindamycin (0.03 to 2 ug/ml), dalfopristin (2 to >32 ug/ml), and tiamulin (0.12 to 32 ug/ml), whereas no change in MICs of erythromycin (0.06 ug/ml), azithromycin (0.03 ug/ml), spiramycin (0.25 ug/ml), telithromycin (0.03 ug/ml), and quinupristin (8 ug/ml) was observed. The phenotype was renamed the LS(A)P phenotype on the basis of cross-resistance to lincosamides, streptogramins A, and pleuromutilins. | |||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
