Drug Information
Drug (ID: DG01264) and It's Reported Resistant Information
| Name |
LY2835219
|
||||
|---|---|---|---|---|---|
| Synonyms |
Abemaciclib; 1231929-97-7; LY2835219; LY2835219 free base; Verzenio; LY-2835219; UNII-60UAB198HK; N-(5-((4-ethylpiperazin-1-yl)methyl)pyridin-2-yl)-5-fluoro-4-(4-fluoro-1-isopropyl-2-methyl-1H-benzo[d]imidazol-6-yl)pyrimidin-2-amine; Abemaciclib (ly2835219); 60UAB198HK; N-[5-[(4-ethylpiperazin-1-yl)methyl]pyridin-2-yl]-5-fluoro-4-(7-fluoro-2-methyl-3-propan-2-ylbenzimidazol-5-yl)pyrimidin-2-amine; LY 2835219; HY-16297A; CS-1230; N-{5-[(4-ethylpiperazin-1-yl)methyl]pyridin-2-yl}-5-fluoro-4-(7-fluoro-3-isopropyl-2-methyl-1,3-benzodiazol-5-yl)pyrimidin-2-amine; N-{5-[(4-Ethylpiperazin-1-yl)methyl]pyridin-2-yl}-5-fluoro-4-[4-fluoro-2-methyl-1-(propan-2-yl)-1H-benzimidazol-6-yl]pyrimidin-2-amine; LY2835219 (free base); Abemaciclib [USAN:INN]; Abemaciclib,LY2835219; Verzenios; rimidin-2-amine; Verzenio (TN); 6ZV; N-{5-[(4-ethylpiperazin-1-yl)methyl]pyridin-2-yl}-5-fluoro-4-[4-fluoro-2-methyl-1-(propan-2-yl)-1H-benzimidazol-6-yl]py rimidin-2-amine; CDK4/6 dual inhibitor; LY2835210; Abemaciclib (JAN/USAN); GTPL7382; SCHEMBL2487229; CHEMBL3301610; DTXSID20673119; EX-A521; LY 2835219 (free base); C27H32F2N8; HMS3673I05; BCP13079; EX-A1588; 3798AH; BDBM50110183; MFCD22665744; NSC768073; NSC783671; s5716; ZINC72318121; 1231929-97-7, Verzenio,; AKOS025404907; LY2835219 free base (Abemaciclib); CCG-269750; DB12001; NSC-768073; NSC-783671; SB16476; NCGC00351599-02; NCGC00351599-06; 2-Pyrimidinamine, N-(5-((4-ethyl-1-piperazinyl)methyl)-2-pyridinyl)-5-fluoro-4-(4-fluoro-2-methyl-1-(1-methylethyl)-1H-benzimidazol-6-yl); 5-(4-ethylpiperazin-1-ylmethyl)pyridin-2-yl)-(5-fluoro-4-(7-fluoro-3-isopropyl-2-methyl-3H-benzimidazol-5-yl)pyrimidin-2-yl)amine; 5-(4-ethylpiperazin-1-ylmethyl)pyridin-2-yl)-(5-fluoro-4-(7-fluoro-3-isopropyl-2-methyl-3H-benzoimidazol-5-yl)pyrimidin-2-yl)amine; AC-30666; AS-10230; DA-33422; QC-11713; LY2835219 Ms salt, Abemaciclib Ms salt; FT-0700134; LY 2835210; A12989; D10688; J-690083; Q23901483; [5-(4-ethyl-piperazin-1-ylmethyl)-pyridin-2-yl]-[5-fluoro-4-(7-fluoro-3-isopropyl-2-methyl-3H-benzoimidazol-5-yl)-pyrimidin-2-yl]-amine; 2-Pyrimidinamine, N-[5-[(4-ethyl-1-piperazinyl)methyl]-2-pyridinyl]-5-fluoro-4-[4-fluoro-2-methyl-1-(1-methylethyl)-1H-benzimidazol-6-yl]-; 2-Pyrimidinamine,N-[5-[(4-ethyl-1-piperazinyl)methyl]-2-pyridinyl]-5-fluoro-4-[4-fluoro-2-methyl-1-(1-methylethyl)-1H-benzimidazol-6-yl]-; LY-2835219; ; ; N-[5-[(4-Ethylpiperazin-1-yl)methyl]pyridin-2-yl]-5-fluoro-4-(7-fluoro-2-methyl-3-propan-2-ylbenzimidazol-5-yl)pyrimidin-2-amine; N-{5-[(4-ethylpiperazin-1-yl)methyl]pyridin-2-yl}-5-fluoro-4-[4-fluoro-2-methyl-1-(propan-2-yl)-1H-benzimidazol-6-yl]py
Click to Show/Hide
|
||||
| Indication |
In total 2 Indication(s)
|
||||
| Structure |
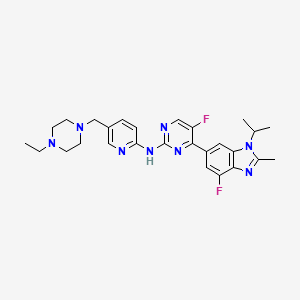
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Resistance Information Validated by in-vivo Model for This Drug
(1 diseases)
[2]
Disease(s) with Resistance Information Discovered by Cell Line Test for This Drug
(1 diseases)
[1]
|
||||
| Target | Cyclin-dependent kinase 4 (CDK4) | CDK4_HUMAN | [1] | ||
| Cyclin-dependent kinase 6 (CDK6) | CDK6_HUMAN | [1] | |||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C27H32F2N8
|
||||
| IsoSMILES |
CCN1CCN(CC1)CC2=CN=C(C=C2)NC3=NC=C(C(=N3)C4=CC5=C(C(=C4)F)N=C(N5C(C)C)C)F
|
||||
| InChI |
1S/C27H32F2N8/c1-5-35-8-10-36(11-9-35)16-19-6-7-24(30-14-19)33-27-31-15-22(29)25(34-27)20-12-21(28)26-23(13-20)37(17(2)3)18(4)32-26/h6-7,12-15,17H,5,8-11,16H2,1-4H3,(H,30,31,33,34)
|
||||
| InChIKey |
UZWDCWONPYILKI-UHFFFAOYSA-N
|
||||
| PubChem CID | |||||
| TTD Drug ID | |||||
| VARIDT ID | |||||
| INTEDE ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-02: Benign/in-situ/malignant neoplasm
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Key Molecule: GTPase KRas (KRAS) | [3] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Missense mutation | p.A146V (c.437C>T) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.70 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.18 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
0
|
G
-
M
M
T
T
E
E
Y
Y
K
K
L
L
V
V
V
V
V
V
10
|
G
G
A
A
G
G
G
G
V
V
G
G
K
K
S
S
A
A
L
L
20
|
T
T
I
I
Q
Q
L
L
I
I
Q
Q
N
N
H
H
F
F
V
V
30
|
D
D
E
E
Y
Y
D
D
P
P
T
T
I
I
E
E
D
D
S
S
40
|
Y
Y
R
R
K
K
Q
Q
V
V
V
V
I
I
D
D
G
G
E
E
50
|
T
T
C
C
L
L
L
L
D
D
I
I
L
L
D
D
T
T
A
A
60
|
G
G
Q
Q
E
E
E
E
Y
Y
S
S
A
A
M
M
R
R
D
D
70
|
Q
Q
Y
Y
M
M
R
R
T
T
G
G
E
E
G
G
F
F
L
L
80
|
C
C
V
V
F
F
A
A
I
I
N
N
N
N
T
T
K
K
S
S
90
|
F
F
E
E
D
D
I
I
H
H
H
H
Y
Y
R
R
E
E
Q
Q
100
|
I
I
K
K
R
R
V
V
K
K
D
D
S
S
E
E
D
D
V
V
110
|
P
P
M
M
V
V
L
L
V
V
G
G
N
N
K
K
C
C
D
D
120
|
L
L
P
P
S
S
R
R
T
T
V
V
D
D
T
T
K
K
Q
Q
130
|
A
A
Q
Q
D
D
L
L
A
A
R
R
S
S
Y
Y
G
G
I
I
140
|
P
P
F
F
I
I
E
E
T
T
S
S
A
V
K
K
T
T
R
R
150
|
Q
Q
R
G
V
V
E
D
D
D
A
A
F
F
Y
Y
T
T
L
L
160
|
V
V
R
R
E
E
I
I
R
R
Q
K
Y
H
R
K
L
E
K
K
170
|
K
M
I
S
S
K
K
D
E
G
E
K
K
K
T
K
P
K
G
K
180
|
C
K
V
S
K
K
I
T
K
K
K
S
C
-
I
-
I
-
M
-
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |||||||||
| Experiment for Molecule Alteration |
Rapid formalin-fixed assay; Paraffin-embedded sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Cell counting assay | ||||||||||||
| Key Molecule: Transcription activator BRG1 (BRG1) | [4] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Copy number loss | . |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | NCI-H1703 cells | Lung | Homo sapiens (Human) | CVCL_1490 | |||||||||
| NCI-H1299 cells | Lymph node | Homo sapiens (Human) | CVCL_0060 | ||||||||||
| In Vivo Model | Female YFP/SCID mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-blue assay | ||||||||||||
| Mechanism Description | SMARCA4/2 loss reduces cyclin D1 expression by a combination of restricting CCND1 chromatin accessibility and suppressing c-Jun, a transcription activator of CCND1. Reduced cyclin D1 in SMARCA4-deficient NSCLC causes sensitivities to CDK4/6 inhibitors, abemaciclib or palbociclib. | ||||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Cyclin-dependent kinase 6 (CDK6) | [1] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Mechanism Description | In some studies, CDK6 overexpression was reported to promote resistance to CDK4/6 inhibitors in preclinical models. Possible mechanisms how CDK6 amplification confers resistance to CDK4/6 inhibitor might be due to kinase-independent function of CDK6, which involves VEGF-A or p16. | |||
| Key Molecule: Cyclin-dependent kinase 4 (CDK4) | [1] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Mechanism Description | Various mechanisms, such as gene amplification, mutations and epigenetic alterations, serve to activate the cyclin D-CDK4/6-RB pathway. Overexpression of CDK4, which has been described in several cancers, may limit the efficacy of CDK4/6 inhibitors. | |||
| Key Molecule: Epithelial discoidin domain-containing receptor 1 (DDR1) | [5] | |||
| Resistant Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Molecule Alteration | Expressiom | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | TFAP2C-DDR1 signaling pathway | Regulation | N.A. | |
| In Vitro Model | MCF7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Drug Resistance |
IC50 assay | |||
| Mechanism Description | The comprehensive analyses revealed DDR1 as a potential factor implicated in mediating resistance to CDK4/6i. Specifically, DDR1 inhibition in combination with palbociclib exhibited remarkable synergistic effects, reducing cell survival signaling and promoting apoptosis in resistant cells. In-vivo xenograft model further validated the synergistic effects, showing a significant reduction in the resistant tumor growth. Exploration into DDR1 activation uncovered TFAP2C as a key transcription factor regulating DDR1 expression in palbociclib resistant cells and inhibition of TFAP2C re-sensitized resistant cells to palbociclib. Gene set enrichment analysis (GSEA) in the NeoPalAna trial demonstrated a significant enrichment of the TFAP2C-DDR1 gene set from patitens after palbociclib treatment, suggesting the possible activation of the TFAP2C-DDR1 axis following palbociclib exposure. | |||
| Key Molecule: Estrogen receptor alpha (ESR1) | [2] | |||
| Resistant Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Molecule Alteration | Mutantion | p.Y537N |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vivo Model | ESR1-WT breast cancer xenograft model; ESR1-WT mutant breast cancer xenograft model | Mus musculus | ||
| Experiment for Drug Resistance |
Tumor volume assay | |||
| Mechanism Description | Imlunestrant degraded ERalpha and decreased ERalpha-mediated gene expression both in vitro and in vivo. Cell proliferation and tumor growth in ESR1 wild-type (WT) and mutant models were significantly inhibited by imlunestrant. Combining imlunestrant with abemaciclib (CDK4/6 inhibitor), alpelisib (PI3K inhibitor), or everolimus (mTOR inhibitor) further enhanced tumor growth inhibition, regardless of ESR1 mutational status. | |||
|
|
||||
| Key Molecule: Histone deacetylase 1 (HDAC1) | [1] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Mechanism Description | Although the involvement of HDAC in resistance to CDK4/6 inhibitors is currently unknown, inhibition of HDAC may increase the efficacy of CDK4/6 inhibitors in CDK4/6 inhibitor-resistant cells by activating p21, resulting in cell cycle arrest at the G1 and G2/M phases, as demonstrated in CDK4/6 inhibitor-sensitive cells. | |||
| Key Molecule: E3 ubiquitin-protein ligase Mdm2 (MDM2) | [1] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Mechanism Description | Approximately 20%-30% of breast cancer patients show overexpression of MDM2, and this overexpression contributes particularly to the progression of HR-positive breast cancer. It is reported that CDK4/6 inhibitor-resistant cells have disrupted senescence pathways and insensitivity to the induction of senescence. Therefore, interruption of the senescence pathway by MDM2 in a p53-dependent manner may cause resistance to CDK4/6 inhibitors. | |||
| Key Molecule: Cyclin dependent kinase 7 (CDK7) | [1] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Mechanism Description | CDK7 is a cell cycle regulator. In addition, it also acts as a transcription factor, after complexation with cyclin H and MAT1. Increased expression of CDK7 is reported to confer resistance to CDK4/6 inhibitors. It acts as a CDK-activating kinase (CAK) and is involved in the G2/M phase by maintaining CDK1 and CDK2 activity. | |||
| Key Molecule: Cyclin-dependent kinase inhibitor 2A (CDKN2A) | [1] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Mechanism Description | Overexpression of p16 occurs during oncogenic stress, with or without the loss of RB. Loss of RB with concurrent p16 overexpression resulted in failure to respond to CDK4/6 inhibitors because of the absence of RB function. Alternatively, p16 overexpression in the presence of functional RB, also confers resistance to CDK4/6 inhibitors as a result of diminished CDK4, indicating depletion of a target of CDK4/6 inhibitors. Although the loss of RB and p16 overexpression seems to occur consequently together, further studies revealing the mechanistic association of RB loss and p16 overexpression might be beneficial in designing the strategies to overcome acquired resistance to CDK4/6 inhibitors. | |||
| Key Molecule: Mothers against decapentaplegic homolog 3 (SMAD3) | [1] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Mechanism Description | Smad3 is a component of the TGF-beta signaling pathway, having antiproliferative effects that contribute to G1 cell cycle arrest. From this perspective, it was demonstrated that the suppression of Smad3 was involved in mechanisms responsible for resistance to certain anticancer drugs, such as trastzumab. Furthermore, some evidences suggested that Smad3 may be correlated with resistance to CDK4/6 inhibitors. Mechanistically, Smad3 was reported to be suppressed through phosphorylation by the cyclin E-CDK2 or cyclin D1-CDK4/6 complexes. | |||
| Key Molecule: Fibroblast growth factor receptor (FGFR) | [1] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | FGFR signaling pathway | Activation | hsa01521 | |
| PI3K/AKT signaling pathway | Inhibition | hsa05235 | ||
| RAS/MEK/ERK signaling pathway | Activation | hsa04010 | ||
| Mechanism Description | The FGFR pathway is frequently activated in several types of cancer, including breast cancer. Of the five FGFRs, FGFR 1-4 have been reported to play an important role in cancer progression. Furthermore, FGFR1 and FGFR2 also appear to be associated with resistance to CDK4/6 inhibitors, as well as with endocrine resistance. Mechanistic investigation showed that FGFR1 amplification activated the PI3K/AKT and RAS/MEK/ERK signaling pathways in endocrine-resistant breast cancer cells. | |||
| Key Molecule: Transcription factor E2F2 (E2F2) | [1] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Mechanism Description | The overexpression of E2F causes the cell to circumvent CDK4/6 inhibition and rely upon signaling pathways other than the cyclin D-CDK4/6 axis for cell cycle progression. Further studies are required to explore the detailed mechanism of this escape pathway. Moreover, inhibition of proteins downstream of E2F, in concert with CDK4/6 inhibition, may increase the efficacy of CDK4/6 inhibitors, overcoming resistance. | |||
| Key Molecule: Retinoblastoma-like protein 1 (RBL1) | [1] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Mechanism Description | The tumor suppressor RB is the aforementioned key checkpoint in the cell cycle. As the primary target of CDK4/6 inhibitors, RB was considered to be one of the most important biomarkers of sensitivity to therapy. In this scenario, loss of RB is the evident cause of resistance to CDK4/6 inhibitors, and various preclinical studies have supported this hypothesis. In addition, some preclinical and clinical studies have also reported that mutations in RB are responsible for the resistance. A study using glioblastoma xenograft cells, a missense mutation in exon 2 of RB(A193T) resulted in resistance to CDK4/6 inhibitors. | |||
| Key Molecule: Retinoblastoma-like protein 1 (RBL1) | [1] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Missense mutation | p.A193T |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Mechanism Description | The tumor suppressor RB is the aforementioned key checkpoint in the cell cycle. As the primary target of CDK4/6 inhibitors, RB was considered to be one of the most important biomarkers of sensitivity to therapy. In this scenario, loss of RB is the evident cause of resistance to CDK4/6 inhibitors, and various preclinical studies have supported this hypothesis. In addition, some preclinical and clinical studies have also reported that mutations in RB are responsible for the resistance. A study using glioblastoma xenograft cells, a missense mutation in exon 2 of RB(A193T) resulted in resistance to CDK4/6 inhibitors. | |||
| Key Molecule: Fizzy and cell division cycle 20 related 1 (FZR1) | [1] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Mechanism Description | The ubiquitin (Ub) ligase APC/C, which is activated via the co-activator FZR1, interacts with RB during the G1 phase of cell cycle. More notably, APC/CFZR1 complex degrades S-phase kinase associated protein 2 (SKP2), which inhibits p27, natural CDK inhibitors, resulting in decreased CDK2, CDK4 and CDK6. Accordingly, the loss of FZR1 results in uncontrolled cell cycle progression from G1 to S phase. | |||
| Key Molecule: Wee1-like protein kinase (WEE1) | [1] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Mechanism Description | WEE1 plays an important role in the G2/M checkpoint. It inhibits the entry of DNA-damaged cells into mitosis in coordination with CDK1. Though the involvement of WEE1 in inducing resistance to CDK4/6 inhibitors is unknown, inhibition of WEE1 has been shown to increase sensitivity to CDK4/6 inhibitors in resistant cells. As WEE1 is associated with a resistant phenotype in preclinical models, targeting the G2/M phase via the inhibition of WEE1 in combination with CDK4/6 inhibition could be a therapeutic option in overcoming resistance. | |||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
