Drug Information
Drug (ID: DG00186) and It's Reported Resistant Information
| Name |
Rifampin
|
||||
|---|---|---|---|---|---|
| Synonyms |
Abrifam; Archidyn; Arficin; Arzide; Benemicin; Benemycin; Dipicin; Doloresum; Eremfat; Famcin; Fenampicin; RFP; RMP; Ramp; Rifa; Rifadin; Rifadine; Rifagen; Rifaldazin; Rifaldazine; Rifaldin; Rifam; Rifamor; Rifampicin; Rifampicina; Rifampicine; Rifampicinum; Rifamsolin; Rifaprodin; Rifcin; Rifinah; Rifobac; Rifoldin; Rifoldine; Riforal; Rimactan; Rimactane; Rimactazid; Rimactizid; Rimazid; Rimycin; Sinerdol; Tubocin; Rifamicin AMP; Rifampicin SV; Rifampicine [French]; Rifampin [USAN]; Rifamycin AMP; Ba 41166; AZT + Rifampin; BA-41166E; Ba 41166/E; DRG-0109; Dione 21-acetate; L-5103; L-5103 Lepetit; Piperine & Rifampicin; R-Cin; R/AMP; Reserpine & Rifampicin; Rifadin (TN); Rifadin I.V; Rifampicin & EEP; Rifampicin & Propolis; Rifampicina [INN-Spanish]; Rifampicinum [INN-Latin]; Rifampin (USP); Rimactan (TN); Rimactane (TN); Rimycin (TN); Sinerdol (TN); Tubocin (TN); Rifadin I.V.; Rifampicin (JP15/INN); Rifampicin[INN:BAN:JAN]; Rifadin, Rimactane, Rifampicin, Rifampin; 1-b]furan-21-yl acetate; 3-(((4-Methyl-1-piperazinyl)imino)-methyl)rifamycin; 3-(((4-Methyl-1-piperazinyl)imino)methyl)rifamycin SV; 3-(4-Methylpiperazinyliminomethyl)-rifamycin SV; 3-(4-Methylpiperazinyliminomethyl)rifamycin SV; 3-([(4-Methyl-1-piperazinyl)imino]methyl)rifamycin SV; 3-[(4-Methyl-1-piperazinyl)iminomethyl]rifamycin SV; 3-[[(4-Methyl-1-piperazinyl)imino]-methyl]rifamycin; 8-(((4-Methyl-1-piperazinyl)imino)methyl)rifamycin SV; 8-(4-Methylpiperazinyliminomethyl) rifamycin SV; 8-[[(4-Methyl-1-piperazinyl)imino[methyl]rifamycin; 8-[[(4-Methyl-1-piperazinyl)imino]methyl]rifamycin sv; 8-[[(4-Methylpiperazinyl)imino]methyl]rifamycin sv; 8CI)
Click to Show/Hide
|
||||
| Indication |
In total 1 Indication(s)
|
||||
| Structure |
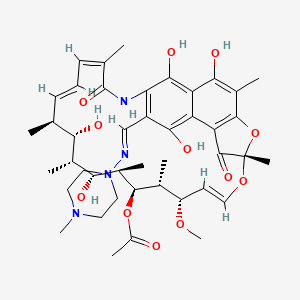
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Clinically Reported Resistance for This Drug
(13 diseases)
[6]
[10]
[11]
[12]
[13]
[14]
[15]
[16]
[18]
[19]
Disease(s) with Resistance Information Discovered by Cell Line Test for This Drug
(2 diseases)
[5]
[17]
Disease(s) with Resistance Information Validated by in-vivo Model for This Drug
(2 diseases)
[20]
[21]
|
||||
| Target | Bacterial RNA polymerase switch region (Bact RNAP-SR) | NOUNIPROTAC | [1] | ||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C43H58N4O12
|
||||
| IsoSMILES |
C[C@H]1/C=C/C=C(\\C(=O)NC2=C(C(=C3C(=C2O)C(=C(C4=C3C(=O)[C@](O4)(O/C=C/[C@@H]([C@H]([C@H]([C@@H]([C@@H]([C@@H]([C@H]1O)C)O)C)OC(=O)C)C)OC)C)C)O)O)/C=N/N5CCN(CC5)C)/C
|
||||
| InChI |
1S/C43H58N4O12/c1-21-12-11-13-22(2)42(55)45-33-28(20-44-47-17-15-46(9)16-18-47)37(52)30-31(38(33)53)36(51)26(6)40-32(30)41(54)43(8,59-40)57-19-14-29(56-10)23(3)39(58-27(7)48)25(5)35(50)24(4)34(21)49/h11-14,19-21,23-25,29,34-35,39,49-53H,15-18H2,1-10H3,(H,45,55)/b12-11+,19-14+,22-13-,44-20+/t21-,23+,24+,25+,29-,34-,35+,39+,43-/m0/s1
|
||||
| InChIKey |
JQXXHWHPUNPDRT-WLSIYKJHSA-N
|
||||
| PubChem CID | |||||
| ChEBI ID | |||||
| TTD Drug ID | |||||
| VARIDT ID | |||||
| INTEDE ID | |||||
| DrugBank ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-01: Infectious/parasitic diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Rifampin phosphotransferase (RPHB) | [22] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Paenibacillus sp. LC231 | 1120679 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | RphB inactivates rifampin by Phosphorylation. | |||
| Key Molecule: rgt1438 (Unclear) | [20] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Streptomyces albus J1074 | 457425 | ||
| Streptomyces speibonae WAC1438 | 195801 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | Rgt1438R encode a rifampin-inactivating glycosyltransferase,as a rifampin resistance determinant from WAC1438 capable of inactivating an assortment of rifamycins. | |||
| Key Molecule: Rifampin monooxygenase (IRI) | [23] | |||
| Resistant Disease | Rhodococcus equi infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli strain MM294 | 562 | ||
| Rhodococcus equi strain ATCC 14887 | 43767 | |||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Monitored by zones of inhibition assay | |||
| Mechanism Description | The original 8-kb clone and all subclones with the intact iri gene conferred similar 25-fold increases in rifampin resistance in rhodococcal strain Ri8. Clones growing on rifampin-containing selective plates all possessed an insert of about 8 kb, and retransformation into strain Ri8 demonstrated that this segment of DNA increased the rifampin MIC about 25-fold and conferred the ability to inactivate the antibiotic: rifampin at a concentration of 20 mg/ml was completely inactivated in about 6 h (as monitored by zones of inhibition on plates spread with a tester strain). inactivation gene cloned from the R.equi type strain, ATCC 14887, can confer a 10-fold increase in resistance to rifampin in E.coli as well as a 25-fold increase in Rhodococcus. | |||
|
|
||||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [3], [4] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | c.ins1593C |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli MG1655 | 511145 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | Frameshift mutations have been reported in rpoB, an essential gene encoding the beta-subunit of RNA polymerase, in rifampicin-resistant clinical isolates of Mycobacterium tuberculosis. Escherichia coli with a +1-nt frameshift mutation centrally located in rpoB is viable and highly resistant to rifampicin. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [21] | |||
| Resistant Disease | Clostridium difficile infection [ICD-11: 1A04.0] | |||
| Molecule Alteration | Mutation | p.R505K |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| Mechanism Description | RIFs (rifampicin and rifaximin) have recently been used as another option for CDI treatment. Nevertheless, the resistance to RIFs in C. difficile has been reported. These drugs target on a DNA-dependent RNA polymerase (RNAP), resulting in the extension of short transcript blockage. Point mutations within the rpoB gene encoding for beta-subunit of RNAP cause resistance to RIFs. Among identified amino acid substitutions, the R505K substitution has been mostly evident to promote the high level of resistance. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: DNA-directed RNA polymerase beta-subunit (rpoB) | [24] | |||
| Resistant Disease | Tuberculosis [ICD-11: 1B10.0] | |||
| Molecule Alteration | Mutations | Q24K+L28M+R30E+A92K |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | THP-1 cells | monocytic | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Microarray assay | |||
| Experiment for Drug Resistance |
Functional enrichment assay | |||
| Mechanism Description | The overexpression of many interferon-stimulated genes (ISGs) in cells infected with the isoniazid-resistant strain, compared to the rifampin-resistant and the drug-sensitive strains. | |||
| Key Molecule: Outer membrane protein A (OmpA) | [17] | |||
| Resistant Disease | Tuberculosis [ICD-11: 1B10.0] | |||
| Molecule Alteration | Expressiom | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Mycobacterium tuberculosis | 1773 | ||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | These results support the model that the roles of OmpA as a porin protein overexpressing in mycobacteria can increase the hydrophilic ability of the cell wall which can facilitate the streptomycin uptakes and increase the mycobacteria's sensitivity to aminoglycosides. | |||
|
|
||||
| Key Molecule: Enoyl-[acyl-carrier-protein] reductase [NADH] (INHA) | [16] | |||
| Resistant Disease | Tuberculosis [ICD-11: 1B10.0] | |||
| Molecule Alteration | Mutation | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis H37Rv | 83332 | ||
| Mycobacterium tuberculosis isolates | 1773 | |||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Mechanism Description | Monoresistance to rifampicin and isoniazid was found in 11% (95% CI: 0.077-0.150; p, 0.087) and 8.5% (95% CI: 0.056-0.123; p, 0.692) of all the patients, respectively. Resistance to RIF and INH among newly diagnosed patients was 10.2% and 8.6%, while among previously treated patients, resistance to RIF and INH was 23.5% and 5.9% respectively. Furthermore, 4.9% of the samples from newly diagnosed with INH monoresistance, were found to have mutations in the InhA region while 8.6% had mutations in the katG region, a condition that can lead to phenotypic isoniazid drug resistance. | |||
|
|
||||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [16] | |||
| Resistant Disease | Tuberculosis [ICD-11: 1B10.0] | |||
| Molecule Alteration | Mutation | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis H37Rv | 83332 | ||
| Mycobacterium tuberculosis isolates | 1773 | |||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Mechanism Description | Monoresistance to rifampicin and isoniazid was found in 11% (95% CI: 0.077-0.150; p, 0.087) and 8.5% (95% CI: 0.056-0.123; p, 0.692) of all the patients, respectively. Resistance to RIF and INH among newly diagnosed patients was 10.2% and 8.6%, while among previously treated patients, resistance to RIF and INH was 23.5% and 5.9% respectively. Furthermore, 4.9% of the samples from newly diagnosed with INH monoresistance, were found to have mutations in the InhA region while 8.6% had mutations in the katG region, a condition that can lead to phenotypic isoniazid drug resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta' (RPOC) | [25] | |||
| Resistant Disease | Tuberculosis [ICD-11: 1B10.0] | |||
| Molecule Alteration | Mutation | Q24K+L28M+R30E+A92K |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Experiment for Molecule Alteration |
GeneSeq assay; Bioinformatics assay | |||
| Mechanism Description | Out of total 112 mycobacterial positive cultures, five?M. bovis?were isolated and underwent WGS. All sequenced strains belonged to?Mycobacterium tuberculosis var bovis, spoligotype BOV_1; BOV_11. Resistance gene mutations were determined in 100% of strains to pyrazinamide (pncA?and?rpsA), isoniazid (KatG?and?ahpC), ethambutol (embB,?embC,?embR?and?ubiA), streptomycin (rpsl) and fluoroquinolones (gyrA?and?gyrB). Rifampin (rpoB?and?rpoC) and delamanid (fbiC) resistance genes were found in 80% of strains. The major represented virulence classes were the secretion system, cell surface components and regulation system. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [25] | |||
| Resistant Disease | Tuberculosis [ICD-11: 1B10.0] | |||
| Molecule Alteration | Mutation | R173C |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Experiment for Molecule Alteration |
GeneSeq assay; Bioinformatics assay | |||
| Mechanism Description | Out of total 112 mycobacterial positive cultures, five?M. bovis?were isolated and underwent WGS. All sequenced strains belonged to?Mycobacterium tuberculosis var bovis, spoligotype BOV_1; BOV_11. Resistance gene mutations were determined in 100% of strains to pyrazinamide (pncA?and?rpsA), isoniazid (KatG?and?ahpC), ethambutol (embB,?embC,?embR?and?ubiA), streptomycin (rpsl) and fluoroquinolones (gyrA?and?gyrB). Rifampin (rpoB?and?rpoC) and delamanid (fbiC) resistance genes were found in 80% of strains. The major represented virulence classes were the secretion system, cell surface components and regulation system. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Dihydrofolate reductase/DNA-directed RNA polymerase subunit beta (DHFR/RPOB) | [11] | |||
| Resistant Disease | Leprosy [ICD-11: 1B20.0] | |||
| Molecule Alteration | Missense mutation | folP p.P55L+poB p.S531L |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium leprae isolates | 1769 | ||
| In Vivo Model | Footpad granuloma from M. leprae-infected nude mice model | Mus musculus | ||
| Experiment for Molecule Alteration |
PCR and single-stranded conformational polymorphism (SSCP) assay | |||
| Experiment for Drug Resistance |
Mouse footpad assay | |||
| Mechanism Description | The mutations genes reported in this study have been demonstrated to be responsible for drug resistance by mouse footpad assay. | |||
| Key Molecule: Dihydrofolate reductase/DNA-directed RNA polymerase subunit beta (DHFR/RPOB) | [11] | |||
| Resistant Disease | Leprosy [ICD-11: 1B20.0] | |||
| Molecule Alteration | Missense mutation | folP p.P55S+rpoB p.S531L+rpoB p.V547I |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium leprae isolates | 1769 | ||
| In Vivo Model | Footpad granuloma from M. leprae-infected nude mice model | Mus musculus | ||
| Experiment for Molecule Alteration |
PCR and single-stranded conformational polymorphism (SSCP) assay | |||
| Experiment for Drug Resistance |
Mouse footpad assay | |||
| Mechanism Description | The mutations genes reported in this study have been demonstrated to be responsible for drug resistance by mouse footpad assay. | |||
| Key Molecule: Dihydrofolate reductase/DNA gyrase subunit A/DNA gyrase subunit B (DHFR/GYRA/GYRB) | [11] | |||
| Resistant Disease | Leprosy [ICD-11: 1B20.0] | |||
| Molecule Alteration | Missense mutation | folP p.P55L+gyrA p.A91V+gyrB p.A91V |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium leprae isolates | 1769 | ||
| In Vivo Model | Footpad granuloma from M. leprae-infected nude mice model | Mus musculus | ||
| Experiment for Molecule Alteration |
PCR and single-stranded conformational polymorphism (SSCP) assay | |||
| Experiment for Drug Resistance |
Mouse footpad assay | |||
| Mechanism Description | The mutations genes reported in this study have been demonstrated to be responsible for drug resistance by mouse footpad assay. | |||
| Key Molecule: Dihydrofolate reductase/DNA gyrase subunit A/DNA gyrase subunit B (DHFR/GYRA/GYRB) | [11] | |||
| Resistant Disease | Leprosy [ICD-11: 1B20.0] | |||
| Molecule Alteration | Missense mutation | folP p.P55L+gyrA p.D205N+gyrB p.D205N |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium leprae isolates | 1769 | ||
| In Vivo Model | Footpad granuloma from M. leprae-infected nude mice model | Mus musculus | ||
| Experiment for Molecule Alteration |
PCR and single-stranded conformational polymorphism (SSCP) assay | |||
| Experiment for Drug Resistance |
Mouse footpad assay | |||
| Mechanism Description | The mutations genes reported in this study have been demonstrated to be responsible for drug resistance by mouse footpad assay. | |||
|
|
||||
| Key Molecule: Dihydrofolate reductase (DHFR) | [11] | |||
| Resistant Disease | Leprosy [ICD-11: 1B20.0] | |||
| Molecule Alteration | Missense mutation | p.T53A |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium leprae isolates | 1769 | ||
| In Vivo Model | Footpad granuloma from M. leprae-infected nude mice model | Mus musculus | ||
| Experiment for Molecule Alteration |
PCR and single-stranded conformational polymorphism (SSCP) assay | |||
| Experiment for Drug Resistance |
Mouse footpad assay | |||
| Mechanism Description | The mutations genes reported in this study have been demonstrated to be responsible for drug resistance by mouse footpad assay. | |||
| Key Molecule: Dihydrofolate reductase (DHFR) | [11] | |||
| Resistant Disease | Leprosy [ICD-11: 1B20.0] | |||
| Molecule Alteration | Missense mutation | p.P55R |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium leprae isolates | 1769 | ||
| In Vivo Model | Footpad granuloma from M. leprae-infected nude mice model | Mus musculus | ||
| Experiment for Molecule Alteration |
PCR and single-stranded conformational polymorphism (SSCP) assay | |||
| Experiment for Drug Resistance |
Mouse footpad assay | |||
| Mechanism Description | The mutations genes reported in this study have been demonstrated to be responsible for drug resistance by mouse footpad assay. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Multidrug efflux pump Tap (TAP) | [1], [2] | |||
| Resistant Disease | Mycobacterium tuberculosis infection [ICD-11: 1B2Z.5] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis H37Rv | 83332 | ||
| Mycobacterium tuberculosis ICC154 | 1773 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | One mechanism proposed for drug resistance in Mycobacterium tuberculosis (MTB) is by efflux of the drugs by membrane located pumps.Mycobacterium tuberculosis isolate with a distinct genomic identity overexpresses a tap-like efflux pump,which confers resistance to Rifampin and Ofloxacin. | |||
| Key Molecule: Multidrug efflux pump Tap (TAP) | [1], [2] | |||
| Resistant Disease | Mycobacterium fortuitum infection [ICD-11: 1B2Z.2] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis H37Rv | 83332 | ||
| Mycobacterium tuberculosis ICC154 | 1773 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | One mechanism proposed for drug resistance in Mycobacterium tuberculosis (MTB) is by efflux of the drugs by membrane located pumps.Mycobacterium tuberculosis isolate with a distinct genomic identity overexpresses a tap-like efflux pump,which confers resistance to Rifampin and Ofloxacin. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [15] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Missense mutation | p.H481N |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Staphylococcus aureus strain T109 | 1280 | ||
| Staphylococcus aureus strain T112 | 1280 | |||
| Staphylococcus aureus strain T113 | 1280 | |||
| Staphylococcus aureus strain T115 | 1280 | |||
| Staphylococcus aureus strain T118 | 1280 | |||
| Staphylococcus aureus strain T124 | 1280 | |||
| Staphylococcus aureus strain T161 | 1280 | |||
| Staphylococcus aureus strain T166 | 1280 | |||
| Staphylococcus aureus strain T20 | 1280 | |||
| Staphylococcus aureus strain T211 | 1280 | |||
| Staphylococcus aureus strain T212 | 1280 | |||
| Staphylococcus aureus strain T23 | 1280 | |||
| Staphylococcus aureus strain T236 | 1280 | |||
| Staphylococcus aureus strain T23aa | 1280 | |||
| Staphylococcus aureus strain T23aac | 1280 | |||
| Staphylococcus aureus strain T23bb | 1280 | |||
| Staphylococcus aureus strain T248 | 1280 | |||
| Staphylococcus aureus strain T249 | 1280 | |||
| Staphylococcus aureus strain T25 | 1280 | |||
| Staphylococcus aureus strain T250 | 1280 | |||
| Staphylococcus aureus strain T262 | 1280 | |||
| Staphylococcus aureus strain T264 | 1280 | |||
| Staphylococcus aureus strain T295 | 1280 | |||
| Staphylococcus aureus strain T296 | 1280 | |||
| Staphylococcus aureus strain T297 | 1280 | |||
| Staphylococcus aureus strain T36 | 1280 | |||
| Staphylococcus aureus strain T38 | 1280 | |||
| Staphylococcus aureus strain T382 | 1280 | |||
| Staphylococcus aureus strain T38aa | 1280 | |||
| Staphylococcus aureus strain T38bb | 1280 | |||
| Staphylococcus aureus strain T397 | 1280 | |||
| Staphylococcus aureus strain T398 | 1280 | |||
| Staphylococcus aureus strain T399 | 1280 | |||
| Staphylococcus aureus strain T4 | 1280 | |||
| Staphylococcus aureus strain T400 | 1280 | |||
| Staphylococcus aureus strain T401 | 1280 | |||
| Staphylococcus aureus strain T402 | 1280 | |||
| Staphylococcus aureus strain T403 | 1280 | |||
| Staphylococcus aureus strain T404 | 1280 | |||
| Staphylococcus aureus strain T46 | 1280 | |||
| Staphylococcus aureus strain T59 | 1280 | |||
| Staphylococcus aureus strain T66 | 1280 | |||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | Twelve mutational changes at 10 positions were identified, with 473Ala-Thr representing a new mutation site. New amino acid substitutions, 465Gln-Arg, 466Leu-Ser, 468Gln-Lys, and 477Ala-Thr in cluster I and 527Ile-Met and 529Ser-Leu in cluster II, were described, thereby emphasizing the high variability of these amino acid positions. Codon 481 was mutated on 32 separate occasions, which indicates a central role of this amino acid. All in vivo isolates that demonstrated two or three amino acid changes exhibited high-level resistance. Interestingly enough, all of these isolates showed the mutational change 481His-Asn, which is capable of conferring low-level resistance on its own, thereby indicating a two-step resistance mechanism in vivo to high-level resistance within these isolates. High-level resistance in vivo, however, was not demonstrated to occur through multiple mutations alone. The single amino acid substitution 468Gln-Lys also causes high-level resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [15] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Missense mutation | p.A473T |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Staphylococcus aureus strain T109 | 1280 | ||
| Staphylococcus aureus strain T112 | 1280 | |||
| Staphylococcus aureus strain T113 | 1280 | |||
| Staphylococcus aureus strain T115 | 1280 | |||
| Staphylococcus aureus strain T118 | 1280 | |||
| Staphylococcus aureus strain T124 | 1280 | |||
| Staphylococcus aureus strain T161 | 1280 | |||
| Staphylococcus aureus strain T166 | 1280 | |||
| Staphylococcus aureus strain T20 | 1280 | |||
| Staphylococcus aureus strain T211 | 1280 | |||
| Staphylococcus aureus strain T212 | 1280 | |||
| Staphylococcus aureus strain T23 | 1280 | |||
| Staphylococcus aureus strain T236 | 1280 | |||
| Staphylococcus aureus strain T23aa | 1280 | |||
| Staphylococcus aureus strain T23aac | 1280 | |||
| Staphylococcus aureus strain T23bb | 1280 | |||
| Staphylococcus aureus strain T248 | 1280 | |||
| Staphylococcus aureus strain T249 | 1280 | |||
| Staphylococcus aureus strain T25 | 1280 | |||
| Staphylococcus aureus strain T250 | 1280 | |||
| Staphylococcus aureus strain T262 | 1280 | |||
| Staphylococcus aureus strain T264 | 1280 | |||
| Staphylococcus aureus strain T295 | 1280 | |||
| Staphylococcus aureus strain T296 | 1280 | |||
| Staphylococcus aureus strain T297 | 1280 | |||
| Staphylococcus aureus strain T36 | 1280 | |||
| Staphylococcus aureus strain T38 | 1280 | |||
| Staphylococcus aureus strain T382 | 1280 | |||
| Staphylococcus aureus strain T38aa | 1280 | |||
| Staphylococcus aureus strain T38bb | 1280 | |||
| Staphylococcus aureus strain T397 | 1280 | |||
| Staphylococcus aureus strain T398 | 1280 | |||
| Staphylococcus aureus strain T399 | 1280 | |||
| Staphylococcus aureus strain T4 | 1280 | |||
| Staphylococcus aureus strain T400 | 1280 | |||
| Staphylococcus aureus strain T401 | 1280 | |||
| Staphylococcus aureus strain T402 | 1280 | |||
| Staphylococcus aureus strain T403 | 1280 | |||
| Staphylococcus aureus strain T404 | 1280 | |||
| Staphylococcus aureus strain T46 | 1280 | |||
| Staphylococcus aureus strain T59 | 1280 | |||
| Staphylococcus aureus strain T66 | 1280 | |||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | Twelve mutational changes at 10 positions were identified, with 473Ala-Thr representing a new mutation site. New amino acid substitutions, 465Gln-Arg, 466Leu-Ser, 468Gln-Lys, and 477Ala-Thr in cluster I and 527Ile-Met and 529Ser-Leu in cluster II, were described, thereby emphasizing the high variability of these amino acid positions. Codon 481 was mutated on 32 separate occasions, which indicates a central role of this amino acid. All in vivo isolates that demonstrated two or three amino acid changes exhibited high-level resistance. Interestingly enough, all of these isolates showed the mutational change 481His-Asn, which is capable of conferring low-level resistance on its own, thereby indicating a two-step resistance mechanism in vivo to high-level resistance within these isolates. High-level resistance in vivo, however, was not demonstrated to occur through multiple mutations alone. The single amino acid substitution 468Gln-Lys also causes high-level resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [15] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Missense mutation | p.Q465R |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Staphylococcus aureus strain T109 | 1280 | ||
| Staphylococcus aureus strain T112 | 1280 | |||
| Staphylococcus aureus strain T113 | 1280 | |||
| Staphylococcus aureus strain T115 | 1280 | |||
| Staphylococcus aureus strain T118 | 1280 | |||
| Staphylococcus aureus strain T124 | 1280 | |||
| Staphylococcus aureus strain T161 | 1280 | |||
| Staphylococcus aureus strain T166 | 1280 | |||
| Staphylococcus aureus strain T20 | 1280 | |||
| Staphylococcus aureus strain T211 | 1280 | |||
| Staphylococcus aureus strain T212 | 1280 | |||
| Staphylococcus aureus strain T23 | 1280 | |||
| Staphylococcus aureus strain T236 | 1280 | |||
| Staphylococcus aureus strain T23aa | 1280 | |||
| Staphylococcus aureus strain T23aac | 1280 | |||
| Staphylococcus aureus strain T23bb | 1280 | |||
| Staphylococcus aureus strain T248 | 1280 | |||
| Staphylococcus aureus strain T249 | 1280 | |||
| Staphylococcus aureus strain T25 | 1280 | |||
| Staphylococcus aureus strain T250 | 1280 | |||
| Staphylococcus aureus strain T262 | 1280 | |||
| Staphylococcus aureus strain T264 | 1280 | |||
| Staphylococcus aureus strain T295 | 1280 | |||
| Staphylococcus aureus strain T296 | 1280 | |||
| Staphylococcus aureus strain T297 | 1280 | |||
| Staphylococcus aureus strain T36 | 1280 | |||
| Staphylococcus aureus strain T38 | 1280 | |||
| Staphylococcus aureus strain T382 | 1280 | |||
| Staphylococcus aureus strain T38aa | 1280 | |||
| Staphylococcus aureus strain T38bb | 1280 | |||
| Staphylococcus aureus strain T397 | 1280 | |||
| Staphylococcus aureus strain T398 | 1280 | |||
| Staphylococcus aureus strain T399 | 1280 | |||
| Staphylococcus aureus strain T4 | 1280 | |||
| Staphylococcus aureus strain T400 | 1280 | |||
| Staphylococcus aureus strain T401 | 1280 | |||
| Staphylococcus aureus strain T402 | 1280 | |||
| Staphylococcus aureus strain T403 | 1280 | |||
| Staphylococcus aureus strain T404 | 1280 | |||
| Staphylococcus aureus strain T46 | 1280 | |||
| Staphylococcus aureus strain T59 | 1280 | |||
| Staphylococcus aureus strain T66 | 1280 | |||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | Twelve mutational changes at 10 positions were identified, with 473Ala-Thr representing a new mutation site. New amino acid substitutions, 465Gln-Arg, 466Leu-Ser, 468Gln-Lys, and 477Ala-Thr in cluster I and 527Ile-Met and 529Ser-Leu in cluster II, were described, thereby emphasizing the high variability of these amino acid positions. Codon 481 was mutated on 32 separate occasions, which indicates a central role of this amino acid. All in vivo isolates that demonstrated two or three amino acid changes exhibited high-level resistance. Interestingly enough, all of these isolates showed the mutational change 481His-Asn, which is capable of conferring low-level resistance on its own, thereby indicating a two-step resistance mechanism in vivo to high-level resistance within these isolates. High-level resistance in vivo, however, was not demonstrated to occur through multiple mutations alone. The single amino acid substitution 468Gln-Lys also causes high-level resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [15] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Missense mutation | p.L466S |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Staphylococcus aureus strain T109 | 1280 | ||
| Staphylococcus aureus strain T112 | 1280 | |||
| Staphylococcus aureus strain T113 | 1280 | |||
| Staphylococcus aureus strain T115 | 1280 | |||
| Staphylococcus aureus strain T118 | 1280 | |||
| Staphylococcus aureus strain T124 | 1280 | |||
| Staphylococcus aureus strain T161 | 1280 | |||
| Staphylococcus aureus strain T166 | 1280 | |||
| Staphylococcus aureus strain T20 | 1280 | |||
| Staphylococcus aureus strain T211 | 1280 | |||
| Staphylococcus aureus strain T212 | 1280 | |||
| Staphylococcus aureus strain T23 | 1280 | |||
| Staphylococcus aureus strain T236 | 1280 | |||
| Staphylococcus aureus strain T23aa | 1280 | |||
| Staphylococcus aureus strain T23aac | 1280 | |||
| Staphylococcus aureus strain T23bb | 1280 | |||
| Staphylococcus aureus strain T248 | 1280 | |||
| Staphylococcus aureus strain T249 | 1280 | |||
| Staphylococcus aureus strain T25 | 1280 | |||
| Staphylococcus aureus strain T250 | 1280 | |||
| Staphylococcus aureus strain T262 | 1280 | |||
| Staphylococcus aureus strain T264 | 1280 | |||
| Staphylococcus aureus strain T295 | 1280 | |||
| Staphylococcus aureus strain T296 | 1280 | |||
| Staphylococcus aureus strain T297 | 1280 | |||
| Staphylococcus aureus strain T36 | 1280 | |||
| Staphylococcus aureus strain T38 | 1280 | |||
| Staphylococcus aureus strain T382 | 1280 | |||
| Staphylococcus aureus strain T38aa | 1280 | |||
| Staphylococcus aureus strain T38bb | 1280 | |||
| Staphylococcus aureus strain T397 | 1280 | |||
| Staphylococcus aureus strain T398 | 1280 | |||
| Staphylococcus aureus strain T399 | 1280 | |||
| Staphylococcus aureus strain T4 | 1280 | |||
| Staphylococcus aureus strain T400 | 1280 | |||
| Staphylococcus aureus strain T401 | 1280 | |||
| Staphylococcus aureus strain T402 | 1280 | |||
| Staphylococcus aureus strain T403 | 1280 | |||
| Staphylococcus aureus strain T404 | 1280 | |||
| Staphylococcus aureus strain T46 | 1280 | |||
| Staphylococcus aureus strain T59 | 1280 | |||
| Staphylococcus aureus strain T66 | 1280 | |||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | Twelve mutational changes at 10 positions were identified, with 473Ala-Thr representing a new mutation site. New amino acid substitutions, 465Gln-Arg, 466Leu-Ser, 468Gln-Lys, and 477Ala-Thr in cluster I and 527Ile-Met and 529Ser-Leu in cluster II, were described, thereby emphasizing the high variability of these amino acid positions. Codon 481 was mutated on 32 separate occasions, which indicates a central role of this amino acid. All in vivo isolates that demonstrated two or three amino acid changes exhibited high-level resistance. Interestingly enough, all of these isolates showed the mutational change 481His-Asn, which is capable of conferring low-level resistance on its own, thereby indicating a two-step resistance mechanism in vivo to high-level resistance within these isolates. High-level resistance in vivo, however, was not demonstrated to occur through multiple mutations alone. The single amino acid substitution 468Gln-Lys also causes high-level resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [15] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Missense mutation | p.Q468K |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Staphylococcus aureus strain T109 | 1280 | ||
| Staphylococcus aureus strain T112 | 1280 | |||
| Staphylococcus aureus strain T113 | 1280 | |||
| Staphylococcus aureus strain T115 | 1280 | |||
| Staphylococcus aureus strain T118 | 1280 | |||
| Staphylococcus aureus strain T124 | 1280 | |||
| Staphylococcus aureus strain T161 | 1280 | |||
| Staphylococcus aureus strain T166 | 1280 | |||
| Staphylococcus aureus strain T20 | 1280 | |||
| Staphylococcus aureus strain T211 | 1280 | |||
| Staphylococcus aureus strain T212 | 1280 | |||
| Staphylococcus aureus strain T23 | 1280 | |||
| Staphylococcus aureus strain T236 | 1280 | |||
| Staphylococcus aureus strain T23aa | 1280 | |||
| Staphylococcus aureus strain T23aac | 1280 | |||
| Staphylococcus aureus strain T23bb | 1280 | |||
| Staphylococcus aureus strain T248 | 1280 | |||
| Staphylococcus aureus strain T249 | 1280 | |||
| Staphylococcus aureus strain T25 | 1280 | |||
| Staphylococcus aureus strain T250 | 1280 | |||
| Staphylococcus aureus strain T262 | 1280 | |||
| Staphylococcus aureus strain T264 | 1280 | |||
| Staphylococcus aureus strain T295 | 1280 | |||
| Staphylococcus aureus strain T296 | 1280 | |||
| Staphylococcus aureus strain T297 | 1280 | |||
| Staphylococcus aureus strain T36 | 1280 | |||
| Staphylococcus aureus strain T38 | 1280 | |||
| Staphylococcus aureus strain T382 | 1280 | |||
| Staphylococcus aureus strain T38aa | 1280 | |||
| Staphylococcus aureus strain T38bb | 1280 | |||
| Staphylococcus aureus strain T397 | 1280 | |||
| Staphylococcus aureus strain T398 | 1280 | |||
| Staphylococcus aureus strain T399 | 1280 | |||
| Staphylococcus aureus strain T4 | 1280 | |||
| Staphylococcus aureus strain T400 | 1280 | |||
| Staphylococcus aureus strain T401 | 1280 | |||
| Staphylococcus aureus strain T402 | 1280 | |||
| Staphylococcus aureus strain T403 | 1280 | |||
| Staphylococcus aureus strain T404 | 1280 | |||
| Staphylococcus aureus strain T46 | 1280 | |||
| Staphylococcus aureus strain T59 | 1280 | |||
| Staphylococcus aureus strain T66 | 1280 | |||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | Twelve mutational changes at 10 positions were identified, with 473Ala-Thr representing a new mutation site. New amino acid substitutions, 465Gln-Arg, 466Leu-Ser, 468Gln-Lys, and 477Ala-Thr in cluster I and 527Ile-Met and 529Ser-Leu in cluster II, were described, thereby emphasizing the high variability of these amino acid positions. Codon 481 was mutated on 32 separate occasions, which indicates a central role of this amino acid. All in vivo isolates that demonstrated two or three amino acid changes exhibited high-level resistance. Interestingly enough, all of these isolates showed the mutational change 481His-Asn, which is capable of conferring low-level resistance on its own, thereby indicating a two-step resistance mechanism in vivo to high-level resistance within these isolates. High-level resistance in vivo, however, was not demonstrated to occur through multiple mutations alone. The single amino acid substitution 468Gln-Lys also causes high-level resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [15] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Missense mutation | p.D471Y |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Staphylococcus aureus strain T109 | 1280 | ||
| Staphylococcus aureus strain T112 | 1280 | |||
| Staphylococcus aureus strain T113 | 1280 | |||
| Staphylococcus aureus strain T115 | 1280 | |||
| Staphylococcus aureus strain T118 | 1280 | |||
| Staphylococcus aureus strain T124 | 1280 | |||
| Staphylococcus aureus strain T161 | 1280 | |||
| Staphylococcus aureus strain T166 | 1280 | |||
| Staphylococcus aureus strain T20 | 1280 | |||
| Staphylococcus aureus strain T211 | 1280 | |||
| Staphylococcus aureus strain T212 | 1280 | |||
| Staphylococcus aureus strain T23 | 1280 | |||
| Staphylococcus aureus strain T236 | 1280 | |||
| Staphylococcus aureus strain T23aa | 1280 | |||
| Staphylococcus aureus strain T23aac | 1280 | |||
| Staphylococcus aureus strain T23bb | 1280 | |||
| Staphylococcus aureus strain T248 | 1280 | |||
| Staphylococcus aureus strain T249 | 1280 | |||
| Staphylococcus aureus strain T25 | 1280 | |||
| Staphylococcus aureus strain T250 | 1280 | |||
| Staphylococcus aureus strain T262 | 1280 | |||
| Staphylococcus aureus strain T264 | 1280 | |||
| Staphylococcus aureus strain T295 | 1280 | |||
| Staphylococcus aureus strain T296 | 1280 | |||
| Staphylococcus aureus strain T297 | 1280 | |||
| Staphylococcus aureus strain T36 | 1280 | |||
| Staphylococcus aureus strain T38 | 1280 | |||
| Staphylococcus aureus strain T382 | 1280 | |||
| Staphylococcus aureus strain T38aa | 1280 | |||
| Staphylococcus aureus strain T38bb | 1280 | |||
| Staphylococcus aureus strain T397 | 1280 | |||
| Staphylococcus aureus strain T398 | 1280 | |||
| Staphylococcus aureus strain T399 | 1280 | |||
| Staphylococcus aureus strain T4 | 1280 | |||
| Staphylococcus aureus strain T400 | 1280 | |||
| Staphylococcus aureus strain T401 | 1280 | |||
| Staphylococcus aureus strain T402 | 1280 | |||
| Staphylococcus aureus strain T403 | 1280 | |||
| Staphylococcus aureus strain T404 | 1280 | |||
| Staphylococcus aureus strain T46 | 1280 | |||
| Staphylococcus aureus strain T59 | 1280 | |||
| Staphylococcus aureus strain T66 | 1280 | |||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | Twelve mutational changes at 10 positions were identified, with 473Ala-Thr representing a new mutation site. New amino acid substitutions, 465Gln-Arg, 466Leu-Ser, 468Gln-Lys, and 477Ala-Thr in cluster I and 527Ile-Met and 529Ser-Leu in cluster II, were described, thereby emphasizing the high variability of these amino acid positions. Codon 481 was mutated on 32 separate occasions, which indicates a central role of this amino acid. All in vivo isolates that demonstrated two or three amino acid changes exhibited high-level resistance. Interestingly enough, all of these isolates showed the mutational change 481His-Asn, which is capable of conferring low-level resistance on its own, thereby indicating a two-step resistance mechanism in vivo to high-level resistance within these isolates. High-level resistance in vivo, however, was not demonstrated to occur through multiple mutations alone. The single amino acid substitution 468Gln-Lys also causes high-level resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [15] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Missense mutation | p.A477T |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Staphylococcus aureus strain T109 | 1280 | ||
| Staphylococcus aureus strain T112 | 1280 | |||
| Staphylococcus aureus strain T113 | 1280 | |||
| Staphylococcus aureus strain T115 | 1280 | |||
| Staphylococcus aureus strain T118 | 1280 | |||
| Staphylococcus aureus strain T124 | 1280 | |||
| Staphylococcus aureus strain T161 | 1280 | |||
| Staphylococcus aureus strain T166 | 1280 | |||
| Staphylococcus aureus strain T20 | 1280 | |||
| Staphylococcus aureus strain T211 | 1280 | |||
| Staphylococcus aureus strain T212 | 1280 | |||
| Staphylococcus aureus strain T23 | 1280 | |||
| Staphylococcus aureus strain T236 | 1280 | |||
| Staphylococcus aureus strain T23aa | 1280 | |||
| Staphylococcus aureus strain T23aac | 1280 | |||
| Staphylococcus aureus strain T23bb | 1280 | |||
| Staphylococcus aureus strain T248 | 1280 | |||
| Staphylococcus aureus strain T249 | 1280 | |||
| Staphylococcus aureus strain T25 | 1280 | |||
| Staphylococcus aureus strain T250 | 1280 | |||
| Staphylococcus aureus strain T262 | 1280 | |||
| Staphylococcus aureus strain T264 | 1280 | |||
| Staphylococcus aureus strain T295 | 1280 | |||
| Staphylococcus aureus strain T296 | 1280 | |||
| Staphylococcus aureus strain T297 | 1280 | |||
| Staphylococcus aureus strain T36 | 1280 | |||
| Staphylococcus aureus strain T38 | 1280 | |||
| Staphylococcus aureus strain T382 | 1280 | |||
| Staphylococcus aureus strain T38aa | 1280 | |||
| Staphylococcus aureus strain T38bb | 1280 | |||
| Staphylococcus aureus strain T397 | 1280 | |||
| Staphylococcus aureus strain T398 | 1280 | |||
| Staphylococcus aureus strain T399 | 1280 | |||
| Staphylococcus aureus strain T4 | 1280 | |||
| Staphylococcus aureus strain T400 | 1280 | |||
| Staphylococcus aureus strain T401 | 1280 | |||
| Staphylococcus aureus strain T402 | 1280 | |||
| Staphylococcus aureus strain T403 | 1280 | |||
| Staphylococcus aureus strain T404 | 1280 | |||
| Staphylococcus aureus strain T46 | 1280 | |||
| Staphylococcus aureus strain T59 | 1280 | |||
| Staphylococcus aureus strain T66 | 1280 | |||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | Twelve mutational changes at 10 positions were identified, with 473Ala-Thr representing a new mutation site. New amino acid substitutions, 465Gln-Arg, 466Leu-Ser, 468Gln-Lys, and 477Ala-Thr in cluster I and 527Ile-Met and 529Ser-Leu in cluster II, were described, thereby emphasizing the high variability of these amino acid positions. Codon 481 was mutated on 32 separate occasions, which indicates a central role of this amino acid. All in vivo isolates that demonstrated two or three amino acid changes exhibited high-level resistance. Interestingly enough, all of these isolates showed the mutational change 481His-Asn, which is capable of conferring low-level resistance on its own, thereby indicating a two-step resistance mechanism in vivo to high-level resistance within these isolates. High-level resistance in vivo, however, was not demonstrated to occur through multiple mutations alone. The single amino acid substitution 468Gln-Lys also causes high-level resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [15] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Missense mutation | p.I527M |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Staphylococcus aureus strain T109 | 1280 | ||
| Staphylococcus aureus strain T112 | 1280 | |||
| Staphylococcus aureus strain T113 | 1280 | |||
| Staphylococcus aureus strain T115 | 1280 | |||
| Staphylococcus aureus strain T118 | 1280 | |||
| Staphylococcus aureus strain T124 | 1280 | |||
| Staphylococcus aureus strain T161 | 1280 | |||
| Staphylococcus aureus strain T166 | 1280 | |||
| Staphylococcus aureus strain T20 | 1280 | |||
| Staphylococcus aureus strain T211 | 1280 | |||
| Staphylococcus aureus strain T212 | 1280 | |||
| Staphylococcus aureus strain T23 | 1280 | |||
| Staphylococcus aureus strain T236 | 1280 | |||
| Staphylococcus aureus strain T23aa | 1280 | |||
| Staphylococcus aureus strain T23aac | 1280 | |||
| Staphylococcus aureus strain T23bb | 1280 | |||
| Staphylococcus aureus strain T248 | 1280 | |||
| Staphylococcus aureus strain T249 | 1280 | |||
| Staphylococcus aureus strain T25 | 1280 | |||
| Staphylococcus aureus strain T250 | 1280 | |||
| Staphylococcus aureus strain T262 | 1280 | |||
| Staphylococcus aureus strain T264 | 1280 | |||
| Staphylococcus aureus strain T295 | 1280 | |||
| Staphylococcus aureus strain T296 | 1280 | |||
| Staphylococcus aureus strain T297 | 1280 | |||
| Staphylococcus aureus strain T36 | 1280 | |||
| Staphylococcus aureus strain T38 | 1280 | |||
| Staphylococcus aureus strain T382 | 1280 | |||
| Staphylococcus aureus strain T38aa | 1280 | |||
| Staphylococcus aureus strain T38bb | 1280 | |||
| Staphylococcus aureus strain T397 | 1280 | |||
| Staphylococcus aureus strain T398 | 1280 | |||
| Staphylococcus aureus strain T399 | 1280 | |||
| Staphylococcus aureus strain T4 | 1280 | |||
| Staphylococcus aureus strain T400 | 1280 | |||
| Staphylococcus aureus strain T401 | 1280 | |||
| Staphylococcus aureus strain T402 | 1280 | |||
| Staphylococcus aureus strain T403 | 1280 | |||
| Staphylococcus aureus strain T404 | 1280 | |||
| Staphylococcus aureus strain T46 | 1280 | |||
| Staphylococcus aureus strain T59 | 1280 | |||
| Staphylococcus aureus strain T66 | 1280 | |||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | Twelve mutational changes at 10 positions were identified, with 473Ala-Thr representing a new mutation site. New amino acid substitutions, 465Gln-Arg, 466Leu-Ser, 468Gln-Lys, and 477Ala-Thr in cluster I and 527Ile-Met and 529Ser-Leu in cluster II, were described, thereby emphasizing the high variability of these amino acid positions. Codon 481 was mutated on 32 separate occasions, which indicates a central role of this amino acid. All in vivo isolates that demonstrated two or three amino acid changes exhibited high-level resistance. Interestingly enough, all of these isolates showed the mutational change 481His-Asn, which is capable of conferring low-level resistance on its own, thereby indicating a two-step resistance mechanism in vivo to high-level resistance within these isolates. High-level resistance in vivo, however, was not demonstrated to occur through multiple mutations alone. The single amino acid substitution 468Gln-Lys also causes high-level resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [15] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Missense mutation | p.S529L |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Staphylococcus aureus strain T109 | 1280 | ||
| Staphylococcus aureus strain T112 | 1280 | |||
| Staphylococcus aureus strain T113 | 1280 | |||
| Staphylococcus aureus strain T115 | 1280 | |||
| Staphylococcus aureus strain T118 | 1280 | |||
| Staphylococcus aureus strain T124 | 1280 | |||
| Staphylococcus aureus strain T161 | 1280 | |||
| Staphylococcus aureus strain T166 | 1280 | |||
| Staphylococcus aureus strain T20 | 1280 | |||
| Staphylococcus aureus strain T211 | 1280 | |||
| Staphylococcus aureus strain T212 | 1280 | |||
| Staphylococcus aureus strain T23 | 1280 | |||
| Staphylococcus aureus strain T236 | 1280 | |||
| Staphylococcus aureus strain T23aa | 1280 | |||
| Staphylococcus aureus strain T23aac | 1280 | |||
| Staphylococcus aureus strain T23bb | 1280 | |||
| Staphylococcus aureus strain T248 | 1280 | |||
| Staphylococcus aureus strain T249 | 1280 | |||
| Staphylococcus aureus strain T25 | 1280 | |||
| Staphylococcus aureus strain T250 | 1280 | |||
| Staphylococcus aureus strain T262 | 1280 | |||
| Staphylococcus aureus strain T264 | 1280 | |||
| Staphylococcus aureus strain T295 | 1280 | |||
| Staphylococcus aureus strain T296 | 1280 | |||
| Staphylococcus aureus strain T297 | 1280 | |||
| Staphylococcus aureus strain T36 | 1280 | |||
| Staphylococcus aureus strain T38 | 1280 | |||
| Staphylococcus aureus strain T382 | 1280 | |||
| Staphylococcus aureus strain T38aa | 1280 | |||
| Staphylococcus aureus strain T38bb | 1280 | |||
| Staphylococcus aureus strain T397 | 1280 | |||
| Staphylococcus aureus strain T398 | 1280 | |||
| Staphylococcus aureus strain T399 | 1280 | |||
| Staphylococcus aureus strain T4 | 1280 | |||
| Staphylococcus aureus strain T400 | 1280 | |||
| Staphylococcus aureus strain T401 | 1280 | |||
| Staphylococcus aureus strain T402 | 1280 | |||
| Staphylococcus aureus strain T403 | 1280 | |||
| Staphylococcus aureus strain T404 | 1280 | |||
| Staphylococcus aureus strain T46 | 1280 | |||
| Staphylococcus aureus strain T59 | 1280 | |||
| Staphylococcus aureus strain T66 | 1280 | |||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | Twelve mutational changes at 10 positions were identified, with 473Ala-Thr representing a new mutation site. New amino acid substitutions, 465Gln-Arg, 466Leu-Ser, 468Gln-Lys, and 477Ala-Thr in cluster I and 527Ile-Met and 529Ser-Leu in cluster II, were described, thereby emphasizing the high variability of these amino acid positions. Codon 481 was mutated on 32 separate occasions, which indicates a central role of this amino acid. All in vivo isolates that demonstrated two or three amino acid changes exhibited high-level resistance. Interestingly enough, all of these isolates showed the mutational change 481His-Asn, which is capable of conferring low-level resistance on its own, thereby indicating a two-step resistance mechanism in vivo to high-level resistance within these isolates. High-level resistance in vivo, however, was not demonstrated to occur through multiple mutations alone. The single amino acid substitution 468Gln-Lys also causes high-level resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [15] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Missense mutation | p.H481N+p.L466S |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Staphylococcus aureus strain T109 | 1280 | ||
| Staphylococcus aureus strain T112 | 1280 | |||
| Staphylococcus aureus strain T113 | 1280 | |||
| Staphylococcus aureus strain T115 | 1280 | |||
| Staphylococcus aureus strain T118 | 1280 | |||
| Staphylococcus aureus strain T124 | 1280 | |||
| Staphylococcus aureus strain T161 | 1280 | |||
| Staphylococcus aureus strain T166 | 1280 | |||
| Staphylococcus aureus strain T20 | 1280 | |||
| Staphylococcus aureus strain T211 | 1280 | |||
| Staphylococcus aureus strain T212 | 1280 | |||
| Staphylococcus aureus strain T23 | 1280 | |||
| Staphylococcus aureus strain T236 | 1280 | |||
| Staphylococcus aureus strain T23aa | 1280 | |||
| Staphylococcus aureus strain T23aac | 1280 | |||
| Staphylococcus aureus strain T23bb | 1280 | |||
| Staphylococcus aureus strain T248 | 1280 | |||
| Staphylococcus aureus strain T249 | 1280 | |||
| Staphylococcus aureus strain T25 | 1280 | |||
| Staphylococcus aureus strain T250 | 1280 | |||
| Staphylococcus aureus strain T262 | 1280 | |||
| Staphylococcus aureus strain T264 | 1280 | |||
| Staphylococcus aureus strain T295 | 1280 | |||
| Staphylococcus aureus strain T296 | 1280 | |||
| Staphylococcus aureus strain T297 | 1280 | |||
| Staphylococcus aureus strain T36 | 1280 | |||
| Staphylococcus aureus strain T38 | 1280 | |||
| Staphylococcus aureus strain T382 | 1280 | |||
| Staphylococcus aureus strain T38aa | 1280 | |||
| Staphylococcus aureus strain T38bb | 1280 | |||
| Staphylococcus aureus strain T397 | 1280 | |||
| Staphylococcus aureus strain T398 | 1280 | |||
| Staphylococcus aureus strain T399 | 1280 | |||
| Staphylococcus aureus strain T4 | 1280 | |||
| Staphylococcus aureus strain T400 | 1280 | |||
| Staphylococcus aureus strain T401 | 1280 | |||
| Staphylococcus aureus strain T402 | 1280 | |||
| Staphylococcus aureus strain T403 | 1280 | |||
| Staphylococcus aureus strain T404 | 1280 | |||
| Staphylococcus aureus strain T46 | 1280 | |||
| Staphylococcus aureus strain T59 | 1280 | |||
| Staphylococcus aureus strain T66 | 1280 | |||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | Twelve mutational changes at 10 positions were identified, with 473Ala-Thr representing a new mutation site. New amino acid substitutions, 465Gln-Arg, 466Leu-Ser, 468Gln-Lys, and 477Ala-Thr in cluster I and 527Ile-Met and 529Ser-Leu in cluster II, were described, thereby emphasizing the high variability of these amino acid positions. Codon 481 was mutated on 32 separate occasions, which indicates a central role of this amino acid. All in vivo isolates that demonstrated two or three amino acid changes exhibited high-level resistance. Interestingly enough, all of these isolates showed the mutational change 481His-Asn, which is capable of conferring low-level resistance on its own, thereby indicating a two-step resistance mechanism in vivo to high-level resistance within these isolates. High-level resistance in vivo, however, was not demonstrated to occur through multiple mutations alone. The single amino acid substitution 468Gln-Lys also causes high-level resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [15] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Missense mutation | p.H481N+p.S529L |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Staphylococcus aureus strain T109 | 1280 | ||
| Staphylococcus aureus strain T112 | 1280 | |||
| Staphylococcus aureus strain T113 | 1280 | |||
| Staphylococcus aureus strain T115 | 1280 | |||
| Staphylococcus aureus strain T118 | 1280 | |||
| Staphylococcus aureus strain T124 | 1280 | |||
| Staphylococcus aureus strain T161 | 1280 | |||
| Staphylococcus aureus strain T166 | 1280 | |||
| Staphylococcus aureus strain T20 | 1280 | |||
| Staphylococcus aureus strain T211 | 1280 | |||
| Staphylococcus aureus strain T212 | 1280 | |||
| Staphylococcus aureus strain T23 | 1280 | |||
| Staphylococcus aureus strain T236 | 1280 | |||
| Staphylococcus aureus strain T23aa | 1280 | |||
| Staphylococcus aureus strain T23aac | 1280 | |||
| Staphylococcus aureus strain T23bb | 1280 | |||
| Staphylococcus aureus strain T248 | 1280 | |||
| Staphylococcus aureus strain T249 | 1280 | |||
| Staphylococcus aureus strain T25 | 1280 | |||
| Staphylococcus aureus strain T250 | 1280 | |||
| Staphylococcus aureus strain T262 | 1280 | |||
| Staphylococcus aureus strain T264 | 1280 | |||
| Staphylococcus aureus strain T295 | 1280 | |||
| Staphylococcus aureus strain T296 | 1280 | |||
| Staphylococcus aureus strain T297 | 1280 | |||
| Staphylococcus aureus strain T36 | 1280 | |||
| Staphylococcus aureus strain T38 | 1280 | |||
| Staphylococcus aureus strain T382 | 1280 | |||
| Staphylococcus aureus strain T38aa | 1280 | |||
| Staphylococcus aureus strain T38bb | 1280 | |||
| Staphylococcus aureus strain T397 | 1280 | |||
| Staphylococcus aureus strain T398 | 1280 | |||
| Staphylococcus aureus strain T399 | 1280 | |||
| Staphylococcus aureus strain T4 | 1280 | |||
| Staphylococcus aureus strain T400 | 1280 | |||
| Staphylococcus aureus strain T401 | 1280 | |||
| Staphylococcus aureus strain T402 | 1280 | |||
| Staphylococcus aureus strain T403 | 1280 | |||
| Staphylococcus aureus strain T404 | 1280 | |||
| Staphylococcus aureus strain T46 | 1280 | |||
| Staphylococcus aureus strain T59 | 1280 | |||
| Staphylococcus aureus strain T66 | 1280 | |||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | Twelve mutational changes at 10 positions were identified, with 473Ala-Thr representing a new mutation site. New amino acid substitutions, 465Gln-Arg, 466Leu-Ser, 468Gln-Lys, and 477Ala-Thr in cluster I and 527Ile-Met and 529Ser-Leu in cluster II, were described, thereby emphasizing the high variability of these amino acid positions. Codon 481 was mutated on 32 separate occasions, which indicates a central role of this amino acid. All in vivo isolates that demonstrated two or three amino acid changes exhibited high-level resistance. Interestingly enough, all of these isolates showed the mutational change 481His-Asn, which is capable of conferring low-level resistance on its own, thereby indicating a two-step resistance mechanism in vivo to high-level resistance within these isolates. High-level resistance in vivo, however, was not demonstrated to occur through multiple mutations alone. The single amino acid substitution 468Gln-Lys also causes high-level resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [15] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Missense mutation | p.H481N+p.I527M |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Staphylococcus aureus strain T109 | 1280 | ||
| Staphylococcus aureus strain T112 | 1280 | |||
| Staphylococcus aureus strain T113 | 1280 | |||
| Staphylococcus aureus strain T115 | 1280 | |||
| Staphylococcus aureus strain T118 | 1280 | |||
| Staphylococcus aureus strain T124 | 1280 | |||
| Staphylococcus aureus strain T161 | 1280 | |||
| Staphylococcus aureus strain T166 | 1280 | |||
| Staphylococcus aureus strain T20 | 1280 | |||
| Staphylococcus aureus strain T211 | 1280 | |||
| Staphylococcus aureus strain T212 | 1280 | |||
| Staphylococcus aureus strain T23 | 1280 | |||
| Staphylococcus aureus strain T236 | 1280 | |||
| Staphylococcus aureus strain T23aa | 1280 | |||
| Staphylococcus aureus strain T23aac | 1280 | |||
| Staphylococcus aureus strain T23bb | 1280 | |||
| Staphylococcus aureus strain T248 | 1280 | |||
| Staphylococcus aureus strain T249 | 1280 | |||
| Staphylococcus aureus strain T25 | 1280 | |||
| Staphylococcus aureus strain T250 | 1280 | |||
| Staphylococcus aureus strain T262 | 1280 | |||
| Staphylococcus aureus strain T264 | 1280 | |||
| Staphylococcus aureus strain T295 | 1280 | |||
| Staphylococcus aureus strain T296 | 1280 | |||
| Staphylococcus aureus strain T297 | 1280 | |||
| Staphylococcus aureus strain T36 | 1280 | |||
| Staphylococcus aureus strain T38 | 1280 | |||
| Staphylococcus aureus strain T382 | 1280 | |||
| Staphylococcus aureus strain T38aa | 1280 | |||
| Staphylococcus aureus strain T38bb | 1280 | |||
| Staphylococcus aureus strain T397 | 1280 | |||
| Staphylococcus aureus strain T398 | 1280 | |||
| Staphylococcus aureus strain T399 | 1280 | |||
| Staphylococcus aureus strain T4 | 1280 | |||
| Staphylococcus aureus strain T400 | 1280 | |||
| Staphylococcus aureus strain T401 | 1280 | |||
| Staphylococcus aureus strain T402 | 1280 | |||
| Staphylococcus aureus strain T403 | 1280 | |||
| Staphylococcus aureus strain T404 | 1280 | |||
| Staphylococcus aureus strain T46 | 1280 | |||
| Staphylococcus aureus strain T59 | 1280 | |||
| Staphylococcus aureus strain T66 | 1280 | |||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | Twelve mutational changes at 10 positions were identified, with 473Ala-Thr representing a new mutation site. New amino acid substitutions, 465Gln-Arg, 466Leu-Ser, 468Gln-Lys, and 477Ala-Thr in cluster I and 527Ile-Met and 529Ser-Leu in cluster II, were described, thereby emphasizing the high variability of these amino acid positions. Codon 481 was mutated on 32 separate occasions, which indicates a central role of this amino acid. All in vivo isolates that demonstrated two or three amino acid changes exhibited high-level resistance. Interestingly enough, all of these isolates showed the mutational change 481His-Asn, which is capable of conferring low-level resistance on its own, thereby indicating a two-step resistance mechanism in vivo to high-level resistance within these isolates. High-level resistance in vivo, however, was not demonstrated to occur through multiple mutations alone. The single amino acid substitution 468Gln-Lys also causes high-level resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [15] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Missense mutation | p.H481N+p.S529L+p.Q465R |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Staphylococcus aureus strain T109 | 1280 | ||
| Staphylococcus aureus strain T112 | 1280 | |||
| Staphylococcus aureus strain T113 | 1280 | |||
| Staphylococcus aureus strain T115 | 1280 | |||
| Staphylococcus aureus strain T118 | 1280 | |||
| Staphylococcus aureus strain T124 | 1280 | |||
| Staphylococcus aureus strain T161 | 1280 | |||
| Staphylococcus aureus strain T166 | 1280 | |||
| Staphylococcus aureus strain T20 | 1280 | |||
| Staphylococcus aureus strain T211 | 1280 | |||
| Staphylococcus aureus strain T212 | 1280 | |||
| Staphylococcus aureus strain T23 | 1280 | |||
| Staphylococcus aureus strain T236 | 1280 | |||
| Staphylococcus aureus strain T23aa | 1280 | |||
| Staphylococcus aureus strain T23aac | 1280 | |||
| Staphylococcus aureus strain T23bb | 1280 | |||
| Staphylococcus aureus strain T248 | 1280 | |||
| Staphylococcus aureus strain T249 | 1280 | |||
| Staphylococcus aureus strain T25 | 1280 | |||
| Staphylococcus aureus strain T250 | 1280 | |||
| Staphylococcus aureus strain T262 | 1280 | |||
| Staphylococcus aureus strain T264 | 1280 | |||
| Staphylococcus aureus strain T295 | 1280 | |||
| Staphylococcus aureus strain T296 | 1280 | |||
| Staphylococcus aureus strain T297 | 1280 | |||
| Staphylococcus aureus strain T36 | 1280 | |||
| Staphylococcus aureus strain T38 | 1280 | |||
| Staphylococcus aureus strain T382 | 1280 | |||
| Staphylococcus aureus strain T38aa | 1280 | |||
| Staphylococcus aureus strain T38bb | 1280 | |||
| Staphylococcus aureus strain T397 | 1280 | |||
| Staphylococcus aureus strain T398 | 1280 | |||
| Staphylococcus aureus strain T399 | 1280 | |||
| Staphylococcus aureus strain T4 | 1280 | |||
| Staphylococcus aureus strain T400 | 1280 | |||
| Staphylococcus aureus strain T401 | 1280 | |||
| Staphylococcus aureus strain T402 | 1280 | |||
| Staphylococcus aureus strain T403 | 1280 | |||
| Staphylococcus aureus strain T404 | 1280 | |||
| Staphylococcus aureus strain T46 | 1280 | |||
| Staphylococcus aureus strain T59 | 1280 | |||
| Staphylococcus aureus strain T66 | 1280 | |||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | Twelve mutational changes at 10 positions were identified, with 473Ala-Thr representing a new mutation site. New amino acid substitutions, 465Gln-Arg, 466Leu-Ser, 468Gln-Lys, and 477Ala-Thr in cluster I and 527Ile-Met and 529Ser-Leu in cluster II, were described, thereby emphasizing the high variability of these amino acid positions. Codon 481 was mutated on 32 separate occasions, which indicates a central role of this amino acid. All in vivo isolates that demonstrated two or three amino acid changes exhibited high-level resistance. Interestingly enough, all of these isolates showed the mutational change 481His-Asn, which is capable of conferring low-level resistance on its own, thereby indicating a two-step resistance mechanism in vivo to high-level resistance within these isolates. High-level resistance in vivo, however, was not demonstrated to occur through multiple mutations alone. The single amino acid substitution 468Gln-Lys also causes high-level resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [15] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Missense mutation | p.H481N+p.A473T+p.A477T |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Staphylococcus aureus strain T109 | 1280 | ||
| Staphylococcus aureus strain T112 | 1280 | |||
| Staphylococcus aureus strain T113 | 1280 | |||
| Staphylococcus aureus strain T115 | 1280 | |||
| Staphylococcus aureus strain T118 | 1280 | |||
| Staphylococcus aureus strain T124 | 1280 | |||
| Staphylococcus aureus strain T161 | 1280 | |||
| Staphylococcus aureus strain T166 | 1280 | |||
| Staphylococcus aureus strain T20 | 1280 | |||
| Staphylococcus aureus strain T211 | 1280 | |||
| Staphylococcus aureus strain T212 | 1280 | |||
| Staphylococcus aureus strain T23 | 1280 | |||
| Staphylococcus aureus strain T236 | 1280 | |||
| Staphylococcus aureus strain T23aa | 1280 | |||
| Staphylococcus aureus strain T23aac | 1280 | |||
| Staphylococcus aureus strain T23bb | 1280 | |||
| Staphylococcus aureus strain T248 | 1280 | |||
| Staphylococcus aureus strain T249 | 1280 | |||
| Staphylococcus aureus strain T25 | 1280 | |||
| Staphylococcus aureus strain T250 | 1280 | |||
| Staphylococcus aureus strain T262 | 1280 | |||
| Staphylococcus aureus strain T264 | 1280 | |||
| Staphylococcus aureus strain T295 | 1280 | |||
| Staphylococcus aureus strain T296 | 1280 | |||
| Staphylococcus aureus strain T297 | 1280 | |||
| Staphylococcus aureus strain T36 | 1280 | |||
| Staphylococcus aureus strain T38 | 1280 | |||
| Staphylococcus aureus strain T382 | 1280 | |||
| Staphylococcus aureus strain T38aa | 1280 | |||
| Staphylococcus aureus strain T38bb | 1280 | |||
| Staphylococcus aureus strain T397 | 1280 | |||
| Staphylococcus aureus strain T398 | 1280 | |||
| Staphylococcus aureus strain T399 | 1280 | |||
| Staphylococcus aureus strain T4 | 1280 | |||
| Staphylococcus aureus strain T400 | 1280 | |||
| Staphylococcus aureus strain T401 | 1280 | |||
| Staphylococcus aureus strain T402 | 1280 | |||
| Staphylococcus aureus strain T403 | 1280 | |||
| Staphylococcus aureus strain T404 | 1280 | |||
| Staphylococcus aureus strain T46 | 1280 | |||
| Staphylococcus aureus strain T59 | 1280 | |||
| Staphylococcus aureus strain T66 | 1280 | |||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | Twelve mutational changes at 10 positions were identified, with 473Ala-Thr representing a new mutation site. New amino acid substitutions, 465Gln-Arg, 466Leu-Ser, 468Gln-Lys, and 477Ala-Thr in cluster I and 527Ile-Met and 529Ser-Leu in cluster II, were described, thereby emphasizing the high variability of these amino acid positions. Codon 481 was mutated on 32 separate occasions, which indicates a central role of this amino acid. All in vivo isolates that demonstrated two or three amino acid changes exhibited high-level resistance. Interestingly enough, all of these isolates showed the mutational change 481His-Asn, which is capable of conferring low-level resistance on its own, thereby indicating a two-step resistance mechanism in vivo to high-level resistance within these isolates. High-level resistance in vivo, however, was not demonstrated to occur through multiple mutations alone. The single amino acid substitution 468Gln-Lys also causes high-level resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [15] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Missense mutation | p.D471Y+p.S486L |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Staphylococcus aureus strain T109 | 1280 | ||
| Staphylococcus aureus strain T112 | 1280 | |||
| Staphylococcus aureus strain T113 | 1280 | |||
| Staphylococcus aureus strain T115 | 1280 | |||
| Staphylococcus aureus strain T118 | 1280 | |||
| Staphylococcus aureus strain T124 | 1280 | |||
| Staphylococcus aureus strain T161 | 1280 | |||
| Staphylococcus aureus strain T166 | 1280 | |||
| Staphylococcus aureus strain T20 | 1280 | |||
| Staphylococcus aureus strain T211 | 1280 | |||
| Staphylococcus aureus strain T212 | 1280 | |||
| Staphylococcus aureus strain T23 | 1280 | |||
| Staphylococcus aureus strain T236 | 1280 | |||
| Staphylococcus aureus strain T23aa | 1280 | |||
| Staphylococcus aureus strain T23aac | 1280 | |||
| Staphylococcus aureus strain T23bb | 1280 | |||
| Staphylococcus aureus strain T248 | 1280 | |||
| Staphylococcus aureus strain T249 | 1280 | |||
| Staphylococcus aureus strain T25 | 1280 | |||
| Staphylococcus aureus strain T250 | 1280 | |||
| Staphylococcus aureus strain T262 | 1280 | |||
| Staphylococcus aureus strain T264 | 1280 | |||
| Staphylococcus aureus strain T295 | 1280 | |||
| Staphylococcus aureus strain T296 | 1280 | |||
| Staphylococcus aureus strain T297 | 1280 | |||
| Staphylococcus aureus strain T36 | 1280 | |||
| Staphylococcus aureus strain T38 | 1280 | |||
| Staphylococcus aureus strain T382 | 1280 | |||
| Staphylococcus aureus strain T38aa | 1280 | |||
| Staphylococcus aureus strain T38bb | 1280 | |||
| Staphylococcus aureus strain T397 | 1280 | |||
| Staphylococcus aureus strain T398 | 1280 | |||
| Staphylococcus aureus strain T399 | 1280 | |||
| Staphylococcus aureus strain T4 | 1280 | |||
| Staphylococcus aureus strain T400 | 1280 | |||
| Staphylococcus aureus strain T401 | 1280 | |||
| Staphylococcus aureus strain T402 | 1280 | |||
| Staphylococcus aureus strain T403 | 1280 | |||
| Staphylococcus aureus strain T404 | 1280 | |||
| Staphylococcus aureus strain T46 | 1280 | |||
| Staphylococcus aureus strain T59 | 1280 | |||
| Staphylococcus aureus strain T66 | 1280 | |||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | Twelve mutational changes at 10 positions were identified, with 473Ala-Thr representing a new mutation site. New amino acid substitutions, 465Gln-Arg, 466Leu-Ser, 468Gln-Lys, and 477Ala-Thr in cluster I and 527Ile-Met and 529Ser-Leu in cluster II, were described, thereby emphasizing the high variability of these amino acid positions. Codon 481 was mutated on 32 separate occasions, which indicates a central role of this amino acid. All in vivo isolates that demonstrated two or three amino acid changes exhibited high-level resistance. Interestingly enough, all of these isolates showed the mutational change 481His-Asn, which is capable of conferring low-level resistance on its own, thereby indicating a two-step resistance mechanism in vivo to high-level resistance within these isolates. High-level resistance in vivo, however, was not demonstrated to occur through multiple mutations alone. The single amino acid substitution 468Gln-Lys also causes high-level resistance. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Serine/threonine-protein kinase (STPK) | [5] | |||
| Resistant Disease | brucellosis [ICD-11: 1B95] | |||
| Molecule Alteration | Deletion mutation | . |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | STPK gene deletion cells | N.A. | Homo sapiens (Human) | N.A. |
| Experiment for Drug Resistance |
MIC assay; MBC assay; RpoB?gene testing | |||
| Mechanism Description | All these findings indicate that the absence of the STPK could increase sulfur metabolism and GSH levels, and decrease the NADPH oxidase activity and NADP+/NADPH ratio, which promotes the antioxidant capacity of B. melitensis. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: DNA-directed RNA polymerase subunit beta 2 (RPOB2) | [13] | |||
| Resistant Disease | Nocardiosis [ICD-11: 1C1B.0] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Nocardia farcinica IFM 10152 | 247156 | ||
| Experiment for Molecule Alteration |
PCR; Southern analysis | |||
| Experiment for Drug Resistance |
Brain heart infusion (BHI) broth assay | |||
| Mechanism Description | RpoB2 may have reduced binding affinity for RIF due to amino acid substitutions and is capable of functioning in the presence of RIF instead of RpoB. Besides, RNAP with RpoB2 may elicit expression of a latent RIF resistance gene which may be present in the genome. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Ribonuclease PH (RPH) | [26], [27], [28] | |||
| Resistant Disease | HIV-infected patients with tuberculosis [ICD-11: 1C60.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli TOP10 | 83333 | ||
| Bacillus cereus RPH-Bc | 1396 | |||
| Escherichia coli Rosetta(DE3) pLysS | 866768 | |||
| L. monocytogenes | 1639 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | RIF phosphotransferase (rph) led to the identification of a new resistance gene and associated enzyme responsible for inactivating rifamycin antibiotics by phosphorylation. | |||
|
|
||||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [7], [8], [9] | |||
| Resistant Disease | HIV-infected patients with tuberculosis [ICD-11: 1C60.0] | |||
| Molecule Alteration | Missense mutation | p.L511P |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis isolates | 1773 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | More than 96% of rifampicin-resistant strains show mutations in a portion of the RNA polymerase B subunit gene (rpoB), called the hot-spot region, encompassing codons 507-533.Mutations L533P, H526L, D516Y and L511P and the double mutation E562G/P564L conferred a low level of resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [7], [8], [9] | |||
| Resistant Disease | HIV-infected patients with tuberculosis [ICD-11: 1C60.0] | |||
| Molecule Alteration | Missense mutation | p.Q513P |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis isolates | 1773 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | More than 96% of rifampicin-resistant strains show mutations in a portion of the RNA polymerase B subunit gene (rpoB), called the hot-spot region, encompassing codons 507-533.Mutations L533P, H526L, D516Y and L511P and the double mutation E562G/P564L conferred a low level of resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [7], [8], [9] | |||
| Resistant Disease | HIV-infected patients with tuberculosis [ICD-11: 1C60.0] | |||
| Molecule Alteration | Missense mutation | p.Q513K |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis isolates | 1773 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | More than 96% of rifampicin-resistant strains show mutations in a portion of the RNA polymerase B subunit gene (rpoB), called the hot-spot region, encompassing codons 507-533.Mutations L533P, H526L, D516Y and L511P and the double mutation E562G/P564L conferred a low level of resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [7], [8], [9] | |||
| Resistant Disease | HIV-infected patients with tuberculosis [ICD-11: 1C60.0] | |||
| Molecule Alteration | Missense mutation | p.D516V |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis isolates | 1773 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | More than 96% of rifampicin-resistant strains show mutations in a portion of the RNA polymerase B subunit gene (rpoB), called the hot-spot region, encompassing codons 507-533.Mutations L533P, H526L, D516Y and L511P and the double mutation E562G/P564L conferred a low level of resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [7], [8], [9] | |||
| Resistant Disease | HIV-infected patients with tuberculosis [ICD-11: 1C60.0] | |||
| Molecule Alteration | Missense mutation | p.H526N+p.L533P |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis isolates | 1773 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | More than 96% of rifampicin-resistant strains show mutations in a portion of the RNA polymerase B subunit gene (rpoB), called the hot-spot region, encompassing codons 507-533.Mutations L533P, H526L, D516Y and L511P and the double mutation E562G/P564L conferred a low level of resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [7], [8], [9] | |||
| Resistant Disease | HIV-infected patients with tuberculosis [ICD-11: 1C60.0] | |||
| Molecule Alteration | Missense mutation | p.H526Y |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis isolates | 1773 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | More than 96% of rifampicin-resistant strains show mutations in a portion of the RNA polymerase B subunit gene (rpoB), called the hot-spot region, encompassing codons 507-533.Mutations L533P, H526L, D516Y and L511P and the double mutation E562G/P564L conferred a low level of resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [7], [8], [9] | |||
| Resistant Disease | HIV-infected patients with tuberculosis [ICD-11: 1C60.0] | |||
| Molecule Alteration | Missense mutation | p.H526R |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis isolates | 1773 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | More than 96% of rifampicin-resistant strains show mutations in a portion of the RNA polymerase B subunit gene (rpoB), called the hot-spot region, encompassing codons 507-533.Mutations L533P, H526L, D516Y and L511P and the double mutation E562G/P564L conferred a low level of resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [7], [8], [9] | |||
| Resistant Disease | HIV-infected patients with tuberculosis [ICD-11: 1C60.0] | |||
| Molecule Alteration | Missense mutation | p.H526D |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis isolates | 1773 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | More than 96% of rifampicin-resistant strains show mutations in a portion of the RNA polymerase B subunit gene (rpoB), called the hot-spot region, encompassing codons 507-533.Mutations L533P, H526L, D516Y and L511P and the double mutation E562G/P564L conferred a low level of resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [7], [8], [9] | |||
| Resistant Disease | HIV-infected patients with tuberculosis [ICD-11: 1C60.0] | |||
| Molecule Alteration | Missense mutation | p.H526N |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis isolates | 1773 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | More than 96% of rifampicin-resistant strains show mutations in a portion of the RNA polymerase B subunit gene (rpoB), called the hot-spot region, encompassing codons 507-533.Mutations L533P, H526L, D516Y and L511P and the double mutation E562G/P564L conferred a low level of resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [7], [8], [9] | |||
| Resistant Disease | HIV-infected patients with tuberculosis [ICD-11: 1C60.0] | |||
| Molecule Alteration | Missense mutation | p.H526L |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis isolates | 1773 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | More than 96% of rifampicin-resistant strains show mutations in a portion of the RNA polymerase B subunit gene (rpoB), called the hot-spot region, encompassing codons 507-533.Mutations L533P, H526L, D516Y and L511P and the double mutation E562G/P564L conferred a low level of resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [7], [8], [9] | |||
| Resistant Disease | HIV-infected patients with tuberculosis [ICD-11: 1C60.0] | |||
| Molecule Alteration | Missense mutation | p.H526C |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis isolates | 1773 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | More than 96% of rifampicin-resistant strains show mutations in a portion of the RNA polymerase B subunit gene (rpoB), called the hot-spot region, encompassing codons 507-533.Mutations L533P, H526L, D516Y and L511P and the double mutation E562G/P564L conferred a low level of resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [7], [8], [9] | |||
| Resistant Disease | HIV-infected patients with tuberculosis [ICD-11: 1C60.0] | |||
| Molecule Alteration | Missense mutation | p.S531W |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis isolates | 1773 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | More than 96% of rifampicin-resistant strains show mutations in a portion of the RNA polymerase B subunit gene (rpoB), called the hot-spot region, encompassing codons 507-533.Mutations L533P, H526L, D516Y and L511P and the double mutation E562G/P564L conferred a low level of resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [7], [8], [9] | |||
| Resistant Disease | HIV-infected patients with tuberculosis [ICD-11: 1C60.0] | |||
| Molecule Alteration | Missense mutation | p.S531L |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis isolates | 1773 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | More than 96% of rifampicin-resistant strains show mutations in a portion of the RNA polymerase B subunit gene (rpoB), called the hot-spot region, encompassing codons 507-533.Mutations L533P, H526L, D516Y and L511P and the double mutation E562G/P564L conferred a low level of resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [7], [8], [9] | |||
| Resistant Disease | HIV-infected patients with tuberculosis [ICD-11: 1C60.0] | |||
| Molecule Alteration | Missense mutation | p.L533P |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis isolates | 1773 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | More than 96% of rifampicin-resistant strains show mutations in a portion of the RNA polymerase B subunit gene (rpoB), called the hot-spot region, encompassing codons 507-533.Mutations L533P, H526L, D516Y and L511P and the double mutation E562G/P564L conferred a low level of resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [7], [8], [9] | |||
| Resistant Disease | HIV-infected patients with tuberculosis [ICD-11: 1C60.0] | |||
| Molecule Alteration | Missense mutation | p.E562G+p.P564L |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis isolates | 1773 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | More than 96% of rifampicin-resistant strains show mutations in a portion of the RNA polymerase B subunit gene (rpoB), called the hot-spot region, encompassing codons 507-533.Mutations L533P, H526L, D516Y and L511P and the double mutation E562G/P564L conferred a low level of resistance. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [7], [8], [9] | |||
| Resistant Disease | HIV-infected patients with tuberculosis [ICD-11: 1C60.0] | |||
| Molecule Alteration | Frameshift mutation | c.513_516del*AA TTC ATG G* |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis isolates | 1773 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | More than 96% of rifampicin-resistant strains show mutations in a portion of the RNA polymerase B subunit gene (rpoB), called the hot-spot region, encompassing codons 507-533.Mutations L533P, H526L, D516Y and L511P and the double mutation E562G/P564L conferred a low level of resistance. | |||
|
|
||||
| Key Molecule: Uncharacterized MFS-type transporter EfpA (EFPA) | [29], [30], [31] | |||
| Resistant Disease | HIV-infected patients with tuberculosis [ICD-11: 1C60.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis H37Rv | 83332 | ||
| Mycobacterium tuberculosis MTB1 | 1773 | |||
| Mycobacterium tuberculosis MTB2 | 1773 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | The induced strains presented an increased efflux activity that was inhibited by the efflux inhibitors (EIs) and showed overexpression of the efflux pump genes efpA, mmpL7, mmr, p55 and the Tap-like gene Rv1258c. Altogether, these results correlate efflux activity with INH resistance and demonstrate that efflux pumps play an important role in acquired INH resistance in M. tuberculosis complex. | |||
|
|
||||
| Key Molecule: Probable arabinosyltransferase B (EMBB) | [32] | |||
| Resistant Disease | HIV-infected patients with tuberculosis [ICD-11: 1C60.0] | |||
| Molecule Alteration | Missense mutation | p.M306I |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis isolates | 1773 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Test for drug susceptibility in L-J medium assay | |||
| Mechanism Description | An embB mutation has a strong relationship to rifampin resistance. Inhibition of cell wall biosynthesis may not play an important role, and inhibition of RNA metabolism may be partly responsible. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [19] | |||
| Resistant Disease | Urinary tuberculosis [ICD-11: 1G80.0] | |||
| Molecule Alteration | Missense mutation | p.S531L |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis isolates | 1773 | ||
| Experiment for Molecule Alteration |
Gene sequencing assay | |||
| Mechanism Description | Regarding drug-resistance mutation profiles, the most prevalent mutation sites were katG S315T1 and rpoB S531L. | |||
| Key Molecule: DNA-directed RNA polymerase subunit beta (RPOB) | [19] | |||
| Resistant Disease | Urinary tuberculosis [ICD-11: 1G80.0] | |||
| Molecule Alteration | Missense mutation | p.S315T1 |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis isolates | 1773 | ||
| Experiment for Molecule Alteration |
Gene sequencing assay | |||
| Mechanism Description | Regarding drug-resistance mutation profiles, the most prevalent mutation sites were katG S315T1 and rpoB S531L. | |||
|
|
||||
| Key Molecule: Catalase-peroxidase (KATG) | [19] | |||
| Resistant Disease | Urinary tuberculosis [ICD-11: 1G80.0] | |||
| Molecule Alteration | Missense mutation | p.S531L |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis isolates | 1773 | ||
| Experiment for Molecule Alteration |
Gene sequencing assay | |||
| Mechanism Description | Regarding drug-resistance mutation profiles, the most prevalent mutation sites were katG S315T1 and rpoB S531L. | |||
| Key Molecule: Catalase-peroxidase (KATG) | [19] | |||
| Resistant Disease | Urinary tuberculosis [ICD-11: 1G80.0] | |||
| Molecule Alteration | Missense mutation | p.S315T1 |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis isolates | 1773 | ||
| Experiment for Molecule Alteration |
Gene sequencing assay | |||
| Mechanism Description | Regarding drug-resistance mutation profiles, the most prevalent mutation sites were katG S315T1 and rpoB S531L. | |||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
