Drug Information
Drug (ID: DG00037) and It's Reported Resistant Information
| Name |
Cefoxitin
|
||||
|---|---|---|---|---|---|
| Synonyms |
Mefoxin; Mefoxitin; CEFOXITIN SODIUM; Cefoxitin sodium salt; Mefoxin (TN); Cefoxitin (USAN/INN); Cefoxitin sodium (JAN/USP); (6R,7S)-3-(carbamoyloxymethyl)-7-methoxy-8-oxo-7-[(2-thiophen-2-ylacetyl)amino]-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylate; (6R,7S)-3-(carbamoyloxymethyl)-7-methoxy-8-oxo-7-[(2-thiophen-2-ylacetyl)amino]-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid; (6R,7S)-3-[(carbamoyloxy)methyl]-7-methoxy-8-oxo-7-[(2-thienylacetyl)amino]-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylate; (6R,7S)-3-{[(aminocarbonyl)oxy]methyl}-7-(methyloxy)-8-oxo-7-[(2-thienylacetyl)amino]-5-thia-1-azabicyclo[4.2.0]oct-2-ene-2-carboxylic acid; 3-[(carbamoyloxy)methyl]-7alpha-methoxy-7beta-[(2-thienylacetyl)amino]-3,4-didehydrocepham-4-carboxylate; 3-[(carbamoyloxy)methyl]-7alpha-methoxy-7beta-[(thiophen-2-yl)acetamido]-3,4-didehydrocepham-4-carboxylic acid
Click to Show/Hide
|
||||
| Indication |
In total 1 Indication(s)
|
||||
| Structure |
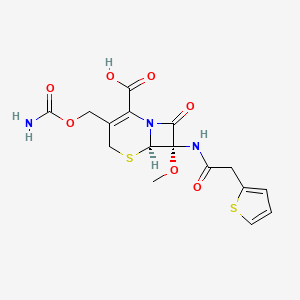
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Clinically Reported Resistance for This Drug
(5 diseases)
[2]
[3]
[1]
[6]
Disease(s) with Resistance Information Validated by in-vivo Model for This Drug
(1 diseases)
[7]
|
||||
| Target | Bacterial Penicillin binding protein (Bact PBP) | NOUNIPROTAC | [1] | ||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C16H17N3O7S2
|
||||
| IsoSMILES |
CO[C@@]1([C@@H]2N(C1=O)C(=C(CS2)COC(=O)N)C(=O)O)NC(=O)CC3=CC=CS3
|
||||
| InChI |
1S/C16H17N3O7S2/c1-25-16(18-10(20)5-9-3-2-4-27-9)13(23)19-11(12(21)22)8(6-26-15(17)24)7-28-14(16)19/h2-4,14H,5-7H2,1H3,(H2,17,24)(H,18,20)(H,21,22)/t14-,16+/m1/s1
|
||||
| InChIKey |
WZOZEZRFJCJXNZ-ZBFHGGJFSA-N
|
||||
| PubChem CID | |||||
| ChEBI ID | |||||
| TTD Drug ID | |||||
| VARIDT ID | |||||
| INTEDE ID | |||||
| DrugBank ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-01: Infectious/parasitic diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Beta-lactamase (BLA) | [8] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.V88L+p.M154L |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli TOP10 | 83333 | ||
| Escherichia coli ST648 | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
Etest assay | |||
| Mechanism Description | NDM-5 differed from existing enzymes due to substitutions at positions 88 (Val - Leu) and 154 (Met - Leu) and reduced the susceptibility of Escherichia coli TOP10 transformants to expanded-spectrum cephalosporins and carbapenems when expressed under its native promoter. | |||
| Key Molecule: Penicillin binding protein PBP 2 (PBP2) | [9] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Staphylococcus aureus RN4220 | 1280 | ||
| Staphylococcus aureus M10/0061 | 1280 | |||
| Staphylococcus aureus M10/0148 | 1280 | |||
| Staphylococcus aureus WGB8404 | 1280 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
Disk diffusion test assay; Etest assay | |||
| Mechanism Description | Methicillin resistance in staphylococci is mediated by penicillin binding protein 2a (PBP 2a), encoded by mecA on mobile staphylococcal cassette chromosome mec (SCCmec) elements.Whole-genome sequencing of one isolate (M10/0061) revealed a 30-kb SCCmec element encoding a class E mec complex with highly divergent blaZ-mecA-mecR1-mecI, a type 8 cassette chromosome recombinase (ccr) complex consisting of ccrA1-ccrB3, an arsenic resistance operon, and flanking direct repeats (DRs). | |||
| Key Molecule: Beta-lactamase (BLA) | [3] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Mutantion | p.V231S |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli DH10B | 316385 | ||
| Escherichia coli VA1171/10 | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
Quadruple disc test assay | |||
| Mechanism Description | Molecular methods revealed a novel, plasmid-localized variant of CMY-2 with a substitution of valine 231 for serine (V231S), which was designated CMY-42. Like the CMY-2-like AmpC beta-lactamase CMY-30, carrying the substitution V231G, CMY-42 displayed increased activity toward expanded spectrum cephalosporins. | |||
| Key Molecule: Beta-lactamase (BLA) | [10], [11] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.V77A+p.D114N+p.S140A+p.N288D |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Citrobacter freundii strain 2524/96 | 546 | ||
| Citrobacter freundii strain 2525/96 | 546 | |||
| Citrobacter freundii strain 2526/96 | 546 | |||
| Escherichia coli strain 2527/96 | 562 | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | Sequencing has revealed that C. freundii isolates produced a new CTX-M-3 enzyme which is very closely related to the CTX-M-1/MEN-1 Beta-lactamase. | |||
| Key Molecule: Beta-lactamase (BLA) | [10], [12] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli strain DH5a | 668369 | ||
| Klebsiella pneumoniae strain HEL-1 | 573 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | The phenotype of Klebsiella pneumoniae HEL-1 indicates a plasmidic cephamycinase gene (blaCMY-2),which is responsible for cephamycin resistance. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Outer membrane porin (OMP38) | [4], [5] | |||
| Resistant Disease | Melioidosis [ICD-11: 1C42.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli | 668369 | ||
| Escherichia coli BL21(DE3) | 469008 | |||
| Burkholderia pseudomallei isolates | 28450 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Bps is highly resistant to many antimicrobial agents and this resistance may result from the low drug permeability of outer membrane proteins, known as porins.An Escherichia coli strain defective in most porins, but expressing BpsOmp38, exhibited considerably lower antimicrobial susceptibility than the control strain. In addition, mutation of Tyr119, the most prominent pore-lining residue in BpsOmp38, markedly altered membrane permeability, substitution with Ala (mutant BpsOmp38Y119A) enhanced uptake of the antimicrobial agents, while substitution with Phe (mutant BpsOmp38Y119F) inhibited uptake. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: ErmR rRNA adenine N6-methyltransferase (ERMR) | [7] | |||
| Resistant Disease | Bacteroides fragilis infection [ICD-11: 1C4Y.6] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Bacteroides distasonis strains | 823 | ||
| Bacteroides distasonis strains V2002 | 823 | |||
| Bacteroides distasonis strains V2003 | 823 | |||
| Bacteroides distasonis strains V2004 | 823 | |||
| Bacteroides fragilis strain | 817 | |||
| Bacteroides fragilis strain V503 | 817 | |||
| Bacteroides ovatus strains | 28116 | |||
| Bacteroides ovatus strains V2008 | 28116 | |||
| Bacteroides thetaiotaomicron strain | 818 | |||
| Bacteroides thetaiotaomicron strain V2005 | 818 | |||
| Bacteroides thetaiotaomicron strain V2006 | 818 | |||
| Bacteroides thetaiotaomicron strain V2007 | 818 | |||
| Bacteroides uniformis strain | 820 | |||
| Bacteroides uniformis strain V1760 | 820 | |||
| Bacteroides uniformis strain V1761 | 820 | |||
| Bacteroides uniformis strain V1918 | 820 | |||
| Bacteroides uniformis strain V1921 | 820 | |||
| Bacteroides uniformis strain V2000 | 820 | |||
| Bacteroides uniformis strain V2001 | 820 | |||
| Bacteroides uniformis strain V528 | 820 | |||
| Bacteroides uniformis strain V844 | 820 | |||
| Experiment for Molecule Alteration |
Southern blotting assay | |||
| Mechanism Description | Clindamycin resistance in Bacteroides spp. is usually macrolide-lincosamide-streptogramin B (MLS) resistance conferred by erm genes which are similar to those seen in gram-positive, facultative anaerobes. Of 13 clinical isolates of the Bacteroides group, all were resistant to tetracycline (>10,ug/ml). | |||
| Key Molecule: ErmR rRNA adenine N6-methyltransferase (ERMR) | [7] | |||
| Resistant Disease | Bacteroides uniformis infection [ICD-11: 1C4Y.11] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Bacteroides distasonis strains | 823 | ||
| Bacteroides distasonis strains V2002 | 823 | |||
| Bacteroides distasonis strains V2003 | 823 | |||
| Bacteroides distasonis strains V2004 | 823 | |||
| Bacteroides fragilis strain | 817 | |||
| Bacteroides fragilis strain V503 | 817 | |||
| Bacteroides ovatus strains | 28116 | |||
| Bacteroides ovatus strains V2008 | 28116 | |||
| Bacteroides thetaiotaomicron strain | 818 | |||
| Bacteroides thetaiotaomicron strain V2005 | 818 | |||
| Bacteroides thetaiotaomicron strain V2006 | 818 | |||
| Bacteroides thetaiotaomicron strain V2007 | 818 | |||
| Bacteroides uniformis strain | 820 | |||
| Bacteroides uniformis strain V1760 | 820 | |||
| Bacteroides uniformis strain V1761 | 820 | |||
| Bacteroides uniformis strain V1918 | 820 | |||
| Bacteroides uniformis strain V1921 | 820 | |||
| Bacteroides uniformis strain V2000 | 820 | |||
| Bacteroides uniformis strain V2001 | 820 | |||
| Bacteroides uniformis strain V528 | 820 | |||
| Bacteroides uniformis strain V844 | 820 | |||
| Experiment for Molecule Alteration |
Southern blotting assay | |||
| Mechanism Description | Clindamycin resistance in Bacteroides spp. is usually macrolide-lincosamide-streptogramin B (MLS) resistance conferred by erm genes which are similar to those seen in gram-positive, facultative anaerobes. Of 13 clinical isolates of the Bacteroides group, all were resistant to tetracycline (>10,ug/ml). | |||
| Key Molecule: ErmR rRNA adenine N6-methyltransferase (ERMR) | [7] | |||
| Resistant Disease | Bacteroides thetaiotaomicron infection [ICD-11: 1C4Y.10] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Bacteroides distasonis strains | 823 | ||
| Bacteroides distasonis strains V2002 | 823 | |||
| Bacteroides distasonis strains V2003 | 823 | |||
| Bacteroides distasonis strains V2004 | 823 | |||
| Bacteroides fragilis strain | 817 | |||
| Bacteroides fragilis strain V503 | 817 | |||
| Bacteroides ovatus strains | 28116 | |||
| Bacteroides ovatus strains V2008 | 28116 | |||
| Bacteroides thetaiotaomicron strain | 818 | |||
| Bacteroides thetaiotaomicron strain V2005 | 818 | |||
| Bacteroides thetaiotaomicron strain V2006 | 818 | |||
| Bacteroides thetaiotaomicron strain V2007 | 818 | |||
| Bacteroides uniformis strain | 820 | |||
| Bacteroides uniformis strain V1760 | 820 | |||
| Bacteroides uniformis strain V1761 | 820 | |||
| Bacteroides uniformis strain V1918 | 820 | |||
| Bacteroides uniformis strain V1921 | 820 | |||
| Bacteroides uniformis strain V2000 | 820 | |||
| Bacteroides uniformis strain V2001 | 820 | |||
| Bacteroides uniformis strain V528 | 820 | |||
| Bacteroides uniformis strain V844 | 820 | |||
| Experiment for Molecule Alteration |
Southern blotting assay | |||
| Mechanism Description | Clindamycin resistance in Bacteroides spp. is usually macrolide-lincosamide-streptogramin B (MLS) resistance conferred by erm genes which are similar to those seen in gram-positive, facultative anaerobes. Of 13 clinical isolates of the Bacteroides group, all were resistant to tetracycline (>10,ug/ml). | |||
| Key Molecule: ErmR rRNA adenine N6-methyltransferase (ERMR) | [7] | |||
| Resistant Disease | Bacteroides ovatus infection [ICD-11: 1C4Y.8] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Bacteroides distasonis strains | 823 | ||
| Bacteroides distasonis strains V2002 | 823 | |||
| Bacteroides distasonis strains V2003 | 823 | |||
| Bacteroides distasonis strains V2004 | 823 | |||
| Bacteroides fragilis strain | 817 | |||
| Bacteroides fragilis strain V503 | 817 | |||
| Bacteroides ovatus strains | 28116 | |||
| Bacteroides ovatus strains V2008 | 28116 | |||
| Bacteroides thetaiotaomicron strain | 818 | |||
| Bacteroides thetaiotaomicron strain V2005 | 818 | |||
| Bacteroides thetaiotaomicron strain V2006 | 818 | |||
| Bacteroides thetaiotaomicron strain V2007 | 818 | |||
| Bacteroides uniformis strain | 820 | |||
| Bacteroides uniformis strain V1760 | 820 | |||
| Bacteroides uniformis strain V1761 | 820 | |||
| Bacteroides uniformis strain V1918 | 820 | |||
| Bacteroides uniformis strain V1921 | 820 | |||
| Bacteroides uniformis strain V2000 | 820 | |||
| Bacteroides uniformis strain V2001 | 820 | |||
| Bacteroides uniformis strain V528 | 820 | |||
| Bacteroides uniformis strain V844 | 820 | |||
| Experiment for Molecule Alteration |
Southern blotting assay | |||
| Mechanism Description | Clindamycin resistance in Bacteroides spp. is usually macrolide-lincosamide-streptogramin B (MLS) resistance conferred by erm genes which are similar to those seen in gram-positive, facultative anaerobes. Of 13 clinical isolates of the Bacteroides group, all were resistant to tetracycline (>10,ug/ml). Seven of the eight clindamycin-resistant clinical isolates constitutively expressed erythromycin resistance and had a high level of resistance to clindamycin (> 10ug/ml). V2002 was susceptible to erythromycin. | |||
|
|
||||
| Key Molecule: Beta-lactamase (BLA) | [2] | |||
| Resistant Disease | Bacteroides infection [ICD-11: 1C4Y.7] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli strain HB101 | 634468 | ||
| Escherichia coli strain DH5a | 668369 | |||
| Bacteroides uniformis strain V528 | 820 | |||
| Bacteroides fragilis strain 638 | 862962 | |||
| Bacteroides fragilis strain IB246 | 817 | |||
| Bacteroides fragilis strain IB246flpSUC2C | 817 | |||
| Bacteroides fragilis strain IB247 | 817 | |||
| Bacteroides vulgatus strain CLA341 | 821 | |||
| Experiment for Molecule Alteration |
Nucleotide sequence assay | |||
| Experiment for Drug Resistance |
Standard broth microdilution method assay | |||
| Mechanism Description | The beta-lactamase gene (cfxA) was subcloned on a 2.2-kb DraI-HindIII fragment, and the nucleotide sequence was determined. These results showed that cfxA encoded a protein of 321 amino acids and 35,375 molecular weight. Mutant strains in which the cfxA structural gene was disrupted by insertional inactivation lost both Fxr and beta-lactamase activity. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: CAM-1 carbapenemase (CAM1) | [1] | |||
| Resistant Disease | Pseudomonas infection [ICD-11: 1F45.1] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli TOP10 | 83333 | ||
| Pseudomonas aeruginosa N17-01167 | 287 | |||
| Pseudomonas aeruginosa N17-01173 | 287 | |||
| Pseudomonas aeruginosa N17-02436 | 287 | |||
| Pseudomonas aeruginosa N17-02437 | 287 | |||
| Experiment for Molecule Alteration |
Whole genome sequencing assay | |||
| Experiment for Drug Resistance |
Vitek 2 assay; Etest assay | |||
| Mechanism Description | A novel class B Beta-lactamase gene, blaCAM-1, exhibited resistance to imipenem, meropenem, doripenem, cefotaxime, ceftazidime, cefoxitin, piperacillin/tazobactam, ceftazidime/avibactam and ceftolozane/tazobactam. | |||
ICD-12: Respiratory system diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Bcr/CflA family efflux transporter (BCML) | [6] | |||
| Resistant Disease | Klebsiella pneumoniae infection [ICD-11: CA40.1] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli DH10B | 316385 | ||
| Escherichia coli strain NCTC 50192 | 562 | |||
| Klebsiella pneumoniae strain ORI-1 | 573 | |||
| Experiment for Molecule Alteration |
PCR and hybridization experiments assay | |||
| Experiment for Drug Resistance |
Agar dilution technique assay | |||
| Mechanism Description | Klebsiella pneumoniae ORI-1 strain harbored a ca. 140-kb nontransferable plasmid, pTk1, that conferred an extended-spectrum cephalosporin resistance profile antagonized by the addition of clavulanic acid, tazobactam, or imipenem. The gene for GES-1 (Guiana extended-spectrum beta-lactamase) was cloned, and its protein was expressed in Escherichia coli DH10B, where this pI-5. 8 beta-lactamase of a ca. 31-kDa molecular mass conferred resistance to oxyimino cephalosporins (mostly to ceftazidime). GES-1 is weakly related to the other plasmid-located Ambler class A extended-spectrum beta-lactamases (ESBLs). | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Bcr/CflA family efflux transporter (BCML) | [6] | |||
| Sensitive Disease | Klebsiella pneumoniae infection [ICD-11: CA40.1] | |||
| Molecule Alteration | Expression | Antagonism |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli DH10B | 316385 | ||
| Escherichia coli strain NCTC 50192 | 562 | |||
| Klebsiella pneumoniae strain ORI-1 | 573 | |||
| Experiment for Molecule Alteration |
PCR and hybridization experiments assay | |||
| Experiment for Drug Resistance |
Agar dilution technique assay | |||
| Mechanism Description | Inhibition studies, as measured by IC50 values with benzylpenicillin as the substrate, showed that GES-1 was inhibited by clavulanic acid (5 uM) and tazobactam (2.5 uM) and strongly inhibited by imipenem (0.1 uM). Beta-Lactam MICs were always lowered by the addition of clavulanic acid or tazobactam, less so by sulbactam, and uncommonly by imipenem. | |||
| Key Molecule: OmpK37 (OMPK37) | [13] | |||
| Sensitive Disease | Klebsiella pneumoniae infection [ICD-11: CA40.1] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli | 668369 | ||
| Klebsiella pneumoniae strain CSUB10R | 573 | |||
| Klebsiella pneumoniae strain CSUB10S | 573 | |||
| Klebsiella pneumoniae strain LB4 | 573 | |||
| Klebsiella pneumoniae strain LB66 | 573 | |||
| Klebsiella pneumoniae strain SD8 | 573 | |||
| Experiment for Molecule Alteration |
Southern blotting assay | |||
| Experiment for Drug Resistance |
Microdilution method assay | |||
| Mechanism Description | Due to its porin deficiency, strain CSUB10R is more resistant to Beta-lactams than is parental strain CSUB10S. As expected, for k. pneumoniae CSUB10R expressing Ompk36 or Ompk35, the MICs reverted to values similar to those observed for strain CSUB10S. | |||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
