Molecule Information
General Information of the Molecule (ID: Mol00119)
| Name |
Multidrug resistance-associated protein 1 (MRP1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
ATP-binding cassette sub-family C member 1; Glutathione-S-conjugate-translocating ATPase ABCC1; Leukotriene C(4) transporter; LTC4 transporter; MRP; MRP1
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
ABCC1
|
||||
| Gene ID | |||||
| Location |
chr16:15949138-16143257[+]
|
||||
| Sequence |
MALRGFCSADGSDPLWDWNVTWNTSNPDFTKCFQNTVLVWVPCFYLWACFPFYFLYLSRH
DRGYIQMTPLNKTKTALGFLLWIVCWADLFYSFWERSRGIFLAPVFLVSPTLLGITMLLA TFLIQLERRKGVQSSGIMLTFWLVALVCALAILRSKIMTALKEDAQVDLFRDITFYVYFS LLLIQLVLSCFSDRSPLFSETIHDPNPCPESSASFLSRITFWWITGLIVRGYRQPLEGSD LWSLNKEDTSEQVVPVLVKNWKKECAKTRKQPVKVVYSSKDPAQPKESSKVDANEEVEAL IVKSPQKEWNPSLFKVLYKTFGPYFLMSFFFKAIHDLMMFSGPQILKLLIKFVNDTKAPD WQGYFYTVLLFVTACLQTLVLHQYFHICFVSGMRIKTAVIGAVYRKALVITNSARKSSTV GEIVNLMSVDAQRFMDLATYINMIWSAPLQVILALYLLWLNLGPSVLAGVAVMVLMVPVN AVMAMKTKTYQVAHMKSKDNRIKLMNEILNGIKVLKLYAWELAFKDKVLAIRQEELKVLK KSAYLSAVGTFTWVCTPFLVALCTFAVYVTIDENNILDAQTAFVSLALFNILRFPLNILP MVISSIVQASVSLKRLRIFLSHEELEPDSIERRPVKDGGGTNSITVRNATFTWARSDPPT LNGITFSIPEGALVAVVGQVGCGKSSLLSALLAEMDKVEGHVAIKGSVAYVPQQAWIQND SLRENILFGCQLEEPYYRSVIQACALLPDLEILPSGDRTEIGEKGVNLSGGQKQRVSLAR AVYSNADIYLFDDPLSAVDAHVGKHIFENVIGPKGMLKNKTRILVTHSMSYLPQVDVIIV MSGGKISEMGSYQELLARDGAFAEFLRTYASTEQEQDAEENGVTGVSGPGKEAKQMENGM LVTDSAGKQLQRQLSSSSSYSGDISRHHNSTAELQKAEAKKEETWKLMEADKAQTGQVKL SVYWDYMKAIGLFISFLSIFLFMCNHVSALASNYWLSLWTDDPIVNGTQEHTKVRLSVYG ALGISQGIAVFGYSMAVSIGGILASRCLHVDLLHSILRSPMSFFERTPSGNLVNRFSKEL DTVDSMIPEVIKMFMGSLFNVIGACIVILLATPIAAIIIPPLGLIYFFVQRFYVASSRQL KRLESVSRSPVYSHFNETLLGVSVIRAFEEQERFIHQSDLKVDENQKAYYPSIVANRWLA VRLECVGNCIVLFAALFAVISRHSLSAGLVGLSVSYSLQVTTYLNWLVRMSSEMETNIVA VERLKEYSETEKEAPWQIQETAPPSSWPQVGRVEFRNYCLRYREDLDFVLRHINVTINGG EKVGIVGRTGAGKSSLTLGLFRINESAEGEIIIDGINIAKIGLHDLRFKITIIPQDPVLF SGSLRMNLDPFSQYSDEEVWTSLELAHLKDFVSALPDKLDHECAEGGENLSVGQRQLVCL ARALLRKTKILVLDEATAAVDLETDDLIQSTIRTQFEDCTVLTIAHRLNTIMDYTRVIVL DKGEIQEYGAPSDLLQQRGLFYSMAKDAGLV Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Mediates export of organic anions and drugs from the cytoplasm. Mediates ATP-dependent transport of glutathione and glutathione conjugates, leukotriene C4, estradiol-17-beta-o-glucuronide, methotrexate, antiviral drugs and other xenobiotics. Confers resistance to anticancer drugs by decreasing accumulation of drug in cells, and by mediating ATP- and GSH-dependent drug export. Hydrolyzes ATP with low efficiency. Catalyzes the export of sphingosine 1-phosphate from mast cells independently of their degranulation. Participates in inflammatory response by allowing export of leukotriene C4 from leukotriene C4-synthezing cells.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
16 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lung cancer [ICD-11: 2C25.5] | [1] | |||
| Resistant Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | |||
| The Specified Disease | Lung cancer | |||
| The Studied Tissue | Lung tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.90E-30 Fold-change: 7.04E-02 Z-score: 1.19E+01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell invasion | Activation | hsa05200 | ||
| Cell migration | Activation | hsa04670 | ||
| Cell proliferation | Activation | hsa05200 | ||
| TGF-beta/Smad2/STAT3/STAT5 signaling pathway | Activation | hsa04350 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay; Flow cytometry assay | |||
| Mechanism Description | miR-10a had an important role in promoting drug resistance in tumors through enhancing drug efflux and inhibiting apoptosis via upregulation of MDR1, MRP1 and RhoE expression. In addition, miR-10a promoted the expression of TGF-beta as wells as regulated the activity of the Smad2/STAT3/STAT5 pathway and its downstream transcriptional factors of HIF and eIF4E, which may be the potential mechanism of drug resistance in A549 cells. Therefore, miR-10a may be an important drug target for improving cancer treatment; however, further studies are required to explore the clinical applications of miR-10a inhibitors. | |||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [3] | |||
| Resistant Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Ovarian cancer [ICD-11: 2C73] | |||
| The Specified Disease | Ovarian cancer | |||
| The Studied Tissue | Ovarian tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.01E-02 Fold-change: 1.42E-01 Z-score: 3.33E+00 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Ovarian cancer tissue | N.A. | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Efficacy evaluation of chemotherapy | |||
| Mechanism Description | Ovarian cancer tissues had much higher expression levels of MRP1, GST-pai, and GSK3beta mRNA than normal ovarian tissues (P<0.05). The expression levels of MRP1, GST-pai, and GSK3beta mRNA in the Chemotherapy-sensitive group were significantly lower than those in the Chemotherapy-resistant group (P<0.05). Patients with high expression of MRP1, GST-pai, and GSK3beta mRNA had a much lower 3-year survival rate than patients with low expression of the genes (P<0.05). Highly expressed in patients with ovarian cancer, MRP1, GST-pai, and GSK3beta mRNA play an important role in the development and drug resistance of ovarian cancer. | |||
| Disease Class: Oral squamous cell carcinoma [ICD-11: 2B6E.0] | [6] | |||
| Resistant Disease | Oral squamous cell carcinoma [ICD-11: 2B6E.0] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Oral squamous cell carcinoma [ICD-11: 2B6E] | |||
| The Specified Disease | Oral cancer | |||
| The Studied Tissue | Oral tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.17E-05 Fold-change: 1.19E-01 Z-score: 4.53E+00 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Caspase-3 signaling pathway | Activation | hsa04210 | |
| In Vitro Model | CAL27 cells | Oral | Homo sapiens (Human) | CVCL_1107 |
| HSC3 cells | Tongue | Homo sapiens (Human) | CVCL_1288 | |
| HaCaT cells | Tongue | Homo sapiens (Human) | CVCL_0038 | |
| OSCC3 cells | Tongue | Homo sapiens (Human) | CVCL_L894 | |
| SCC4 cells | Tongue | Homo sapiens (Human) | CVCL_1684 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Midkine derived from cancer-associated fibroblasts promotes cisplatin-resistance via up-regulation of the expression of LncRNA ANRIL in tumour cells. ANRIL knockdown overcomes Mk-induced cisplatin resistance via activation of caspase-3-dependent apoptosis. Overexpression of LncRNA ANRIL promots the up-regulation of ABC family proteins MRP1 and ABCC2, which ultimately results in tumour cell resistance to cisplatin. | |||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [7] | |||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| BGC823 cells | Gastric | Homo sapiens (Human) | CVCL_3360 | |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | LncRNA SNHG5 promotes cisplatin resistance in gastric cancer via inhibiting cell apoptosis and upregulating drug resistance-related genes. | |||
| Disease Class: Lung cancer [ICD-11: 2C25.5] | [8] | |||
| Resistant Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell colony | Activation | hsa05200 | ||
| Cell invasion | Activation | hsa05200 | ||
| Cell viability | Activation | hsa05200 | ||
| STAT3 signaling pathway | Activation | hsa04550 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay; TUNEL assay | |||
| Mechanism Description | LncRNA-MALAT1 contributes to the cisplatin-resistance of lung cancer by upregulating MRP1 and MDR1 via STAT3 activation. | |||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [9] | |||
| Resistant Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell viability | Activation | hsa05200 | ||
| Intrinsic apoptotic signaling pathway | Inhibition | hsa04210 | ||
| In Vitro Model | HT29 Cells | Colon | Homo sapiens (Human) | CVCL_A8EZ |
| SW480 cells | Colon | Homo sapiens (Human) | CVCL_0546 | |
| HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |
| LOVO cells | Colon | Homo sapiens (Human) | CVCL_0399 | |
| RkO cells | Colon | Homo sapiens (Human) | CVCL_0504 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; TUNEL assay; Flow cytometry assay | |||
| Mechanism Description | PVT1 involved in cisplatin resistance of CRC cells via upregulation of drug resistance-associated molecules, including multidrug resistance 1 (MDR1) and multidrug resistance protein 1 (MRP1), by blocking the intrinsic apoptotic pathway. | |||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [10] | |||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| mTOR/HIF-1alpha /P-gp/MRP1 signaling pathway | Regulation | N.A. | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| BGC823 cells | Gastric | Homo sapiens (Human) | CVCL_3360 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | Overexpression of long non-coding RNA PVT1 in gastric cancer cells promotes the development of multidrug resistance.PVT-1 was highly expressed in gastric cancer tissues of cisplatin-resistant patients and cisplatin-resistant cells. While, PVT1 overexpression exhibit the anti-apoptotic property in BGC823 and SGC7901 cells transfected with LV-PVT1-GFP and treated with cisplatin. Moreover, qRT-PCR and western blotting revealed that PVT1 up-regulation increased the expression of MDR1, MRP, mTOR and HIF-1alpha. Overexpression of LncRNA PVT1 in gastric carcinoma promotes the development of MDR, suggesting an efficacious target for reversing MDR in gastric cancer therapy. | |||
| Disease Class: Prostate cancer [ICD-11: 2C82.0] | [4] | |||
| Resistant Disease | Prostate cancer [ICD-11: 2C82.0] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | PC3 cells | Prostate | Homo sapiens (Human) | CVCL_0035 |
| 22RV1 cells | Prostate | Homo sapiens (Human) | CVCL_1045 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | However, higher concentrations of probenecid (500 uM) failed to demonstrate a chemosensitizing effect. Consistent with this lower chemosensitizing efficacy in higher-concentration probenecid treatment, we observed that the expression of ABCG2, a drug-efflux transporter, increased in a dose-dependent manner following probenecid treatment. Thus, probenecid could enhance the chemosensitivity of 3D-cultured prostate cancer cells, but not at higher concentr. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [11] | |||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| A549/DDP cells | Lung | Homo sapiens (Human) | CVCL_0023 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; TUNEL assay | |||
| Mechanism Description | miR185-5p exhibited negative correlation with ABCC1 in A549/DDP cells., inhibition of miR185-5p was involved in chemo-resistance of NSCLC cells to cisplatin via down-regulating ABCC1. | |||
| Disease Class: Lung cancer [ICD-11: 2C25.5] | [12] | |||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell viability | Inhibition | hsa05200 | |
| Wnt signaling pathway | Inhibition | hsa04310 | ||
| In Vitro Model | H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Si-HOTAIR interference significantly increased the sensitivity of cells to DDP, the IC50 of cells was decreased from 131.85 to 44.34 M (P<0.05), the expression levels of MRP1 and MDR1 were significantly decreased, and the activation of Wnt signaling pathway was significantly inhibited. | |||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [13] | |||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | miR-21 mainly achieves drug resistance by inhibiting cisplatin-induced apoptosis, and its specific mechanisms include the following: (1) improving the expression level of EGFR and protecting the toxic effect of tumor cells during chemotherapy; (2) Increase the expression of LRP and decrease the effective concentration of the target drug through the barrier of drug transport between nucleus and cell; (3) Enhance the expression of multidrug resistance associated protein (MRP1) and assist in pumping chemotherapeutic drugs from the inside to the outside of the cell. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hepatocellular carcinoma [ICD-11: 2C12.2] | [2] | |||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Resistant Drug | Oxaliplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Liver cancer [ICD-11: 2C12] | |||
| The Specified Disease | Liver cancer | |||
| The Studied Tissue | Liver tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.12E-23 Fold-change: 1.52E-01 Z-score: 1.09E+01 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell viability | Activation | hsa05200 | |
| miR7-5p/ABCC1 signaling pathway | Regulation | N.A. | ||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| SMMC7721 cells | Uterus | Homo sapiens (Human) | CVCL_0534 | |
| Skhep1 cells | Liver | Homo sapiens (Human) | CVCL_0525 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Overexpression of kCNQ1OT1 enhances OXA resistance through downregulating miR-7-5p and upregulating ABCC1 in HCC cells. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Prostate cancer [ICD-11: 2C82.0] | [4] | |||
| Resistant Disease | Prostate cancer [ICD-11: 2C82.0] | |||
| Resistant Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Prostate cancer [ICD-11: 2C82] | |||
| The Specified Disease | Prostate cancer | |||
| The Studied Tissue | Prostate | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.18E-07 Fold-change: 1.22E-01 Z-score: 6.14E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | PC3 cells | Prostate | Homo sapiens (Human) | CVCL_0035 |
| 22RV1 cells | Prostate | Homo sapiens (Human) | CVCL_1045 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | However, higher concentrations of probenecid (500 uM) failed to demonstrate a chemosensitizing effect. Consistent with this lower chemosensitizing efficacy in higher-concentration probenecid treatment, we observed that the expression of ABCG2, a drug-efflux transporter, increased in a dose-dependent manner following probenecid treatment. Thus, probenecid could enhance the chemosensitivity of 3D-cultured prostate cancer cells, but not at higher concentr. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [15] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| PI3K/AKT/mTOR signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Colony formation assay | |||
| Mechanism Description | The protein levels of MDR1, MRP1 and ABCB1 were significantly decreased in DOXR-MCF-7 siR-HOTAIR1 cells compared with the siR-NC DOXR-MCF-7 cells and HOTAIR silencing reduces the sensitivity of drug resistance in drug-resistant MCF-7 cells. | |||
| Disease Class: Osteosarcoma [ICD-11: 2B51.0] | [16] | |||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| SAOS-2 cells | Bone marrow | Homo sapiens (Human) | CVCL_0548 | |
| HOS cells | Bone | Homo sapiens (Human) | CVCL_0312 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometric analysis | |||
| Mechanism Description | LncRNA FENDRR sensitizes doxorubicin-resistance of osteosarcoma cells through down-regulating ABCB1 and ABCC1. | |||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [17] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| miR199a/MRP1 signaling pathway | Regulation | N.A. | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
XTT assay; Flow cytometry assay; Caspase 9 activity assay | |||
| Mechanism Description | Linc00518 downregulation reduced MDR by upregulating miR-199a which downregulates MRP1 in breast cancer. | |||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [18] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| MDA-MB-453 cells | Breast | Homo sapiens (Human) | CVCL_0418 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |
| MDA-kb2 cells | Breast | Homo sapiens (Human) | CVCL_6421 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-145 suppressed MRP1 expression by directly targeting MRP1 3'-untranslated regions. Overexpression of miR-145 sensitized breast cancer cells to doxorubicin in vitro and (+) to doxorubicin chemotherapy in vivo through inducing intracellular doxorubicin accumulation via inhibiting MRP1. | |||
| Disease Class: Lung cancer [ICD-11: 2C25.5] | [19] | |||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | AKT/ERK signaling pathway | Inhibition | hsa04010 | |
| Cell apoptosis | Activation | hsa04210 | ||
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Suppression of miR-155 in this cell line considerably reversed doxorubicin resistance, and doxorubicin-induced apoptosis and cell cycle arrest were recovered. Furthermore, reverse transcription-polymerase chain reaction and western blot analysis revealed that miR-155 suppression downregulated the expression of multidrug resistance protein 1, multidrug resistance-associated protein 1, breast cancer resistance protein, glutathione S-transferase-Pi, Survivin and B-cell lymphoma 2, and upregulated the expression of caspase-3 and caspase-8. In addition, it was found that miR-155 suppression inhibited the activation of AkT and extracellular signal-regulated kinase. The transcriptional activity of nuclear factor-kB and activator protein-1 was also downregulated. | |||
| Disease Class: Glioma [ICD-11: 2A00.1] | [20] | |||
| Sensitive Disease | Glioma [ICD-11: 2A00.1] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | AKT signaling pathway | Inhibition | hsa04151 | |
| Cell apoptosis | Activation | hsa04210 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| U87-MG cells | Brain | Homo sapiens (Human) | CVCL_0022 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay; Flow cytometry assay | |||
| Mechanism Description | microRNA-127 silencing significantly affects cell growth and increases the sensitivity to adriamycin. microRNA-127 silencing arrests the cell cycle, potentiates adriamycin-induced apoptosis, and increases cellular Rh-123 uptake. microRNA-127 silencing down-regulates MDR1, MRP1, Runx2, Bcl-2, Survivin and ErbB4 expression while up-regulates p53 expression. microRNA-127 silencing inhibits AkT phosphorylation. | |||
| Disease Class: Pancreatic carcinoma [ICD-11: 2C10.2] | [21] | |||
| Sensitive Disease | Pancreatic carcinoma [ICD-11: 2C10.2] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell growth | Inhibition | hsa05200 | |
| In Vitro Model | H69 cells | Lung | Homo sapiens (Human) | CVCL_8121 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Hsa-miR-1291-directed downregulation of ABCC1 led to a greater intracellular drug accumulation and sensitized the cells to doxorubicin. | |||
| Disease Class: Lung small cell carcinoma [ICD-11: 2C25.2] | [21] | |||
| Sensitive Disease | Lung small cell carcinoma [ICD-11: 2C25.2] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell growth | Inhibition | hsa05200 | |
| In Vitro Model | PANC-1 cells | Pancreas | Homo sapiens (Human) | CVCL_0480 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Hsa-miR-1291-directed downregulation of ABCC1 led to a greater intracellular drug accumulation and sensitized the cells to doxorubicin. | |||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [22] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell proliferation | Activation | hsa05200 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | The elevated levels of miR-326 in the mimics-transfected VP-16-resistant cell line, MCF-7/VP, downregulated MRP-1 expression and sensitized these cells to VP-16 and doxorubicin. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Malignant glioma [ICD-11: 2A00.2] | [5] | |||
| Resistant Disease | Malignant glioma [ICD-11: 2A00.2] | |||
| Resistant Drug | Temozolomide | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Brain cancer [ICD-11: 2A00] | |||
| The Specified Disease | Brain cancer | |||
| The Studied Tissue | Nervous tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.59E-117 Fold-change: 1.21E-01 Z-score: 2.56E+01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| U87 cells | Brain | Homo sapiens (Human) | CVCL_0022 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Knockdown of long noncoding RNA H19 sensitizes human glioma cells to temozolomide therapy.the expression level of H19 transcripts was increased compared to wild-type or nonresistant clones.Furthermore, the reduced expression of H19 altered major drug resistance genes, such as ABCB1 (MDR1), ABCC (MRP), and ABCG2 (BCRP), both at the mRNA and protein levels. Taken together, these findings suggest that H19 plays an important role in the development of TMZ resistance, and may represent a novel therapeutic target for TMZ-resistant gliomas.Our results suggested that knockdown of H19 significantly downregulated the expression of these drug-resistant genes, both at the mRNA (P<0.001 vs respective control siRNA) and protein levels. These data confirm that the H19-induced TMZ resistance is in part mediated by MDR, MRP, and ABCG2. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Glioblastoma [ICD-11: 2A00.02] | [34] | |||
| Sensitive Disease | Glioblastoma [ICD-11: 2A00.02] | |||
| Sensitive Drug | Temozolomide | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell viability | Inhibition | hsa05200 | |
| Hedgehog signaling pathway | Inhibition | hsa04340 | ||
| MAPK signaling pathway | Inhibition | hsa04010 | ||
| p53 signaling pathway | Inhibition | hsa04115 | ||
| In Vitro Model | LN229 cells | Brain | Homo sapiens (Human) | CVCL_0393 |
| U87 cells | Brain | Homo sapiens (Human) | CVCL_0022 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis; RIP assay; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Overexpression of miR-1268a inhibited protein translation of ABCC1, which enhanced sensitivity of GBM cells to TMZ. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer bone metastasis [ICD-11: 2E03.1] | [14] | |||
| Resistant Disease | Breast cancer bone metastasis [ICD-11: 2E03.1] | |||
| Resistant Drug | Docetaxel | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Flow cytometric | |||
| Mechanism Description | Studies have shown that resistance can occur from altered expression of apoptosis regulatory proteins (p53, Bcl-2), and overexpression of ATP binding cassette (ABC) transporters/ multidrug resistance-related proteins such as multidrug resistance-associated protein 1 (MRP1), ATP-binding cassette super-family G member 2 (ABCG2). | |||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [3] | |||
| Resistant Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Resistant Drug | Docetaxel | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Ovarian cancer tissue | N.A. | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Efficacy evaluation of chemotherapy | |||
| Mechanism Description | Ovarian cancer tissues had much higher expression levels of MRP1, GST-pai, and GSK3beta mRNA than normal ovarian tissues (P<0.05). The expression levels of MRP1, GST-pai, and GSK3beta mRNA in the Chemotherapy-sensitive group were significantly lower than those in the Chemotherapy-resistant group (P<0.05). Patients with high expression of MRP1, GST-pai, and GSK3beta mRNA had a much lower 3-year survival rate than patients with low expression of the genes (P<0.05). Highly expressed in patients with ovarian cancer, MRP1, GST-pai, and GSK3beta mRNA play an important role in the development and drug resistance of ovarian cancer. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hypertrophic scar [ICD-11: EE60.0] | [23] | |||
| Resistant Disease | Hypertrophic scar [ICD-11: EE60.0] | |||
| Resistant Drug | Etoposide | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Hypertrophic scar tissue isolates | N.A. | ||
| Experiment for Molecule Alteration |
Western blotting assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Fibroblasts derived from hypertrophic scar and normal skin tissues were first compared for their resistance to verapamil and etoposide phosphate. Scar fibroblasts showed stronger resistance to both verapamil and etoposide than normal fibroblasts, also scar fibroblasts expressed more P-glycoprotein and MRP1 than normal fibroblasts. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [22] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Etoposide | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell proliferation | Activation | hsa05200 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | The elevated levels of miR-326 in the mimics-transfected VP-16-resistant cell line, MCF-7/VP, downregulated MRP-1 expression and sensitized these cells to VP-16 and doxorubicin. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [24] | |||
| Resistant Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Resistant Drug | Fluorouracil | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell migration | Activation | hsa04670 | ||
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | HT29 Cells | Colon | Homo sapiens (Human) | CVCL_A8EZ |
| SW480 cells | Colon | Homo sapiens (Human) | CVCL_0546 | |
| HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |
| LOVO cells | Colon | Homo sapiens (Human) | CVCL_0399 | |
| RkO cells | Colon | Homo sapiens (Human) | CVCL_0504 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Colony formation assay; Transwell assays and wound healing assay; Flow cytometry assay | |||
| Mechanism Description | ANRIL promotes chemoresistance via disturbing expression of ABCC1 by inhibiting the expression of Let-7a in colorectal cancer. | |||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [25] | |||
| Resistant Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Resistant Drug | Fluorouracil | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell viability | Activation | hsa05200 | ||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| HCT-8 cells | Colon | Homo sapiens (Human) | CVCL_2478 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | The overexpression of PVT1 increased the mRNA and protein expression levels of multidrug resistance associated protein 1, P glycoprotein, serine/threonine protein kinase mTOR and apoptosis regulator Bcl2. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Chronic myeloid leukemia [ICD-11: 2A20.0] | [26] | |||
| Resistant Disease | Chronic myeloid leukemia [ICD-11: 2A20.0] | |||
| Resistant Drug | Imatinib | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | PI3K/AKT signaling pathway | Activation | hsa04151 | |
| In Vitro Model | K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 |
| K562-R cells | Pleural effusion | Homo sapiens (Human) | CVCL_5950 | |
| Experiment for Molecule Alteration |
qRT-PCR; Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay; Annexin V/propidium iodide staining assay | |||
| Mechanism Description | Knockdown of HOTAIR expression downregulated the MRP1 expression levels in the k562-imatinib cells and resulted in higher sensitivity to the imatinib treatment. In addition, the activation of PI3k/Akt was greatly attenuated when HOTAIR was knocked down in k562-imatinib cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Chronic myeloid leukemia [ICD-11: 2A20.0] | [27] | |||
| Sensitive Disease | Chronic myeloid leukemia [ICD-11: 2A20.0] | |||
| Sensitive Drug | Imatinib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Annexin V-FITC/PI Apoptosis Detection assay | |||
| Mechanism Description | Overexpression of MEG3 in imatinib-resistant k562 cells markedly decreased cell proliferation, increased cell apoptosis, reversed imatinib resistance, and reduced the expression of MRP1, MDR1, and ABCG2. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [28] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Indomethacin | |||
| Molecule Alteration | Function | Inhibition |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Mechanism Description | Vitamin E d-alpha-tocopheryl poly(ethylene glycol) 1000 succinate (TPGS) and indomethacin (IDM) can reverse multidrug resistance (MDR) via inhibiting P-glycoprotein (P-gp) and multidrug resistance associated protein 1 (MRP1) respectively, but their drawbacks in physicochemical properties limit their clinical application. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Mature B-cell neoplasms [.] | [29] | |||
| Sensitive Disease | Mature B-cell neoplasms [.] | |||
| Sensitive Drug | Inotuzumab ozogamicin | |||
| Molecule Alteration | Function | C2617T/A; A2063G |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | pre-InO and/or post-InO tumor cells | N.A. | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
GeneSeq assay; Mutation assay | |||
| Mechanism Description | Multiple mechanisms drive CD22 antigen escape, including epitope loss (protein truncation and destabilization) and epitope alteration.Hypermutation caused by error-prone DNA damage repair may serve as a driver of CD22 mutation and escape. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Chronic myeloid leukemia [ICD-11: 2A20.0] | [30] | |||
| Sensitive Disease | Chronic myeloid leukemia [ICD-11: 2A20.0] | |||
| Sensitive Drug | Ivermectin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | K562/FLM cells | Blood | Homo sapiens (Human) | CVCL_E7CM |
| Experiment for Molecule Alteration |
Western blot assay | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | It was found that ivermectin effectively suppressed the expression of autophagy and transport proteins in K562/FLM cells, reduced the activity of the aforementioned phosphoproteins, and promoted apoptotic cell death. The significant effects of ivermectin might offer a novel therapeutic strategy to overcome flumatinib resistance and optimize the treatment outcomes of CML. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Retinoblastoma [ICD-11: 2D02.2] | [31] | |||
| Resistant Disease | Retinoblastoma [ICD-11: 2D02.2] | |||
| Resistant Drug | Melphalan | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Similar Genomic Alterations but Distinctive Expression of Influx/Efflux Transporters Between Chemoresistant and Parental Cells | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Epithelial ovarian cancer [ICD-11: 2B5D.0] | [32] | |||
| Resistant Disease | Epithelial ovarian cancer [ICD-11: 2B5D.0] | |||
| Resistant Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell invasion | Activation | hsa05200 | ||
| Cell migration | Activation | hsa04670 | ||
| In Vitro Model | SkOV3 cells | Ovary | Homo sapiens (Human) | CVCL_0532 |
| A2780 cells | Ovary | Homo sapiens (Human) | CVCL_0134 | |
| COC1 cells | Ovary | Homo sapiens (Human) | CVCL_6891 | |
| SkOV3-TR30 cells | Ovary | Homo sapiens (Human) | CVCL_0532 | |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
MTS assay; Flow cytometry assay | |||
| Mechanism Description | LINC01118 Can enhance ABCC1 expression by suppressing miR-134 expression to promote paclitaxel resistance in epithelial ovarian cancer. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [33] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7/PR cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Sulforhodamine B assay | |||
| Mechanism Description | Down-regulation of LncRNA RP11-770J1.3 and TMEM25 enhanced the sensitivity of MCF-7/PR cells to paclitaxel, and inhibited the expression of MRP, BCRP and MDR1/P-gp. | |||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [17] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| miR199a/MRP1 signaling pathway | Regulation | N.A. | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
XTT assay; Flow cytometry assay; Caspase 9 activity assay | |||
| Mechanism Description | Linc00518 downregulation reduced MDR by upregulating miR-199a which downregulates MRP1 in breast cancer. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Prostate cancer [ICD-11: 2C82.0] | [4] | |||
| Sensitive Disease | Prostate cancer [ICD-11: 2C82.0] | |||
| Sensitive Drug | Probenecid | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell colony | Inhibition | hsa05200 | |
| In Vitro Model | PC3 cells | Prostate | Homo sapiens (Human) | CVCL_0035 |
| 22RV1 cells | Prostate | Homo sapiens (Human) | CVCL_1045 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | However, probenecid only weakly inhibits ABCG2. Thus, probenecid enhanced the efficacy of anticancer drugs against 22Rv1 spheroids by inhibiting drug resistance-related transporters such as MRP; at high probenecid concentrations, the chemosensitization effect may be reduced owing to promotion of alternate drug excretion pathways via upregulated ABCG2 expression. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hypertrophic scar [ICD-11: EE60.0] | [23] | |||
| Resistant Disease | Hypertrophic scar [ICD-11: EE60.0] | |||
| Resistant Drug | Verapamil | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Hypertrophic scar tissue isolates | N.A. | ||
| Experiment for Molecule Alteration |
Western blotting assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Fibroblasts derived from hypertrophic scar and normal skin tissues were first compared for their resistance to verapamil and etoposide phosphate. Scar fibroblasts showed stronger resistance to both verapamil and etoposide than normal fibroblasts, also scar fibroblasts expressed more P-glycoprotein and MRP1 than normal fibroblasts. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [17] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Vincristine | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| miR199a/MRP1 signaling pathway | Regulation | N.A. | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
XTT assay; Flow cytometry assay; Caspase 9 activity assay | |||
| Mechanism Description | Linc00518 downregulation reduced MDR by upregulating miR-199a which downregulates MRP1 in breast cancer. | |||
Clinical Trial Drug(s)
2 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Chronic myeloid leukemia [ICD-11: 2A20.0] | [30] | |||
| Resistant Disease | Chronic myeloid leukemia [ICD-11: 2A20.0] | |||
| Resistant Drug | Flumatinib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | K562/FLM cells | Blood | Homo sapiens (Human) | CVCL_E7CM |
| Experiment for Molecule Alteration |
Western blot assay | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | Through cellular experimentation, we explored the resistance mechanisms, which indicated that K562/FLM cells evade flumatinib cytotoxicity by enhancing autophagy, increasing the expression of membrane transport proteins, particularly P-glycoprotein, ABCC1 and ABCC4, as well as enhancing phosphorylation of p-EGFR, p-ERK and p-STAT3 proteins. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Thyroid gland cancer [ICD-11: 2D10.0] | [35] | |||
| Sensitive Disease | Thyroid gland cancer [ICD-11: 2D10.0] | |||
| Sensitive Drug | BI-847325 | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SW1736 cells | Thyroid | Homo sapiens (Human) | CVCL_3883 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Apoptosis assay | |||
| Mechanism Description | Suppression of MAPK signaling pathway activity by BI-847325 treatment could significantly decrease the expression of?MDR1?and?MRP1?genes in C643 and SW1736 ATC cell lines. BI-847325 decreased multidrug resistance through downregulation of MDR1 and MRP1. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Nervous tissue | |
| The Specified Disease | Brain cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.59E-117; Fold-change: 5.51E-01; Z-score: 1.85E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
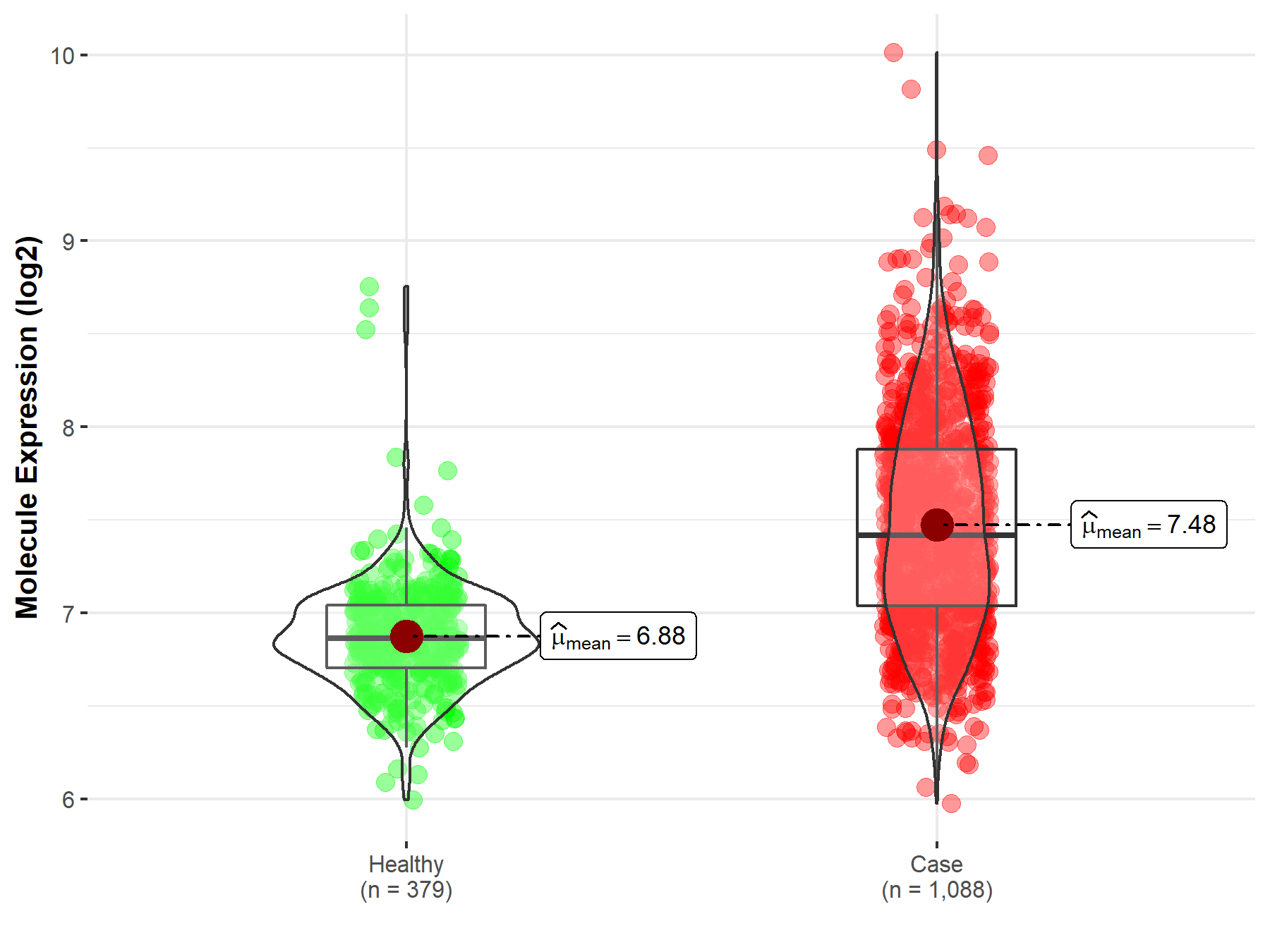
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.13E-02; Fold-change: 3.90E-01; Z-score: 1.94E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| The Studied Tissue | White matter | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.63E-06; Fold-change: 1.04E+00; Z-score: 2.01E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
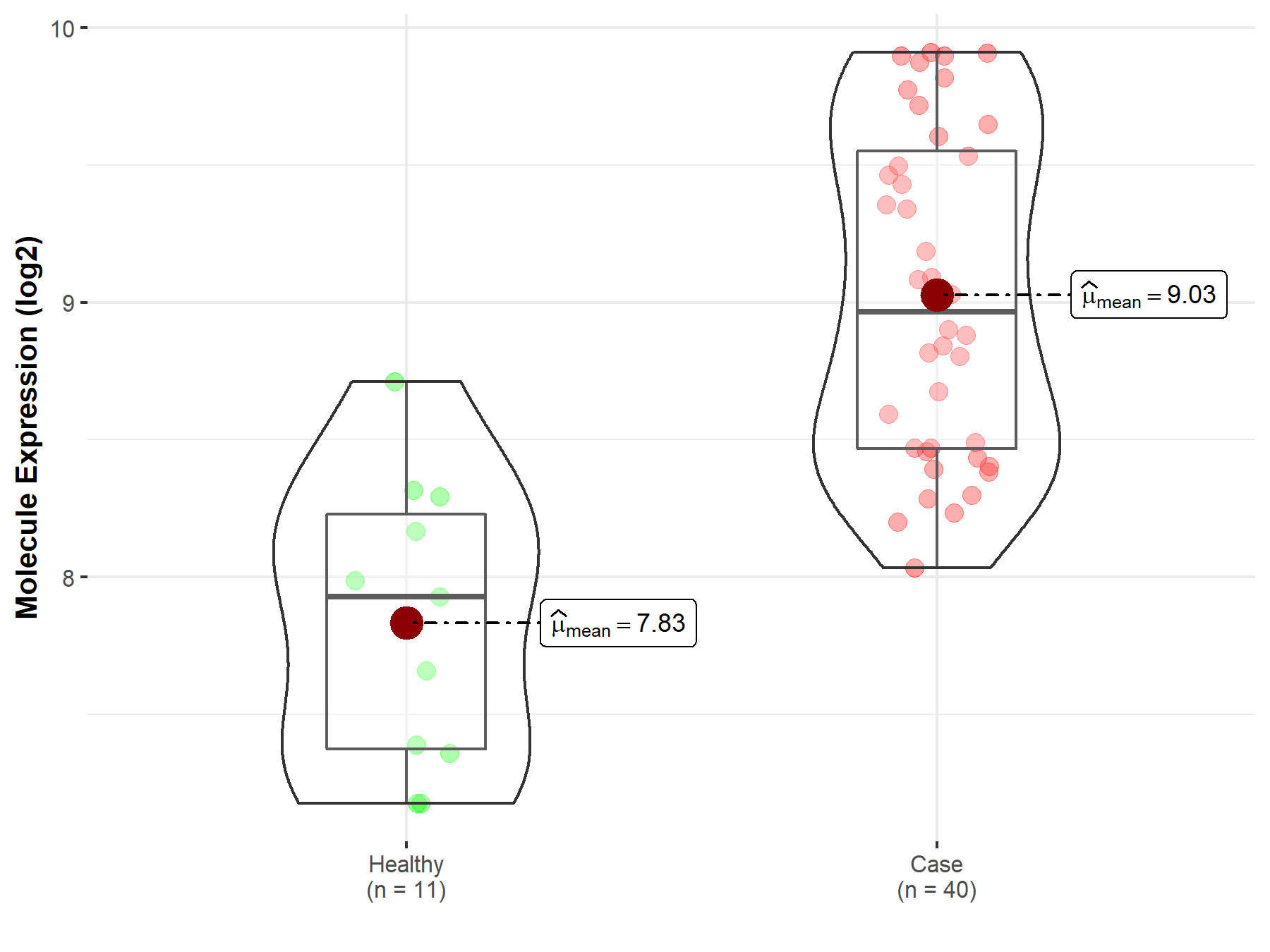
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Neuroectodermal tumor | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.68E-08; Fold-change: 5.65E-01; Z-score: 3.11E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Whole blood | |
| The Specified Disease | Myelofibrosis | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.79E-03; Fold-change: -2.19E-01; Z-score: -1.94E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
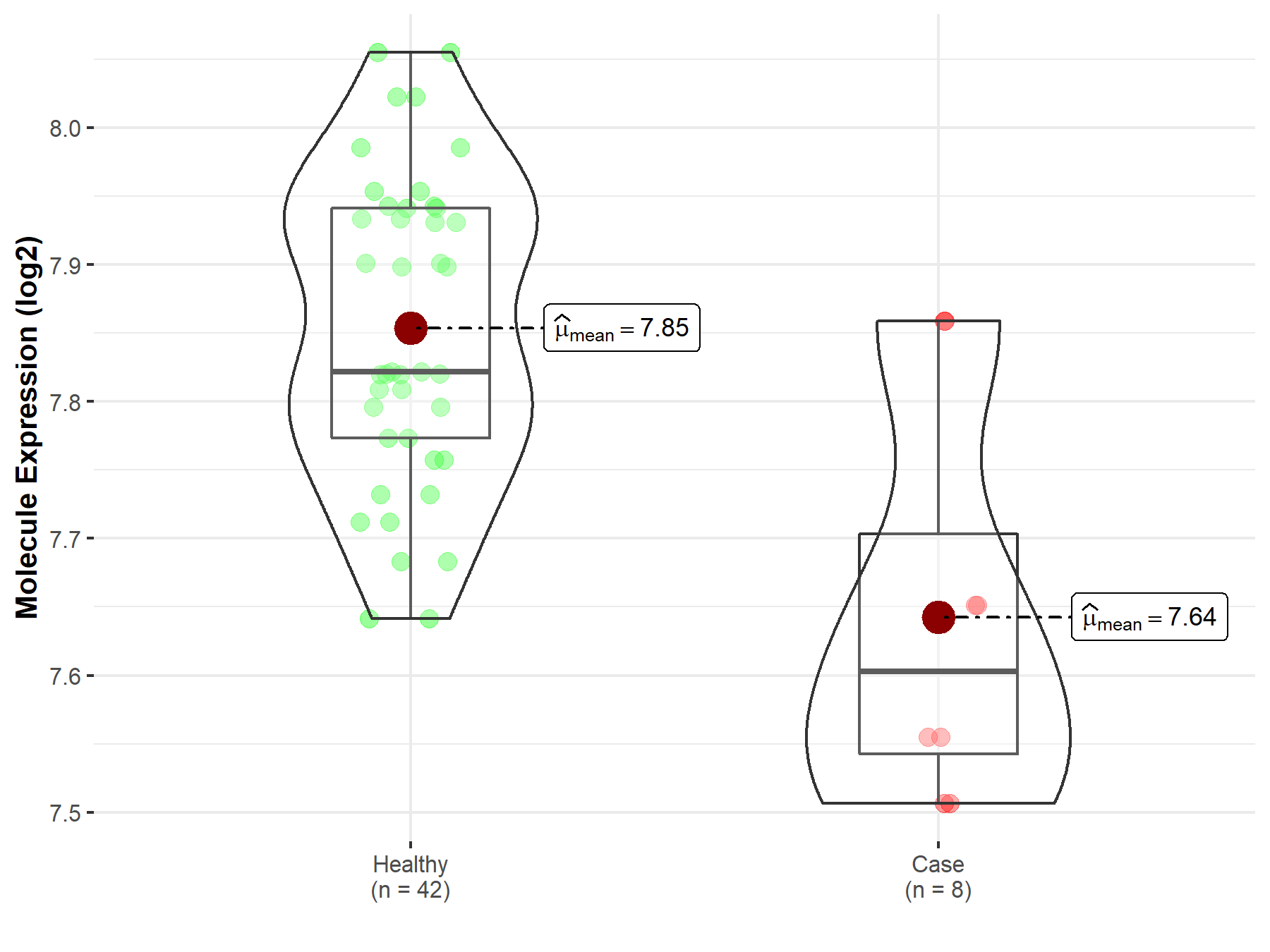
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Whole blood | |
| The Specified Disease | Polycythemia vera | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.62E-18; Fold-change: -2.79E-01; Z-score: -2.34E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
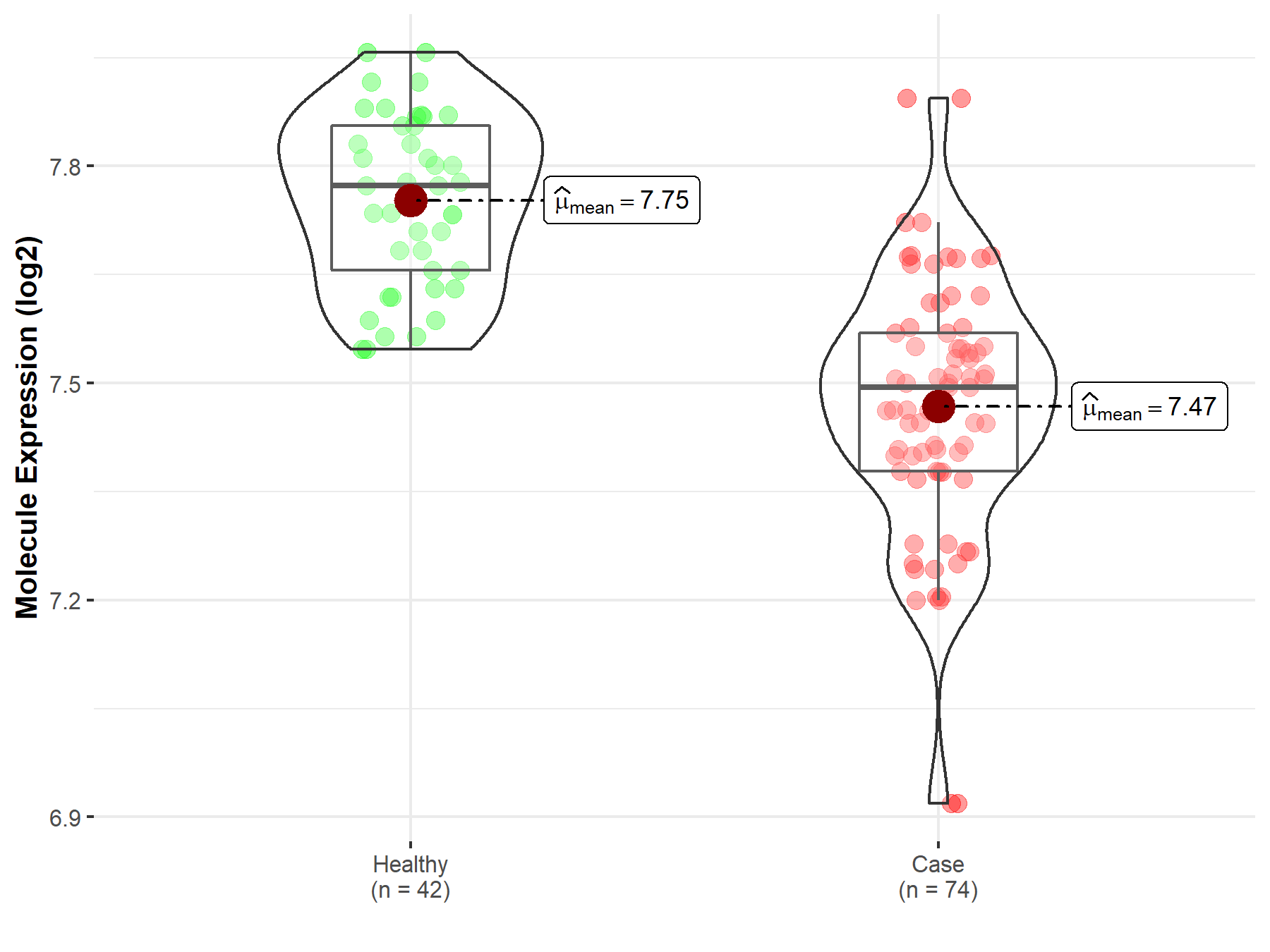
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
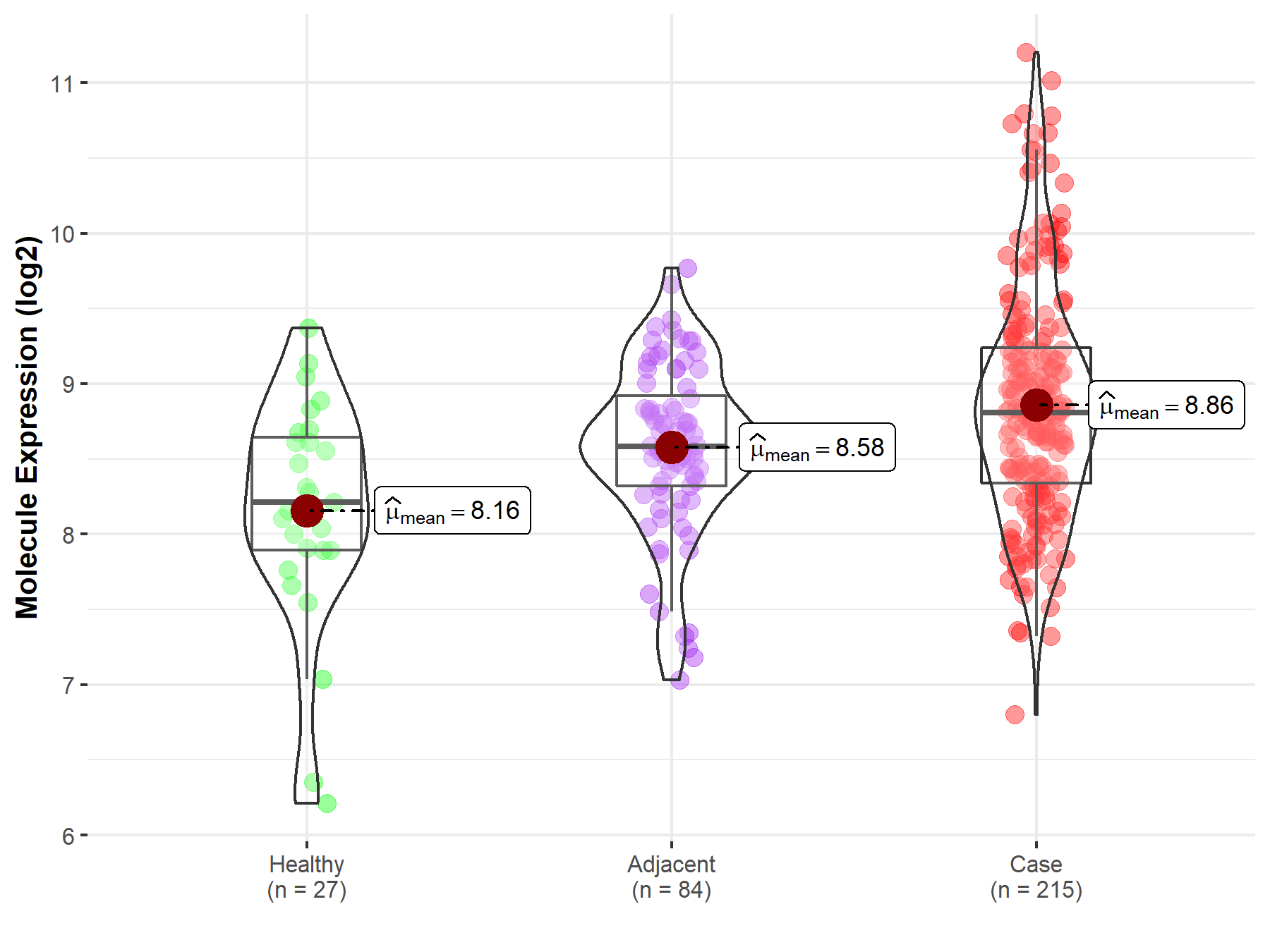
| The Studied Tissue | Oral tissue | |
| The Specified Disease | Oral squamous cell carcinoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.17E-05; Fold-change: 5.93E-01; Z-score: 7.85E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 6.51E-04; Fold-change: 2.24E-01; Z-score: 4.00E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Gastric tissue | |
| The Specified Disease | Gastric cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.46E-01; Fold-change: 1.71E-02; Z-score: 2.50E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 4.93E-01; Fold-change: 7.14E-02; Z-score: 2.80E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
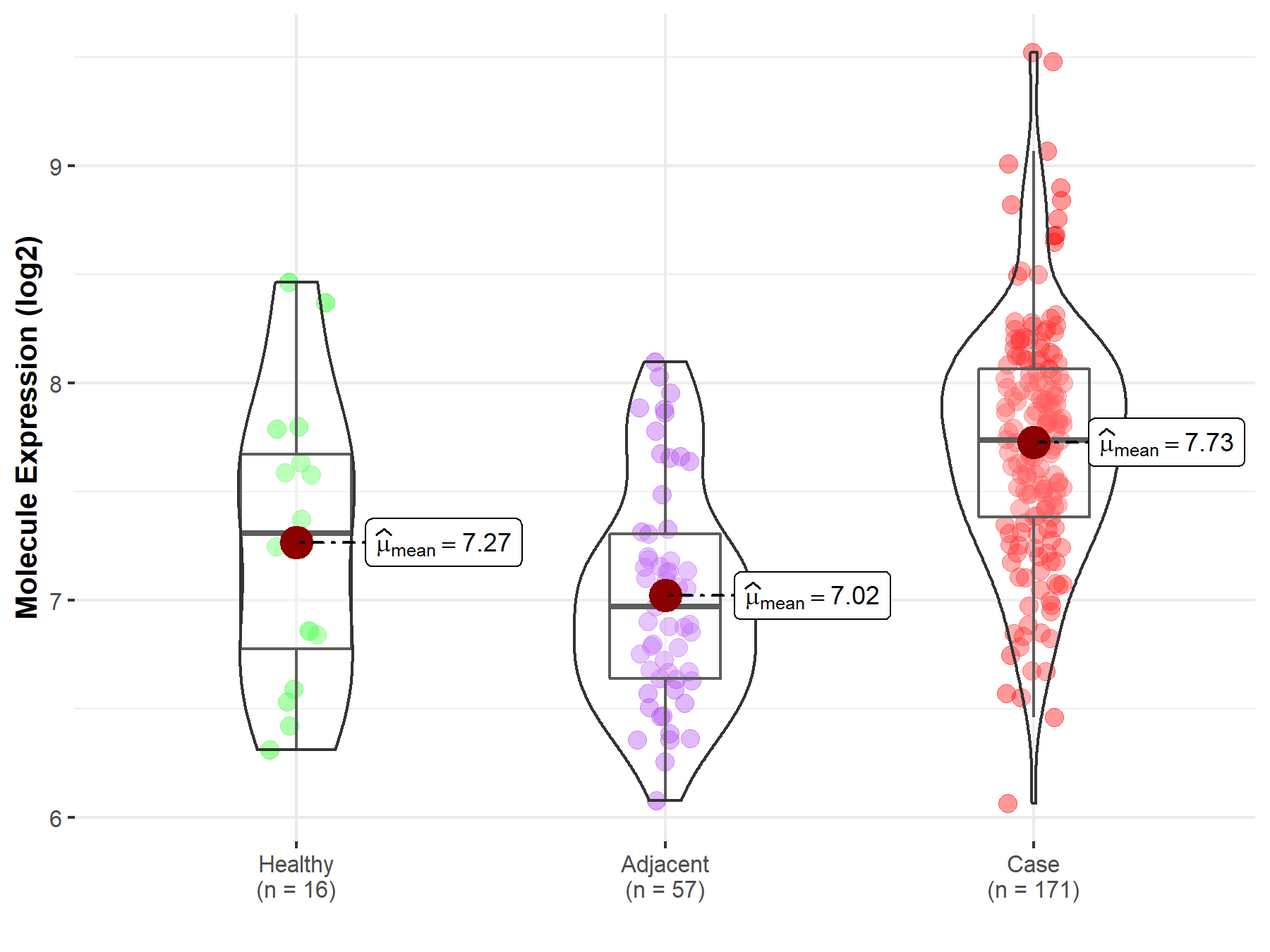
| The Studied Tissue | Pancreas | |
| The Specified Disease | Pancreatic cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.59E-02; Fold-change: 4.29E-01; Z-score: 6.42E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.33E-14; Fold-change: 7.66E-01; Z-score: 1.53E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Liver | |
| The Specified Disease | Liver cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.12E-23; Fold-change: 4.83E-01; Z-score: 2.27E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.88E-07; Fold-change: 1.56E-01; Z-score: 4.09E-01 | |
| The Expression Level of Disease Section Compare with the Other Disease Section | p-value: 8.92E-03; Fold-change: 4.07E-01; Z-score: 1.90E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
Molecule expression in tissue other than the diseased tissue of patients
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Lung | |
| The Specified Disease | Lung cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.90E-30; Fold-change: 1.76E-01; Z-score: 7.31E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.93E-06; Fold-change: 1.39E-03; Z-score: 4.52E-03 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.33E-51; Fold-change: 5.67E-01; Z-score: 1.25E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.38E-08; Fold-change: 4.14E-01; Z-score: 8.45E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
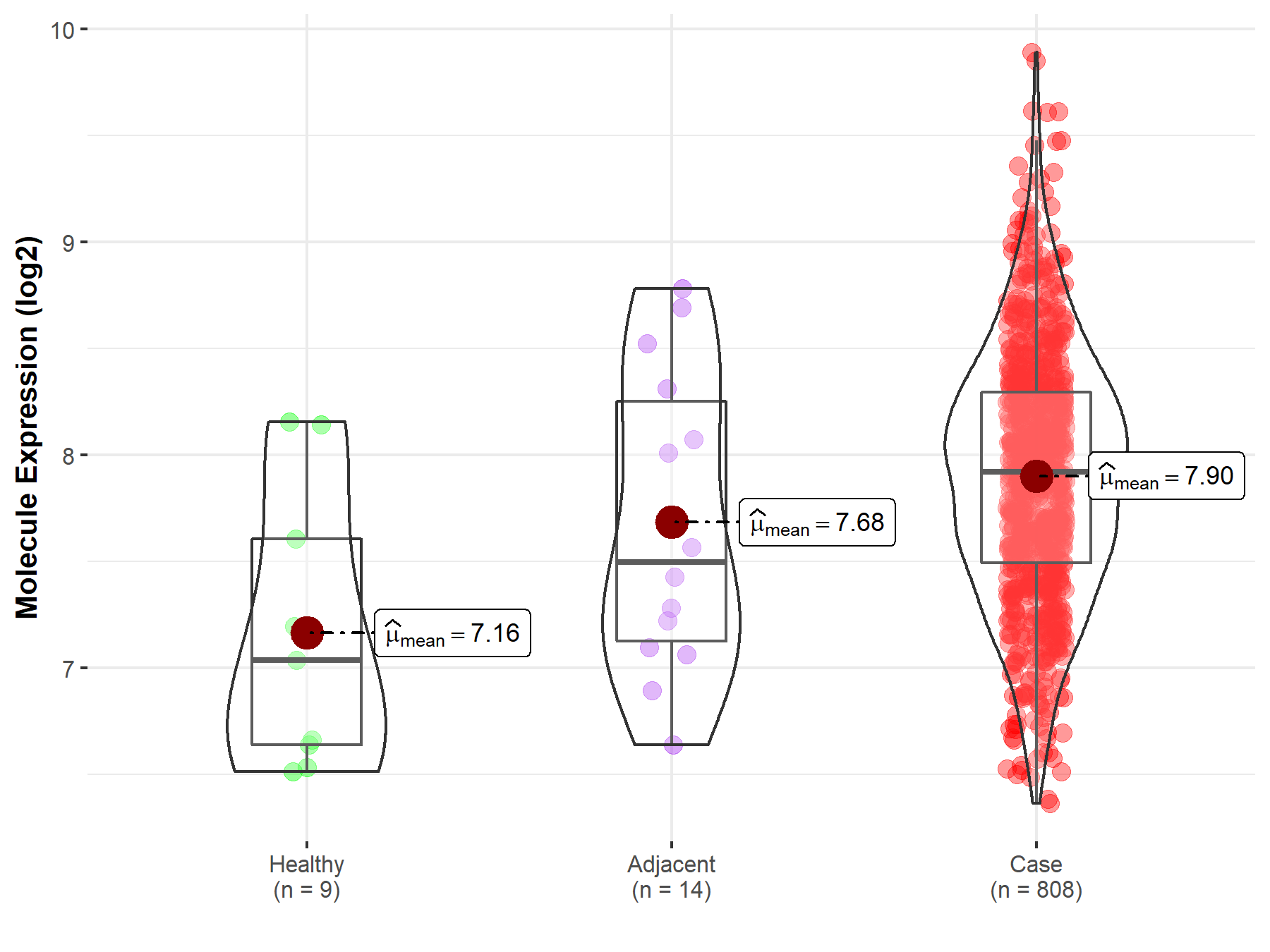
| The Studied Tissue | Ovary | |
| The Specified Disease | Ovarian cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.01E-02; Fold-change: 8.86E-01; Z-score: 1.34E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.70E-01; Fold-change: 4.25E-01; Z-score: 6.01E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
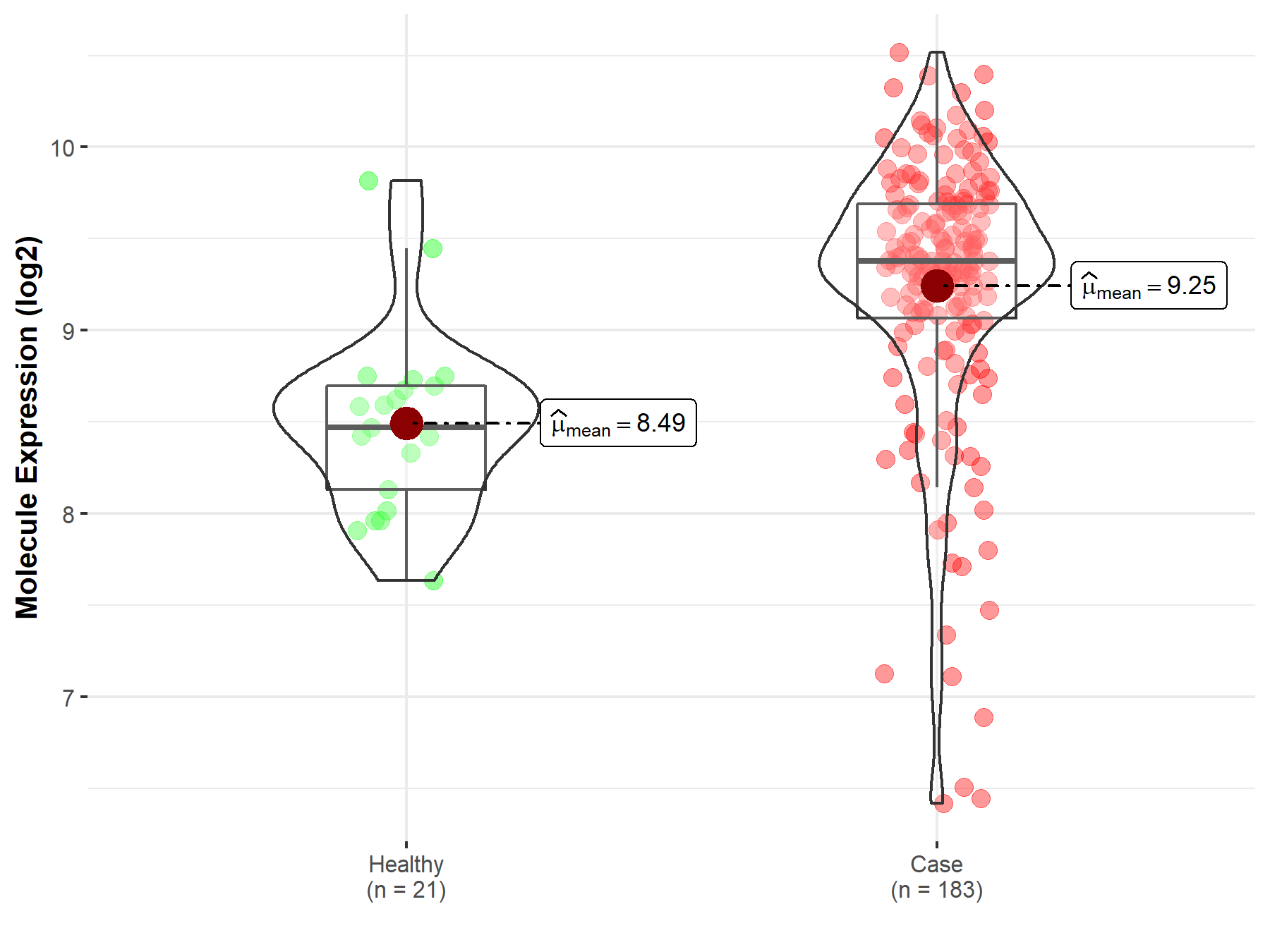
| The Studied Tissue | Prostate | |
| The Specified Disease | Prostate cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.18E-07; Fold-change: 9.06E-01; Z-score: 1.81E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

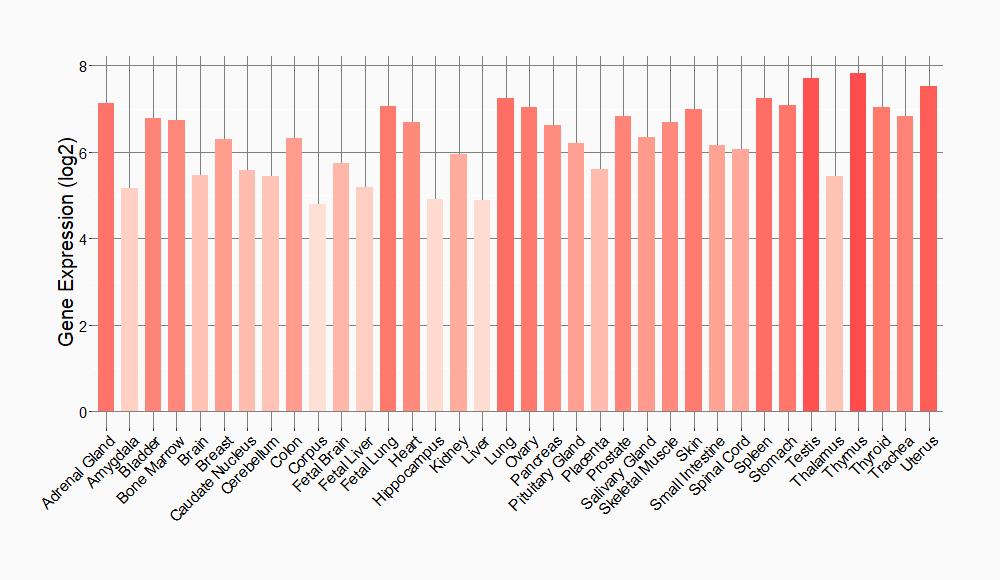
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
