Molecule Information
General Information of the Molecule (ID: Mol00102)
| Name |
Tyrosine-protein kinase JAK2 (JAK3)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Janus kinase 2; JAK-2
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
JAK2
|
||||
| Gene ID | |||||
| Location |
chr9:4984390-5129948[+]
|
||||
| Sequence |
MGMACLTMTEMEGTSTSSIYQNGDISGNANSMKQIDPVLQVYLYHSLGKSEADYLTFPSG
EYVAEEICIAASKACGITPVYHNMFALMSETERIWYPPNHVFHIDESTRHNVLYRIRFYF PRWYCSGSNRAYRHGISRGAEAPLLDDFVMSYLFAQWRHDFVHGWIKVPVTHETQEECLG MAVLDMMRIAKENDQTPLAIYNSISYKTFLPKCIRAKIQDYHILTRKRIRYRFRRFIQQF SQCKATARNLKLKYLINLETLQSAFYTEKFEVKEPGSGPSGEEIFATIIITGNGGIQWSR GKHKESETLTEQDLQLYCDFPNIIDVSIKQANQEGSNESRVVTIHKQDGKNLEIELSSLR EALSFVSLIDGYYRLTADAHHYLCKEVAPPAVLENIQSNCHGPISMDFAISKLKKAGNQT GLYVLRCSPKDFNKYFLTFAVERENVIEYKHCLITKNENEEYNLSGTKKNFSSLKDLLNC YQMETVRSDNIIFQFTKCCPPKPKDKSNLLVFRTNGVSDVPTSPTLQRPTHMNQMVFHKI RNEDLIFNESLGQGTFTKIFKGVRREVGDYGQLHETEVLLKVLDKAHRNYSESFFEAASM MSKLSHKHLVLNYGVCVCGDENILVQEFVKFGSLDTYLKKNKNCINILWKLEVAKQLAWA MHFLEENTLIHGNVCAKNILLIREEDRKTGNPPFIKLSDPGISITVLPKDILQERIPWVP PECIENPKNLNLATDKWSFGTTLWEICSGGDKPLSALDSQRKLQFYEDRHQLPAPKWAEL ANLINNCMDYEPDFRPSFRAIIRDLNSLFTPDYELLTENDMLPNMRIGALGFSGAFEDRD PTQFEERHLKFLQQLGKGNFGSVEMCRYDPLQDNTGEVVAVKKLQHSTEEHLRDFEREIE ILKSLQHDNIVKYKGVCYSAGRRNLKLIMEYLPYGSLRDYLQKHKERIDHIKLLQYTSQI CKGMEYLGTKRYIHRDLATRNILVENENRVKIGDFGLTKVLPQDKEYYKVKEPGESPIFW YAPESLTESKFSVASDVWSFGVVLYELFTYIEKSKSPPAEFMRMIGNDKQGQMIVFHLIE LLKNNGRLPRPDGCPDEIYMIMTECWNNNVNQRPSFRDLALRVDQIRDNMAG Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Non-receptor tyrosine kinase involved in various processes such as cell growth, development, differentiation or histone modifications. Mediates essential signaling events in both innate and adaptive immunity. In the cytoplasm, plays a pivotal role in signal transduction via its association with type I receptors such as growth hormone (GHR), prolactin (PRLR), leptin (LEPR), erythropoietin (EPOR), thrombopoietin (THPO); or type II receptors including IFN-alpha, IFN-beta, IFN-gamma and multiple interleukins. Following ligand-binding to cell surface receptors, phosphorylates specific tyrosine residues on the cytoplasmic tails of the receptor, creating docking sites for STATs proteins. Subsequently, phosphorylates the STATs proteins once they are recruited to the receptor. Phosphorylated STATs then form homodimer or heterodimers and translocate to the nucleus to activate gene transcription. For example, cell stimulation with erythropoietin (EPO) during erythropoiesis leads to JAK2 autophosphorylation, activation, and its association with erythropoietin receptor (EPOR) that becomes phosphorylated in its cytoplasmic domain. Then, STAT5 (STAT5A or STAT5B) is recruited, phosphorylated and activated by JAK2. Once activated, dimerized STAT5 translocates into the nucleus and promotes the transcription of several essential genes involved in the modulation of erythropoiesis. Part of a signaling cascade that is activated by increased cellular retinol and that leads to the activation of STAT5 (STAT5A or STAT5B). In addition, JAK2 mediates angiotensin-2-induced ARHGEF1 phosphorylation. Plays a role in cell cycle by phosphorylating CDKN1B. Cooperates with TEC through reciprocal phosphorylation to mediate cytokine-driven activation of FOS transcription. In the nucleus, plays a key role in chromatin by specifically mediating phosphorylation of 'Tyr-41' of histone H3 (H3Y41ph), a specific tag that promotes exclusion of CBX5 (HP1 alpha) from chromatin.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
6 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Epithelial ovarian cancer [ICD-11: 2B5D.0] | [1] | |||
| Resistant Disease | Epithelial ovarian cancer [ICD-11: 2B5D.0] | |||
| Resistant Drug | Carboplatin | |||
| Molecule Alteration | Phosphorylation | . |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SKOV-3 cells | Ovary | Homo sapiens (Human) | CVCL_0532 |
| OVCAR-3 cells | Ascites | Homo sapiens (Human) | CVCL_0465 | |
| OVCAR5 cells | Ovary | Homo sapiens (Human) | CVCL_1628 | |
| OVCAR8 cells | Ovary | Homo sapiens (Human) | CVCL_1629 | |
| Mechanism Description | We show that the addition of AAFs to the culture media of EOC cell lines has the potential to induce resistance to standard-of-care drugs (SCDs). We also show that AAFs induce time- and concentration-dependent activation of downstream signalling to signal transducer and activator of transcription 3 (STAT3), and concomitantly altered phosphorylation of mitogen-activated protein kinase kinase (MEK), phosphoinositide 3-kinase (PI3K)-protein kinase B (AKT) and nuclear factor NF-kappa-B (NFkappaB). Antibodies targeting the interleukin-6 receptor (IL6R) effectively blocked phosphorylation of STAT3 and STAT1. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Head and neck squamous cell carcinoma [ICD-11: 2D42.1] | [2] | |||
| Sensitive Disease | Head and neck squamous cell carcinoma [ICD-11: 2D42.1] | |||
| Sensitive Drug | Cetuximab | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | JAKT2/STAT3 signaling pathway | Inhibition | hsa04030 | |
| In Vitro Model | 5-8F cells | Nasopharynx | Homo sapiens (Human) | CVCL_C528 |
| CNE2 cells | Nasopharynx | Homo sapiens (Human) | CVCL_6889 | |
| Experiment for Molecule Alteration |
qRT-PCR; Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR204 inhibits angiogenesis and promotes sensitivity to cetuximab in head and neck squamous cell carcinoma cells by blocking JAk2-STAT3 signaling. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Primary myelofibrosis [ICD-11: 2A20.2] | [3] | |||
| Sensitive Disease | Primary myelofibrosis [ICD-11: 2A20.2] | |||
| Sensitive Drug | Danazol | |||
| Molecule Alteration | Function | Inhibition |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Mechanism Description | In non-transplant candidates, conventional treatment for anemia includes androgens, prednisone, thalidomide, and danazol; for symptomatic splenomegaly, hydroxyurea and ruxolitinib; and for constitutional symptoms, ruxolitinib. Fedratinib, another JAK2 inhibitor, has now been FDA-approved for use in ruxolitinib failures. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hematologic Cancer [ICD-11: MG24.Y] | [4] | |||
| Resistant Disease | Hematologic Cancer [ICD-11: MG24.Y] | |||
| Resistant Drug | Fedratinib | |||
| Molecule Alteration | Missense mutation | p.R867Q (c.2600G>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
WST-1 cell proliferation assay | |||
| Disease Class: Myeloproliferative neoplasm [ICD-11: 2A22.0] | [5] | |||
| Resistant Disease | Myeloproliferative neoplasm [ICD-11: 2A22.0] | |||
| Resistant Drug | Fedratinib | |||
| Molecule Alteration | Mutation | V617F+L902Q+E1028K |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Sanger sequencing assay | |||
| Experiment for Drug Resistance |
MTS-based assay | |||
| Mechanism Description | In this study, we have recovered seven residues in the kinase domain of JAK2 that affect ruxolitinib sensitivity. All these mutations confer cross-resistance across the panel of JAK2 kinase inhibitors except JAK2-L983F. JAK2-L983F reduces the sensitivity towards ruxolitinib. However, it is sensitive towards fedratinib indicating that our screen identifies the drug-specific resistance profiles. These results suggest that fedratinib might be effective in the suppression of ATP site mutations generated by ruxolitinib due to its ability to bind additional substrate binding sites. | |||
| Disease Class: Myeloproliferative neoplasm [ICD-11: 2A22.0] | [5] | |||
| Resistant Disease | Myeloproliferative neoplasm [ICD-11: 2A22.0] | |||
| Resistant Drug | Fedratinib | |||
| Molecule Alteration | Mutation | V617F+L902Q+R938E |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Sanger sequencing assay | |||
| Experiment for Drug Resistance |
MTS-based assay | |||
| Mechanism Description | In this study, we have recovered seven residues in the kinase domain of JAK2 that affect ruxolitinib sensitivity. All these mutations confer cross-resistance across the panel of JAK2 kinase inhibitors except JAK2-L983F. JAK2-L983F reduces the sensitivity towards ruxolitinib. However, it is sensitive towards fedratinib indicating that our screen identifies the drug-specific resistance profiles. These results suggest that fedratinib might be effective in the suppression of ATP site mutations generated by ruxolitinib due to its ability to bind additional substrate binding sites. | |||
| Disease Class: Myeloproliferative neoplasm [ICD-11: 2A22.0] | [5] | |||
| Resistant Disease | Myeloproliferative neoplasm [ICD-11: 2A22.0] | |||
| Resistant Drug | Fedratinib | |||
| Molecule Alteration | Mutation | V617F+L902Q+R947Q |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Sanger sequencing assay | |||
| Experiment for Drug Resistance |
MTS-based assay | |||
| Mechanism Description | In this study, we have recovered seven residues in the kinase domain of JAK2 that affect ruxolitinib sensitivity. All these mutations confer cross-resistance across the panel of JAK2 kinase inhibitors except JAK2-L983F. JAK2-L983F reduces the sensitivity towards ruxolitinib. However, it is sensitive towards fedratinib indicating that our screen identifies the drug-specific resistance profiles. These results suggest that fedratinib might be effective in the suppression of ATP site mutations generated by ruxolitinib due to its ability to bind additional substrate binding sites. | |||
| Disease Class: Myeloproliferative neoplasm [ICD-11: 2A22.0] | [5] | |||
| Resistant Disease | Myeloproliferative neoplasm [ICD-11: 2A22.0] | |||
| Resistant Drug | Fedratinib | |||
| Molecule Alteration | Mutation | V617F+Y931C |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Sanger sequencing assay | |||
| Experiment for Drug Resistance |
MTS-based assay | |||
| Mechanism Description | In this study, we have recovered seven residues in the kinase domain of JAK2 that affect ruxolitinib sensitivity. All these mutations confer cross-resistance across the panel of JAK2 kinase inhibitors except JAK2-L983F. JAK2-L983F reduces the sensitivity towards ruxolitinib. However, it is sensitive towards fedratinib indicating that our screen identifies the drug-specific resistance profiles. These results suggest that fedratinib might be effective in the suppression of ATP site mutations generated by ruxolitinib due to its ability to bind additional substrate binding sites. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Primary myelofibrosis [ICD-11: 2A20.2] | [3] | |||
| Sensitive Disease | Primary myelofibrosis [ICD-11: 2A20.2] | |||
| Sensitive Drug | Fedratinib | |||
| Molecule Alteration | Function | Inhibition |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Mechanism Description | In non-transplant candidates, conventional treatment for anemia includes androgens, prednisone, thalidomide, and danazol; for symptomatic splenomegaly, hydroxyurea and ruxolitinib; and for constitutional symptoms, ruxolitinib. Fedratinib, another JAK2 inhibitor, has now been FDA-approved for use in ruxolitinib failures. | |||
| Disease Class: Chronic myeloid leukemia [ICD-11: 2A20.0] | [6] | |||
| Sensitive Disease | Chronic myeloid leukemia [ICD-11: 2A20.0] | |||
| Sensitive Drug | Fedratinib | |||
| Molecule Alteration | Missense mutation | p.V617F (c.1849G>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HEL cells | Blood | Homo sapiens (Human) | CVCL_0001 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
XTT assay | |||
| Mechanism Description | The missense mutation p.V617F (c.1849G>T) in gene JAK2 cause the sensitivity of Fedratinib by aberration of the drug's therapeutic target | |||
| Disease Class: Myeloproliferative neoplasm [ICD-11: 2A22.0] | [5] | |||
| Sensitive Disease | Myeloproliferative neoplasm [ICD-11: 2A22.0] | |||
| Sensitive Drug | Fedratinib | |||
| Molecule Alteration | Mutation | V617F+L902Q+E1028K |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Sanger sequencing assay | |||
| Experiment for Drug Resistance |
MTS-based assay | |||
| Mechanism Description | In this study, we have recovered seven residues in the kinase domain of JAK2 that affect ruxolitinib sensitivity. All these mutations confer cross-resistance across the panel of JAK2 kinase inhibitors except JAK2-L983F. JAK2-L983F reduces the sensitivity towards ruxolitinib. However, it is sensitive towards fedratinib indicating that our screen identifies the drug-specific resistance profiles. These results suggest that fedratinib might be effective in the suppression of ATP site mutations generated by ruxolitinib due to its ability to bind additional substrate binding sites. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Epithelial ovarian cancer [ICD-11: 2B5D.0] | [1] | |||
| Resistant Disease | Epithelial ovarian cancer [ICD-11: 2B5D.0] | |||
| Resistant Drug | Paclitaxel | |||
| Molecule Alteration | Phosphorylation | . |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SKOV-3 cells | Ovary | Homo sapiens (Human) | CVCL_0532 |
| OVCAR-3 cells | Ascites | Homo sapiens (Human) | CVCL_0465 | |
| OVCAR5 cells | Ovary | Homo sapiens (Human) | CVCL_1628 | |
| OVCAR8 cells | Ovary | Homo sapiens (Human) | CVCL_1629 | |
| Mechanism Description | We show that the addition of AAFs to the culture media of EOC cell lines has the potential to induce resistance to standard-of-care drugs (SCDs). We also show that AAFs induce time- and concentration-dependent activation of downstream signalling to signal transducer and activator of transcription 3 (STAT3), and concomitantly altered phosphorylation of mitogen-activated protein kinase kinase (MEK), phosphoinositide 3-kinase (PI3K)-protein kinase B (AKT) and nuclear factor NF-kappa-B (NFkappaB). Antibodies targeting the interleukin-6 receptor (IL6R) effectively blocked phosphorylation of STAT3 and STAT1. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Polycythaemia vera [ICD-11: 2A20.4] | [7] | |||
| Resistant Disease | Polycythaemia vera [ICD-11: 2A20.4] | |||
| Resistant Drug | Ruxolitinib | |||
| Molecule Alteration | Missense mutation | p.V617F |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Mechanism Description | JAK2 mutation confers resistance to Ruxolitinib. | |||
| Disease Class: Acute lymphocytic leukemia [ICD-11: 2B33.0] | [8] | |||
| Resistant Disease | Acute lymphocytic leukemia [ICD-11: 2B33.0] | |||
| Resistant Drug | Ruxolitinib | |||
| Molecule Alteration | Missense mutation | p.R938Q (c.2813G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Experiment for Drug Resistance |
FACS assay | |||
| Mechanism Description | Mutations within the kinase domain of JAK2 are associated with resistance to type I JAK inhibitors. | |||
| Disease Class: Hematologic Cancer [ICD-11: MG24.Y] | [4] | |||
| Resistant Disease | Hematologic Cancer [ICD-11: MG24.Y] | |||
| Resistant Drug | Ruxolitinib | |||
| Molecule Alteration | Missense mutation | p.R867Q (c.2600G>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
WST-1 cell proliferation assay | |||
| Disease Class: Myeloproliferative neoplasm [ICD-11: 2A22.0] | [5] | |||
| Resistant Disease | Myeloproliferative neoplasm [ICD-11: 2A22.0] | |||
| Resistant Drug | Ruxolitinib | |||
| Molecule Alteration | Mutation | V617F+L902Q |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Sanger sequencing assay | |||
| Experiment for Drug Resistance |
MTS-based assay | |||
| Mechanism Description | In this study, we have recovered seven residues in the kinase domain of JAK2 that affect ruxolitinib sensitivity. All these mutations confer cross-resistance across the panel of JAK2 kinase inhibitors except JAK2-L983F. JAK2-L983F reduces the sensitivity towards ruxolitinib. However, it is sensitive towards fedratinib indicating that our screen identifies the drug-specific resistance profiles. | |||
| Disease Class: Myeloproliferative neoplasm [ICD-11: 2A22.0] | [5] | |||
| Resistant Disease | Myeloproliferative neoplasm [ICD-11: 2A22.0] | |||
| Resistant Drug | Ruxolitinib | |||
| Molecule Alteration | Mutation | V617F+L902Q+R938E |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Sanger sequencing assay | |||
| Experiment for Drug Resistance |
MTS-based assay | |||
| Mechanism Description | In this study, we have recovered seven residues in the kinase domain of JAK2 that affect ruxolitinib sensitivity. All these mutations confer cross-resistance across the panel of JAK2 kinase inhibitors except JAK2-L983F. JAK2-L983F reduces the sensitivity towards ruxolitinib. However, it is sensitive towards fedratinib indicating that our screen identifies the drug-specific resistance profiles. | |||
| Disease Class: Myeloproliferative neoplasm [ICD-11: 2A22.0] | [5] | |||
| Resistant Disease | Myeloproliferative neoplasm [ICD-11: 2A22.0] | |||
| Resistant Drug | Ruxolitinib | |||
| Molecule Alteration | Mutation | V617F+L902Q+R947Q |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Sanger sequencing assay | |||
| Experiment for Drug Resistance |
MTS-based assay | |||
| Mechanism Description | In this study, we have recovered seven residues in the kinase domain of JAK2 that affect ruxolitinib sensitivity. All these mutations confer cross-resistance across the panel of JAK2 kinase inhibitors except JAK2-L983F. JAK2-L983F reduces the sensitivity towards ruxolitinib. However, it is sensitive towards fedratinib indicating that our screen identifies the drug-specific resistance profiles. | |||
| Disease Class: Myeloproliferative neoplasm [ICD-11: 2A22.0] | [5] | |||
| Resistant Disease | Myeloproliferative neoplasm [ICD-11: 2A22.0] | |||
| Resistant Drug | Ruxolitinib | |||
| Molecule Alteration | Mutation | V617F+L983F |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Sanger sequencing assay | |||
| Experiment for Drug Resistance |
MTS-based assay | |||
| Mechanism Description | In this study, we have recovered seven residues in the kinase domain of JAK2 that affect ruxolitinib sensitivity. All these mutations confer cross-resistance across the panel of JAK2 kinase inhibitors except JAK2-L983F. JAK2-L983F reduces the sensitivity towards ruxolitinib. However, it is sensitive towards fedratinib indicating that our screen identifies the drug-specific resistance profiles. | |||
| Disease Class: Myeloproliferative neoplasm [ICD-11: 2A22.0] | [5] | |||
| Resistant Disease | Myeloproliferative neoplasm [ICD-11: 2A22.0] | |||
| Resistant Drug | Ruxolitinib | |||
| Molecule Alteration | Mutation | V617F+L983F+Q959H |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Sanger sequencing assay | |||
| Experiment for Drug Resistance |
MTS-based assay | |||
| Mechanism Description | In this study, we have recovered seven residues in the kinase domain of JAK2 that affect ruxolitinib sensitivity. All these mutations confer cross-resistance across the panel of JAK2 kinase inhibitors except JAK2-L983F. JAK2-L983F reduces the sensitivity towards ruxolitinib. However, it is sensitive towards fedratinib indicating that our screen identifies the drug-specific resistance profiles. | |||
| Disease Class: Myeloproliferative neoplasm [ICD-11: 2A22.0] | [5] | |||
| Resistant Disease | Myeloproliferative neoplasm [ICD-11: 2A22.0] | |||
| Resistant Drug | Ruxolitinib | |||
| Molecule Alteration | Mutation | V617F+Y931C |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Sanger sequencing assay | |||
| Experiment for Drug Resistance |
MTS-based assay | |||
| Mechanism Description | In this study, we have recovered seven residues in the kinase domain of JAK2 that affect ruxolitinib sensitivity. All these mutations confer cross-resistance across the panel of JAK2 kinase inhibitors except JAK2-L983F. JAK2-L983F reduces the sensitivity towards ruxolitinib. However, it is sensitive towards fedratinib indicating that our screen identifies the drug-specific resistance profiles. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Primary myelofibrosis [ICD-11: 2A20.2] | [3] | |||
| Sensitive Disease | Primary myelofibrosis [ICD-11: 2A20.2] | |||
| Sensitive Drug | Ruxolitinib | |||
| Molecule Alteration | Function | Inhibition |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Mechanism Description | In non-transplant candidates, conventional treatment for anemia includes androgens, prednisone, thalidomide, and danazol; for symptomatic splenomegaly, hydroxyurea and ruxolitinib; and for constitutional symptoms, ruxolitinib. Fedratinib, another JAK2 inhibitor, has now been FDA-approved for use in ruxolitinib failures. | |||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [9] | |||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Sensitive Drug | Ruxolitinib | |||
| Molecule Alteration | Missense mutation | p.V617F (c.1849G>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Bone marrow | N.A. | ||
| Mechanism Description | The missense mutation p.V617F (c.1849G>T) in gene JAK2 cause the sensitivity of Ruxolitinib by aberration of the drug's therapeutic target | |||
| Disease Class: Acute lymphocytic leukemia [ICD-11: 2B33.0] | [10] | |||
| Sensitive Disease | Acute lymphocytic leukemia [ICD-11: 2B33.0] | |||
| Sensitive Drug | Ruxolitinib | |||
| Molecule Alteration | Missense mutation | p.R683G (c.2047A>G) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
|
|
||||
| Disease Class: Acute lymphocytic leukemia [ICD-11: 2B33.0] | [11] | |||
| Sensitive Disease | Acute lymphocytic leukemia [ICD-11: 2B33.0] | |||
| Sensitive Drug | Ruxolitinib | |||
| Molecule Alteration | Missense mutation | p.F694L (c.2080T>C) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | JAKT2/STAT3 signaling pathway | Inhibition | hsa04030 | |
Clinical Trial Drug(s)
7 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hematologic Cancer [ICD-11: MG24.Y] | [4] | |||
| Resistant Disease | Hematologic Cancer [ICD-11: MG24.Y] | |||
| Resistant Drug | Momelotinib | |||
| Molecule Alteration | Missense mutation | p.R867Q (c.2600G>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
WST-1 cell proliferation assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Chronic myeloid leukemia [ICD-11: 2A20.0] | [12] | |||
| Sensitive Disease | Chronic myeloid leukemia [ICD-11: 2A20.0] | |||
| Sensitive Drug | Ropeginterferon alfa-2b | |||
| Molecule Alteration | Missense mutation | p.V617F (c.1849G>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | HEL cells | Blood | Homo sapiens (Human) | CVCL_0001 |
| UKE-1 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0104 | |
| Experiment for Drug Resistance |
Trypan blue staining assay | |||
| Disease Class: Myeloproliferative neoplasm [ICD-11: 2A22.0] | [12] | |||
| Sensitive Disease | Myeloproliferative neoplasm [ICD-11: 2A22.0] | |||
| Sensitive Drug | Ropeginterferon alfa-2b | |||
| Molecule Alteration | Missense mutation | p.V617F (c.1849G>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | HEL cells | Blood | Homo sapiens (Human) | CVCL_0001 |
| UKE-1 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0104 | |
| Experiment for Drug Resistance |
Trypan blue staining assay | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hematologic Cancer [ICD-11: MG24.Y] | [4] | |||
| Resistant Disease | Hematologic Cancer [ICD-11: MG24.Y] | |||
| Resistant Drug | AUY922 | |||
| Molecule Alteration | Missense mutation | p.R867Q (c.2600G>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
WST-1 cell proliferation assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Myeloproliferative neoplasm [ICD-11: 2A22.0] | [13] | |||
| Sensitive Disease | Myeloproliferative neoplasm [ICD-11: 2A22.0] | |||
| Sensitive Drug | BMS-911543 | |||
| Molecule Alteration | Missense mutation | p.V617F (c.1849G>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
[3H] thymidine incorporation assay | |||
| Mechanism Description | The missense mutation p.V617F (c.1849G>T) in gene JAK2 cause the sensitivity of BMS-911543 by aberration of the drug's therapeutic target | |||
| Disease Class: Essential thrombocythemia [ICD-11: 3B63.Z] | [13] | |||
| Sensitive Disease | Essential thrombocythemia [ICD-11: 3B63.Z] | |||
| Sensitive Drug | BMS-911543 | |||
| Molecule Alteration | Missense mutation | p.V617F (c.1849G>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
[3H] thymidine incorporation assay | |||
| Mechanism Description | The missense mutation p.V617F (c.1849G>T) in gene JAK2 cause the sensitivity of BMS-911543 by aberration of the drug's therapeutic target | |||
| Disease Class: Hematologic Cancer [ICD-11: MG24.Y] | [13] | |||
| Sensitive Disease | Hematologic Cancer [ICD-11: MG24.Y] | |||
| Sensitive Drug | BMS-911543 | |||
| Molecule Alteration | Missense mutation | p.V617F (c.1849G>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
[3H] thymidine incorporation assay | |||
| Mechanism Description | The missense mutation p.V617F (c.1849G>T) in gene JAK2 cause the sensitivity of BMS-911543 by aberration of the drug's therapeutic target | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Myeloproliferative neoplasm [ICD-11: 2A22.0] | [14] | |||
| Sensitive Disease | Myeloproliferative neoplasm [ICD-11: 2A22.0] | |||
| Sensitive Drug | Gandotinib | |||
| Molecule Alteration | Missense mutation | p.V617F (c.1849G>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Myeloproliferative neoplasm [ICD-11: 2A22.0] | [5] | |||
| Sensitive Disease | Myeloproliferative neoplasm [ICD-11: 2A22.0] | |||
| Sensitive Drug | Tanespimycin | |||
| Molecule Alteration | Mutation | V617F+L902Q |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Sanger sequencing assay | |||
| Experiment for Drug Resistance |
MTS-based assay | |||
| Mechanism Description | These results indicate that these mutants are dependent on the HSP90 for their folding. To know that downregulation of JAK2 protein leads to the decrease of cell proliferation, we performed biochemical analysis on these mutant JAK2 cells and found that ruxolitinib-resistant variants are sensitive towards 17-AAG and treatment of the cells with 17-AAG leads to the downregulation of JAK2 protein and decrease of STAT5 activation. This study shows that HSP90 inhibitors are potent against ruxolitinib-resistant variants through the JAK2 degradation and provides the rationale for clinical evaluation of potent HSP90 inhibitors in genetic resistance driven by JAK2 inhibitors. | |||
| Disease Class: Myeloproliferative neoplasm [ICD-11: 2A22.0] | [5] | |||
| Sensitive Disease | Myeloproliferative neoplasm [ICD-11: 2A22.0] | |||
| Sensitive Drug | Tanespimycin | |||
| Molecule Alteration | Mutation | V617F+L902Q+E1028K |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Sanger sequencing assay | |||
| Experiment for Drug Resistance |
MTS-based assay | |||
| Mechanism Description | These results indicate that these mutants are dependent on the HSP90 for their folding. To know that downregulation of JAK2 protein leads to the decrease of cell proliferation, we performed biochemical analysis on these mutant JAK2 cells and found that ruxolitinib-resistant variants are sensitive towards 17-AAG and treatment of the cells with 17-AAG leads to the downregulation of JAK2 protein and decrease of STAT5 activation. This study shows that HSP90 inhibitors are potent against ruxolitinib-resistant variants through the JAK2 degradation and provides the rationale for clinical evaluation of potent HSP90 inhibitors in genetic resistance driven by JAK2 inhibitors. | |||
| Disease Class: Myeloproliferative neoplasm [ICD-11: 2A22.0] | [5] | |||
| Sensitive Disease | Myeloproliferative neoplasm [ICD-11: 2A22.0] | |||
| Sensitive Drug | Tanespimycin | |||
| Molecule Alteration | Mutation | V617F+L902Q+R938E |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Sanger sequencing assay | |||
| Experiment for Drug Resistance |
MTS-based assay | |||
| Mechanism Description | These results indicate that these mutants are dependent on the HSP90 for their folding. To know that downregulation of JAK2 protein leads to the decrease of cell proliferation, we performed biochemical analysis on these mutant JAK2 cells and found that ruxolitinib-resistant variants are sensitive towards 17-AAG and treatment of the cells with 17-AAG leads to the downregulation of JAK2 protein and decrease of STAT5 activation. This study shows that HSP90 inhibitors are potent against ruxolitinib-resistant variants through the JAK2 degradation and provides the rationale for clinical evaluation of potent HSP90 inhibitors in genetic resistance driven by JAK2 inhibitors. | |||
| Disease Class: Myeloproliferative neoplasm [ICD-11: 2A22.0] | [5] | |||
| Sensitive Disease | Myeloproliferative neoplasm [ICD-11: 2A22.0] | |||
| Sensitive Drug | Tanespimycin | |||
| Molecule Alteration | Mutation | V617F+L902Q+R947Q |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Sanger sequencing assay | |||
| Experiment for Drug Resistance |
MTS-based assay | |||
| Mechanism Description | These results indicate that these mutants are dependent on the HSP90 for their folding. To know that downregulation of JAK2 protein leads to the decrease of cell proliferation, we performed biochemical analysis on these mutant JAK2 cells and found that ruxolitinib-resistant variants are sensitive towards 17-AAG and treatment of the cells with 17-AAG leads to the downregulation of JAK2 protein and decrease of STAT5 activation. This study shows that HSP90 inhibitors are potent against ruxolitinib-resistant variants through the JAK2 degradation and provides the rationale for clinical evaluation of potent HSP90 inhibitors in genetic resistance driven by JAK2 inhibitors. | |||
| Disease Class: Myeloproliferative neoplasm [ICD-11: 2A22.0] | [5] | |||
| Sensitive Disease | Myeloproliferative neoplasm [ICD-11: 2A22.0] | |||
| Sensitive Drug | Tanespimycin | |||
| Molecule Alteration | Mutation | V617F+L983F |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Sanger sequencing assay | |||
| Experiment for Drug Resistance |
MTS-based assay | |||
| Mechanism Description | These results indicate that these mutants are dependent on the HSP90 for their folding. To know that downregulation of JAK2 protein leads to the decrease of cell proliferation, we performed biochemical analysis on these mutant JAK2 cells and found that ruxolitinib-resistant variants are sensitive towards 17-AAG and treatment of the cells with 17-AAG leads to the downregulation of JAK2 protein and decrease of STAT5 activation. This study shows that HSP90 inhibitors are potent against ruxolitinib-resistant variants through the JAK2 degradation and provides the rationale for clinical evaluation of potent HSP90 inhibitors in genetic resistance driven by JAK2 inhibitors. | |||
| Disease Class: Myeloproliferative neoplasm [ICD-11: 2A22.0] | [5] | |||
| Sensitive Disease | Myeloproliferative neoplasm [ICD-11: 2A22.0] | |||
| Sensitive Drug | Tanespimycin | |||
| Molecule Alteration | Mutation | V617F+L983F+Q959H |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Sanger sequencing assay | |||
| Experiment for Drug Resistance |
MTS-based assay | |||
| Mechanism Description | These results indicate that these mutants are dependent on the HSP90 for their folding. To know that downregulation of JAK2 protein leads to the decrease of cell proliferation, we performed biochemical analysis on these mutant JAK2 cells and found that ruxolitinib-resistant variants are sensitive towards 17-AAG and treatment of the cells with 17-AAG leads to the downregulation of JAK2 protein and decrease of STAT5 activation. This study shows that HSP90 inhibitors are potent against ruxolitinib-resistant variants through the JAK2 degradation and provides the rationale for clinical evaluation of potent HSP90 inhibitors in genetic resistance driven by JAK2 inhibitors. | |||
| Disease Class: Myeloproliferative neoplasm [ICD-11: 2A22.0] | [5] | |||
| Sensitive Disease | Myeloproliferative neoplasm [ICD-11: 2A22.0] | |||
| Sensitive Drug | Tanespimycin | |||
| Molecule Alteration | Mutation | V617F+Y931C |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Sanger sequencing assay | |||
| Experiment for Drug Resistance |
MTS-based assay | |||
| Mechanism Description | These results indicate that these mutants are dependent on the HSP90 for their folding. To know that downregulation of JAK2 protein leads to the decrease of cell proliferation, we performed biochemical analysis on these mutant JAK2 cells and found that ruxolitinib-resistant variants are sensitive towards 17-AAG and treatment of the cells with 17-AAG leads to the downregulation of JAK2 protein and decrease of STAT5 activation. This study shows that HSP90 inhibitors are potent against ruxolitinib-resistant variants through the JAK2 degradation and provides the rationale for clinical evaluation of potent HSP90 inhibitors in genetic resistance driven by JAK2 inhibitors. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hematologic Cancer [ICD-11: MG24.Y] | [15] | |||
| Sensitive Disease | Hematologic Cancer [ICD-11: MG24.Y] | |||
| Sensitive Drug | NS-018 | |||
| Molecule Alteration | Missense mutation | p.V617F (c.1849G>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Sf9 cells | Ovary | Homo sapiens (Human) | CVCL_0549 | |
| Experiment for Molecule Alteration |
Colony formation assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The missense mutation p.V617F (c.1849G>T) in gene JAK2 cause the sensitivity of NS-018 by aberration of the drug's therapeutic target | |||
Preclinical Drug(s)
7 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [16] | |||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Sensitive Drug | A-1155463 | |||
| Molecule Alteration | Missense mutation | p.V617F (c.1849G>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SCLC cells | N.A. | Homo sapiens (Human) | N.A. |
| NHL cells | N.A. | Homo sapiens (Human) | N.A. | |
| In Vivo Model | SCID and SCID-bg mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay; Colony formation assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Metastatic bone cancer [ICD-11: 2E03.0] | [17] | |||
| Sensitive Disease | Metastatic bone cancer [ICD-11: 2E03.0] | |||
| Sensitive Drug | AZD1208/Ruxolitinib | |||
| Molecule Alteration | Missense mutation | p.V617F (c.1849G>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HEL cells | Blood | Homo sapiens (Human) | CVCL_0001 |
| HEL cells | Blood | Homo sapiens (Human) | CVCL_0001 | |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| UKE-1 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0104 | |
| UKE-1 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0104 | |
| SET2 cells | Peripheral blood | Homo sapiens (Human) | CVCL_2187 | |
| SET2 cells | Peripheral blood | Homo sapiens (Human) | CVCL_2187 | |
| JAK2-V617F cells | N.A. | N.A. | N.A. | |
| Experiment for Molecule Alteration |
Western blot analysis; Microarray gene expression analysis; Crystallization and structure determination assay | |||
| Experiment for Drug Resistance |
MTS assay; Colony formation assay | |||
| Disease Class: Metastatic bone cancer [ICD-11: 2E03.0] | [17] | |||
| Sensitive Disease | Metastatic bone cancer [ICD-11: 2E03.0] | |||
| Sensitive Drug | AZD1208/Ruxolitinib | |||
| Molecule Alteration | Missense mutation | p.V617F (c.1849G>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HEL cells | Blood | Homo sapiens (Human) | CVCL_0001 |
| HEL cells | Blood | Homo sapiens (Human) | CVCL_0001 | |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| UKE-1 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0104 | |
| UKE-1 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0104 | |
| SET2 cells | Peripheral blood | Homo sapiens (Human) | CVCL_2187 | |
| SET2 cells | Peripheral blood | Homo sapiens (Human) | CVCL_2187 | |
| JAK2-V617F cells | N.A. | N.A. | N.A. | |
| Experiment for Molecule Alteration |
Western blot analysis; Microarray gene expression analysis; Crystallization and structure determination assay | |||
| Experiment for Drug Resistance |
MTS assay; Colony formation assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Metastatic bone cancer [ICD-11: 2E03.0] | [18] | |||
| Sensitive Disease | Metastatic bone cancer [ICD-11: 2E03.0] | |||
| Sensitive Drug | CHZ868 | |||
| Molecule Alteration | Missense mutation | p.V617F (c.1849G>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | TF-1 cells | Bone marrow | Homo sapiens (Human) | CVCL_0559 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| W515L cells | Blood | Homo sapiens (Human) | N.A. | |
| SET2 cells | Peripheral blood | Homo sapiens (Human) | CVCL_2187 | |
| In Vivo Model | CD45.2 Jak2V617F mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Cell viability luminescent assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [19] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Pictilisib/Ruxolitinib | |||
| Molecule Alteration | Missense mutation | p.V617F (c.1849G>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| In Vivo Model | JAK2 mutant Ba/F3 tumour mouse model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay; Colony assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Metastatic bone cancer [ICD-11: 2E03.0] | [17] | |||
| Sensitive Disease | Metastatic bone cancer [ICD-11: 2E03.0] | |||
| Sensitive Drug | Ruxolitinib/SGI-1776 | |||
| Molecule Alteration | Missense mutation | p.V617F (c.1849G>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HEL cells | Blood | Homo sapiens (Human) | CVCL_0001 |
| HEL cells | Blood | Homo sapiens (Human) | CVCL_0001 | |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| UKE-1 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0104 | |
| UKE-1 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0104 | |
| SET2 cells | Peripheral blood | Homo sapiens (Human) | CVCL_2187 | |
| SET2 cells | Peripheral blood | Homo sapiens (Human) | CVCL_2187 | |
| JAK2-V617F cells | N.A. | N.A. | N.A. | |
| Experiment for Molecule Alteration |
Western blot analysis; Microarray gene expression analysis; Crystallization and structure determination assay | |||
| Experiment for Drug Resistance |
MTS assay; Colony formation assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [19] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Ruxolitinib/ZSTK474 | |||
| Molecule Alteration | Missense mutation | p.V617F (c.1849G>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| In Vivo Model | JAK2 mutant Ba/F3 tumour mouse model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay; Colony assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hematologic Cancer [ICD-11: MG24.Y] | [20] | |||
| Sensitive Disease | Hematologic Cancer [ICD-11: MG24.Y] | |||
| Sensitive Drug | SHP099 | |||
| Molecule Alteration | Missense mutation | p.V617F (c.1849G>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | RAS/ERK signaling pathway | Inhibition | hsa01521 | |
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| NCI-H2228 cells | Lung | Homo sapiens (Human) | CVCL_1543 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| A2058 cells | Skin | Homo sapiens (Human) | CVCL_1059 | |
| KYSE520 cells | Esophagus | Homo sapiens (Human) | CVCL_1355 | |
| KATO-3 cells | Gastric | Homo sapiens (Human) | CVCL_0371 | |
| Sum52 cells | Pleural effusion | Homo sapiens (Human) | CVCL_3425 | |
| NCI-H2170 cells | Lung | Homo sapiens (Human) | CVCL_1535 | |
| NCI-H2170 cells | Lung | Homo sapiens (Human) | CVCL_1535 | |
| H293 cells | Kidney | Homo sapiens (Human) | N.A. | |
| In Vivo Model | Athymic nude mouse PDX model | Mus musculus | ||
| Experiment for Drug Resistance |
CellTitre-Glo assay; Crystal violet staining assay | |||
| Mechanism Description | SHP099 suppresses RAS-ERK signalling to inhibit the proliferation of receptor-tyrosine-kinase-driven human cancer cells in vitro and is efficacious in mouse tumour xenograft models. | |||
Investigative Drug(s)
4 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hematologic Cancer [ICD-11: MG24.Y] | [4] | |||
| Resistant Disease | Hematologic Cancer [ICD-11: MG24.Y] | |||
| Resistant Drug | AZ960 | |||
| Molecule Alteration | Missense mutation | p.R867Q (c.2600G>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
WST-1 cell proliferation assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [21] | |||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Sensitive Drug | GO6976 | |||
| Molecule Alteration | Missense mutation | p.V617F (c.1849G>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | TF-1 cells | Bone marrow | Homo sapiens (Human) | CVCL_0559 |
| HEL cells | Blood | Homo sapiens (Human) | CVCL_0001 | |
| Mo7E cells | Peripheral blood | Homo sapiens (Human) | CVCL_2106 | |
| FDCP1 cells | Bone marrow | Mus musculus (Mouse) | CVCL_2039 | |
| 32D cells | Bone marrow | Homo sapiens (Human) | CVCL_0118 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | The missense mutation p.V617F (c.1849G>T) in gene JAK2 cause the sensitivity of Go6976 by unusual activation of pro-survival pathway | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Chronic myeloid leukemia [ICD-11: 2A20.0] | [22] | |||
| Sensitive Disease | Chronic myeloid leukemia [ICD-11: 2A20.0] | |||
| Sensitive Drug | Peginterferon alfa-2a | |||
| Molecule Alteration | Missense mutation | p.V617F (c.1849G>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Bone marrow | N.A. | ||
| Mechanism Description | The missense mutation p.V617F (c.1849G>T) in gene JAK2 cause the sensitivity of Peginterferon alfa-2a by unusual activation of pro-survival pathway | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [23] | |||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Sensitive Drug | Pyridone 6 | |||
| Molecule Alteration | Copy number gain | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | JAKT/STAT signaling pathway | Inhibition | hsa04630 | |
| In Vitro Model | HCC70 cells | Breast | Homo sapiens (Human) | CVCL_1270 |
| HCC1954 cells | Breast | Homo sapiens (Human) | CVCL_1259 | |
| MDA-231 cells | Pleural effusion | Homo sapiens (Human) | CVCL_0062 | |
| MDA-MB-436 cells | Breast | Homo sapiens (Human) | CVCL_0623 | |
| SUM159PT cells | Breast | Homo sapiens (Human) | CVCL_5423/CVCL_5590 | |
| HCC38 cells | Breast | Homo sapiens (Human) | CVCL_1267 | |
| HCC1143 cells | Breast | Homo sapiens (Human) | CVCL_1245 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
SRB assay | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Whole blood | |
| The Specified Disease | Myelofibrosis | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.29E-03; Fold-change: 1.98E-01; Z-score: 5.31E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
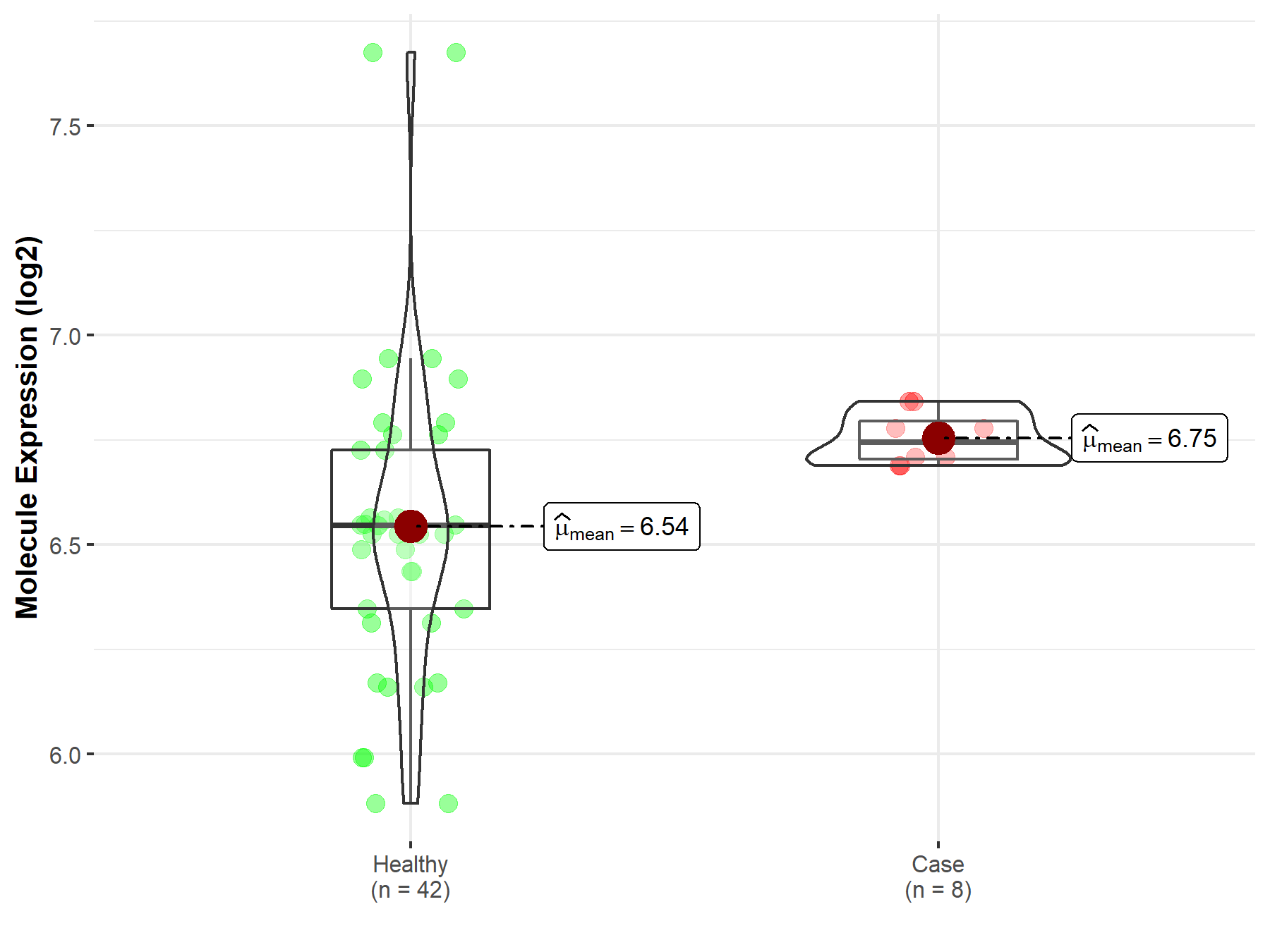
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Whole blood | |
| The Specified Disease | Polycythemia vera | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.41E-07; Fold-change: 3.61E-01; Z-score: 1.03E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Bone marrow | |
| The Specified Disease | Acute myeloid leukemia | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.46E-10; Fold-change: -4.97E-01; Z-score: -8.17E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
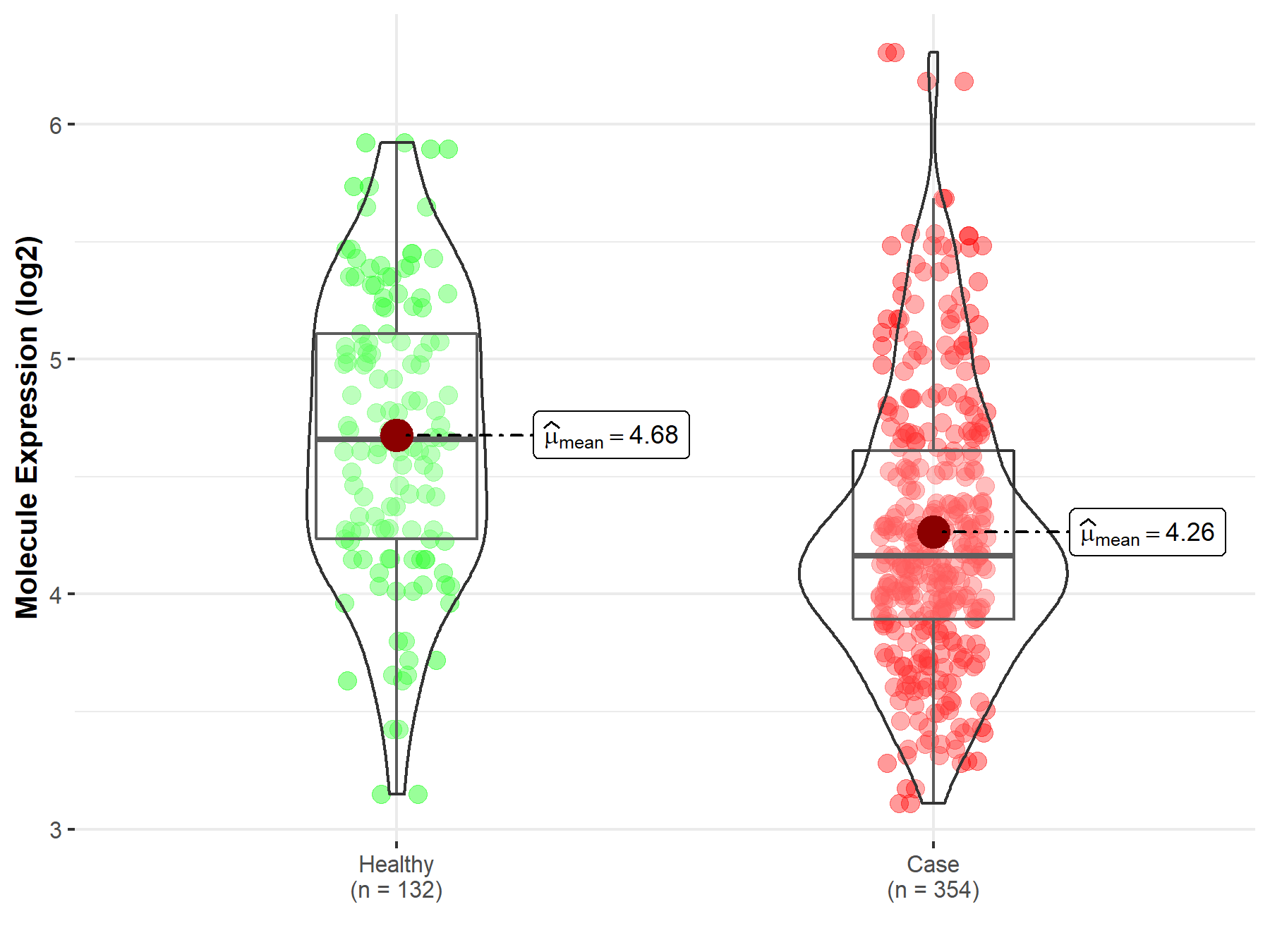
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.64E-02; Fold-change: 3.95E-02; Z-score: 5.89E-02 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 8.64E-01; Fold-change: -4.74E-02; Z-score: -6.30E-02 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
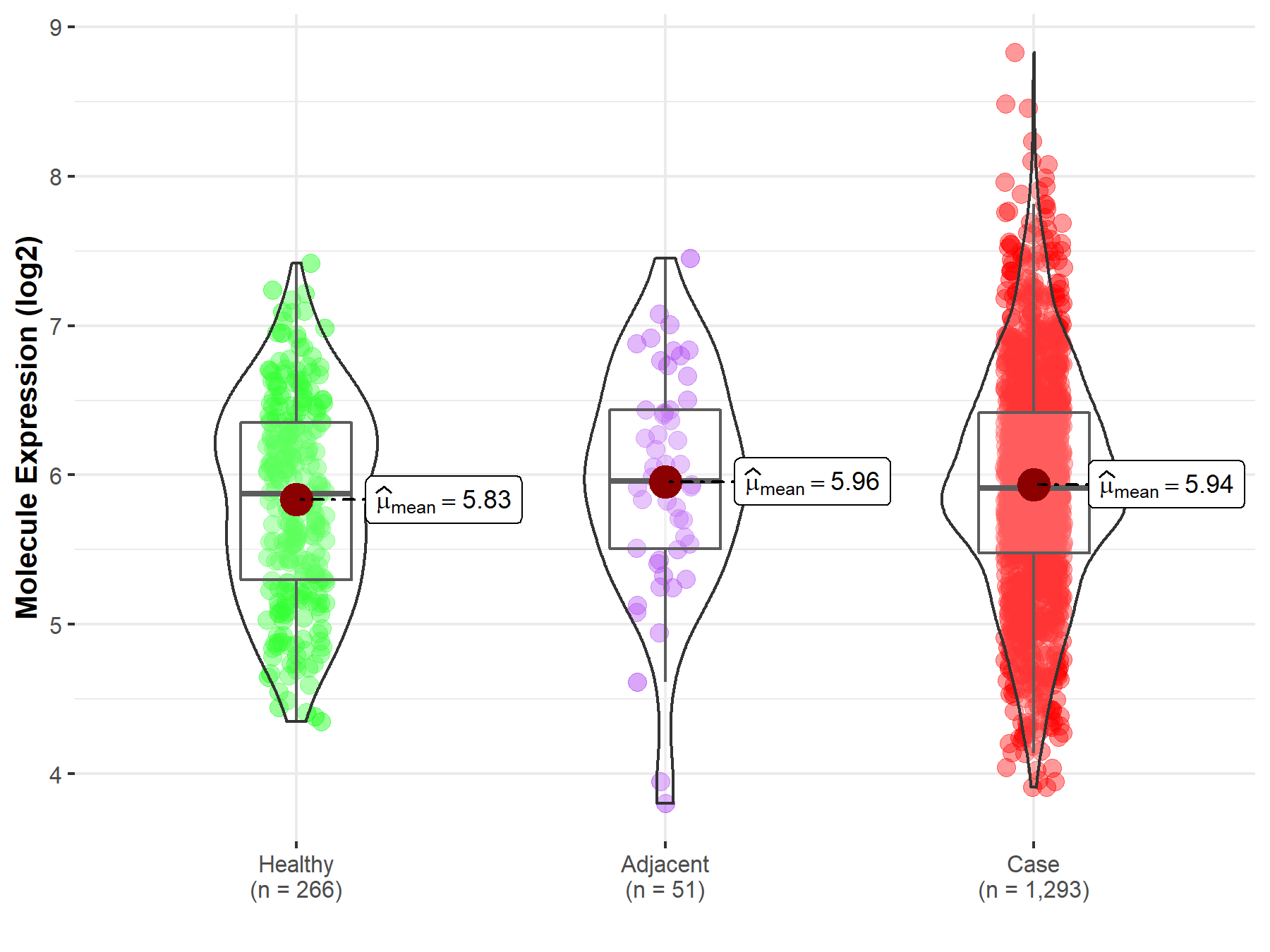
|
Click to View the Clearer Original Diagram |
ICD Disease Classification 03

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Whole blood | |
| The Specified Disease | Essential thrombocythemia | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.90E-02; Fold-change: 5.70E-02; Z-score: 1.54E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
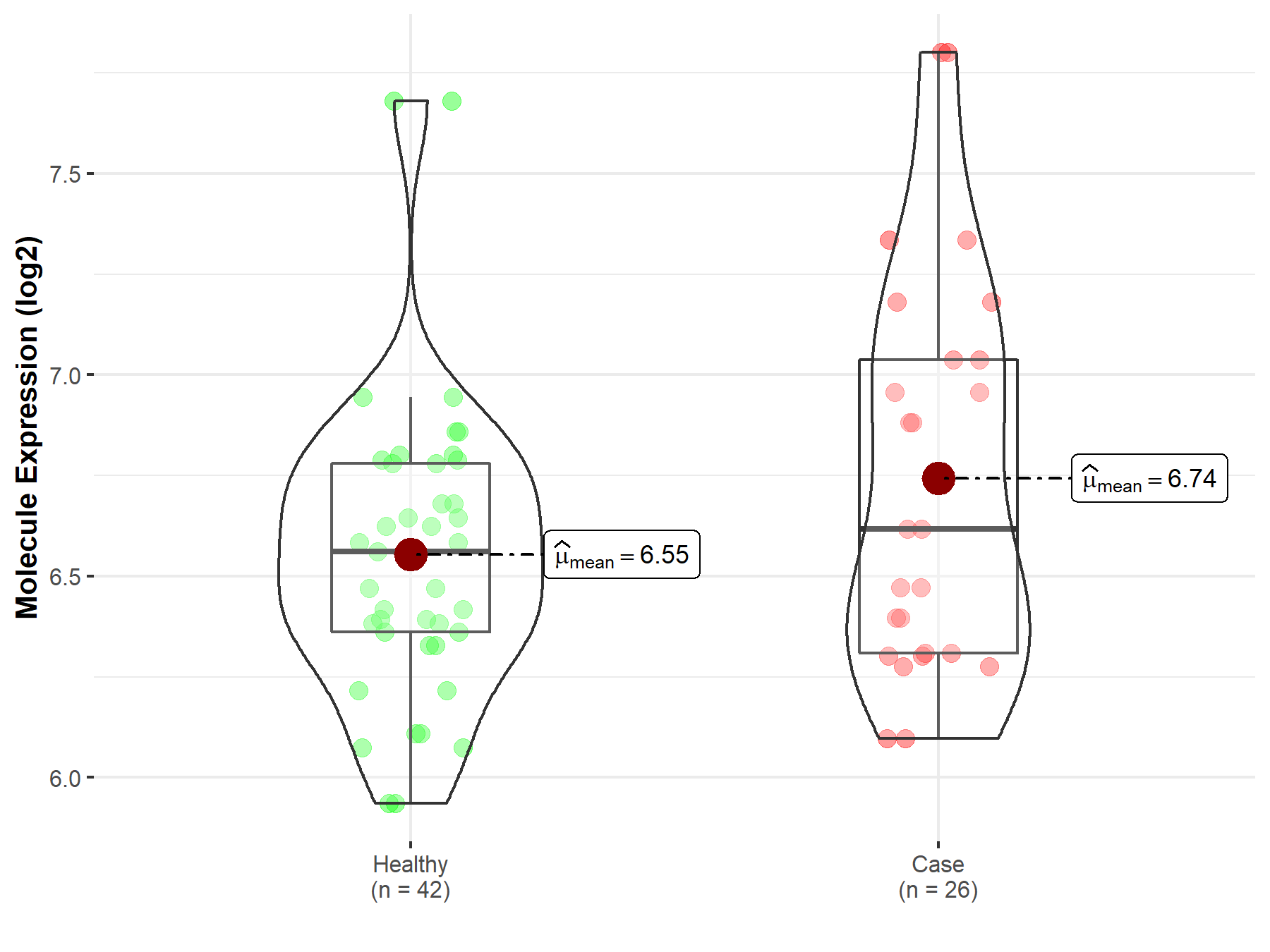
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

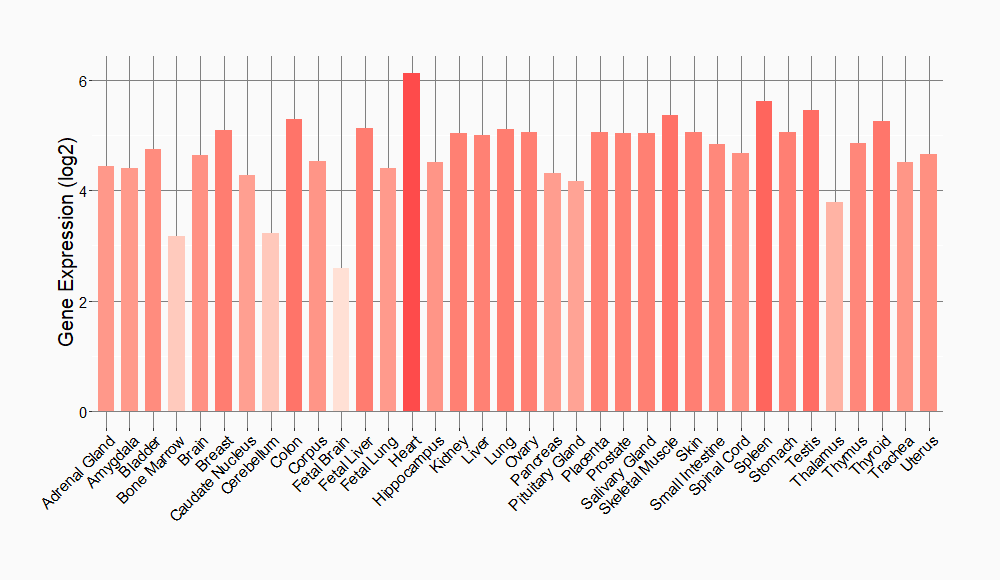
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
