Drug Information
Drug (ID: DG00575) and It's Reported Resistant Information
| Name |
Metronidazole
|
||||
|---|---|---|---|---|---|
| Synonyms |
Metronidazole; 443-48-1; Flagyl; Metronidazol; 2-Methyl-5-nitroimidazole-1-ethanol; Anagiardil; Gineflavir; Trichazol; MetroGel; Bayer 5360; Deflamon; Meronidal; Metronidaz; Novonidazol; Trichopol; Trivazol; Danizol; Mexibol; Orvagil; Vagilen; Clont; Flagemona; Giatricol; Metronidazolo; Sanatrichom; Takimetol; Trichocide; Trichomol; Trikacide; Acromona; Atrivyl; Efloran; Entizol; Flagesol; Monagyl; Monasin; Trichex; Tricocet; Trikamon; Trikojol; Trikozol; Trimeks; Vagimid; Vertisal; Wagitran; Arilin; Bexon; Elyzol; Eumin; Flagil; Klion; Klont; Nalox; Tricom; 2-(2-Methyl-5-nitro-1H-imidazol-1-yl)ethanol; neo-Tric; Tricowas B; Deflamon-wirkstoff; Protostat; Satric; MetroCream; MetroLotion; MetroGel-Vaginal; CONT; NIDA; Methronidazole; Metromidol; Trichopal; Flegyl; Fossyol; 1H-Imidazole-1-ethanol, 2-methyl-5-nitro-; Flagyl Er; Metronidazolum; Metro I.V.; Metrolyl; 2-(2-methyl-5-nitroimidazol-1-yl)ethanol; Metric 21; Trichomonacid 'pharmachim'; 1-(2-Hydroxyethyl)-2-methyl-5-nitroimidazole; RP 8823; NSC-50364; Metronidazole in Plastic Container; 2-Methyl-1-(2-hydroxyethyl)-5-nitroimidazole; 2-Methyl-3-(2-hydroxyethyl)-4-nitroimidazole; SC 10295; 1-(beta-Ethylol)-2-methyl-5-nitro-3-azapyrrole; 1-(2-Hydroxy-1-ethyl)-2-methyl-5-nitroimidazole; 2-(2-methyl-5-nitro-1H-imidazol-1-yl)ethan-1-ol; 2-(2-methyl-5-nitro-1-imidazolyl)ethanol; MFCD00009750; 1-(beta-Hydroxyethyl)-2-methyl-5-nitroimidazole; 1-Hydroxyethyl-2-methyl-5-nitroimidazole; Imidazole-1-ethanol, 2-methyl-5-nitro-; FLAGYL I.V. RTU IN PLASTIC CONTAINER; 1-(beta-Oxyethyl)-2-methyl-5-nitroimidazole; NSC 50364; UNII-140QMO216E; 2-(2-methyl-5-nitro-imidazol-1-yl)ethanol; BAY-5360; NSC69587; Noritate; MLS000028590; CHEBI:6909; 140QMO216E; Metro Gel; NSC50364; NSC-69587; NCGC00016446-06; CAS-443-48-1; Metrolag; Metrotop; Rathimed; SMR000058175; Vandazole; Zadstat; Tricho cordes; DSSTox_CID_892; Metronidazolo [DCIT]; Tricho-gynaedron; DSSTox_RID_75848; DSSTox_GSID_20892; Mexibol 'silanes'; Metro I.V. In Plastic Container; 1-(.beta.-Ethylol)-2-methyl-5-nitro-3-azapyrrole; 1-(.beta.-Hydroxyethyl)-2-methyl-5-nitroimidazole; Metronidazol [INN-Spanish]; Metronidazolum [INN-Latin]; Flagyl I.V. RTU; Flagyl 375; Trichobrol; Florazole; Mepagyl; Nidagyl; Rosased; Zidoval; Caswell No. 579AA; WLN: T5N CNJ A2Q B1 ENW; Noritate (TN); CCRIS 410; Metro cream & gel; Flagyl (TN); HSDB 3129; WLN: T6NTJ DQ ANU1- ET5N CNJ A1 BNW; SR-01000000244; EINECS 207-136-1; NSC 69587; EPA Pesticide Chemical Code 120401; BRN 0611683; Polibiotic; Trikhopol; Donnan; Flazol; CB-01-14 MMX; Metro IV; Vandazole (TN); Metronidazole,(S); Prestwick_334; Nuvessa (TN); IDR-90105; Cimetrol 500LPCI; Metronidazole solution; RP-8823; Metronidazole, BioXtra; Metronidazole (Flagyl); Spectrum_001035; Metronidazole [USAN:USP:INN:BAN:JAN]; HELIDAC (Salt/Mix); 2-(2-methyl-5-nitroimidazolyl)ethan-1-ol; Maybridge1_001999; Opera_ID_1585; Prestwick0_000081; Prestwick1_000081; Prestwick2_000081; Prestwick3_000081; Spectrum2_000883; Spectrum3_000506; Spectrum4_000060; Spectrum5_001289; M0924; CHEMBL137; NCIOpen2_000337; SCHEMBL23042; BSPBio_000002; BSPBio_002031; KBioGR_000559; KBioSS_001515; 5-23-05-00063 (Beilstein Handbook Reference); MLS000758286; MLS001424018; BIDD:GT0107; DivK1c_000007; SPECTRUM1500412; SPBio_000666; SPBio_001941; BAYER-5360; BPBio1_000004; DTXSID2020892; Flagyl I.V. RTU (Salt/Mix); BCBcMAP01_000184; GTPL10914; HMS500A09; HMS547C19; KBio1_000007; KBio2_001515; KBio2_004083; KBio2_006651; KBio3_001531; Metronidazole (JP17/USP/INN); Metronidazole, analytical standard; NINDS_000007; HMS1568A04; HMS1920N19; HMS2051G07; HMS2090B19; HMS2091F14; HMS2095A04; HMS2231E11; HMS3373O05; HMS3393G07; HMS3655E22; HMS3712A04; Pharmakon1600-01500412; ZINC113442; BCP13757; HY-B0318; Tox21_110441; Tox21_202413; Tox21_302794; BBL005452; BDBM50375309; CCG-40016; FP-250; NSC757118; s1907; STK177359; Metronidazole 2.0 mg/ml in Methanol; AKOS000269646; AKOS005169650; Tox21_110441_1; DB00916; KS-5140; MCULE-6891596695; NC00020; NSC-757118; IDI1_000007; SMP1_000189; NCGC00016446-01; NCGC00016446-02; NCGC00016446-03; NCGC00016446-04; NCGC00016446-05; NCGC00016446-07; NCGC00016446-08; NCGC00016446-09; NCGC00016446-11; NCGC00016446-12; NCGC00016446-17; NCGC00022059-03; NCGC00022059-04; NCGC00022059-05; NCGC00256513-01; NCGC00259962-01; AC-23968; SY002821; SBI-0051447.P003; DB-051212; Metronidazole, SAJ first grade, >=99.0%; AB00052046; BB 0218386; FT-0603394; SW196613-4; C07203; D00409; AB00052046-17; AB00052046_18; AB00052046_19; A826552; Metronidazole, VETRANAL(TM), analytical standard; Q169569; 2-(2-methyl-5-nitro-1H-imidazol-1-yl)-1-ethanol; 2-(2-Methyl-5-nitro-1H-imidazol-1-yl)ethanol #; Metronidazole, Antibiotic for Culture Media Use Only; Q-201403; SR-01000000244-4; SR-01000000244-5; BRD-K52020312-001-05-2; BRD-K52020312-001-15-1; Z87001124; F1773-0073; Metronidazole, certified reference material, TraceCERT(R); Metronidazole, British Pharmacopoeia (BP) Reference Standard; Metronidazole, European Pharmacopoeia (EP) Reference Standard; Metronidazole, United States Pharmacopeia (USP) Reference Standard; Metronidazole solution, 2.0 mg/mL in methanol, ampule of 1 mL, certified reference material; Metronidazole, Pharmaceutical Secondary Standard; Certified Reference Material
Click to Show/Hide
|
||||
| Indication |
In total 3 Indication(s)
|
||||
| Structure |
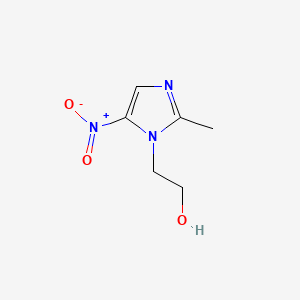
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Clinically Reported Resistance for This Drug
(6 diseases)
[2]
[3]
[4]
[5]
[6]
[7]
Disease(s) with Resistance Information Validated by in-vivo Model for This Drug
(4 diseases)
[8]
[9]
[8]
[1]
|
||||
| Target | Bacterial Deoxyribonucleic acid (Bact DNA) | NOUNIPROTAC | [8] | ||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C6H9N3O3
|
||||
| IsoSMILES |
CC1=NC=C(N1CCO)[N+](=O)[O-]
|
||||
| InChI |
1S/C6H9N3O3/c1-5-7-4-6(9(11)12)8(5)2-3-10/h4,10H,2-3H2,1H3
|
||||
| InChIKey |
VAOCPAMSLUNLGC-UHFFFAOYSA-N
|
||||
| PubChem CID | |||||
| ChEBI ID | |||||
| TTD Drug ID | |||||
| INTEDE ID | |||||
| DrugBank ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-01: Infectious/parasitic diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: 2-Nitroimidazole nitrohydrolase (NNHA) | [8] | |||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | THP1 cells | Pleural effusion | Homo sapiens (Human) | CVCL_0006 |
| Mechanism Description | The oxygen-insensitive major and minor (nsfA and nfsB, respectively) flavoprotein nitroreductases of E. coli, were shown to reduce nitrofurans and later, metronidazole. These nitroreductases share a similar structure to the nim genes described below. Mutants of both nfsA/nfsB in E. coli show reduced susceptibility to metronidazole. A 2-nitroimidazole nitrohydrolase (NnhA) was also shown to render E. coli resistant to 2-nitroimidazoles. | |||
|
|
||||
| Key Molecule: G2-specific protein kinase (NIMA) | [8] | |||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | |||
| Molecule Alteration | Expression | Acquired |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | THP1 cells | Pleural effusion | Homo sapiens (Human) | CVCL_0006 |
| Mechanism Description | In B. fragilis, nimA encodes a 5-nitroimidazole reductase reducing the metronidazole analogue dimetronidazole (1,2-dimethyl-5-nitroimidazole) to the amino derivative, preventing ring fission and associated toxicity. The role of nim genes in metronidazole resistance is controversial. It has been established that overexpression of a NimA homologue from B. fragilis induces a 3-fold increase in metronidazole resistance in E. coli. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Thioredoxin (TRX) | [9] | |||
| Resistant Disease | Clostridium difficile infection [ICD-11: 1A04.0] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| Mechanism Description | The decrement of redox proteins including pyruvate:ferredoxin oxidoreductase (OR), pyruvate:flavodoxin OR, thioredoxin and thioredoxin reductase are thought to be responsible for MTZ resistance as they are required for drug activation. | |||
| Key Molecule: Thioredoxin (TRX) | [9] | |||
| Resistant Disease | Clostridium difficile infection [ICD-11: 1A04.0] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| Mechanism Description | The decrement of redox proteins including pyruvate:ferredoxin oxidoreductase (OR), pyruvate:flavodoxin OR, thioredoxin and thioredoxin reductase are thought to be responsible for MTZ resistance as they are required for drug activation. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: ABC transporter family protein (ABC) | [1] | |||
| Resistant Disease | Trichomoniasis [ICD-11: 1A92.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Trichomonas vaginalis ATCC 30001 | 5722 | ||
| Trichomonas vaginalis ATCC 30236 | 5722 | |||
| Trichomonas vaginalis ATCC50148 | 5722 | |||
| Trichomonas vaginalis ATCC 50142 | 5722 | |||
| Trichomonas vaginalis ATCC 50143 | 5722 | |||
| Trichomonas vaginalis ATCC 30238 | 5722 | |||
| Experiment for Molecule Alteration |
Gene sequencing assay | |||
| Experiment for Drug Resistance |
Trypan blue exclusion assay | |||
| Mechanism Description | Comparative transcriptomes analysis identified that several putative drug-resistant genes were exclusively upregulated in different MTZ-R strains, such as ATP-binding cassette (ABC) transporters and multidrug resistance pumps. | |||
| Key Molecule: Multidrug resistance protein MdtE (MDTE) | [1] | |||
| Resistant Disease | Trichomoniasis [ICD-11: 1A92.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Trichomonas vaginalis ATCC 30001 | 5722 | ||
| Trichomonas vaginalis ATCC 30236 | 5722 | |||
| Trichomonas vaginalis ATCC50148 | 5722 | |||
| Trichomonas vaginalis ATCC 50142 | 5722 | |||
| Trichomonas vaginalis ATCC 50143 | 5722 | |||
| Trichomonas vaginalis ATCC 30238 | 5722 | |||
| Experiment for Molecule Alteration |
Gene sequencing assay | |||
| Experiment for Drug Resistance |
Trypan blue exclusion assay | |||
| Mechanism Description | Comparative transcriptomes analysis identified that several putative drug-resistant genes were exclusively upregulated in different MTZ-R strains, such as ATP-binding cassette (ABC) transporters and multidrug resistance pumps. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Glycosyltransferase Bme (BME) | [8] | |||
| Resistant Disease | Bacillus clausii infection [ICD-11: 1C4Y.1] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Vero cells | Kidney | Chlorocebus sabaeus (Green monkey) (Cercopithecus sabaeus) | CVCL_0059 |
| Mechanism Description | E. coli mutants lacking the ability to reduce nitrate and chlorate, presumably reducing the rate of metronidazole uptake, are also resistant. A reduced growth rate is also associated with metronidazole resistance in C. perfringens, C. difficile and B. fragilis. Bacteroides resistance-nodulation-division (RND) efflux pumps (bme) also reduce susceptibility to metronidazole. | |||
|
|
||||
| Key Molecule: HTH-type transcriptional activator (RHAR) | [8] | |||
| Resistant Disease | Bacillus clausii infection [ICD-11: 1C4Y.1] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Vero/TMPRSS2 cells | Kidney | Chlorocebus sabaeus (Green monkey) (Cercopithecus sabaeus) | CVCL_YQ48 |
| Mechanism Description | In B. thetaiotaomicron, the upregulation of rhamnose catabolism regulatory protein (RhaR) results in a similar rerouting and increased resistance to metronidazole. | |||
| Key Molecule: G2-specific protein kinase (NIMA) | [8] | |||
| Resistant Disease | Bacillus clausii infection [ICD-11: 1C4Y.1] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Vero cells | Kidney | Chlorocebus sabaeus (Green monkey) (Cercopithecus sabaeus) | CVCL_0059 |
| Mechanism Description | In B. fragilis, nimA encodes a 5-nitroimidazole reductase reducing the metronidazole analogue dimetronidazole (1,2-dimethyl-5-nitroimidazole) to the amino derivative, preventing ring fission and associated toxicity. The role of nim genes in metronidazole resistance is controversial. It has been established that overexpression of a NimA homologue from B. fragilis induces a 3-fold increase in metronidazole resistance in E. coli. | |||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
