Drug Information
Drug (ID: DG00047) and It's Reported Resistant Information
| Name |
Ofloxacin
|
||||
|---|---|---|---|---|---|
| Synonyms |
Bactocin; DEXTROFLOXACINE; Danoflox; Effexin; Exocin; Exocine; Flobacin; Flodemex; Flotavid; Flovid; Floxal; Floxil; Floxin; Floxstat; Fugacin; Inoflox; Kinflocin; Kinoxacin; Liflox; Loxinter; Marfloxacin; Medofloxine; Mergexin; Novecin; Nufafloqo; OFLX; OFX; Obide; Occidal; Ocuflox; Ofcin; Oflin; Oflocee; Oflocet; Oflocin; Oflodal; Oflodex; Oflodura; Oflox; Ofloxacina; Ofloxacine; Ofloxacino; Ofloxacinum; Ofloxin; Ofus; Onexacin; Operan; Orocin; Otonil; Oxaldin; Pharflox; Praxin; Puiritol; Qinolon; Qipro; Quinolon; Quotavil; Rilox; Sinflo; Tabrin; Taravid; Tariflox; Tarivid; Telbit; Tructum; Viotisone; Visiren; XED; Zanocin; Floxin otic; Ofloxacin Otic; Ofloxacina [DCIT]; Ofloxacine [French]; Ofloxacino [Spanish]; Ofloxacinum [Latin]; Uro Tarivid; DL 8280; HOE 280; O 8757; ORF 18489; PT 01; DL-8280; FLOXIN IN DEXTROSE 5%; FLOXIN IN DEXTROSE 5% IN PLASTIC CONTAINER; Floxin Otic (TN); HOE-280; Hoe-280; Marfloxacin (TN); O-Flox; ORF-28489; Ocuflox (TN); Ru-43280; WP-0405; Ofloxacin (JP15/USP/INN); Ofloxacin [USAN:BAN:INN:JAN]; Ofloxacin, (S)-Isomer; DL-8280, HOE-280, Exocin, Flobacin, Floxin, Floxil, Monoflocet, Ofloxacin; (+-)-9-Fluoro-2,3-dihydro-3-methyl-10-(4-methyl-1-piperazinyl)-7-oxo-7H-pyrido(1,2,3-de)-1,4-benzoxazine-6-carboxylic acid; (+/-)-9-Fluoro-2,3-dihydro-3-methyl-10-(4-methyl-1-piperaz inyl)-7-oxo-7H-pyrido(1,2,3-de)-1,4-benzoxazine-6-carboxylic acid; (+/-)-9-Fluoro-2,3-dihydro-3-methyl-10-(4-methyl-1-piperazinyl)-7-oxo-7H-pyrido(1,2,3-de)-1,4-benzoxazine-6-carboxylic acid; (+/-)-9-Fluoro-2,3-dihydro-3-methyl-10-(4-methyl-1-piperazinyl)-7-oxo-7H-pyrido[1,2,3-de]-1,4-benzoxazine-6-carboxylic acid; (+/-)-Floxin; (-)-9-Fluoro-2,3-dihydro-3-methyl-10-(4-methyl-1-piperazinyl)-7-oxo-7H-pyrido(1,2,3-de)(1,4)benzoxazin-6-carbonsaeure; 9-fluoro-3-methyl-10-(4-methylpiperazin-1-yl)-7-oxo-2,3-dihydro-7H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid
Click to Show/Hide
|
||||
| Indication |
In total 1 Indication(s)
|
||||
| Structure |
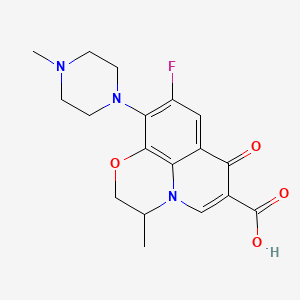
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Clinically Reported Resistance for This Drug
(8 diseases)
[3]
[6]
[7]
[9]
[7]
[3]
Disease(s) with Resistance Information Discovered by Cell Line Test for This Drug
(2 diseases)
[8]
[10]
Disease(s) with Resistance Information Validated by in-vivo Model for This Drug
(2 diseases)
[11]
[12]
|
||||
| Target | Bacterial DNA gyrase (Bact gyrase) |
GYRA_STAAU
; GYRB_STAAU |
[1] | ||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C18H20FN3O4
|
||||
| IsoSMILES |
CC1COC2=C3N1C=C(C(=O)C3=CC(=C2N4CCN(CC4)C)F)C(=O)O
|
||||
| InChI |
1S/C18H20FN3O4/c1-10-9-26-17-14-11(16(23)12(18(24)25)8-22(10)14)7-13(19)15(17)21-5-3-20(2)4-6-21/h7-8,10H,3-6,9H2,1-2H3,(H,24,25)
|
||||
| InChIKey |
GSDSWSVVBLHKDQ-UHFFFAOYSA-N
|
||||
| PubChem CID | |||||
| ChEBI ID | |||||
| TTD Drug ID | |||||
| VARIDT ID | |||||
| INTEDE ID | |||||
| DrugBank ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-01: Infectious/parasitic diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: DNA gyrase subunit A (GYRA) | [13], [14] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.T83I |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Pseudomonas aeruginosa isolates | 287 | ||
| Pseudomonas aeruginosa ATCC10145 | 287 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Etest assay | |||
| Mechanism Description | The major mechanism of the resistance of this Pseudomonas aeruginosa to fluoroquinolones is the modification of type II topoisomerases (DNA gyrase and topoisomerase IV). | |||
| Key Molecule: DNA gyrase subunit A (GYRA) | [13], [14] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.H83R |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Pseudomonas aeruginosa isolates | 287 | ||
| Pseudomonas aeruginosa ATCC10145 | 287 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Etest assay | |||
| Mechanism Description | The major mechanism of the resistance of this Pseudomonas aeruginosa to fluoroquinolones is the modification of type II topoisomerases (DNA gyrase and topoisomerase IV). | |||
| Key Molecule: DNA gyrase subunit A (GYRA) | [15], [16], [17] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.S83L |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli strain kL16 | 1425342 | ||
| Escherichia coli strain N-112 | 562 | |||
| Escherichia coli strain N-118 | 562 | |||
| Escherichia coli strain N-119 | 562 | |||
| Escherichia coli strain N-51 | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Mechanism Description | Quinolones are considered to exert antibacterial activity by inhibiting DNA gyrase (EC 5.99.1.3), which catalyzes topological changes of DNA.DNA gyrase of Escherichia coli consists of subunits A and B, which are the products of the gyrA and gyrB genes, respectively. Mutations in either gene can cause quinolone resistance. | |||
| Key Molecule: DNA gyrase subunit A (GYRA) | [15], [16], [17] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.S83W |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli strain kL16 | 1425342 | ||
| Escherichia coli strain P-18 | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Mechanism Description | Quinolones are considered to exert antibacterial activity by inhibiting DNA gyrase (EC 5.99.1.3), which catalyzes topological changes of DNA.DNA gyrase of Escherichia coli consists of subunits A and B, which are the products of the gyrA and gyrB genes, respectively. Mutations in either gene can cause quinolone resistance. | |||
| Key Molecule: DNA gyrase subunit A (GYRA) | [15], [16], [17] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.D87N |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli strain kL16 | 1425342 | ||
| Escherichia coli strain N-113 | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Mechanism Description | Quinolones are considered to exert antibacterial activity by inhibiting DNA gyrase (EC 5.99.1.3), which catalyzes topological changes of DNA.DNA gyrase of Escherichia coli consists of subunits A and B, which are the products of the gyrA and gyrB genes, respectively. Mutations in either gene can cause quinolone resistance. | |||
| Key Molecule: DNA gyrase subunit A (GYRA) | [15], [16], [17] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.G81C |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli strain kL16 | 1425342 | ||
| Escherichia coli strain N-97 | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Mechanism Description | Quinolones are considered to exert antibacterial activity by inhibiting DNA gyrase (EC 5.99.1.3), which catalyzes topological changes of DNA.DNA gyrase of Escherichia coli consists of subunits A and B, which are the products of the gyrA and gyrB genes, respectively. Mutations in either gene can cause quinolone resistance. | |||
| Key Molecule: DNA gyrase subunit A (GYRA) | [15], [16], [17] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.A84P |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli strain kL16 | 1425342 | ||
| Escherichia coli strain P-5 | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Mechanism Description | Quinolones are considered to exert antibacterial activity by inhibiting DNA gyrase (EC 5.99.1.3), which catalyzes topological changes of DNA.DNA gyrase of Escherichia coli consists of subunits A and B, which are the products of the gyrA and gyrB genes, respectively. Mutations in either gene can cause quinolone resistance. | |||
| Key Molecule: DNA gyrase subunit A (GYRA) | [15], [16], [17] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.A67S |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli strain kL16 | 1425342 | ||
| Escherichia coli strain P-10 | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Mechanism Description | Quinolones are considered to exert antibacterial activity by inhibiting DNA gyrase (EC 5.99.1.3), which catalyzes topological changes of DNA.DNA gyrase of Escherichia coli consists of subunits A and B, which are the products of the gyrA and gyrB genes, respectively. Mutations in either gene can cause quinolone resistance. | |||
| Key Molecule: DNA gyrase subunit A (GYRA) | [15], [16], [17] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.Q106H |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli strain kL16 | 1425342 | ||
| Escherichia coli strain N-89 | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Mechanism Description | Quinolones are considered to exert antibacterial activity by inhibiting DNA gyrase (EC 5.99.1.3), which catalyzes topological changes of DNA.DNA gyrase of Escherichia coli consists of subunits A and B, which are the products of the gyrA and gyrB genes, respectively. Mutations in either gene can cause quinolone resistance. | |||
|
|
||||
| Key Molecule: DNA topoisomerase 4 subunit B (PARE) | [18] | |||
| Resistant Disease | Morganella morganii infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.S463A |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Morganella morganii isolate | 582 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | The mutations in DNA gyrase (gyrA and gyrB) and topoisomerase IV (parC,parE) genes result in quinolone susceptibility. | |||
| Key Molecule: DNA topoisomerase 4 subunit B (PARE) | [18] | |||
| Resistant Disease | Morganella morganii infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.S464Y |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Morganella morganii isolate | 582 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | The mutations in DNA gyrase (gyrA and gyrB) and topoisomerase IV (parC,parE) genes result in quinolone susceptibility. | |||
| Key Molecule: DNA topoisomerase 4 subunit A (PARC) | [18] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.S80I |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Morganella morganii isolate | 582 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | The mutations in DNA gyrase (gyrA and gyrB) and topoisomerase IV (parC,parE) genes result in quinolone susceptibility. | |||
|
|
||||
| Key Molecule: Multidrug efflux pump Tap (TAP) | [1], [2] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis H37Rv | 83332 | ||
| Mycobacterium tuberculosis ICC154 | 1773 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | One mechanism proposed for drug resistance in Mycobacterium tuberculosis (MTB) is by efflux of the drugs by membrane located pumps.Mycobacterium tuberculosis isolate with a distinct genomic identity overexpresses a tap-like efflux pump,which confers resistance to Rifampin and Ofloxacin. | |||
| Key Molecule: Multidrug efflux pump Tap (TAP) | [1], [2] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium tuberculosis H37Rv | 83332 | ||
| Mycobacterium tuberculosis ICC154 | 1773 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | One mechanism proposed for drug resistance in Mycobacterium tuberculosis (MTB) is by efflux of the drugs by membrane located pumps.Mycobacterium tuberculosis isolate with a distinct genomic identity overexpresses a tap-like efflux pump,which confers resistance to Rifampin and Ofloxacin. | |||
| Key Molecule: Putative ABC transporter ATP-binding component (OTRC) | [19] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli BL21 (DE3) | 469008 | ||
| Escherichia coli | 668369 | |||
| Escherichia coli ET12567 (pUZ8002) | 562 | |||
| Streptomyces rimosus M4018 | 1927 | |||
| Streptomyces rimosus SR16 | 1927 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | OtrC is a multidrug resistance protein based on an ATP hydrolysis-dependent active efflux mechanism.OtrC is a multidrug resistance protein based on an ATP hydrolysis-dependent active efflux mechanism. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: DNA topoisomerase 4 subunit A (PARC) | [12] | |||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | |||
| Molecule Alteration | Missense mutation | p.S80l |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli ECIS803 | 562 | ||
| Escherichia coli ATCC 43869 | 562 | |||
| Experiment for Molecule Alteration |
PCR; DNA sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Mutational substitutions in the quinolone target enzymes, namely DNA topoisomerase II (GyrA) and topoisomerase IV (ParC), are recognised to be the major mechanisms through which resistance develops. | |||
| Key Molecule: DNA topoisomerase 4 subunit A (PARC) | [12] | |||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | |||
| Molecule Alteration | Missense mutation | p.E84G |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli ECIS803 | 562 | ||
| Escherichia coli ATCC 43869 | 562 | |||
| Experiment for Molecule Alteration |
PCR; DNA sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Mutational substitutions in the quinolone target enzymes, namely DNA topoisomerase II (GyrA) and topoisomerase IV (ParC), are recognised to be the major mechanisms through which resistance develops. | |||
| Key Molecule: DNA topoisomerase 4 subunit B (PARE) | [12] | |||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | |||
| Molecule Alteration | Missense mutation | p.D476N |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli ECIS803 | 562 | ||
| Escherichia coli ATCC 43869 | 562 | |||
| Experiment for Molecule Alteration |
PCR; DNA sequence assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Mutational substitutions in the quinolone target enzymes, namely DNA topoisomerase II (GyrA) and topoisomerase IV (ParC), are recognised to be the major mechanisms through which resistance develops. | |||
|
|
||||
| Key Molecule: Quinolone resistance protein NorA (NORA) | [3] | |||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | |||
| Molecule Alteration | Expression | Acquired |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli HB101 | 634468 | ||
| Staphylococcus aureus strain SA113 | 1280 | |||
| Experiment for Molecule Alteration |
Dideoxy chain-termination method assay | |||
| Mechanism Description | The norA gene cloned from chromosomal DNA of quinolone-resistant Staphylococcus aureus Tk2566 conferred relatively high resistance to hydrophilic quinolones such as norfloxacin, enoxacin, ofloxacin, and ciprofloxacin, but only low or no resistance at all to hydrophobic ones such as nalidixic acid, oxolinic acid, and sparfloxacin in S. aureus and Escherichia coli. Escherichia coli strains containing one of the plasmids carrying the norA gene (pTUS1, pTUS180, pTUS829, and pTUS206) were 8 to 64 times more resistant to the hydrophilic quinolones than the parent quinolone-susceptible strain. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: DNA gyrase subunit A (GYRA) | [11] | |||
| Resistant Disease | Clostridium difficile infection [ICD-11: 1A04.0] | |||
| Molecule Alteration | Mutation | p.T82I |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| Mechanism Description | Mutations in the gyrA or gyrB gene within quinolone resistance-determining region lead to the reduction in fidelity or prevention of drug binding via the target conformation change. Although several amino acid substitutions have been noted in GyrA and/or GyrB, the most frequent amino acid change has been recognized at T82I in GyrA subunit. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Outer membrane protein A (OmpA) | [10] | |||
| Resistant Disease | Tuberculosis [ICD-11: 1B10.0] | |||
| Molecule Alteration | Expressiom | D1194A+R1181K+D1194G |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Mycobacterium tuberculosis | 1773 | ||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | These results support the model that the roles of OmpA as a porin protein overexpressing in mycobacteria can increase the hydrophilic ability of the cell wall which can facilitate the streptomycin uptakes and increase the mycobacteria's sensitivity to aminoglycosides. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Dihydrofolate reductase/DNA-directed RNA polymerase subunit beta (DHFR/RPOB) | [6] | |||
| Resistant Disease | Leprosy [ICD-11: 1B20.0] | |||
| Molecule Alteration | Missense mutation | folP p.P55L+poB p.S531L |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium leprae isolates | 1769 | ||
| In Vivo Model | Footpad granuloma from M. leprae-infected nude mice model | Mus musculus | ||
| Experiment for Molecule Alteration |
PCR and single-stranded conformational polymorphism (SSCP) assay | |||
| Experiment for Drug Resistance |
Mouse footpad assay | |||
| Mechanism Description | The mutations genes reported in this study have been demonstrated to be responsible for drug resistance by mouse footpad assay. | |||
| Key Molecule: Dihydrofolate reductase/DNA-directed RNA polymerase subunit beta (DHFR/RPOB) | [6] | |||
| Resistant Disease | Leprosy [ICD-11: 1B20.0] | |||
| Molecule Alteration | Missense mutation | folP p.P55S+rpoB p.S531L+rpoB p.V547I |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium leprae isolates | 1769 | ||
| In Vivo Model | Footpad granuloma from M. leprae-infected nude mice model | Mus musculus | ||
| Experiment for Molecule Alteration |
PCR and single-stranded conformational polymorphism (SSCP) assay | |||
| Experiment for Drug Resistance |
Mouse footpad assay | |||
| Mechanism Description | The mutations genes reported in this study have been demonstrated to be responsible for drug resistance by mouse footpad assay. | |||
| Key Molecule: Dihydrofolate reductase/DNA gyrase subunit A/DNA gyrase subunit B (DHFR/GYRA/GYRB) | [6] | |||
| Resistant Disease | Leprosy [ICD-11: 1B20.0] | |||
| Molecule Alteration | Missense mutation | folP p.P55L+gyrA p.A91V+gyrB p.A91V |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium leprae isolates | 1769 | ||
| In Vivo Model | Footpad granuloma from M. leprae-infected nude mice model | Mus musculus | ||
| Experiment for Molecule Alteration |
PCR and single-stranded conformational polymorphism (SSCP) assay | |||
| Experiment for Drug Resistance |
Mouse footpad assay | |||
| Mechanism Description | The mutations genes reported in this study have been demonstrated to be responsible for drug resistance by mouse footpad assay. | |||
| Key Molecule: Dihydrofolate reductase/DNA gyrase subunit A/DNA gyrase subunit B (DHFR/GYRA/GYRB) | [6] | |||
| Resistant Disease | Leprosy [ICD-11: 1B20.0] | |||
| Molecule Alteration | Missense mutation | folP p.P55L+gyrA p.D205N+gyrB p.D205N |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium leprae isolates | 1769 | ||
| In Vivo Model | Footpad granuloma from M. leprae-infected nude mice model | Mus musculus | ||
| Experiment for Molecule Alteration |
PCR and single-stranded conformational polymorphism (SSCP) assay | |||
| Experiment for Drug Resistance |
Mouse footpad assay | |||
| Mechanism Description | The mutations genes reported in this study have been demonstrated to be responsible for drug resistance by mouse footpad assay. | |||
|
|
||||
| Key Molecule: DNA topoisomerase 4 subunit B (PARE) | [20] | |||
| Resistant Disease | Leprosy [ICD-11: 1B20.0] | |||
| Molecule Alteration | Missense mutation | p.D464N |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli BL21 (DE3) | 469008 | ||
| Escherichia coli Rosetta-gami 2 | 562 | |||
| Escherichia coli TOP-10 | 83333 | |||
| Mycobacterium leprae Thai-53 | 1769 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
DNA supercoiling assay; DNA cleavage assay | |||
| Mechanism Description | FQs are known to interact with both A and B subunits of DNA gyrase and inhibit supercoiling activity of this enzyme.The FQ-inhibited supercoiling assay and FQ-induced cleavage assay demonstrated the important roles of these amino acid substitutions in reduced sensitivity to FQ with marked influence by amino acid substitution, especially at position 502. | |||
| Key Molecule: DNA topoisomerase 4 subunit B (PARE) | [20] | |||
| Resistant Disease | Leprosy [ICD-11: 1B20.0] | |||
| Molecule Alteration | Missense mutation | p.N502D |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli BL21 (DE3) | 469008 | ||
| Escherichia coli Rosetta-gami 2 | 562 | |||
| Escherichia coli TOP-10 | 83333 | |||
| Mycobacterium leprae Thai-53 | 1769 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
DNA supercoiling assay; DNA cleavage assay | |||
| Mechanism Description | FQs are known to interact with both A and B subunits of DNA gyrase and inhibit supercoiling activity of this enzyme.The FQ-inhibited supercoiling assay and FQ-induced cleavage assay demonstrated the important roles of these amino acid substitutions in reduced sensitivity to FQ with marked influence by amino acid substitution, especially at position 502. | |||
| Key Molecule: DNA topoisomerase 4 subunit B (PARE) | [20] | |||
| Resistant Disease | Leprosy [ICD-11: 1B20.0] | |||
| Molecule Alteration | Missense mutation | p.E504V |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli BL21 (DE3) | 469008 | ||
| Escherichia coli Rosetta-gami 2 | 562 | |||
| Escherichia coli TOP-10 | 83333 | |||
| Mycobacterium leprae Thai-53 | 1769 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay; Allelic frequency measurement assay | |||
| Experiment for Drug Resistance |
DNA supercoiling assay; DNA cleavage assay | |||
| Mechanism Description | FQs are known to interact with both A and B subunits of DNA gyrase and inhibit supercoiling activity of this enzyme.The FQ-inhibited supercoiling assay and FQ-induced cleavage assay demonstrated the important roles of these amino acid substitutions in reduced sensitivity to FQ with marked influence by amino acid substitution, especially at position 502. | |||
|
|
||||
| Key Molecule: Dihydrofolate reductase (DHFR) | [6] | |||
| Resistant Disease | Leprosy [ICD-11: 1B20.0] | |||
| Molecule Alteration | Missense mutation | p.T53A |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium leprae isolates | 1769 | ||
| In Vivo Model | Footpad granuloma from M. leprae-infected nude mice model | Mus musculus | ||
| Experiment for Molecule Alteration |
PCR and single-stranded conformational polymorphism (SSCP) assay | |||
| Experiment for Drug Resistance |
Mouse footpad assay | |||
| Mechanism Description | The mutations genes reported in this study have been demonstrated to be responsible for drug resistance by mouse footpad assay. | |||
| Key Molecule: Dihydrofolate reductase (DHFR) | [6] | |||
| Resistant Disease | Leprosy [ICD-11: 1B20.0] | |||
| Molecule Alteration | Missense mutation | p.P55R |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycobacterium leprae isolates | 1769 | ||
| In Vivo Model | Footpad granuloma from M. leprae-infected nude mice model | Mus musculus | ||
| Experiment for Molecule Alteration |
PCR and single-stranded conformational polymorphism (SSCP) assay | |||
| Experiment for Drug Resistance |
Mouse footpad assay | |||
| Mechanism Description | The mutations genes reported in this study have been demonstrated to be responsible for drug resistance by mouse footpad assay. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: DNA topoisomerase (ATP-hydrolyzing) (PARC) | [7] | |||
| Resistant Disease | Mycoplasma hominis genital infection [ICD-11: 1B2Z.7] | |||
| Molecule Alteration | Missense mutation | p.K134R |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycoplasma hominis ATCC 23114(PG21) | 347256 | ||
| Mycoplasma hominis isolate | 2098 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Mechanism Description | The single amino acid mutation in ParC of MH may relate to the resistance to OFX and LVX and the high-level resistance to fluoroquinolones for MH is associated with mutations in both DNA gyrase and the ParC subunit of topoisomerase IV. | |||
| Key Molecule: DNA topoisomerase (ATP-hydrolyzing) (PARC) | [7] | |||
| Resistant Disease | Mycoplasma hominis mycoplasma infection [ICD-11: 1B2Z.4] | |||
| Molecule Alteration | Missense mutation | p.K134R |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycoplasma hominis ATCC 23114(PG21) | 347256 | ||
| Mycoplasma hominis isolate | 2098 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Mechanism Description | The single amino acid mutation in ParC of MH may relate to the resistance to OFX and LVX and the high-level resistance to fluoroquinolones for MH is associated with mutations in both DNA gyrase and the ParC subunit of topoisomerase IV. | |||
|
|
||||
| Key Molecule: P-type ATPase zinc transporter Rv3270 | [8] | |||
| Resistant Disease | Bone infection [ICD-11: 1B2Z.9] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | E. coli XL1-Blue | 562 | ||
| E. coli CS109 | 562 | |||
| M. smegmatis MC2 156 | 1772 | |||
| Experiment for Molecule Alteration |
Gene expression analysis | |||
| Experiment for Drug Resistance |
Antimicrobial susceptibility assay; Intracellular drug accumulation activity assay | |||
| Mechanism Description | Metal homeostasis is maintained by the uptake, storage and efflux of metal ions that are necessary for the survival of the bacterium. Homeostasis is mostly regulated by a group of transporters categorized as ABC transporters and P-type ATPases. On the other hand, efflux pumps often play a role in drug-metal cross-resistance. Here, with the help of antibiotic sensitivity, antibiotic/dye accumulation and semi-quantitative biofilm formation assessments we report the ability of Rv3270, a P-type ATPase known for its role in combating Mn2+ and Zn2+ metal ion toxicity in Mycobacterium tuberculosis, in influencing the extrusion of multiple structurally unrelated drugs and enhancing the biofilm formation of Escherichia coli and Mycobacterium smegmatis. Overexpression of Rv3270 increased the tolerance of host cells to norfloxacin, ofloxacin, sparfloxacin, ampicillin, oxacillin, amikacin and isoniazid. A significantly lower accumulation of norfloxacin, ethidium bromide, bocillin FL and levofloxacin in cells harbouring Rv3270 as compared to host cells indicated its role in enhancing efflux activity. Although over-expression of Rv3270 did not alter the susceptibility levels of levofloxacin, rifampicin and apramycin, the presence of a sub-inhibitory concentration of Zn2+ resulted in low-level tolerance towards these drugs. Of note, the expression of Rv3270 enhanced the biofilm-forming ability of the host cells strengthening its role in antimicrobial resistance. Therefore, the study indicated that the over-expression of Rv3270 enhances the drug efflux activity of the micro-organism where zinc might facilitate drug-metal cross-resistance for some antibiotics. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Quinolone resistance protein NorA (NORA) | [3] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli HB101 | 634468 | ||
| Staphylococcus aureus strain SA113 | 1280 | |||
| Experiment for Molecule Alteration |
Dideoxy chain-termination method assay | |||
| Mechanism Description | The norA gene cloned from chromosomal DNA of quinolone-resistant Staphylococcus aureus Tk2566 conferred relatively high resistance to hydrophilic quinolones such as norfloxacin, enoxacin, ofloxacin, and ciprofloxacin, but only low or no resistance at all to hydrophobic ones such as nalidixic acid, oxolinic acid, and sparfloxacin in S. aureus and Escherichia coli. | |||
| Key Molecule: Quinolone resistance protein NorA (NORA) | [3] | |||
| Resistant Disease | Staphylococcus aureus infection [ICD-11: 1B54.0] | |||
| Molecule Alteration | Expression | Acquired |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli HB101 | 634468 | ||
| Staphylococcus aureus strain SA113 | 1280 | |||
| Experiment for Molecule Alteration |
Dideoxy chain-termination method assay | |||
| Mechanism Description | The norA gene cloned from chromosomal DNA of quinolone-resistant Staphylococcus aureus Tk2566 conferred relatively high resistance to hydrophilic quinolones such as norfloxacin, enoxacin, ofloxacin, and ciprofloxacin, but only low or no resistance at all to hydrophobic ones such as nalidixic acid, oxolinic acid, and sparfloxacin in S. aureus and Escherichia coli. S. aureus SA113 (pTUS20) harboring a plasmid carrying the staphylococcal norA gene was 16 to 64 times more resistant to relatively hydrophilic quinolones. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: DNA topoisomerase 4 subunit B (PARE) | [4], [5] | |||
| Resistant Disease | HIV-infected patients with tuberculosis [ICD-11: 1C60.0] | |||
| Molecule Alteration | Missense mutation | p.N538D |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli | 668369 | ||
| Escherichia coli HB101 | 634468 | |||
| Mycobacterium smegmatis LR222 | 1772 | |||
| Mycobacterium tuberculosis MLB 262 | 1773 | |||
| Mycobacterium tuberculosis isolates | 1773 | |||
| Mycobacterium tuberculosis liquid | 1773 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay; disk diffusion test assay | |||
| Mechanism Description | DNA gyrase consists of two GyrA and two GyrB subunits encoded by gyrA and gyrB, respectively.Fluoroquinolone belong to the quinolone class of antibiotics which inhibit bacterial DNA gyrase and topoisomerase IV.Certain gyrA and gyrB mutations reported to confer cross-resistance to different FQ antibiotics based on clinical data have not yet been characterized in well-studied M. tuberculosis backgrounds. | |||
ICD-02: Benign/in-situ/malignant neoplasm
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: DNA topoisomerase (ATP-hydrolyzing) (PARC) | [7] | |||
| Resistant Disease | Mycoplasma hominis prostate cancer [ICD-11: 2C82.Y] | |||
| Molecule Alteration | Missense mutation | p.K134R |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Mycoplasma hominis ATCC 23114(PG21) | 347256 | ||
| Mycoplasma hominis isolate | 2098 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Mechanism Description | The single amino acid mutation in ParC of MH may relate to the resistance to OFX and LVX and the high-level resistance to fluoroquinolones for MH is associated with mutations in both DNA gyrase and the ParC subunit of topoisomerase IV. | |||
ICD-12: Respiratory system diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: MATE family efflux transporter (ABEM) | [9] | |||
| Resistant Disease | Acinetobacter baumannii infection [ICD-11: CA40.4] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli kAM32 | 562 | ||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | AbeM was found to be an H+-coupled multidrug efflux pump and a unique member of the MATE family which lead to drug resistance. | |||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
