Molecule Information
General Information of the Molecule (ID: Mol00509)
| Name |
MAPK/ERK kinase 1 (MEK1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
MAP kinase kinase 1; MAPKK 1; MKK1; ERK activator kinase 1; MAPK/ERK kinase 1; MEK 1; MEK1; PRKMK1
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
MAP2K1
|
||||
| Gene ID | |||||
| Location |
chr15:66386837-66491656[+]
|
||||
| Sequence |
MPKKKPTPIQLNPAPDGSAVNGTSSAETNLEALQKKLEELELDEQQRKRLEAFLTQKQKV
GELKDDDFEKISELGAGNGGVVFKVSHKPSGLVMARKLIHLEIKPAIRNQIIRELQVLHE CNSPYIVGFYGAFYSDGEISICMEHMDGGSLDQVLKKAGRIPEQILGKVSIAVIKGLTYL REKHKIMHRDVKPSNILVNSRGEIKLCDFGVSGQLIDSMANSFVGTRSYMSPERLQGTHY SVQSDIWSMGLSLVEMAVGRYPIPPPDAKELELMFGCQVEGDAAETPPRPRTPGRPLSSY GMDSRPPMAIFELLDYIVNEPPPKLPSGVFSLEFQDFVNKCLIKNPAERADLKQLMVHAF IKRSDAEEVDFAGWLCSTIGLNQPSTPTHAAGV Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Dual specificity protein kinase which acts as an essential component of the MAP kinase signal transduction pathway. Binding of extracellular ligands such as growth factors, cytokines and hormones to their cell-surface receptors activates RAS and this initiates RAF1 activation. RAF1 then further activates the dual-specificity protein kinases MAP2K1/MEK1 and MAP2K2/MEK2. Both MAP2K1/MEK1 and MAP2K2/MEK2 function specifically in the MAPK/ERK cascade, and catalyze the concomitant phosphorylation of a threonine and a tyrosine residue in a Thr-Glu-Tyr sequence located in the extracellular signal-regulated kinases MAPK3/ERK1 and MAPK1/ERK2, leading to their activation and further transduction of the signal within the MAPK/ERK cascade. Activates BRAF in a KSR1 or KSR2-dependent manner; by binding to KSR1 or KSR2 releases the inhibitory intramolecular interaction between KSR1 or KSR2 protein kinase and N-terminal domains which promotes KSR1 or KSR2-BRAF dimerization and BRAF activation. Depending on the cellular context, this pathway mediates diverse biological functions such as cell growth, adhesion, survival and differentiation, predominantly through the regulation of transcription, metabolism and cytoskeletal rearrangements. One target of the MAPK/ERK cascade is peroxisome proliferator-activated receptor gamma (PPARG), a nuclear receptor that promotes differentiation and apoptosis. MAP2K1/MEK1 has been shown to export PPARG from the nucleus. The MAPK/ERK cascade is also involved in the regulation of endosomal dynamics, including lysosome processing and endosome cycling through the perinuclear recycling compartment (PNRC), as well as in the fragmentation of the Golgi apparatus during mitosis.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
5 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Glioma [ICD-11: 2A00.1] | [1] | |||
| Sensitive Disease | Glioma [ICD-11: 2A00.1] | |||
| Sensitive Drug | Temozolomide | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Brain cancer [ICD-11: 2A00] | |||
| The Specified Disease | Brain cancer | |||
| The Studied Tissue | Nervous tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.08E-99 Fold-change: -1.64E-01 Z-score: -2.63E+01 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| MAPK signaling pathway | Inhibition | hsa04010 | ||
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| U87 cells | Brain | Homo sapiens (Human) | CVCL_0022 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | miR-181b independently predicted chemoresponse to temozolomide and enhanced temozolomide sensitivity via MEk1 downregulation. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [2] | |||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Resistant Drug | Binimetinib | |||
| Molecule Alteration | Missense mutation | p.V211D (c.632T>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | NIH-3T3 cells | Embryo | Mus musculus (Mouse) | CVCL_0594 |
| Phoenix AMPHO cells | Fetal kidney | Homo sapiens (Human) | CVCL_H716 | |
| In Vivo Model | NOD scid gamma xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Single cell sequencing assay | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | Regorafenib | ||||||||||||
| Molecule Alteration | Missense mutation | p.C121S (c.361T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.70 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.01 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
20
|
M
-
A
-
H
-
H
-
H
-
H
-
H
-
H
-
A
-
A
-
30
|
A
-
E
-
N
-
L
-
Y
-
F
-
Q
-
L
-
E
-
E
-
40
|
L
-
E
-
L
M
D
T
E
L
Q
Q
Q
Q
R
R
K
K
R
R
50
|
L
L
E
E
A
A
F
F
L
L
T
T
Q
Q
K
K
Q
Q
K
K
60
|
V
V
G
G
E
E
L
L
K
K
D
D
D
D
D
D
F
F
E
E
70
|
K
K
I
I
S
S
E
E
L
L
G
G
A
A
G
G
N
N
G
G
80
|
G
G
V
V
V
V
F
F
K
K
V
V
S
S
H
H
K
K
P
P
90
|
S
S
G
G
L
L
V
V
M
M
A
A
R
R
K
K
L
L
I
I
100
|
H
H
L
L
E
E
I
I
K
K
P
P
A
A
I
I
R
R
N
N
110
|
Q
Q
I
I
I
I
R
R
E
E
L
L
Q
Q
V
V
L
L
H
H
120
|
E
E
C
S
N
N
S
S
P
P
Y
Y
I
I
V
V
G
G
F
F
130
|
Y
Y
G
G
A
A
F
F
Y
Y
S
S
D
D
G
G
E
E
I
I
140
|
S
S
I
I
C
C
M
M
E
E
H
H
M
M
D
D
G
G
G
G
150
|
S
S
L
L
D
D
Q
Q
V
V
L
L
K
K
K
K
A
A
G
G
160
|
R
R
I
I
P
P
E
E
Q
Q
I
I
L
L
G
G
K
K
V
V
170
|
S
S
I
I
A
A
V
V
I
I
K
K
G
G
L
L
T
T
Y
Y
180
|
L
L
R
R
E
E
K
K
H
H
K
K
I
I
M
M
H
H
R
R
190
|
D
D
V
V
K
K
P
P
S
S
N
N
I
I
L
L
V
V
N
N
200
|
S
S
R
R
G
G
E
E
I
I
K
K
L
L
C
C
D
D
F
F
210
|
G
G
V
V
S
S
G
G
Q
Q
L
L
I
I
D
D
S
S
M
M
220
|
A
A
N
N
S
S
F
F
V
V
G
G
T
T
R
R
S
S
Y
Y
230
|
M
M
S
S
P
P
E
E
R
R
L
L
Q
Q
G
G
T
T
H
H
240
|
Y
Y
S
S
V
V
Q
Q
S
S
D
D
I
I
W
W
S
S
M
M
250
|
G
G
L
L
S
S
L
L
V
V
E
E
M
M
A
A
V
V
G
G
260
|
R
R
Y
Y
P
P
I
I
-
G
-
S
-
G
-
S
-
G
-
S
270
|
-
M
-
A
-
I
-
F
-
E
-
L
-
L
-
D
-
Y
-
I
280
|
-
V
-
N
-
E
-
P
-
P
-
P
-
K
-
L
-
P
-
S
290
|
-
G
-
V
-
F
-
S
-
L
-
E
-
F
-
Q
-
D
-
F
300
|
-
V
-
N
G
K
S
C
G
L
S
I
G
K
S
N
M
P
A
A
310
|
I
E
F
R
E
A
L
D
L
L
D
K
Y
Q
I
L
V
M
N
V
320
|
E
H
P
A
P
F
P
I
K
K
L
R
P
S
S
D
G
A
V
E
330
|
F
E
S
V
L
D
E
F
F
A
Q
G
D
W
F
L
V
C
N
S
340
|
K
T
C
I
L
G
I
L
K
N
N
Q
P
P
A
S
E
T
R
P
350
|
A
T
D
H
L
A
K
A
Q
G
L
E
M
G
V
H
H
H
A
H
360
|
F
H
I
H
K
H
R
-
S
-
D
-
A
-
E
-
E
-
V
-
370
|
D
-
F
-
A
-
G
-
W
-
L
-
C
-
S
-
T
-
I
-
380
|
G
-
L
-
N
-
Q
-
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.C121S (c.361T>A) in gene MAP2K1 cause the resistance of Regorafenib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | Regorafenib | ||||||||||||
| Molecule Alteration | Missense mutation | p.F53L (c.157T>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.F53L (c.157T>C) in gene MAP2K1 cause the resistance of Regorafenib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | Regorafenib | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q56P (c.167A>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.Q56P (c.167A>C) in gene MAP2K1 cause the resistance of Regorafenib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | Regorafenib | ||||||||||||
| Molecule Alteration | Missense mutation | p.K57N (c.171G>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.K57N (c.171G>C) in gene MAP2K1 cause the resistance of Regorafenib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | Regorafenib | ||||||||||||
| Molecule Alteration | Missense mutation | p.I111S (c.332T>G) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.I111S (c.332T>G) in gene MAP2K1 cause the resistance of Regorafenib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | Regorafenib | ||||||||||||
| Molecule Alteration | Missense mutation | p.F129L (c.385T>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.F129L (c.385T>C) in gene MAP2K1 cause the resistance of Regorafenib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | Regorafenib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V211D (c.632T>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.V211D (c.632T>A) in gene MAP2K1 cause the resistance of Regorafenib by unusual activation of pro-survival pathway | ||||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Salmonella enterica infection [ICD-11: 1A09.0] | [4] | |||
| Resistant Disease | Salmonella enterica infection [ICD-11: 1A09.0] | |||
| Resistant Drug | Reserpine | |||
| Molecule Alteration | Function | Activation |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| Cell Pathway Regulation | T cell receptor signaling pathway | Regulation | N.A. | |
| EGFR signaling pathway | Inhibition | hsa01521 | ||
| mTOR signaling pathway | Inhibition | hsa04150 | ||
| MEK1/2 signaling pathway | Activation | hsa04010 | ||
| In Vivo Model | Chicken ceca explant model | Gallus gallus | ||
| Experiment for Molecule Alteration |
Chicken-specific kinome peptide array | |||
| Experiment for Drug Resistance |
Bactericidal assays against Salmonella | |||
| Mechanism Description | Reserpine treatment induced T cell activation, reduced CTLA-4 gene expression, and deactivated metabolic pathways like epidermal growth factor receptor (EGFR) signaling and mammalian target of rapamycin (mTOR) signaling, which were linked to antimicrobial responses. MEK1/2 activation plays a central role in reserpine-induced antimicrobial activities. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [5] | |||
| Resistant Disease | Melanoma [ICD-11: 2C30.0] | |||
| Resistant Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.V60E (c.179T>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 |
| A2058 cells | Skin | Homo sapiens (Human) | CVCL_1059 | |
| WM2664 cells | Skin | Homo sapiens (Human) | CVCL_2765 | |
| SkMEL28 cells | Skin | Homo sapiens (Human) | CVCL_0526 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [6] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.Q56P (c.167A>C) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| AGS cells | Gastric | Homo sapiens (Human) | CVCL_0139 | |
| NCI-N87 cells | Gastric | Homo sapiens (Human) | CVCL_1603 | |
| NCI-H508 cells | Colon | Homo sapiens (Human) | CVCL_1564 | |
| SW48 cells | Colon | Homo sapiens (Human) | CVCL_1724 | |
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |
| NCI-H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | |
| NCI-H1650 cells | Lung | Homo sapiens (Human) | CVCL_1483 | |
| SW1573 cells | Lung | Homo sapiens (Human) | CVCL_1720 | |
| SNU-C1 cells | Peritoneum | Homo sapiens (Human) | CVCL_1708 | |
| OCUM-1 cells | Pleural effusion | Homo sapiens (Human) | CVCL_3084 | |
| NCI-H226 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1544 | |
| NCI-H196 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1509 | |
| NCI-H1437 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1472 | |
| NCI-H1355 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1464 | |
| MKN7 cells | Lymph node | Homo sapiens (Human) | CVCL_1417 | |
| NCI-H1299 cells | Lymph node | Homo sapiens (Human) | CVCL_0060 | |
| HCC366 cells | Lung | Homo sapiens (Human) | CVCL_2059 | |
| NCI-H2126 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1532 | |
| In Vivo Model | Female nu/nu mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay | |||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [6] | |||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.F53L (c.157T>C) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |
| AGS cells | Gastric | Homo sapiens (Human) | CVCL_0139 | |
| NCI-H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | |
| NCI-N87 cells | Gastric | Homo sapiens (Human) | CVCL_1603 | |
| NCI-H1650 cells | Lung | Homo sapiens (Human) | CVCL_1483 | |
| NCI-H508 cells | Colon | Homo sapiens (Human) | CVCL_1564 | |
| SW48 cells | Colon | Homo sapiens (Human) | CVCL_1724 | |
| SW1573 cells | Lung | Homo sapiens (Human) | CVCL_1720 | |
| SNU-C1 cells | Peritoneum | Homo sapiens (Human) | CVCL_1708 | |
| OCUM-1 cells | Pleural effusion | Homo sapiens (Human) | CVCL_3084 | |
| NCI-H226 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1544 | |
| NCI-H196 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1509 | |
| NCI-H1437 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1472 | |
| NCI-H1355 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1464 | |
| NCI-H1299 cells | Lymph node | Homo sapiens (Human) | CVCL_0060 | |
| MKN7 cells | Lymph node | Homo sapiens (Human) | CVCL_1417 | |
| HCC366 cells | Lung | Homo sapiens (Human) | CVCL_2059 | |
| NCI-H2126 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1532 | |
| In Vivo Model | Female nu/nu mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay | |||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [6] | |||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.Q56P (c.167A>C) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| AGS cells | Gastric | Homo sapiens (Human) | CVCL_0139 | |
| NCI-N87 cells | Gastric | Homo sapiens (Human) | CVCL_1603 | |
| NCI-H508 cells | Colon | Homo sapiens (Human) | CVCL_1564 | |
| SW48 cells | Colon | Homo sapiens (Human) | CVCL_1724 | |
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |
| NCI-H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | |
| NCI-H1650 cells | Lung | Homo sapiens (Human) | CVCL_1483 | |
| SW1573 cells | Lung | Homo sapiens (Human) | CVCL_1720 | |
| SNU-C1 cells | Peritoneum | Homo sapiens (Human) | CVCL_1708 | |
| OCUM-1 cells | Pleural effusion | Homo sapiens (Human) | CVCL_3084 | |
| NCI-H226 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1544 | |
| NCI-H196 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1509 | |
| NCI-H1437 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1472 | |
| NCI-H1355 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1464 | |
| MKN7 cells | Lymph node | Homo sapiens (Human) | CVCL_1417 | |
| NCI-H1299 cells | Lymph node | Homo sapiens (Human) | CVCL_0060 | |
| HCC366 cells | Lung | Homo sapiens (Human) | CVCL_2059 | |
| NCI-H2126 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1532 | |
| In Vivo Model | Female nu/nu mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay | |||
| Disease Class: Gastric adenocarcinoma [ICD-11: 2B72.0] | [6] | |||
| Sensitive Disease | Gastric adenocarcinoma [ICD-11: 2B72.0] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.Q56P (c.167A>C) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| AGS cells | Gastric | Homo sapiens (Human) | CVCL_0139 | |
| NCI-N87 cells | Gastric | Homo sapiens (Human) | CVCL_1603 | |
| NCI-H508 cells | Colon | Homo sapiens (Human) | CVCL_1564 | |
| SW48 cells | Colon | Homo sapiens (Human) | CVCL_1724 | |
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |
| NCI-H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | |
| NCI-H1650 cells | Lung | Homo sapiens (Human) | CVCL_1483 | |
| SW1573 cells | Lung | Homo sapiens (Human) | CVCL_1720 | |
| SNU-C1 cells | Peritoneum | Homo sapiens (Human) | CVCL_1708 | |
| OCUM-1 cells | Pleural effusion | Homo sapiens (Human) | CVCL_3084 | |
| NCI-H226 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1544 | |
| NCI-H196 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1509 | |
| NCI-H1437 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1472 | |
| NCI-H1355 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1464 | |
| MKN7 cells | Lymph node | Homo sapiens (Human) | CVCL_1417 | |
| NCI-H1299 cells | Lymph node | Homo sapiens (Human) | CVCL_0060 | |
| HCC366 cells | Lung | Homo sapiens (Human) | CVCL_2059 | |
| NCI-H2126 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1532 | |
| In Vivo Model | Female nu/nu mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay | |||
|
|
||||
| Disease Class: Lymphatic system cancer [ICD-11: 2E81.1] | [7] | |||
| Sensitive Disease | Lymphatic system cancer [ICD-11: 2E81.1] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.K57N (c.171G>C) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Skin | N.A. | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay | |||
Clinical Trial Drug(s)
5 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [2] | |||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Resistant Drug | Cobimetinib | |||
| Molecule Alteration | Missense mutation | p.V211D (c.632T>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | NIH-3T3 cells | Embryo | Mus musculus (Mouse) | CVCL_0594 |
| Phoenix AMPHO cells | Fetal kidney | Homo sapiens (Human) | CVCL_H716 | |
| In Vivo Model | NOD scid gamma xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Single cell sequencing assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lymphatic system cancer [ICD-11: 2E81.1] | [8] | |||
| Sensitive Disease | Lymphatic system cancer [ICD-11: 2E81.1] | |||
| Sensitive Drug | Cobimetinib | |||
| Molecule Alteration | Missense mutation | p.P124L (c.371C>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Whole exome sequencing; Targeted Exon Sequencing | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay; IC50 assay | |||
| Disease Class: Lymphatic system cancer [ICD-11: 2E81.1] | [7] | |||
| Sensitive Disease | Lymphatic system cancer [ICD-11: 2E81.1] | |||
| Sensitive Drug | Cobimetinib | |||
| Molecule Alteration | Missense mutation | p.Q56P (c.167A>C) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Disease Class: Lymphatic system cancer [ICD-11: 2E81.1] | [8] | |||
| Sensitive Disease | Lymphatic system cancer [ICD-11: 2E81.1] | |||
| Sensitive Drug | Cobimetinib | |||
| Molecule Alteration | IF-deletion | p.P105_I107delPAI (c.314_322delCCGCAATCC) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Whole exome sequencing; Targeted Exon Sequencing | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay; IC50 assay | |||
| Disease Class: Lymphatic system cancer [ICD-11: 2E81.1] | [8] | |||
| Sensitive Disease | Lymphatic system cancer [ICD-11: 2E81.1] | |||
| Sensitive Drug | Cobimetinib | |||
| Molecule Alteration | Missense mutation | p.P124Q (c.371C>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Experiment for Molecule Alteration |
Whole exome sequencing; Targeted Exon Sequencing | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay; IC50 assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [6] | |||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Sensitive Drug | Refametinib | |||
| Molecule Alteration | Missense mutation | p.Q56P (c.167A>C) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| AGS cells | Gastric | Homo sapiens (Human) | CVCL_0139 | |
| NCI-N87 cells | Gastric | Homo sapiens (Human) | CVCL_1603 | |
| NCI-H508 cells | Colon | Homo sapiens (Human) | CVCL_1564 | |
| SW48 cells | Colon | Homo sapiens (Human) | CVCL_1724 | |
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |
| NCI-H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | |
| NCI-H1650 cells | Lung | Homo sapiens (Human) | CVCL_1483 | |
| SW1573 cells | Lung | Homo sapiens (Human) | CVCL_1720 | |
| SNU-C1 cells | Peritoneum | Homo sapiens (Human) | CVCL_1708 | |
| OCUM-1 cells | Pleural effusion | Homo sapiens (Human) | CVCL_3084 | |
| NCI-H226 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1544 | |
| NCI-H196 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1509 | |
| NCI-H1437 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1472 | |
| NCI-H1355 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1464 | |
| MKN7 cells | Lymph node | Homo sapiens (Human) | CVCL_1417 | |
| NCI-H1299 cells | Lymph node | Homo sapiens (Human) | CVCL_0060 | |
| HCC366 cells | Lung | Homo sapiens (Human) | CVCL_2059 | |
| NCI-H2126 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1532 | |
| In Vivo Model | Female nu/nu mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [9] | |||
| Resistant Disease | Melanoma [ICD-11: 2C30.0] | |||
| Resistant Drug | Selumetinib | |||
| Molecule Alteration | Missense mutation | p.Q56P (c.167A>C) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 |
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Colony formation assay | |||
| Mechanism Description | The missense mutation p.Q56P (c.167A>C) in gene MAP2K1 cause the resistance of Selumetinib by aberration of the drug's therapeutic target | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [9] | |||
| Resistant Disease | Melanoma [ICD-11: 2C30.0] | |||
| Resistant Drug | Selumetinib | |||
| Molecule Alteration | Missense mutation | p.I103N (c.308T>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 |
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Colony formation assay | |||
| Mechanism Description | The missense mutation p.I103N (c.308T>A) in gene MAP2K1 cause the resistance of Selumetinib by aberration of the drug's therapeutic target | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [9] | |||
| Resistant Disease | Melanoma [ICD-11: 2C30.0] | |||
| Resistant Drug | Selumetinib | |||
| Molecule Alteration | Missense mutation | p.L115P (c.344T>C) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 |
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Colony formation assay | |||
| Mechanism Description | The missense mutation p.L115P (c.344T>C) in gene MAP2K1 cause the resistance of Selumetinib by aberration of the drug's therapeutic target | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [9] | |||
| Resistant Disease | Melanoma [ICD-11: 2C30.0] | |||
| Resistant Drug | Selumetinib | |||
| Molecule Alteration | Missense mutation | p.P124S (c.370C>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 |
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Colony formation assay | |||
| Mechanism Description | The missense mutation p.P124S (c.370C>T) in gene MAP2K1 cause the resistance of Selumetinib by aberration of the drug's therapeutic target | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [9] | |||
| Resistant Disease | Melanoma [ICD-11: 2C30.0] | |||
| Resistant Drug | Selumetinib | |||
| Molecule Alteration | Missense mutation | p.P124L (c.371C>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 |
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Colony formation assay | |||
| Mechanism Description | The missense mutation p.P124L (c.371C>T) in gene MAP2K1 cause the resistance of Selumetinib by aberration of the drug's therapeutic target | |||
|
|
||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | |||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Resistant Drug | Selumetinib | |||
| Molecule Alteration | Missense mutation | p.V211D (c.632T>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 |
| Experiment for Molecule Alteration |
SDS-PAGE assay | |||
| Mechanism Description | The missense mutation p.V211D (c.632T>A) in gene MAP2K1 cause the resistance of Selumetinib by unusual activation of pro-survival pathway | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [6] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Sensitive Drug | Selumetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q56P (c.167A>C) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| AGS cells | Gastric | Homo sapiens (Human) | CVCL_0139 | ||||||||||
| NCI-N87 cells | Gastric | Homo sapiens (Human) | CVCL_1603 | ||||||||||
| NCI-H508 cells | Colon | Homo sapiens (Human) | CVCL_1564 | ||||||||||
| SW48 cells | Colon | Homo sapiens (Human) | CVCL_1724 | ||||||||||
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| NCI-H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | ||||||||||
| NCI-H1650 cells | Lung | Homo sapiens (Human) | CVCL_1483 | ||||||||||
| SW1573 cells | Lung | Homo sapiens (Human) | CVCL_1720 | ||||||||||
| SNU-C1 cells | Peritoneum | Homo sapiens (Human) | CVCL_1708 | ||||||||||
| OCUM-1 cells | Pleural effusion | Homo sapiens (Human) | CVCL_3084 | ||||||||||
| NCI-H226 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1544 | ||||||||||
| NCI-H196 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1509 | ||||||||||
| NCI-H1437 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1472 | ||||||||||
| NCI-H1355 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1464 | ||||||||||
| MKN7 cells | Lymph node | Homo sapiens (Human) | CVCL_1417 | ||||||||||
| NCI-H1299 cells | Lymph node | Homo sapiens (Human) | CVCL_0060 | ||||||||||
| HCC366 cells | Lung | Homo sapiens (Human) | CVCL_2059 | ||||||||||
| NCI-H2126 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1532 | ||||||||||
| In Vivo Model | Female nu/nu mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay | ||||||||||||
| Disease Class: Gastric adenocarcinoma [ICD-11: 2B72.0] | [10] | ||||||||||||
| Sensitive Disease | Gastric adenocarcinoma [ICD-11: 2B72.0] | ||||||||||||
| Sensitive Drug | Selumetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q56P (c.167A>C) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | KATO-3 cells | Gastric | Homo sapiens (Human) | CVCL_0371 | |||||||||
| OCUM-1 cells | Pleural effusion | Homo sapiens (Human) | CVCL_3084 | ||||||||||
| NUGC-4 cells | Lymph node | Homo sapiens (Human) | CVCL_3082/CVCL_8372 | ||||||||||
| Experiment for Molecule Alteration |
Multiplex deep sequencing of MAP2K1 cDNAs assay | ||||||||||||
| Experiment for Drug Resistance |
Focus formation assay | ||||||||||||
| Mechanism Description | The missense mutation p.Q56P (c.167A>C) in gene MAP2K1 cause the sensitivity of Selumetinib by aberration of the drug's therapeutic target | ||||||||||||
|
|
|||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | Selumetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.C121S (c.361T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.70 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.01 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
20
|
M
-
A
-
H
-
H
-
H
-
H
-
H
-
H
-
A
-
A
-
30
|
A
-
E
-
N
-
L
-
Y
-
F
-
Q
-
L
-
E
-
E
-
40
|
L
-
E
-
L
M
D
T
E
L
Q
Q
Q
Q
R
R
K
K
R
R
50
|
L
L
E
E
A
A
F
F
L
L
T
T
Q
Q
K
K
Q
Q
K
K
60
|
V
V
G
G
E
E
L
L
K
K
D
D
D
D
D
D
F
F
E
E
70
|
K
K
I
I
S
S
E
E
L
L
G
G
A
A
G
G
N
N
G
G
80
|
G
G
V
V
V
V
F
F
K
K
V
V
S
S
H
H
K
K
P
P
90
|
S
S
G
G
L
L
V
V
M
M
A
A
R
R
K
K
L
L
I
I
100
|
H
H
L
L
E
E
I
I
K
K
P
P
A
A
I
I
R
R
N
N
110
|
Q
Q
I
I
I
I
R
R
E
E
L
L
Q
Q
V
V
L
L
H
H
120
|
E
E
C
S
N
N
S
S
P
P
Y
Y
I
I
V
V
G
G
F
F
130
|
Y
Y
G
G
A
A
F
F
Y
Y
S
S
D
D
G
G
E
E
I
I
140
|
S
S
I
I
C
C
M
M
E
E
H
H
M
M
D
D
G
G
G
G
150
|
S
S
L
L
D
D
Q
Q
V
V
L
L
K
K
K
K
A
A
G
G
160
|
R
R
I
I
P
P
E
E
Q
Q
I
I
L
L
G
G
K
K
V
V
170
|
S
S
I
I
A
A
V
V
I
I
K
K
G
G
L
L
T
T
Y
Y
180
|
L
L
R
R
E
E
K
K
H
H
K
K
I
I
M
M
H
H
R
R
190
|
D
D
V
V
K
K
P
P
S
S
N
N
I
I
L
L
V
V
N
N
200
|
S
S
R
R
G
G
E
E
I
I
K
K
L
L
C
C
D
D
F
F
210
|
G
G
V
V
S
S
G
G
Q
Q
L
L
I
I
D
D
S
S
M
M
220
|
A
A
N
N
S
S
F
F
V
V
G
G
T
T
R
R
S
S
Y
Y
230
|
M
M
S
S
P
P
E
E
R
R
L
L
Q
Q
G
G
T
T
H
H
240
|
Y
Y
S
S
V
V
Q
Q
S
S
D
D
I
I
W
W
S
S
M
M
250
|
G
G
L
L
S
S
L
L
V
V
E
E
M
M
A
A
V
V
G
G
260
|
R
R
Y
Y
P
P
I
I
-
G
-
S
-
G
-
S
-
G
-
S
270
|
-
M
-
A
-
I
-
F
-
E
-
L
-
L
-
D
-
Y
-
I
280
|
-
V
-
N
-
E
-
P
-
P
-
P
-
K
-
L
-
P
-
S
290
|
-
G
-
V
-
F
-
S
-
L
-
E
-
F
-
Q
-
D
-
F
300
|
-
V
-
N
G
K
S
C
G
L
S
I
G
K
S
N
M
P
A
A
310
|
I
E
F
R
E
A
L
D
L
L
D
K
Y
Q
I
L
V
M
N
V
320
|
E
H
P
A
P
F
P
I
K
K
L
R
P
S
S
D
G
A
V
E
330
|
F
E
S
V
L
D
E
F
F
A
Q
G
D
W
F
L
V
C
N
S
340
|
K
T
C
I
L
G
I
L
K
N
N
Q
P
P
A
S
E
T
R
P
350
|
A
T
D
H
L
A
K
A
Q
G
L
E
M
G
V
H
H
H
A
H
360
|
F
H
I
H
K
H
R
-
S
-
D
-
A
-
E
-
E
-
V
-
370
|
D
-
F
-
A
-
G
-
W
-
L
-
C
-
S
-
T
-
I
-
380
|
G
-
L
-
N
-
Q
-
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.C121S (c.361T>A) in gene MAP2K1 cause the sensitivity of Selumetinib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [11] | ||||||||||||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | ||||||||||||
| Sensitive Drug | Selumetinib | ||||||||||||
| Molecule Alteration | IF-deletion | p.Q56_V60delQKQKV (c.166_180del15) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Ovary | N.A. | |||||||||||
| In Vivo Model | Female nude mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Whole transcriptome analysis | ||||||||||||
| Experiment for Drug Resistance |
Colony formation assay | ||||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [12] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Sensitive Drug | Selumetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.K57N (c.171G>T) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | |||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Trypan blue staining assay | ||||||||||||
| Mechanism Description | The missense mutation p.K57N (c.171G>T) in gene MAP2K1 cause the sensitivity of Selumetinib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | Selumetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.F53L (c.157T>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.F53L (c.157T>C) in gene MAP2K1 cause the sensitivity of Selumetinib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | Selumetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q56P (c.167A>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.Q56P (c.167A>C) in gene MAP2K1 cause the sensitivity of Selumetinib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | Selumetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.K57N (c.171G>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.K57N (c.171G>C) in gene MAP2K1 cause the sensitivity of Selumetinib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | Selumetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.I111S (c.332T>G) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.I111S (c.332T>G) in gene MAP2K1 cause the sensitivity of Selumetinib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | Selumetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.F129L (c.385T>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.F129L (c.385T>C) in gene MAP2K1 cause the sensitivity of Selumetinib by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [12] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Sensitive Drug | Selumetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.K57N (c.171G>C) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | |||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Trypan blue staining assay | ||||||||||||
| Mechanism Description | The missense mutation p.K57N (c.171G>C) in gene MAP2K1 cause the sensitivity of Selumetinib by unusual activation of pro-survival pathway | ||||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [13] | |||
| Resistant Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Resistant Drug | PD-0325901 | |||
| Molecule Alteration | Missense mutation | p.F129L |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | RAS/RAF/MEk signaling pathway | Activation | hsa04010 | |
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| HCT-116 MEk-R cells | Colon | Homo sapiens (Human) | CVCL_V401 | |
| Experiment for Molecule Alteration |
Exome sequencing assay | |||
| Experiment for Drug Resistance |
BrdUrd assay | |||
| Mechanism Description | The RAS/RAF/MEk pathway is activated in more than 30% of human cancers, most commonly via mutation in the k-ras oncogene and also via mutations in BRAF. Importantly, in all cases the MEk-resistant cell lines retained their addiction to the mitogen-activated protein kinase (MAPk) pathway, as evidenced by their sensitivity to a selective inhibitor of the ERk1/2 kinases. These data suggest that tumors with acquired MEk inhibitor resistance remain dependent on the MAPk pathway and are therefore sensitive to inhibitors that act downstream of the mutated MEk target. | |||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [13] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | PD-0325901 | |||
| Molecule Alteration | Missense mutation | p.L115P |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | RAS/RAF/MEk signaling pathway | Activation | hsa04010 | |
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| HCT-116 MEk-R cells | Colon | Homo sapiens (Human) | CVCL_V401 | |
| Experiment for Molecule Alteration |
Exome sequencing assay | |||
| Experiment for Drug Resistance |
BrdUrd assay | |||
| Mechanism Description | The RAS/RAF/MEk pathway is activated in more than 30% of human cancers, most commonly via mutation in the k-ras oncogene and also via mutations in BRAF. Importantly, in all cases the MEk-resistant cell lines retained their addiction to the mitogen-activated protein kinase (MAPk) pathway, as evidenced by their sensitivity to a selective inhibitor of the ERk1/2 kinases. These data suggest that tumors with acquired MEk inhibitor resistance remain dependent on the MAPk pathway and are therefore sensitive to inhibitors that act downstream of the mutated MEk target. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [9] | |||
| Resistant Disease | Melanoma [ICD-11: 2C30.0] | |||
| Resistant Drug | PLX4720 | |||
| Molecule Alteration | Missense mutation | p.Q56P (c.167A>C) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 |
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Colony formation assay | |||
| Mechanism Description | The missense mutation p.Q56P (c.167A>C) in gene MAP2K1 cause the resistance of PLX4720 by unusual activation of pro-survival pathway | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | PLX4720 | ||||||||||||
| Molecule Alteration | Missense mutation | p.C121S (c.361T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.70 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.01 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
20
|
M
-
A
-
H
-
H
-
H
-
H
-
H
-
H
-
A
-
A
-
30
|
A
-
E
-
N
-
L
-
Y
-
F
-
Q
-
L
-
E
-
E
-
40
|
L
-
E
-
L
M
D
T
E
L
Q
Q
Q
Q
R
R
K
K
R
R
50
|
L
L
E
E
A
A
F
F
L
L
T
T
Q
Q
K
K
Q
Q
K
K
60
|
V
V
G
G
E
E
L
L
K
K
D
D
D
D
D
D
F
F
E
E
70
|
K
K
I
I
S
S
E
E
L
L
G
G
A
A
G
G
N
N
G
G
80
|
G
G
V
V
V
V
F
F
K
K
V
V
S
S
H
H
K
K
P
P
90
|
S
S
G
G
L
L
V
V
M
M
A
A
R
R
K
K
L
L
I
I
100
|
H
H
L
L
E
E
I
I
K
K
P
P
A
A
I
I
R
R
N
N
110
|
Q
Q
I
I
I
I
R
R
E
E
L
L
Q
Q
V
V
L
L
H
H
120
|
E
E
C
S
N
N
S
S
P
P
Y
Y
I
I
V
V
G
G
F
F
130
|
Y
Y
G
G
A
A
F
F
Y
Y
S
S
D
D
G
G
E
E
I
I
140
|
S
S
I
I
C
C
M
M
E
E
H
H
M
M
D
D
G
G
G
G
150
|
S
S
L
L
D
D
Q
Q
V
V
L
L
K
K
K
K
A
A
G
G
160
|
R
R
I
I
P
P
E
E
Q
Q
I
I
L
L
G
G
K
K
V
V
170
|
S
S
I
I
A
A
V
V
I
I
K
K
G
G
L
L
T
T
Y
Y
180
|
L
L
R
R
E
E
K
K
H
H
K
K
I
I
M
M
H
H
R
R
190
|
D
D
V
V
K
K
P
P
S
S
N
N
I
I
L
L
V
V
N
N
200
|
S
S
R
R
G
G
E
E
I
I
K
K
L
L
C
C
D
D
F
F
210
|
G
G
V
V
S
S
G
G
Q
Q
L
L
I
I
D
D
S
S
M
M
220
|
A
A
N
N
S
S
F
F
V
V
G
G
T
T
R
R
S
S
Y
Y
230
|
M
M
S
S
P
P
E
E
R
R
L
L
Q
Q
G
G
T
T
H
H
240
|
Y
Y
S
S
V
V
Q
Q
S
S
D
D
I
I
W
W
S
S
M
M
250
|
G
G
L
L
S
S
L
L
V
V
E
E
M
M
A
A
V
V
G
G
260
|
R
R
Y
Y
P
P
I
I
-
G
-
S
-
G
-
S
-
G
-
S
270
|
-
M
-
A
-
I
-
F
-
E
-
L
-
L
-
D
-
Y
-
I
280
|
-
V
-
N
-
E
-
P
-
P
-
P
-
K
-
L
-
P
-
S
290
|
-
G
-
V
-
F
-
S
-
L
-
E
-
F
-
Q
-
D
-
F
300
|
-
V
-
N
G
K
S
C
G
L
S
I
G
K
S
N
M
P
A
A
310
|
I
E
F
R
E
A
L
D
L
L
D
K
Y
Q
I
L
V
M
N
V
320
|
E
H
P
A
P
F
P
I
K
K
L
R
P
S
S
D
G
A
V
E
330
|
F
E
S
V
L
D
E
F
F
A
Q
G
D
W
F
L
V
C
N
S
340
|
K
T
C
I
L
G
I
L
K
N
N
Q
P
P
A
S
E
T
R
P
350
|
A
T
D
H
L
A
K
A
Q
G
L
E
M
G
V
H
H
H
A
H
360
|
F
H
I
H
K
H
R
-
S
-
D
-
A
-
E
-
E
-
V
-
370
|
D
-
F
-
A
-
G
-
W
-
L
-
C
-
S
-
T
-
I
-
380
|
G
-
L
-
N
-
Q
-
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.C121S (c.361T>A) in gene MAP2K1 cause the sensitivity of PLX4720 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | PLX4720 | ||||||||||||
| Molecule Alteration | Missense mutation | p.F53L (c.157T>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.F53L (c.157T>C) in gene MAP2K1 cause the sensitivity of PLX4720 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | PLX4720 | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q56P (c.167A>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.Q56P (c.167A>C) in gene MAP2K1 cause the sensitivity of PLX4720 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | PLX4720 | ||||||||||||
| Molecule Alteration | Missense mutation | p.K57N (c.171G>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.K57N (c.171G>C) in gene MAP2K1 cause the sensitivity of PLX4720 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | PLX4720 | ||||||||||||
| Molecule Alteration | Missense mutation | p.I111S (c.332T>G) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.I111S (c.332T>G) in gene MAP2K1 cause the sensitivity of PLX4720 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | PLX4720 | ||||||||||||
| Molecule Alteration | Missense mutation | p.F129L (c.385T>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.F129L (c.385T>C) in gene MAP2K1 cause the sensitivity of PLX4720 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | PLX4720 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V211D (c.632T>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.V211D (c.632T>A) in gene MAP2K1 cause the sensitivity of PLX4720 by unusual activation of pro-survival pathway | ||||||||||||
Discontinued Drug(s)
1 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | AZD-8330 | ||||||||||||
| Molecule Alteration | Missense mutation | p.C121S (c.361T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.70 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.01 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
20
|
M
-
A
-
H
-
H
-
H
-
H
-
H
-
H
-
A
-
A
-
30
|
A
-
E
-
N
-
L
-
Y
-
F
-
Q
-
L
-
E
-
E
-
40
|
L
-
E
-
L
M
D
T
E
L
Q
Q
Q
Q
R
R
K
K
R
R
50
|
L
L
E
E
A
A
F
F
L
L
T
T
Q
Q
K
K
Q
Q
K
K
60
|
V
V
G
G
E
E
L
L
K
K
D
D
D
D
D
D
F
F
E
E
70
|
K
K
I
I
S
S
E
E
L
L
G
G
A
A
G
G
N
N
G
G
80
|
G
G
V
V
V
V
F
F
K
K
V
V
S
S
H
H
K
K
P
P
90
|
S
S
G
G
L
L
V
V
M
M
A
A
R
R
K
K
L
L
I
I
100
|
H
H
L
L
E
E
I
I
K
K
P
P
A
A
I
I
R
R
N
N
110
|
Q
Q
I
I
I
I
R
R
E
E
L
L
Q
Q
V
V
L
L
H
H
120
|
E
E
C
S
N
N
S
S
P
P
Y
Y
I
I
V
V
G
G
F
F
130
|
Y
Y
G
G
A
A
F
F
Y
Y
S
S
D
D
G
G
E
E
I
I
140
|
S
S
I
I
C
C
M
M
E
E
H
H
M
M
D
D
G
G
G
G
150
|
S
S
L
L
D
D
Q
Q
V
V
L
L
K
K
K
K
A
A
G
G
160
|
R
R
I
I
P
P
E
E
Q
Q
I
I
L
L
G
G
K
K
V
V
170
|
S
S
I
I
A
A
V
V
I
I
K
K
G
G
L
L
T
T
Y
Y
180
|
L
L
R
R
E
E
K
K
H
H
K
K
I
I
M
M
H
H
R
R
190
|
D
D
V
V
K
K
P
P
S
S
N
N
I
I
L
L
V
V
N
N
200
|
S
S
R
R
G
G
E
E
I
I
K
K
L
L
C
C
D
D
F
F
210
|
G
G
V
V
S
S
G
G
Q
Q
L
L
I
I
D
D
S
S
M
M
220
|
A
A
N
N
S
S
F
F
V
V
G
G
T
T
R
R
S
S
Y
Y
230
|
M
M
S
S
P
P
E
E
R
R
L
L
Q
Q
G
G
T
T
H
H
240
|
Y
Y
S
S
V
V
Q
Q
S
S
D
D
I
I
W
W
S
S
M
M
250
|
G
G
L
L
S
S
L
L
V
V
E
E
M
M
A
A
V
V
G
G
260
|
R
R
Y
Y
P
P
I
I
-
G
-
S
-
G
-
S
-
G
-
S
270
|
-
M
-
A
-
I
-
F
-
E
-
L
-
L
-
D
-
Y
-
I
280
|
-
V
-
N
-
E
-
P
-
P
-
P
-
K
-
L
-
P
-
S
290
|
-
G
-
V
-
F
-
S
-
L
-
E
-
F
-
Q
-
D
-
F
300
|
-
V
-
N
G
K
S
C
G
L
S
I
G
K
S
N
M
P
A
A
310
|
I
E
F
R
E
A
L
D
L
L
D
K
Y
Q
I
L
V
M
N
V
320
|
E
H
P
A
P
F
P
I
K
K
L
R
P
S
S
D
G
A
V
E
330
|
F
E
S
V
L
D
E
F
F
A
Q
G
D
W
F
L
V
C
N
S
340
|
K
T
C
I
L
G
I
L
K
N
N
Q
P
P
A
S
E
T
R
P
350
|
A
T
D
H
L
A
K
A
Q
G
L
E
M
G
V
H
H
H
A
H
360
|
F
H
I
H
K
H
R
-
S
-
D
-
A
-
E
-
E
-
V
-
370
|
D
-
F
-
A
-
G
-
W
-
L
-
C
-
S
-
T
-
I
-
380
|
G
-
L
-
N
-
Q
-
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.C121S (c.361T>A) in gene MAP2K1 cause the resistance of AZD-8330 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | AZD-8330 | ||||||||||||
| Molecule Alteration | Missense mutation | p.F53L (c.157T>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.F53L (c.157T>C) in gene MAP2K1 cause the resistance of AZD-8330 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | AZD-8330 | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q56P (c.167A>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.Q56P (c.167A>C) in gene MAP2K1 cause the resistance of AZD-8330 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | AZD-8330 | ||||||||||||
| Molecule Alteration | Missense mutation | p.K57N (c.171G>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.K57N (c.171G>C) in gene MAP2K1 cause the resistance of AZD-8330 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | AZD-8330 | ||||||||||||
| Molecule Alteration | Missense mutation | p.I111S (c.332T>G) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.I111S (c.332T>G) in gene MAP2K1 cause the resistance of AZD-8330 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | AZD-8330 | ||||||||||||
| Molecule Alteration | Missense mutation | p.F129L (c.385T>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.F129L (c.385T>C) in gene MAP2K1 cause the resistance of AZD-8330 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | AZD-8330 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V211D (c.632T>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.V211D (c.632T>A) in gene MAP2K1 cause the resistance of AZD-8330 by unusual activation of pro-survival pathway | ||||||||||||
Preclinical Drug(s)
3 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [14] | ||||||||||||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | ||||||||||||
| Sensitive Drug | Cetuximab/Trametinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.C121S (c.361T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.70 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.01 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
20
|
M
-
A
-
H
-
H
-
H
-
H
-
H
-
H
-
A
-
A
-
30
|
A
-
E
-
N
-
L
-
Y
-
F
-
Q
-
L
-
E
-
E
-
40
|
L
-
E
-
L
M
D
T
E
L
Q
Q
Q
Q
R
R
K
K
R
R
50
|
L
L
E
E
A
A
F
F
L
L
T
T
Q
Q
K
K
Q
Q
K
K
60
|
V
V
G
G
E
E
L
L
K
K
D
D
D
D
D
D
F
F
E
E
70
|
K
K
I
I
S
S
E
E
L
L
G
G
A
A
G
G
N
N
G
G
80
|
G
G
V
V
V
V
F
F
K
K
V
V
S
S
H
H
K
K
P
P
90
|
S
S
G
G
L
L
V
V
M
M
A
A
R
R
K
K
L
L
I
I
100
|
H
H
L
L
E
E
I
I
K
K
P
P
A
A
I
I
R
R
N
N
110
|
Q
Q
I
I
I
I
R
R
E
E
L
L
Q
Q
V
V
L
L
H
H
120
|
E
E
C
S
N
N
S
S
P
P
Y
Y
I
I
V
V
G
G
F
F
130
|
Y
Y
G
G
A
A
F
F
Y
Y
S
S
D
D
G
G
E
E
I
I
140
|
S
S
I
I
C
C
M
M
E
E
H
H
M
M
D
D
G
G
G
G
150
|
S
S
L
L
D
D
Q
Q
V
V
L
L
K
K
K
K
A
A
G
G
160
|
R
R
I
I
P
P
E
E
Q
Q
I
I
L
L
G
G
K
K
V
V
170
|
S
S
I
I
A
A
V
V
I
I
K
K
G
G
L
L
T
T
Y
Y
180
|
L
L
R
R
E
E
K
K
H
H
K
K
I
I
M
M
H
H
R
R
190
|
D
D
V
V
K
K
P
P
S
S
N
N
I
I
L
L
V
V
N
N
200
|
S
S
R
R
G
G
E
E
I
I
K
K
L
L
C
C
D
D
F
F
210
|
G
G
V
V
S
S
G
G
Q
Q
L
L
I
I
D
D
S
S
M
M
220
|
A
A
N
N
S
S
F
F
V
V
G
G
T
T
R
R
S
S
Y
Y
230
|
M
M
S
S
P
P
E
E
R
R
L
L
Q
Q
G
G
T
T
H
H
240
|
Y
Y
S
S
V
V
Q
Q
S
S
D
D
I
I
W
W
S
S
M
M
250
|
G
G
L
L
S
S
L
L
V
V
E
E
M
M
A
A
V
V
G
G
260
|
R
R
Y
Y
P
P
I
I
-
G
-
S
-
G
-
S
-
G
-
S
270
|
-
M
-
A
-
I
-
F
-
E
-
L
-
L
-
D
-
Y
-
I
280
|
-
V
-
N
-
E
-
P
-
P
-
P
-
K
-
L
-
P
-
S
290
|
-
G
-
V
-
F
-
S
-
L
-
E
-
F
-
Q
-
D
-
F
300
|
-
V
-
N
G
K
S
C
G
L
S
I
G
K
S
N
M
P
A
A
310
|
I
E
F
R
E
A
L
D
L
L
D
K
Y
Q
I
L
V
M
N
V
320
|
E
H
P
A
P
F
P
I
K
K
L
R
P
S
S
D
G
A
V
E
330
|
F
E
S
V
L
D
E
F
F
A
Q
G
D
W
F
L
V
C
N
S
340
|
K
T
C
I
L
G
I
L
K
N
N
Q
P
P
A
S
E
T
R
P
350
|
A
T
D
H
L
A
K
A
Q
G
L
E
M
G
V
H
H
H
A
H
360
|
F
H
I
H
K
H
R
-
S
-
D
-
A
-
E
-
E
-
V
-
370
|
D
-
F
-
A
-
G
-
W
-
L
-
C
-
S
-
T
-
I
-
380
|
G
-
L
-
N
-
Q
-
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | NCI-H508 cells | Colon | Homo sapiens (Human) | CVCL_1564 | |||||||||
| CCK-81 cells | N.A. | Homo sapiens (Human) | CVCL_2873 | ||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | GDC0879 | ||||||||||||
| Molecule Alteration | Missense mutation | p.C121S (c.361T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.70 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.01 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
20
|
M
-
A
-
H
-
H
-
H
-
H
-
H
-
H
-
A
-
A
-
30
|
A
-
E
-
N
-
L
-
Y
-
F
-
Q
-
L
-
E
-
E
-
40
|
L
-
E
-
L
M
D
T
E
L
Q
Q
Q
Q
R
R
K
K
R
R
50
|
L
L
E
E
A
A
F
F
L
L
T
T
Q
Q
K
K
Q
Q
K
K
60
|
V
V
G
G
E
E
L
L
K
K
D
D
D
D
D
D
F
F
E
E
70
|
K
K
I
I
S
S
E
E
L
L
G
G
A
A
G
G
N
N
G
G
80
|
G
G
V
V
V
V
F
F
K
K
V
V
S
S
H
H
K
K
P
P
90
|
S
S
G
G
L
L
V
V
M
M
A
A
R
R
K
K
L
L
I
I
100
|
H
H
L
L
E
E
I
I
K
K
P
P
A
A
I
I
R
R
N
N
110
|
Q
Q
I
I
I
I
R
R
E
E
L
L
Q
Q
V
V
L
L
H
H
120
|
E
E
C
S
N
N
S
S
P
P
Y
Y
I
I
V
V
G
G
F
F
130
|
Y
Y
G
G
A
A
F
F
Y
Y
S
S
D
D
G
G
E
E
I
I
140
|
S
S
I
I
C
C
M
M
E
E
H
H
M
M
D
D
G
G
G
G
150
|
S
S
L
L
D
D
Q
Q
V
V
L
L
K
K
K
K
A
A
G
G
160
|
R
R
I
I
P
P
E
E
Q
Q
I
I
L
L
G
G
K
K
V
V
170
|
S
S
I
I
A
A
V
V
I
I
K
K
G
G
L
L
T
T
Y
Y
180
|
L
L
R
R
E
E
K
K
H
H
K
K
I
I
M
M
H
H
R
R
190
|
D
D
V
V
K
K
P
P
S
S
N
N
I
I
L
L
V
V
N
N
200
|
S
S
R
R
G
G
E
E
I
I
K
K
L
L
C
C
D
D
F
F
210
|
G
G
V
V
S
S
G
G
Q
Q
L
L
I
I
D
D
S
S
M
M
220
|
A
A
N
N
S
S
F
F
V
V
G
G
T
T
R
R
S
S
Y
Y
230
|
M
M
S
S
P
P
E
E
R
R
L
L
Q
Q
G
G
T
T
H
H
240
|
Y
Y
S
S
V
V
Q
Q
S
S
D
D
I
I
W
W
S
S
M
M
250
|
G
G
L
L
S
S
L
L
V
V
E
E
M
M
A
A
V
V
G
G
260
|
R
R
Y
Y
P
P
I
I
-
G
-
S
-
G
-
S
-
G
-
S
270
|
-
M
-
A
-
I
-
F
-
E
-
L
-
L
-
D
-
Y
-
I
280
|
-
V
-
N
-
E
-
P
-
P
-
P
-
K
-
L
-
P
-
S
290
|
-
G
-
V
-
F
-
S
-
L
-
E
-
F
-
Q
-
D
-
F
300
|
-
V
-
N
G
K
S
C
G
L
S
I
G
K
S
N
M
P
A
A
310
|
I
E
F
R
E
A
L
D
L
L
D
K
Y
Q
I
L
V
M
N
V
320
|
E
H
P
A
P
F
P
I
K
K
L
R
P
S
S
D
G
A
V
E
330
|
F
E
S
V
L
D
E
F
F
A
Q
G
D
W
F
L
V
C
N
S
340
|
K
T
C
I
L
G
I
L
K
N
N
Q
P
P
A
S
E
T
R
P
350
|
A
T
D
H
L
A
K
A
Q
G
L
E
M
G
V
H
H
H
A
H
360
|
F
H
I
H
K
H
R
-
S
-
D
-
A
-
E
-
E
-
V
-
370
|
D
-
F
-
A
-
G
-
W
-
L
-
C
-
S
-
T
-
I
-
380
|
G
-
L
-
N
-
Q
-
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.C121S (c.361T>A) in gene MAP2K1 cause the resistance of GDC0879 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | GDC0879 | ||||||||||||
| Molecule Alteration | Missense mutation | p.F53L (c.157T>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.F53L (c.157T>C) in gene MAP2K1 cause the resistance of GDC0879 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | GDC0879 | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q56P (c.167A>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.Q56P (c.167A>C) in gene MAP2K1 cause the resistance of GDC0879 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | GDC0879 | ||||||||||||
| Molecule Alteration | Missense mutation | p.K57N (c.171G>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.K57N (c.171G>C) in gene MAP2K1 cause the resistance of GDC0879 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | GDC0879 | ||||||||||||
| Molecule Alteration | Missense mutation | p.I111S (c.332T>G) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.I111S (c.332T>G) in gene MAP2K1 cause the resistance of GDC0879 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | GDC0879 | ||||||||||||
| Molecule Alteration | Missense mutation | p.F129L (c.385T>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.F129L (c.385T>C) in gene MAP2K1 cause the resistance of GDC0879 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | GDC0879 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V211D (c.632T>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.V211D (c.632T>A) in gene MAP2K1 cause the resistance of GDC0879 by unusual activation of pro-survival pathway | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [15] | |||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Sensitive Drug | RO4927350 | |||
| Molecule Alteration | Missense mutation | p.F129L (c.385T>C) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HT-29 cells | Colon | Homo sapiens (Human) | CVCL_0320 |
| In Vivo Model | Mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The missense mutation p.F129L (c.385T>C) in gene MAP2K1 cause the sensitivity of RO4927350 by unusual activation of pro-survival pathway | |||
Investigative Drug(s)
4 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [9] | |||
| Resistant Disease | Melanoma [ICD-11: 2C30.0] | |||
| Resistant Drug | CI-1040 | |||
| Molecule Alteration | Missense mutation | p.I103N (c.308T>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 |
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Colony formation assay | |||
| Mechanism Description | The missense mutation p.I103N (c.308T>A) in gene MAP2K1 cause the resistance of CI-1040 by aberration of the drug's therapeutic target | |||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [16] | |||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Resistant Drug | CI-1040 | |||
| Molecule Alteration | Missense mutation | p.I103N (c.308T>A) |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Saccharomyces cerevisiae strain SY2002 | 4932 | ||
| Experiment for Molecule Alteration |
Kinase assay | |||
| Mechanism Description | The missense mutation p.I103N (c.308T>A) in gene MAP2K1 cause the resistance of CI-1040 by unusual activation of pro-survival pathway | |||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [16] | |||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Resistant Drug | CI-1040 | |||
| Molecule Alteration | Missense mutation | p.L115A (c.343_344delCTinsGC) |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Saccharomyces cerevisiae strain SY2002 | 4932 | ||
| Experiment for Molecule Alteration |
Kinase assay | |||
| Mechanism Description | The missense mutation p.L115A (c.343_344delCTinsGC) in gene MAP2K1 cause the resistance of CI-1040 by unusual activation of pro-survival pathway | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [9] | |||
| Resistant Disease | Melanoma [ICD-11: 2C30.0] | |||
| Resistant Drug | CI-1040 | |||
| Molecule Alteration | Missense mutation | p.L115P (c.344T>C) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 |
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Colony formation assay | |||
| Mechanism Description | The missense mutation p.L115P (c.344T>C) in gene MAP2K1 cause the resistance of CI-1040 by aberration of the drug's therapeutic target | |||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [16] | |||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Resistant Drug | CI-1040 | |||
| Molecule Alteration | Missense mutation | p.L115P (c.344T>C) |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Saccharomyces cerevisiae strain SY2002 | 4932 | ||
| Experiment for Molecule Alteration |
Kinase assay | |||
| Mechanism Description | The missense mutation p.L115P (c.344T>C) in gene MAP2K1 cause the resistance of CI-1040 by unusual activation of pro-survival pathway | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [16] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | CI-1040 | |||
| Molecule Alteration | Missense mutation | p.F53S (c.158T>C) |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Saccharomyces cerevisiae strain SY2002 | 4932 | ||
| Experiment for Molecule Alteration |
Kinase assay | |||
| Mechanism Description | The missense mutation p.F53S (c.158T>C) in gene MAP2K1 cause the sensitivity of CI-1040 by unusual activation of pro-survival pathway | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [9] | ||||||||||||
| Resistant Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Resistant Drug | MEK inhibitors | ||||||||||||
| Molecule Alteration | Missense mutation | p.P124L (c.371C>T) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Colony formation assay | ||||||||||||
| Mechanism Description | The missense mutation p.P124L (c.371C>T) in gene MAP2K1 cause the resistance of MEK inhibitors by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [9] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | MEK inhibitors | ||||||||||||
| Molecule Alteration | Missense mutation | p.D67N (c.199G>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Colony formation assay | ||||||||||||
| Mechanism Description | The missense mutation p.D67N (c.199G>A) in gene MAP2K1 cause the resistance of MEK inhibitors by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [9] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | MEK inhibitors | ||||||||||||
| Molecule Alteration | Missense mutation | p.I99T (c.296T>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Colony formation assay | ||||||||||||
| Mechanism Description | The missense mutation p.I99T (c.296T>C) in gene MAP2K1 cause the resistance of MEK inhibitors by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [9] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | MEK inhibitors | ||||||||||||
| Molecule Alteration | Missense mutation | p.I103N (c.308T>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Colony formation assay | ||||||||||||
| Mechanism Description | The missense mutation p.I103N (c.308T>A) in gene MAP2K1 cause the resistance of MEK inhibitors by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [9] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | MEK inhibitors | ||||||||||||
| Molecule Alteration | Missense mutation | p.K104N (c.312A>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Colony formation assay | ||||||||||||
| Mechanism Description | The missense mutation p.K104N (c.312A>C) in gene MAP2K1 cause the resistance of MEK inhibitors by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [9] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | MEK inhibitors | ||||||||||||
| Molecule Alteration | Missense mutation | p.I111N (c.332T>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Colony formation assay | ||||||||||||
| Mechanism Description | The missense mutation p.I111N (c.332T>A) in gene MAP2K1 cause the resistance of MEK inhibitors by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [9] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | MEK inhibitors | ||||||||||||
| Molecule Alteration | Missense mutation | p.L115P (c.344T>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Colony formation assay | ||||||||||||
| Mechanism Description | The missense mutation p.L115P (c.344T>C) in gene MAP2K1 cause the resistance of MEK inhibitors by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [9] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | MEK inhibitors | ||||||||||||
| Molecule Alteration | Missense mutation | p.H119P (c.356A>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Colony formation assay | ||||||||||||
| Mechanism Description | The missense mutation p.H119P (c.356A>C) in gene MAP2K1 cause the resistance of MEK inhibitors by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [9] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | MEK inhibitors | ||||||||||||
| Molecule Alteration | Missense mutation | p.E120D (c.360G>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Colony formation assay | ||||||||||||
| Mechanism Description | The missense mutation p.E120D (c.360G>C) in gene MAP2K1 cause the resistance of MEK inhibitors by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [9] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | MEK inhibitors | ||||||||||||
| Molecule Alteration | Missense mutation | p.G128D (c.383G>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Colony formation assay | ||||||||||||
| Mechanism Description | The missense mutation p.G128D (c.383G>A) in gene MAP2K1 cause the resistance of MEK inhibitors by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [9] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | MEK inhibitors | ||||||||||||
| Molecule Alteration | Missense mutation | p.F129L (c.385T>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Colony formation assay | ||||||||||||
| Mechanism Description | The missense mutation p.F129L (c.385T>C) in gene MAP2K1 cause the resistance of MEK inhibitors by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [9] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | MEK inhibitors | ||||||||||||
| Molecule Alteration | Missense mutation | p.F133L (c.397T>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Colony formation assay | ||||||||||||
| Mechanism Description | The missense mutation p.F133L (c.397T>C) in gene MAP2K1 cause the resistance of MEK inhibitors by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [9] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | MEK inhibitors | ||||||||||||
| Molecule Alteration | Missense mutation | p.V211D (c.632T>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Colony formation assay | ||||||||||||
| Mechanism Description | The missense mutation p.V211D (c.632T>A) in gene MAP2K1 cause the resistance of MEK inhibitors by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [9] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | MEK inhibitors | ||||||||||||
| Molecule Alteration | Missense mutation | p.L215P (c.644T>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |||||||||
| 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Colony formation assay | ||||||||||||
| Mechanism Description | The missense mutation p.L215P (c.644T>C) in gene MAP2K1 cause the resistance of MEK inhibitors by aberration of the drug's therapeutic target | ||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [17] | ||||||||||||
| Resistant Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Resistant Drug | MEK inhibitors | ||||||||||||
| Molecule Alteration | Missense mutation | p.C121S (c.361T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.70 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.01 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
20
|
M
-
A
-
H
-
H
-
H
-
H
-
H
-
H
-
A
-
A
-
30
|
A
-
E
-
N
-
L
-
Y
-
F
-
Q
-
L
-
E
-
E
-
40
|
L
-
E
-
L
M
D
T
E
L
Q
Q
Q
Q
R
R
K
K
R
R
50
|
L
L
E
E
A
A
F
F
L
L
T
T
Q
Q
K
K
Q
Q
K
K
60
|
V
V
G
G
E
E
L
L
K
K
D
D
D
D
D
D
F
F
E
E
70
|
K
K
I
I
S
S
E
E
L
L
G
G
A
A
G
G
N
N
G
G
80
|
G
G
V
V
V
V
F
F
K
K
V
V
S
S
H
H
K
K
P
P
90
|
S
S
G
G
L
L
V
V
M
M
A
A
R
R
K
K
L
L
I
I
100
|
H
H
L
L
E
E
I
I
K
K
P
P
A
A
I
I
R
R
N
N
110
|
Q
Q
I
I
I
I
R
R
E
E
L
L
Q
Q
V
V
L
L
H
H
120
|
E
E
C
S
N
N
S
S
P
P
Y
Y
I
I
V
V
G
G
F
F
130
|
Y
Y
G
G
A
A
F
F
Y
Y
S
S
D
D
G
G
E
E
I
I
140
|
S
S
I
I
C
C
M
M
E
E
H
H
M
M
D
D
G
G
G
G
150
|
S
S
L
L
D
D
Q
Q
V
V
L
L
K
K
K
K
A
A
G
G
160
|
R
R
I
I
P
P
E
E
Q
Q
I
I
L
L
G
G
K
K
V
V
170
|
S
S
I
I
A
A
V
V
I
I
K
K
G
G
L
L
T
T
Y
Y
180
|
L
L
R
R
E
E
K
K
H
H
K
K
I
I
M
M
H
H
R
R
190
|
D
D
V
V
K
K
P
P
S
S
N
N
I
I
L
L
V
V
N
N
200
|
S
S
R
R
G
G
E
E
I
I
K
K
L
L
C
C
D
D
F
F
210
|
G
G
V
V
S
S
G
G
Q
Q
L
L
I
I
D
D
S
S
M
M
220
|
A
A
N
N
S
S
F
F
V
V
G
G
T
T
R
R
S
S
Y
Y
230
|
M
M
S
S
P
P
E
E
R
R
L
L
Q
Q
G
G
T
T
H
H
240
|
Y
Y
S
S
V
V
Q
Q
S
S
D
D
I
I
W
W
S
S
M
M
250
|
G
G
L
L
S
S
L
L
V
V
E
E
M
M
A
A
V
V
G
G
260
|
R
R
Y
Y
P
P
I
I
-
G
-
S
-
G
-
S
-
G
-
S
270
|
-
M
-
A
-
I
-
F
-
E
-
L
-
L
-
D
-
Y
-
I
280
|
-
V
-
N
-
E
-
P
-
P
-
P
-
K
-
L
-
P
-
S
290
|
-
G
-
V
-
F
-
S
-
L
-
E
-
F
-
Q
-
D
-
F
300
|
-
V
-
N
G
K
S
C
G
L
S
I
G
K
S
N
M
P
A
A
310
|
I
E
F
R
E
A
L
D
L
L
D
K
Y
Q
I
L
V
M
N
V
320
|
E
H
P
A
P
F
P
I
K
K
L
R
P
S
S
D
G
A
V
E
330
|
F
E
S
V
L
D
E
F
F
A
Q
G
D
W
F
L
V
C
N
S
340
|
K
T
C
I
L
G
I
L
K
N
N
Q
P
P
A
S
E
T
R
P
350
|
A
T
D
H
L
A
K
A
Q
G
L
E
M
G
V
H
H
H
A
H
360
|
F
H
I
H
K
H
R
-
S
-
D
-
A
-
E
-
E
-
V
-
370
|
D
-
F
-
A
-
G
-
W
-
L
-
C
-
S
-
T
-
I
-
380
|
G
-
L
-
N
-
Q
-
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Human melanoma tissue | N.A. | |||||||||||
| Mechanism Description | The missense mutation p.C121S (c.361T>A) in gene MAP2K1 cause the resistance of MEK inhibitors by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [18] | ||||||||||||
| Resistant Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Resistant Drug | MEK inhibitors | ||||||||||||
| Molecule Alteration | Missense mutation | p.K57N (c.171G>C) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Skin | N.A. | |||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Mechanism Description | The missense mutation p.K57N (c.171G>C) in gene MAP2K1 cause the resistance of MEK inhibitors by unusual activation of pro-survival pathway | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [11] | |||
| Sensitive Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Sensitive Drug | MEK inhibitors | |||
| Molecule Alteration | IF-deletion | p.Q56_V60delQKQKV (c.166_180del15) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Ovary | N.A. | ||
| In Vivo Model | Female nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Whole transcriptome analysis | |||
| Experiment for Drug Resistance |
Colony formation assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [19] | |||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Sensitive Drug | Panitumumab/Trametinib | |||
| Molecule Alteration | Missense mutation | p.K57N (c.171G>C) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HCA46 cells | Colon | Homo sapiens (Human) | CVCL_2468 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay | |||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [19] | |||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Sensitive Drug | Panitumumab/Trametinib | |||
| Molecule Alteration | Missense mutation | p.K57T (c.170A>C) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HCA46 cells | Colon | Homo sapiens (Human) | CVCL_2468 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay | |||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [19] | |||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Sensitive Drug | Panitumumab/Trametinib | |||
| Molecule Alteration | Missense mutation | p.K57T (c.170A>C) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HCA46 cells | Colon | Homo sapiens (Human) | CVCL_2468 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | PD98059 | ||||||||||||
| Molecule Alteration | Missense mutation | p.C121S (c.361T>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.70 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.01 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
20
|
M
-
A
-
H
-
H
-
H
-
H
-
H
-
H
-
A
-
A
-
30
|
A
-
E
-
N
-
L
-
Y
-
F
-
Q
-
L
-
E
-
E
-
40
|
L
-
E
-
L
M
D
T
E
L
Q
Q
Q
Q
R
R
K
K
R
R
50
|
L
L
E
E
A
A
F
F
L
L
T
T
Q
Q
K
K
Q
Q
K
K
60
|
V
V
G
G
E
E
L
L
K
K
D
D
D
D
D
D
F
F
E
E
70
|
K
K
I
I
S
S
E
E
L
L
G
G
A
A
G
G
N
N
G
G
80
|
G
G
V
V
V
V
F
F
K
K
V
V
S
S
H
H
K
K
P
P
90
|
S
S
G
G
L
L
V
V
M
M
A
A
R
R
K
K
L
L
I
I
100
|
H
H
L
L
E
E
I
I
K
K
P
P
A
A
I
I
R
R
N
N
110
|
Q
Q
I
I
I
I
R
R
E
E
L
L
Q
Q
V
V
L
L
H
H
120
|
E
E
C
S
N
N
S
S
P
P
Y
Y
I
I
V
V
G
G
F
F
130
|
Y
Y
G
G
A
A
F
F
Y
Y
S
S
D
D
G
G
E
E
I
I
140
|
S
S
I
I
C
C
M
M
E
E
H
H
M
M
D
D
G
G
G
G
150
|
S
S
L
L
D
D
Q
Q
V
V
L
L
K
K
K
K
A
A
G
G
160
|
R
R
I
I
P
P
E
E
Q
Q
I
I
L
L
G
G
K
K
V
V
170
|
S
S
I
I
A
A
V
V
I
I
K
K
G
G
L
L
T
T
Y
Y
180
|
L
L
R
R
E
E
K
K
H
H
K
K
I
I
M
M
H
H
R
R
190
|
D
D
V
V
K
K
P
P
S
S
N
N
I
I
L
L
V
V
N
N
200
|
S
S
R
R
G
G
E
E
I
I
K
K
L
L
C
C
D
D
F
F
210
|
G
G
V
V
S
S
G
G
Q
Q
L
L
I
I
D
D
S
S
M
M
220
|
A
A
N
N
S
S
F
F
V
V
G
G
T
T
R
R
S
S
Y
Y
230
|
M
M
S
S
P
P
E
E
R
R
L
L
Q
Q
G
G
T
T
H
H
240
|
Y
Y
S
S
V
V
Q
Q
S
S
D
D
I
I
W
W
S
S
M
M
250
|
G
G
L
L
S
S
L
L
V
V
E
E
M
M
A
A
V
V
G
G
260
|
R
R
Y
Y
P
P
I
I
-
G
-
S
-
G
-
S
-
G
-
S
270
|
-
M
-
A
-
I
-
F
-
E
-
L
-
L
-
D
-
Y
-
I
280
|
-
V
-
N
-
E
-
P
-
P
-
P
-
K
-
L
-
P
-
S
290
|
-
G
-
V
-
F
-
S
-
L
-
E
-
F
-
Q
-
D
-
F
300
|
-
V
-
N
G
K
S
C
G
L
S
I
G
K
S
N
M
P
A
A
310
|
I
E
F
R
E
A
L
D
L
L
D
K
Y
Q
I
L
V
M
N
V
320
|
E
H
P
A
P
F
P
I
K
K
L
R
P
S
S
D
G
A
V
E
330
|
F
E
S
V
L
D
E
F
F
A
Q
G
D
W
F
L
V
C
N
S
340
|
K
T
C
I
L
G
I
L
K
N
N
Q
P
P
A
S
E
T
R
P
350
|
A
T
D
H
L
A
K
A
Q
G
L
E
M
G
V
H
H
H
A
H
360
|
F
H
I
H
K
H
R
-
S
-
D
-
A
-
E
-
E
-
V
-
370
|
D
-
F
-
A
-
G
-
W
-
L
-
C
-
S
-
T
-
I
-
380
|
G
-
L
-
N
-
Q
-
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.C121S (c.361T>A) in gene MAP2K1 cause the resistance of PD98059 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | PD98059 | ||||||||||||
| Molecule Alteration | Missense mutation | p.F53L (c.157T>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.F53L (c.157T>C) in gene MAP2K1 cause the resistance of PD98059 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | PD98059 | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q56P (c.167A>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.Q56P (c.167A>C) in gene MAP2K1 cause the resistance of PD98059 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | PD98059 | ||||||||||||
| Molecule Alteration | Missense mutation | p.K57N (c.171G>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.K57N (c.171G>C) in gene MAP2K1 cause the resistance of PD98059 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | PD98059 | ||||||||||||
| Molecule Alteration | Missense mutation | p.I111S (c.332T>G) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.I111S (c.332T>G) in gene MAP2K1 cause the resistance of PD98059 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | PD98059 | ||||||||||||
| Molecule Alteration | Missense mutation | p.F129L (c.385T>C) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.F129L (c.385T>C) in gene MAP2K1 cause the resistance of PD98059 by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | PD98059 | ||||||||||||
| Molecule Alteration | Missense mutation | p.V211D (c.632T>A) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
SDS-PAGE assay | ||||||||||||
| Mechanism Description | The missense mutation p.V211D (c.632T>A) in gene MAP2K1 cause the resistance of PD98059 by unusual activation of pro-survival pathway | ||||||||||||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Nervous tissue | |
| The Specified Disease | Brain cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.08E-99; Fold-change: -1.34E+00; Z-score: -1.74E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
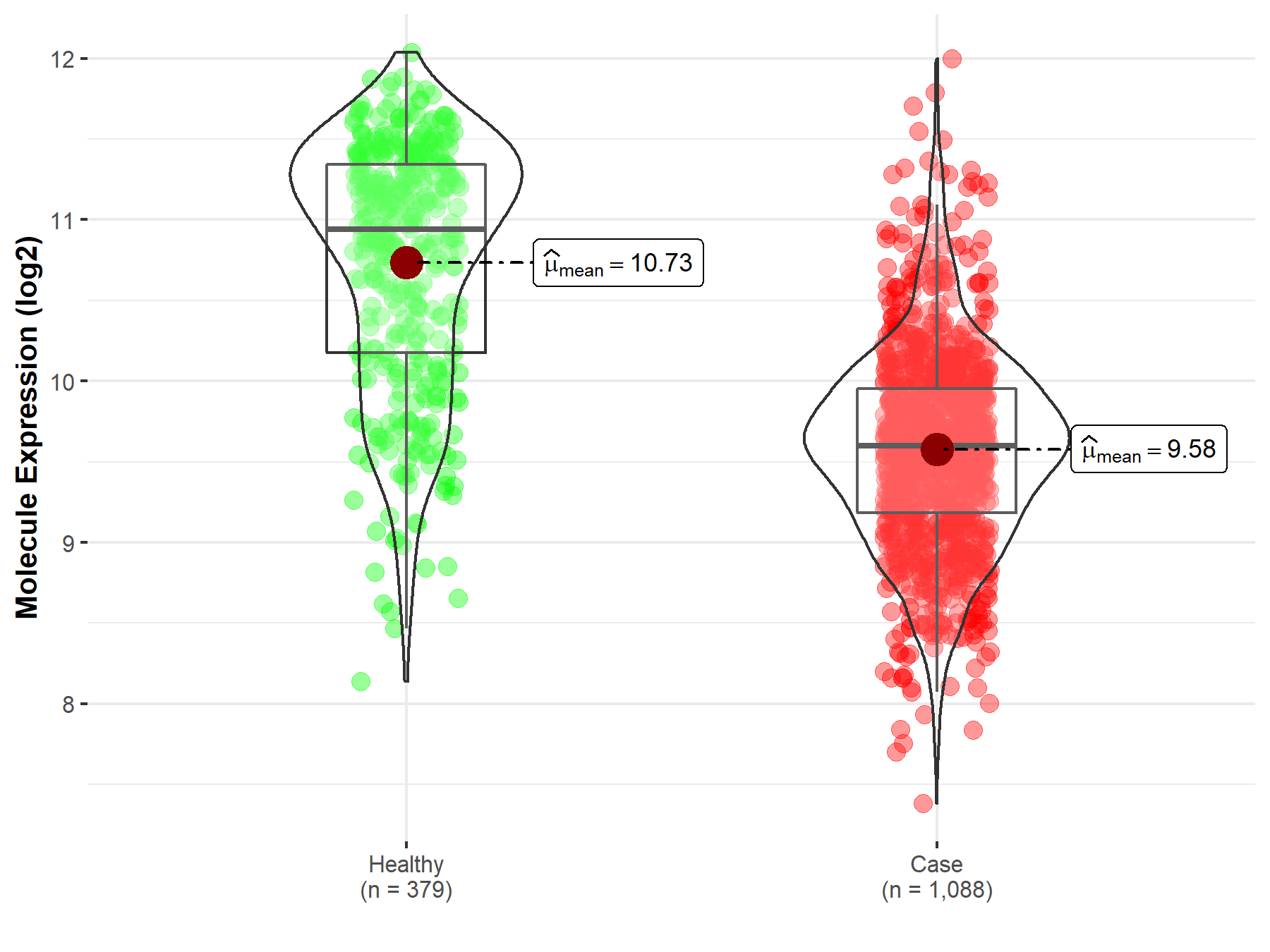
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.33E-01; Fold-change: 1.15E-01; Z-score: 3.55E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| The Studied Tissue | White matter | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.41E-01; Fold-change: 2.73E-01; Z-score: 5.53E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
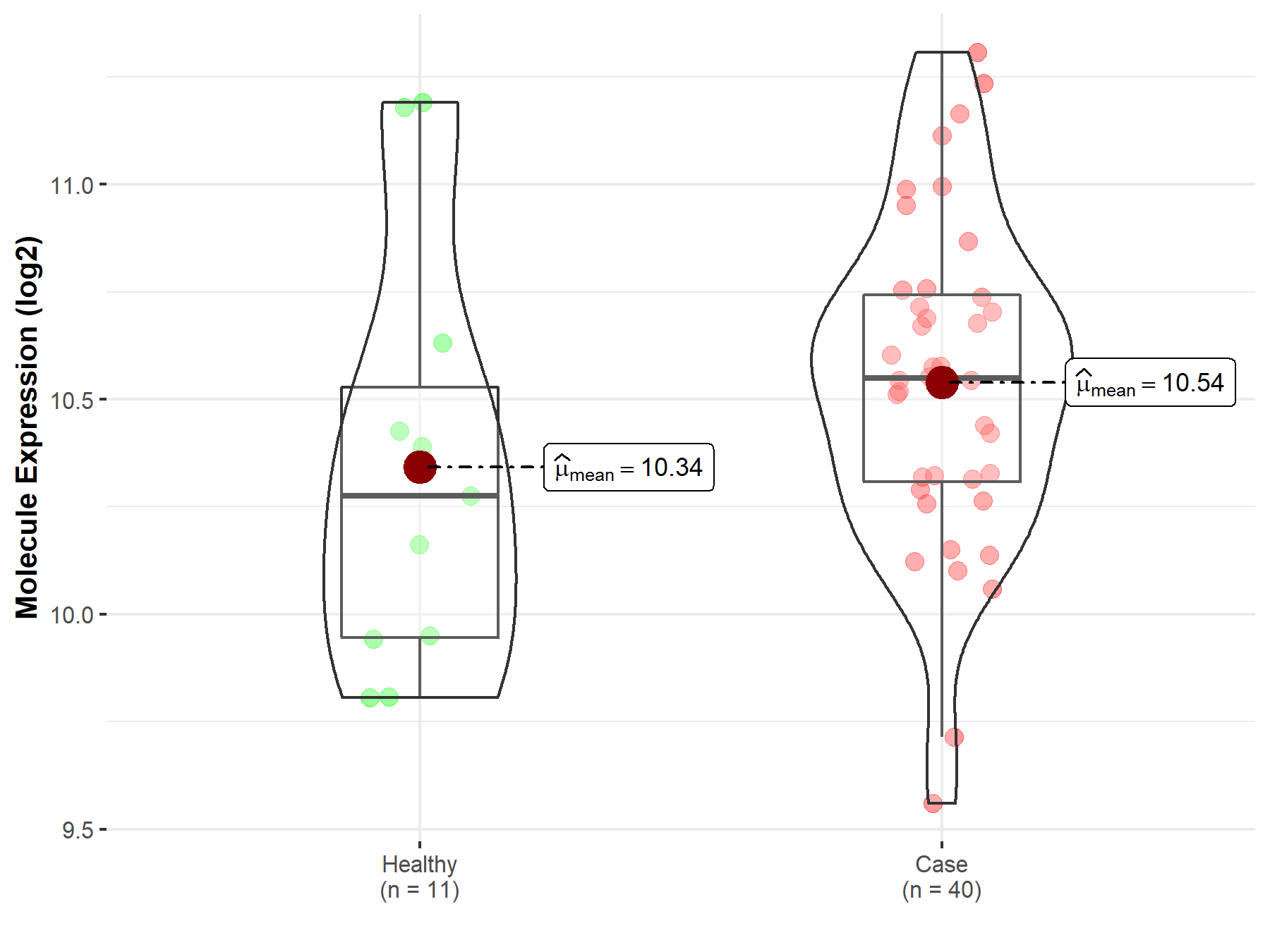
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Neuroectodermal tumor | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.61E-01; Fold-change: 4.74E-02; Z-score: 2.07E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
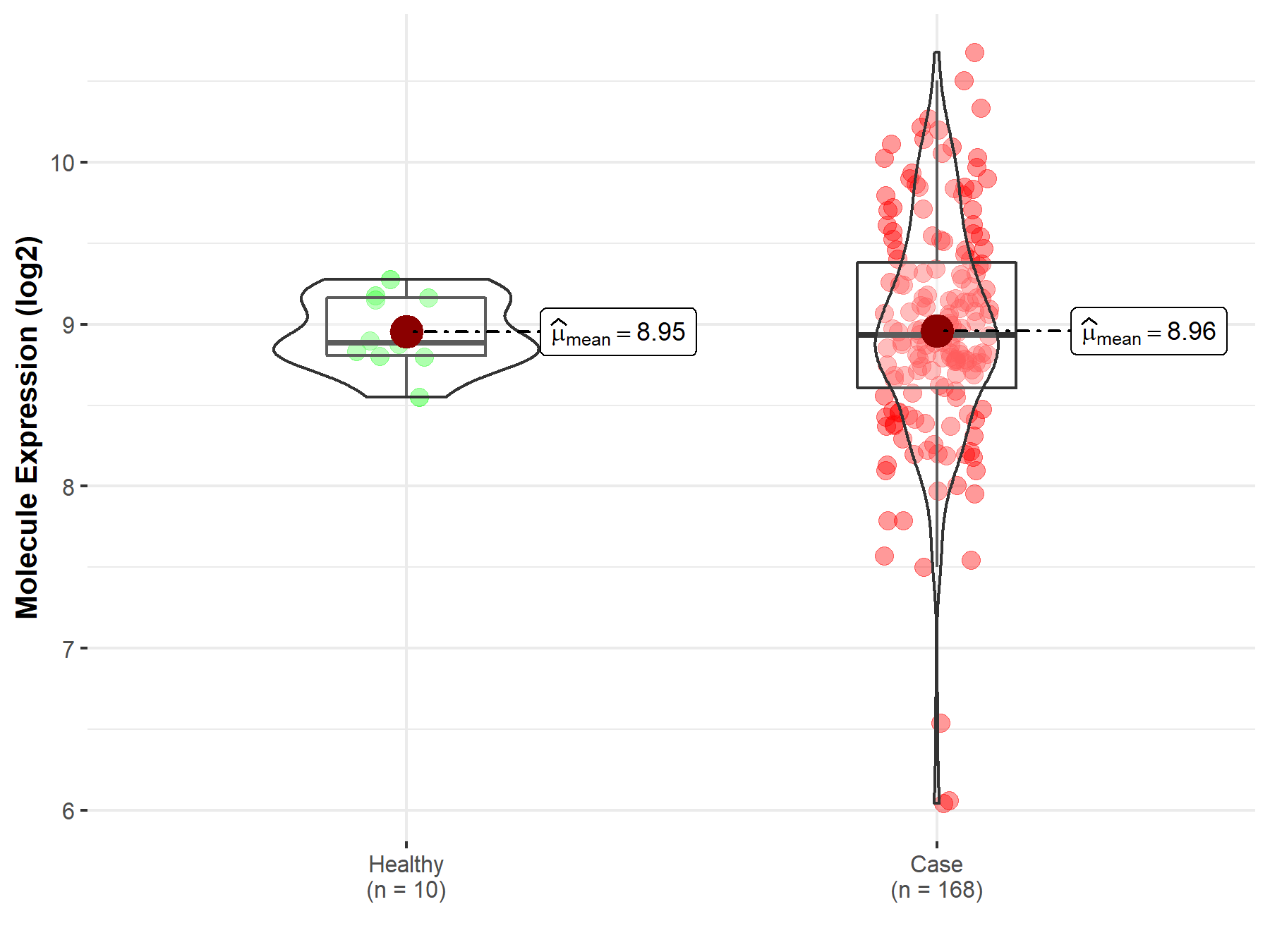
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Gastric tissue | |
| The Specified Disease | Gastric cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.23E-02; Fold-change: 6.72E-01; Z-score: 2.19E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 7.10E-03; Fold-change: 2.28E-01; Z-score: 6.54E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Lung | |
| The Specified Disease | Lung cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.41E-20; Fold-change: 1.95E-01; Z-score: 7.65E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 5.11E-02; Fold-change: 1.17E-01; Z-score: 2.98E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
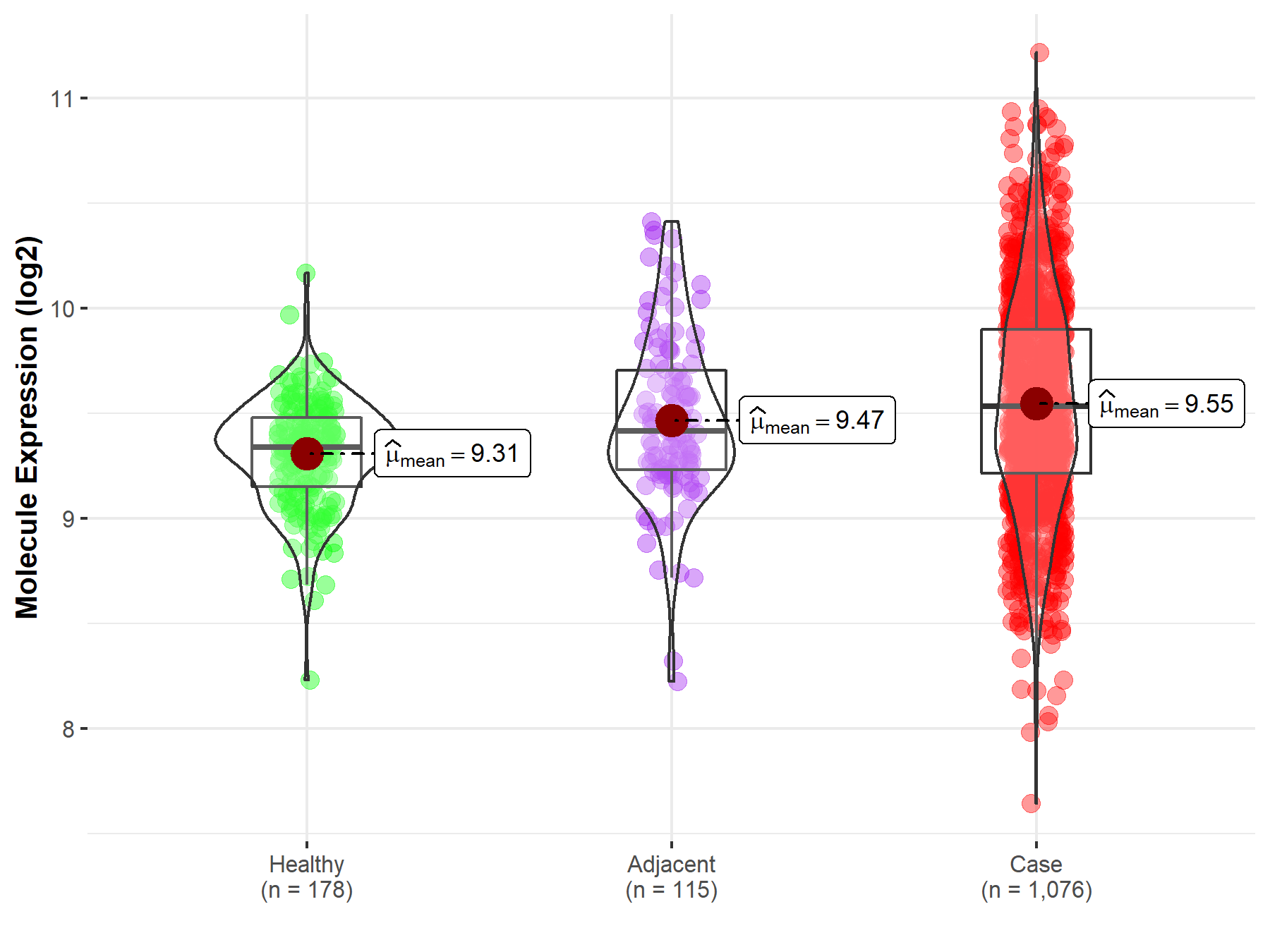
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Skin | |
| The Specified Disease | Melanoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.00E-01; Fold-change: -6.72E-02; Z-score: -1.54E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
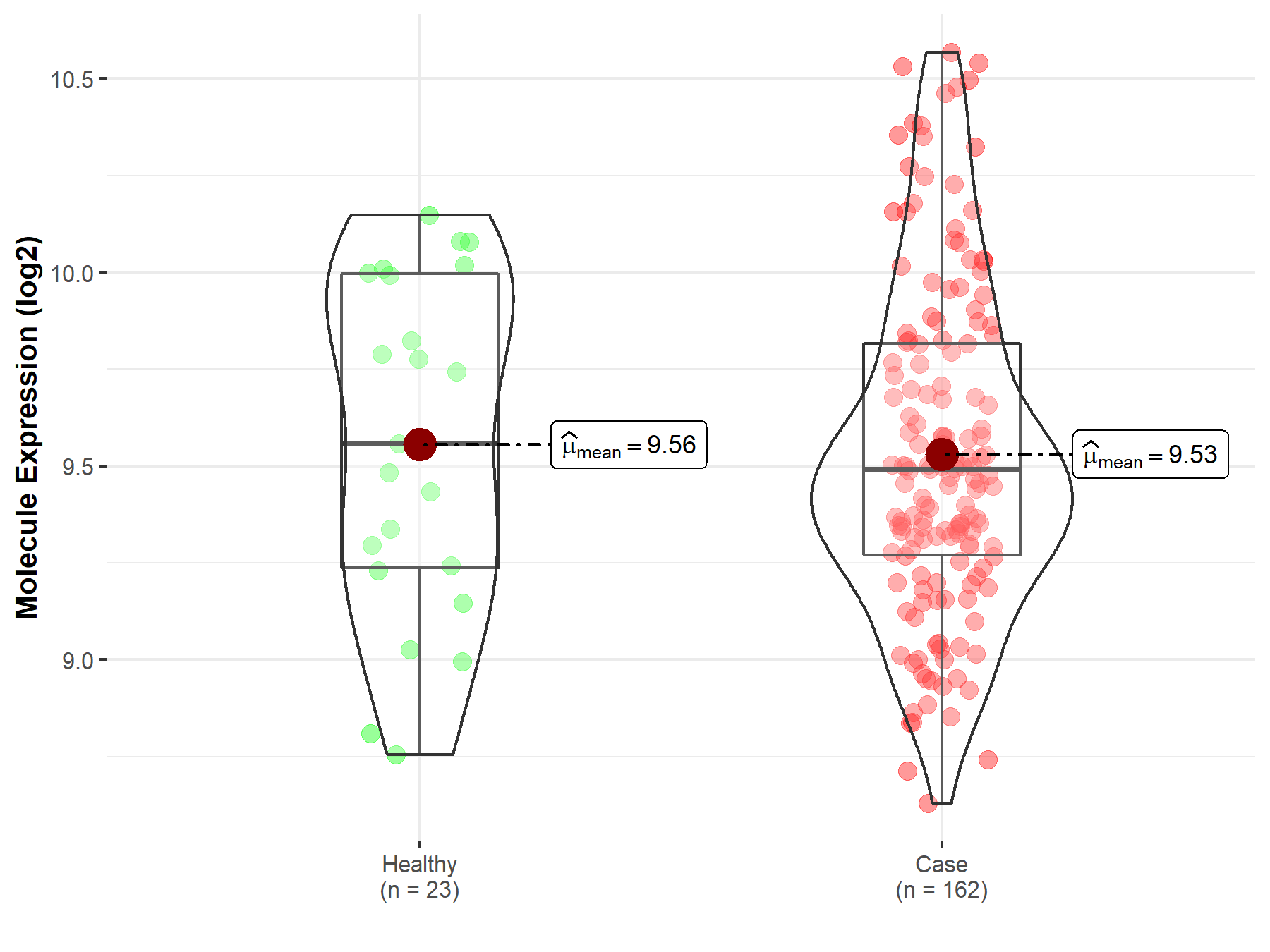
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.25E-04; Fold-change: -2.21E-01; Z-score: -2.84E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 5.77E-01; Fold-change: 2.96E-01; Z-score: 3.02E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Ovary | |
| The Specified Disease | Ovarian cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.61E-02; Fold-change: 6.77E-01; Z-score: 1.31E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 6.55E-03; Fold-change: -4.35E-01; Z-score: -5.88E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
