Molecule Information
General Information of the Molecule (ID: Mol00423)
| Name |
Oxalosuccinate decarboxylase (IDH1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
IDH; Cytosolic NADP-isocitrate dehydrogenase; IDP; NADP(+)-specific ICDH; Oxalosuccinate decarboxylase; PICD
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
IDH1
|
||||
| Gene ID | |||||
| Location |
chr2:208236229-208266074[-]
|
||||
| Sequence |
MSKKISGGSVVEMQGDEMTRIIWELIKEKLIFPYVELDLHSYDLGIENRDATNDQVTKDA
AEAIKKHNVGVKCATITPDEKRVEEFKLKQMWKSPNGTIRNILGGTVFREAIICKNIPRL VSGWVKPIIIGRHAYGDQYRATDFVVPGPGKVEITYTPSDGTQKVTYLVHNFEEGGGVAM GMYNQDKSIEDFAHSSFQMALSKGWPLYLSTKNTILKKYDGRFKDIFQEIYDKQYKSQFE AQKIWYEHRLIDDMVAQAMKSEGGFIWACKNYDGDVQSDSVAQGYGSLGMMTSVLVCPDG KTVEAEAAHGTVTRHYRMYQKGQETSTNPIASIFAWTRGLAHRAKLDNNKELAFFANALE EVSIETIEAGFMTKDLAACIKGLPNVQRSDYLNTFEFMDKLGENLKIKLAQAKL Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
6 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Glioblastoma [ICD-11: 2A00.02] | [1] | |||
| Resistant Disease | Glioblastoma [ICD-11: 2A00.02] | |||
| Resistant Drug | Dichloroacetate | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Brain cancer [ICD-11: 2A00] | |||
| The Specified Disease | Brain cancer | |||
| The Studied Tissue | Nervous tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.42E-176 Fold-change: 2.81E-01 Z-score: 3.90E+01 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell migration | Activation | hsa04670 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | DBTRG cells | Brain | Homo sapiens (Human) | CVCL_1169 |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
Colorimetric SRB assay | |||
| Mechanism Description | The potential of miR-144 overexpression to reduce GB cell malignancy, both by decreasing Cell migration and invasion abilities and by sensitizing resistant tumor cells to chemotherapy, paving the way to a novel and more effective GB therapy. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: FGFR-tacc positive glioblastoma [ICD-11: 2A00.01] | [2] | ||||||||||||
| Sensitive Disease | FGFR-tacc positive glioblastoma [ICD-11: 2A00.01] | ||||||||||||
| Sensitive Drug | Azacitidine | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132H (c.395G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.65 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.88 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
M
S
S
K
K
K
K
I
I
S
S
G
G
G
G
S
S
10
|
V
V
V
V
E
E
M
M
Q
Q
G
G
D
D
E
E
M
M
T
T
20
|
R
R
I
I
I
I
W
W
E
E
L
L
I
I
K
K
E
E
K
K
30
|
L
L
I
I
F
F
P
P
Y
Y
V
V
E
E
L
L
D
D
L
L
40
|
H
H
S
S
Y
Y
D
D
L
L
G
G
I
I
E
E
N
N
R
R
50
|
D
D
A
A
T
T
N
N
D
D
Q
Q
V
V
T
T
K
K
D
D
60
|
A
A
A
A
E
E
A
A
I
I
K
K
K
K
H
H
N
N
V
V
70
|
G
G
V
V
K
K
C
C
A
A
T
T
I
I
T
T
P
P
D
D
80
|
E
E
K
K
R
R
V
V
E
E
E
E
F
F
K
K
L
L
K
K
90
|
Q
Q
M
M
W
W
K
K
S
S
P
P
N
N
G
G
T
T
I
I
100
|
R
R
N
N
I
I
L
L
G
G
G
G
T
T
V
V
F
F
R
R
110
|
E
E
A
A
I
I
I
I
C
C
K
K
N
N
I
I
P
P
R
R
120
|
L
L
V
V
S
S
G
G
W
W
V
V
K
K
P
P
I
I
I
I
130
|
I
I
G
G
R
H
H
H
A
A
Y
Y
G
G
D
D
Q
Q
Y
Y
140
|
R
R
A
A
T
T
D
D
F
F
V
V
V
V
P
P
G
G
P
P
150
|
G
G
K
K
V
V
E
E
I
I
T
T
Y
Y
T
T
P
P
S
S
160
|
D
D
G
G
T
T
Q
Q
K
K
V
V
T
T
Y
Y
L
L
V
V
170
|
H
H
N
N
F
F
E
E
E
E
G
G
G
G
G
G
V
V
A
A
180
|
M
M
G
G
M
M
Y
Y
N
N
Q
Q
D
D
K
K
S
S
I
I
190
|
E
E
D
D
F
F
A
A
H
H
S
S
S
S
F
F
Q
Q
M
M
200
|
A
A
L
L
S
S
K
K
G
G
W
W
P
P
L
L
Y
Y
L
L
210
|
S
S
T
T
K
K
N
N
T
T
I
I
L
L
K
K
K
K
Y
Y
220
|
D
D
G
G
R
R
F
F
K
K
D
D
I
I
F
F
Q
Q
E
E
230
|
I
I
Y
Y
D
D
K
K
Q
Q
Y
Y
K
K
S
S
Q
Q
F
F
240
|
E
E
A
A
Q
Q
K
K
I
I
W
W
Y
Y
E
E
H
H
R
R
250
|
L
L
I
I
D
D
D
D
M
M
V
V
A
A
Q
Q
A
A
M
M
260
|
K
K
S
S
E
E
G
G
G
G
F
F
I
I
W
W
A
A
C
C
270
|
K
K
N
N
Y
Y
D
D
G
G
D
D
V
V
Q
Q
S
S
D
D
280
|
S
S
V
V
A
A
Q
Q
G
G
Y
Y
G
G
S
S
L
L
G
G
290
|
M
M
M
M
T
T
S
S
V
V
L
L
V
V
C
C
P
P
D
D
300
|
G
G
K
K
T
T
V
V
E
E
A
A
E
E
A
A
A
A
H
H
310
|
G
G
T
T
V
V
T
T
R
R
H
H
Y
Y
R
R
M
M
Y
Y
320
|
Q
Q
K
K
G
G
Q
Q
E
E
T
T
S
S
T
T
N
N
P
P
330
|
I
I
A
A
S
S
I
I
F
F
A
A
W
W
T
T
R
R
G
G
340
|
L
L
A
A
H
H
R
R
A
A
K
K
L
L
D
D
N
N
N
N
350
|
K
K
E
E
L
L
A
A
F
F
F
F
A
A
N
N
A
A
L
L
360
|
E
E
E
E
V
V
S
S
I
I
E
E
T
T
I
I
E
E
A
A
370
|
G
G
F
F
M
M
T
T
K
K
D
D
L
L
A
A
A
A
C
C
380
|
I
I
K
K
G
G
L
L
P
P
N
N
V
V
Q
Q
R
R
S
S
390
|
D
D
Y
Y
L
L
N
N
T
T
F
F
E
E
F
F
M
M
D
D
400
|
K
K
L
L
G
G
E
E
N
N
L
L
K
K
I
I
K
K
L
L
410
|
A
A
Q
Q
A
A
K
K
L
L
S
S
L
L
E
E
H
H
H
H
420
|
H
H
H
H
H
H
H
H
H
H
H
H
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Brain | N.A. | |||||||||||
| In Vivo Model | Female athymic nude mouse (NCI-Frederick) model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Tumor volume measurement assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Brain glioma [ICD-11: 2A00.0] | [3] | ||||||||||||
| Sensitive Disease | Brain glioma [ICD-11: 2A00.0] | ||||||||||||
| Sensitive Drug | Bevacizumab | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132C (c.394C>T) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.93 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
M
-
S
-
K
-
K
-
I
-
S
-
G
-
G
-
S
10
|
-
V
-
V
-
E
-
M
-
Q
-
G
-
D
-
E
-
M
-
T
20
|
-
R
-
I
-
I
-
W
-
E
-
L
-
I
-
K
-
E
-
K
30
|
-
L
-
I
-
F
-
P
-
Y
-
V
-
E
-
L
-
D
-
L
40
|
-
H
-
S
-
Y
-
D
-
L
-
G
-
I
-
E
-
N
-
R
50
|
-
D
-
A
-
T
-
N
-
D
-
Q
-
V
-
T
-
K
-
D
60
|
-
A
-
A
-
E
-
A
-
I
-
K
-
K
-
H
-
N
-
V
70
|
-
G
-
V
-
K
-
C
-
A
-
T
-
I
-
T
-
P
-
D
80
|
-
E
-
K
-
R
-
V
-
E
-
E
-
F
-
K
-
L
-
K
90
|
-
Q
-
M
-
W
-
K
-
S
-
P
-
N
-
G
-
T
-
I
100
|
-
R
-
N
-
I
-
L
-
G
-
G
-
T
-
V
-
F
-
R
110
|
-
E
-
A
-
I
-
I
-
C
-
K
-
N
-
I
-
P
-
R
120
|
-
L
-
V
-
S
-
G
-
W
-
V
K
K
P
P
I
I
I
I
130
|
I
I
G
G
S
C
H
H
A
A
Y
Y
G
G
D
D
-
Q
-
Y
140
|
-
R
-
A
-
T
-
D
-
F
-
V
-
V
-
P
-
G
-
P
150
|
-
G
-
K
-
V
-
E
-
I
-
T
-
Y
-
T
-
P
-
S
160
|
-
D
-
G
-
T
-
Q
-
K
-
V
-
T
-
Y
-
L
-
V
170
|
-
H
-
N
-
F
-
E
-
E
-
G
-
G
-
G
-
V
-
A
180
|
-
M
-
G
-
M
-
Y
-
N
-
Q
-
D
-
K
-
S
-
I
190
|
-
E
-
D
-
F
-
A
-
H
-
S
-
S
-
F
-
Q
-
M
200
|
-
A
-
L
-
S
-
K
-
G
-
W
-
P
-
L
-
Y
-
L
210
|
-
S
-
T
-
K
-
N
-
T
-
I
-
L
-
K
-
K
-
Y
220
|
-
D
-
G
-
R
-
F
-
K
-
D
-
I
-
F
-
Q
-
E
230
|
-
I
-
Y
-
D
-
K
-
Q
-
Y
-
K
-
S
-
Q
-
F
240
|
-
E
-
A
-
Q
-
K
-
I
-
W
-
Y
-
E
-
H
-
R
250
|
-
L
-
I
-
D
-
D
-
M
-
V
-
A
-
Q
-
A
-
M
260
|
-
K
-
S
-
E
-
G
-
G
-
F
-
I
-
W
-
A
-
C
270
|
-
K
-
N
-
Y
-
D
-
G
-
D
-
V
-
Q
-
S
-
D
280
|
-
S
-
V
-
A
-
Q
-
G
-
Y
-
G
-
S
-
L
-
G
290
|
-
M
-
M
-
T
-
S
-
V
-
L
-
V
-
C
-
P
-
D
300
|
-
G
-
K
-
T
-
V
-
E
-
A
-
E
-
A
-
A
-
H
310
|
-
G
-
T
-
V
-
T
-
R
-
H
-
Y
-
R
-
M
-
Y
320
|
-
Q
-
K
-
G
-
Q
-
E
-
T
-
S
-
T
-
N
-
P
330
|
-
I
-
A
-
S
-
I
-
F
-
A
-
W
-
T
-
R
-
G
340
|
-
L
-
A
-
H
-
R
-
A
-
K
-
L
-
D
-
N
-
N
350
|
-
K
-
E
-
L
-
A
-
F
-
F
-
A
-
N
-
A
-
L
360
|
-
E
-
E
-
V
-
S
-
I
-
E
-
T
-
I
-
E
-
A
370
|
-
G
-
F
-
M
-
T
-
K
-
D
-
L
-
A
-
A
-
C
380
|
-
I
-
K
-
G
-
L
-
P
-
N
-
V
-
Q
-
R
-
S
390
|
-
D
-
Y
-
L
-
N
-
T
-
F
-
E
-
F
-
M
-
D
400
|
-
K
-
L
-
G
-
E
-
N
-
L
-
K
-
I
-
K
-
L
410
|
-
A
-
Q
-
A
-
K
-
L
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Brain | N.A. | |||||||||||
| Mechanism Description | The missense mutation p.R132C (c.394C>T) in gene IDH1 cause the sensitivity of Bevacizumab by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Brain glioma [ICD-11: 2A00.0] | [3] | ||||||||||||
| Sensitive Disease | Brain glioma [ICD-11: 2A00.0] | ||||||||||||
| Sensitive Drug | Bevacizumab | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132S (c.394C>A) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Brain | N.A. | |||||||||||
| Mechanism Description | The missense mutation p.R132S (c.394C>A) in gene IDH1 cause the sensitivity of Bevacizumab by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Brain glioma [ICD-11: 2A00.0] | [3] | ||||||||||||
| Sensitive Disease | Brain glioma [ICD-11: 2A00.0] | ||||||||||||
| Sensitive Drug | Bevacizumab | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132L (c.395G>T) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Brain | N.A. | |||||||||||
| Mechanism Description | The missense mutation p.R132L (c.395G>T) in gene IDH1 cause the sensitivity of Bevacizumab by aberration of the drug's therapeutic target | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [4] | ||||||||||||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Sensitive Drug | Ivosidenib | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132H (c.395G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.65 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.88 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
M
S
S
K
K
K
K
I
I
S
S
G
G
G
G
S
S
10
|
V
V
V
V
E
E
M
M
Q
Q
G
G
D
D
E
E
M
M
T
T
20
|
R
R
I
I
I
I
W
W
E
E
L
L
I
I
K
K
E
E
K
K
30
|
L
L
I
I
F
F
P
P
Y
Y
V
V
E
E
L
L
D
D
L
L
40
|
H
H
S
S
Y
Y
D
D
L
L
G
G
I
I
E
E
N
N
R
R
50
|
D
D
A
A
T
T
N
N
D
D
Q
Q
V
V
T
T
K
K
D
D
60
|
A
A
A
A
E
E
A
A
I
I
K
K
K
K
H
H
N
N
V
V
70
|
G
G
V
V
K
K
C
C
A
A
T
T
I
I
T
T
P
P
D
D
80
|
E
E
K
K
R
R
V
V
E
E
E
E
F
F
K
K
L
L
K
K
90
|
Q
Q
M
M
W
W
K
K
S
S
P
P
N
N
G
G
T
T
I
I
100
|
R
R
N
N
I
I
L
L
G
G
G
G
T
T
V
V
F
F
R
R
110
|
E
E
A
A
I
I
I
I
C
C
K
K
N
N
I
I
P
P
R
R
120
|
L
L
V
V
S
S
G
G
W
W
V
V
K
K
P
P
I
I
I
I
130
|
I
I
G
G
R
H
H
H
A
A
Y
Y
G
G
D
D
Q
Q
Y
Y
140
|
R
R
A
A
T
T
D
D
F
F
V
V
V
V
P
P
G
G
P
P
150
|
G
G
K
K
V
V
E
E
I
I
T
T
Y
Y
T
T
P
P
S
S
160
|
D
D
G
G
T
T
Q
Q
K
K
V
V
T
T
Y
Y
L
L
V
V
170
|
H
H
N
N
F
F
E
E
E
E
G
G
G
G
G
G
V
V
A
A
180
|
M
M
G
G
M
M
Y
Y
N
N
Q
Q
D
D
K
K
S
S
I
I
190
|
E
E
D
D
F
F
A
A
H
H
S
S
S
S
F
F
Q
Q
M
M
200
|
A
A
L
L
S
S
K
K
G
G
W
W
P
P
L
L
Y
Y
L
L
210
|
S
S
T
T
K
K
N
N
T
T
I
I
L
L
K
K
K
K
Y
Y
220
|
D
D
G
G
R
R
F
F
K
K
D
D
I
I
F
F
Q
Q
E
E
230
|
I
I
Y
Y
D
D
K
K
Q
Q
Y
Y
K
K
S
S
Q
Q
F
F
240
|
E
E
A
A
Q
Q
K
K
I
I
W
W
Y
Y
E
E
H
H
R
R
250
|
L
L
I
I
D
D
D
D
M
M
V
V
A
A
Q
Q
A
A
M
M
260
|
K
K
S
S
E
E
G
G
G
G
F
F
I
I
W
W
A
A
C
C
270
|
K
K
N
N
Y
Y
D
D
G
G
D
D
V
V
Q
Q
S
S
D
D
280
|
S
S
V
V
A
A
Q
Q
G
G
Y
Y
G
G
S
S
L
L
G
G
290
|
M
M
M
M
T
T
S
S
V
V
L
L
V
V
C
C
P
P
D
D
300
|
G
G
K
K
T
T
V
V
E
E
A
A
E
E
A
A
A
A
H
H
310
|
G
G
T
T
V
V
T
T
R
R
H
H
Y
Y
R
R
M
M
Y
Y
320
|
Q
Q
K
K
G
G
Q
Q
E
E
T
T
S
S
T
T
N
N
P
P
330
|
I
I
A
A
S
S
I
I
F
F
A
A
W
W
T
T
R
R
G
G
340
|
L
L
A
A
H
H
R
R
A
A
K
K
L
L
D
D
N
N
N
N
350
|
K
K
E
E
L
L
A
A
F
F
F
F
A
A
N
N
A
A
L
L
360
|
E
E
E
E
V
V
S
S
I
I
E
E
T
T
I
I
E
E
A
A
370
|
G
G
F
F
M
M
T
T
K
K
D
D
L
L
A
A
A
A
C
C
380
|
I
I
K
K
G
G
L
L
P
P
N
N
V
V
Q
Q
R
R
S
S
390
|
D
D
Y
Y
L
L
N
N
T
T
F
F
E
E
F
F
M
M
D
D
400
|
K
K
L
L
G
G
E
E
N
N
L
L
K
K
I
I
K
K
L
L
410
|
A
A
Q
Q
A
A
K
K
L
L
S
S
L
L
E
E
H
H
H
H
420
|
H
H
H
H
H
H
H
H
H
H
H
H
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [4] | ||||||||||||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Sensitive Drug | Ivosidenib | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132C (c.394C>T) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.93 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
M
-
S
-
K
-
K
-
I
-
S
-
G
-
G
-
S
10
|
-
V
-
V
-
E
-
M
-
Q
-
G
-
D
-
E
-
M
-
T
20
|
-
R
-
I
-
I
-
W
-
E
-
L
-
I
-
K
-
E
-
K
30
|
-
L
-
I
-
F
-
P
-
Y
-
V
-
E
-
L
-
D
-
L
40
|
-
H
-
S
-
Y
-
D
-
L
-
G
-
I
-
E
-
N
-
R
50
|
-
D
-
A
-
T
-
N
-
D
-
Q
-
V
-
T
-
K
-
D
60
|
-
A
-
A
-
E
-
A
-
I
-
K
-
K
-
H
-
N
-
V
70
|
-
G
-
V
-
K
-
C
-
A
-
T
-
I
-
T
-
P
-
D
80
|
-
E
-
K
-
R
-
V
-
E
-
E
-
F
-
K
-
L
-
K
90
|
-
Q
-
M
-
W
-
K
-
S
-
P
-
N
-
G
-
T
-
I
100
|
-
R
-
N
-
I
-
L
-
G
-
G
-
T
-
V
-
F
-
R
110
|
-
E
-
A
-
I
-
I
-
C
-
K
-
N
-
I
-
P
-
R
120
|
-
L
-
V
-
S
-
G
-
W
-
V
K
K
P
P
I
I
I
I
130
|
I
I
G
G
S
C
H
H
A
A
Y
Y
G
G
D
D
-
Q
-
Y
140
|
-
R
-
A
-
T
-
D
-
F
-
V
-
V
-
P
-
G
-
P
150
|
-
G
-
K
-
V
-
E
-
I
-
T
-
Y
-
T
-
P
-
S
160
|
-
D
-
G
-
T
-
Q
-
K
-
V
-
T
-
Y
-
L
-
V
170
|
-
H
-
N
-
F
-
E
-
E
-
G
-
G
-
G
-
V
-
A
180
|
-
M
-
G
-
M
-
Y
-
N
-
Q
-
D
-
K
-
S
-
I
190
|
-
E
-
D
-
F
-
A
-
H
-
S
-
S
-
F
-
Q
-
M
200
|
-
A
-
L
-
S
-
K
-
G
-
W
-
P
-
L
-
Y
-
L
210
|
-
S
-
T
-
K
-
N
-
T
-
I
-
L
-
K
-
K
-
Y
220
|
-
D
-
G
-
R
-
F
-
K
-
D
-
I
-
F
-
Q
-
E
230
|
-
I
-
Y
-
D
-
K
-
Q
-
Y
-
K
-
S
-
Q
-
F
240
|
-
E
-
A
-
Q
-
K
-
I
-
W
-
Y
-
E
-
H
-
R
250
|
-
L
-
I
-
D
-
D
-
M
-
V
-
A
-
Q
-
A
-
M
260
|
-
K
-
S
-
E
-
G
-
G
-
F
-
I
-
W
-
A
-
C
270
|
-
K
-
N
-
Y
-
D
-
G
-
D
-
V
-
Q
-
S
-
D
280
|
-
S
-
V
-
A
-
Q
-
G
-
Y
-
G
-
S
-
L
-
G
290
|
-
M
-
M
-
T
-
S
-
V
-
L
-
V
-
C
-
P
-
D
300
|
-
G
-
K
-
T
-
V
-
E
-
A
-
E
-
A
-
A
-
H
310
|
-
G
-
T
-
V
-
T
-
R
-
H
-
Y
-
R
-
M
-
Y
320
|
-
Q
-
K
-
G
-
Q
-
E
-
T
-
S
-
T
-
N
-
P
330
|
-
I
-
A
-
S
-
I
-
F
-
A
-
W
-
T
-
R
-
G
340
|
-
L
-
A
-
H
-
R
-
A
-
K
-
L
-
D
-
N
-
N
350
|
-
K
-
E
-
L
-
A
-
F
-
F
-
A
-
N
-
A
-
L
360
|
-
E
-
E
-
V
-
S
-
I
-
E
-
T
-
I
-
E
-
A
370
|
-
G
-
F
-
M
-
T
-
K
-
D
-
L
-
A
-
A
-
C
380
|
-
I
-
K
-
G
-
L
-
P
-
N
-
V
-
Q
-
R
-
S
390
|
-
D
-
Y
-
L
-
N
-
T
-
F
-
E
-
F
-
M
-
D
400
|
-
K
-
L
-
G
-
E
-
N
-
L
-
K
-
I
-
K
-
L
410
|
-
A
-
Q
-
A
-
K
-
L
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [4] | ||||||||||||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Sensitive Drug | Ivosidenib | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132S (c.394C>A) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [4] | ||||||||||||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Sensitive Drug | Ivosidenib | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132G (c.394C>G) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [4] | ||||||||||||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Sensitive Drug | Ivosidenib | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132L (c.395G>T) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [5] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | Rucaparib | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132H (c.395G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.65 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.88 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
M
S
S
K
K
K
K
I
I
S
S
G
G
G
G
S
S
10
|
V
V
V
V
E
E
M
M
Q
Q
G
G
D
D
E
E
M
M
T
T
20
|
R
R
I
I
I
I
W
W
E
E
L
L
I
I
K
K
E
E
K
K
30
|
L
L
I
I
F
F
P
P
Y
Y
V
V
E
E
L
L
D
D
L
L
40
|
H
H
S
S
Y
Y
D
D
L
L
G
G
I
I
E
E
N
N
R
R
50
|
D
D
A
A
T
T
N
N
D
D
Q
Q
V
V
T
T
K
K
D
D
60
|
A
A
A
A
E
E
A
A
I
I
K
K
K
K
H
H
N
N
V
V
70
|
G
G
V
V
K
K
C
C
A
A
T
T
I
I
T
T
P
P
D
D
80
|
E
E
K
K
R
R
V
V
E
E
E
E
F
F
K
K
L
L
K
K
90
|
Q
Q
M
M
W
W
K
K
S
S
P
P
N
N
G
G
T
T
I
I
100
|
R
R
N
N
I
I
L
L
G
G
G
G
T
T
V
V
F
F
R
R
110
|
E
E
A
A
I
I
I
I
C
C
K
K
N
N
I
I
P
P
R
R
120
|
L
L
V
V
S
S
G
G
W
W
V
V
K
K
P
P
I
I
I
I
130
|
I
I
G
G
R
H
H
H
A
A
Y
Y
G
G
D
D
Q
Q
Y
Y
140
|
R
R
A
A
T
T
D
D
F
F
V
V
V
V
P
P
G
G
P
P
150
|
G
G
K
K
V
V
E
E
I
I
T
T
Y
Y
T
T
P
P
S
S
160
|
D
D
G
G
T
T
Q
Q
K
K
V
V
T
T
Y
Y
L
L
V
V
170
|
H
H
N
N
F
F
E
E
E
E
G
G
G
G
G
G
V
V
A
A
180
|
M
M
G
G
M
M
Y
Y
N
N
Q
Q
D
D
K
K
S
S
I
I
190
|
E
E
D
D
F
F
A
A
H
H
S
S
S
S
F
F
Q
Q
M
M
200
|
A
A
L
L
S
S
K
K
G
G
W
W
P
P
L
L
Y
Y
L
L
210
|
S
S
T
T
K
K
N
N
T
T
I
I
L
L
K
K
K
K
Y
Y
220
|
D
D
G
G
R
R
F
F
K
K
D
D
I
I
F
F
Q
Q
E
E
230
|
I
I
Y
Y
D
D
K
K
Q
Q
Y
Y
K
K
S
S
Q
Q
F
F
240
|
E
E
A
A
Q
Q
K
K
I
I
W
W
Y
Y
E
E
H
H
R
R
250
|
L
L
I
I
D
D
D
D
M
M
V
V
A
A
Q
Q
A
A
M
M
260
|
K
K
S
S
E
E
G
G
G
G
F
F
I
I
W
W
A
A
C
C
270
|
K
K
N
N
Y
Y
D
D
G
G
D
D
V
V
Q
Q
S
S
D
D
280
|
S
S
V
V
A
A
Q
Q
G
G
Y
Y
G
G
S
S
L
L
G
G
290
|
M
M
M
M
T
T
S
S
V
V
L
L
V
V
C
C
P
P
D
D
300
|
G
G
K
K
T
T
V
V
E
E
A
A
E
E
A
A
A
A
H
H
310
|
G
G
T
T
V
V
T
T
R
R
H
H
Y
Y
R
R
M
M
Y
Y
320
|
Q
Q
K
K
G
G
Q
Q
E
E
T
T
S
S
T
T
N
N
P
P
330
|
I
I
A
A
S
S
I
I
F
F
A
A
W
W
T
T
R
R
G
G
340
|
L
L
A
A
H
H
R
R
A
A
K
K
L
L
D
D
N
N
N
N
350
|
K
K
E
E
L
L
A
A
F
F
F
F
A
A
N
N
A
A
L
L
360
|
E
E
E
E
V
V
S
S
I
I
E
E
T
T
I
I
E
E
A
A
370
|
G
G
F
F
M
M
T
T
K
K
D
D
L
L
A
A
A
A
C
C
380
|
I
I
K
K
G
G
L
L
P
P
N
N
V
V
Q
Q
R
R
S
S
390
|
D
D
Y
Y
L
L
N
N
T
T
F
F
E
E
F
F
M
M
D
D
400
|
K
K
L
L
G
G
E
E
N
N
L
L
K
K
I
I
K
K
L
L
410
|
A
A
Q
Q
A
A
K
K
L
L
S
S
L
L
E
E
H
H
H
H
420
|
H
H
H
H
H
H
H
H
H
H
H
H
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | IDH2 cells | Ovary | Homo sapiens (Human) | CVCL_D3DY | |||||||||
| IDH1 cells | Ovary | Homo sapiens (Human) | CVCL_D3DY | ||||||||||
| In Vivo Model | Female athymic nu/nu mouse PDX model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
Promega assay | ||||||||||||
| Mechanism Description | The oncometabolite, 2-hydroxyglutarate, renders IDH1/2 mutant cancer cells deficient in homologous recombination and confers vulnerability to synthetic lethal targeting with PARP inhibitors. | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Colon cancer [ICD-11: 2B90.1] | [5] | ||||||||||||
| Sensitive Disease | Colon cancer [ICD-11: 2B90.1] | ||||||||||||
| Sensitive Drug | Talazoparib | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132H (c.395G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.65 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.88 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
M
S
S
K
K
K
K
I
I
S
S
G
G
G
G
S
S
10
|
V
V
V
V
E
E
M
M
Q
Q
G
G
D
D
E
E
M
M
T
T
20
|
R
R
I
I
I
I
W
W
E
E
L
L
I
I
K
K
E
E
K
K
30
|
L
L
I
I
F
F
P
P
Y
Y
V
V
E
E
L
L
D
D
L
L
40
|
H
H
S
S
Y
Y
D
D
L
L
G
G
I
I
E
E
N
N
R
R
50
|
D
D
A
A
T
T
N
N
D
D
Q
Q
V
V
T
T
K
K
D
D
60
|
A
A
A
A
E
E
A
A
I
I
K
K
K
K
H
H
N
N
V
V
70
|
G
G
V
V
K
K
C
C
A
A
T
T
I
I
T
T
P
P
D
D
80
|
E
E
K
K
R
R
V
V
E
E
E
E
F
F
K
K
L
L
K
K
90
|
Q
Q
M
M
W
W
K
K
S
S
P
P
N
N
G
G
T
T
I
I
100
|
R
R
N
N
I
I
L
L
G
G
G
G
T
T
V
V
F
F
R
R
110
|
E
E
A
A
I
I
I
I
C
C
K
K
N
N
I
I
P
P
R
R
120
|
L
L
V
V
S
S
G
G
W
W
V
V
K
K
P
P
I
I
I
I
130
|
I
I
G
G
R
H
H
H
A
A
Y
Y
G
G
D
D
Q
Q
Y
Y
140
|
R
R
A
A
T
T
D
D
F
F
V
V
V
V
P
P
G
G
P
P
150
|
G
G
K
K
V
V
E
E
I
I
T
T
Y
Y
T
T
P
P
S
S
160
|
D
D
G
G
T
T
Q
Q
K
K
V
V
T
T
Y
Y
L
L
V
V
170
|
H
H
N
N
F
F
E
E
E
E
G
G
G
G
G
G
V
V
A
A
180
|
M
M
G
G
M
M
Y
Y
N
N
Q
Q
D
D
K
K
S
S
I
I
190
|
E
E
D
D
F
F
A
A
H
H
S
S
S
S
F
F
Q
Q
M
M
200
|
A
A
L
L
S
S
K
K
G
G
W
W
P
P
L
L
Y
Y
L
L
210
|
S
S
T
T
K
K
N
N
T
T
I
I
L
L
K
K
K
K
Y
Y
220
|
D
D
G
G
R
R
F
F
K
K
D
D
I
I
F
F
Q
Q
E
E
230
|
I
I
Y
Y
D
D
K
K
Q
Q
Y
Y
K
K
S
S
Q
Q
F
F
240
|
E
E
A
A
Q
Q
K
K
I
I
W
W
Y
Y
E
E
H
H
R
R
250
|
L
L
I
I
D
D
D
D
M
M
V
V
A
A
Q
Q
A
A
M
M
260
|
K
K
S
S
E
E
G
G
G
G
F
F
I
I
W
W
A
A
C
C
270
|
K
K
N
N
Y
Y
D
D
G
G
D
D
V
V
Q
Q
S
S
D
D
280
|
S
S
V
V
A
A
Q
Q
G
G
Y
Y
G
G
S
S
L
L
G
G
290
|
M
M
M
M
T
T
S
S
V
V
L
L
V
V
C
C
P
P
D
D
300
|
G
G
K
K
T
T
V
V
E
E
A
A
E
E
A
A
A
A
H
H
310
|
G
G
T
T
V
V
T
T
R
R
H
H
Y
Y
R
R
M
M
Y
Y
320
|
Q
Q
K
K
G
G
Q
Q
E
E
T
T
S
S
T
T
N
N
P
P
330
|
I
I
A
A
S
S
I
I
F
F
A
A
W
W
T
T
R
R
G
G
340
|
L
L
A
A
H
H
R
R
A
A
K
K
L
L
D
D
N
N
N
N
350
|
K
K
E
E
L
L
A
A
F
F
F
F
A
A
N
N
A
A
L
L
360
|
E
E
E
E
V
V
S
S
I
I
E
E
T
T
I
I
E
E
A
A
370
|
G
G
F
F
M
M
T
T
K
K
D
D
L
L
A
A
A
A
C
C
380
|
I
I
K
K
G
G
L
L
P
P
N
N
V
V
Q
Q
R
R
S
S
390
|
D
D
Y
Y
L
L
N
N
T
T
F
F
E
E
F
F
M
M
D
D
400
|
K
K
L
L
G
G
E
E
N
N
L
L
K
K
I
I
K
K
L
L
410
|
A
A
Q
Q
A
A
K
K
L
L
S
S
L
L
E
E
H
H
H
H
420
|
H
H
H
H
H
H
H
H
H
H
H
H
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | IDH2 cells | Ovary | Homo sapiens (Human) | CVCL_D3DY | |||||||||
| IDH1 cells | Ovary | Homo sapiens (Human) | CVCL_D3DY | ||||||||||
| In Vivo Model | Female athymic nu/nu mouse PDX model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
Promega assay | ||||||||||||
| Mechanism Description | The oncometabolite, 2-hydroxyglutarate, renders IDH1/2 mutant cancer cells deficient in homologous recombination and confers vulnerability to synthetic lethal targeting with PARP inhibitors. | ||||||||||||
| Disease Class: Brain glioma [ICD-11: 2A00.0] | [5] | ||||||||||||
| Sensitive Disease | Brain glioma [ICD-11: 2A00.0] | ||||||||||||
| Sensitive Drug | Talazoparib | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132H (c.395G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.65 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.88 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
M
S
S
K
K
K
K
I
I
S
S
G
G
G
G
S
S
10
|
V
V
V
V
E
E
M
M
Q
Q
G
G
D
D
E
E
M
M
T
T
20
|
R
R
I
I
I
I
W
W
E
E
L
L
I
I
K
K
E
E
K
K
30
|
L
L
I
I
F
F
P
P
Y
Y
V
V
E
E
L
L
D
D
L
L
40
|
H
H
S
S
Y
Y
D
D
L
L
G
G
I
I
E
E
N
N
R
R
50
|
D
D
A
A
T
T
N
N
D
D
Q
Q
V
V
T
T
K
K
D
D
60
|
A
A
A
A
E
E
A
A
I
I
K
K
K
K
H
H
N
N
V
V
70
|
G
G
V
V
K
K
C
C
A
A
T
T
I
I
T
T
P
P
D
D
80
|
E
E
K
K
R
R
V
V
E
E
E
E
F
F
K
K
L
L
K
K
90
|
Q
Q
M
M
W
W
K
K
S
S
P
P
N
N
G
G
T
T
I
I
100
|
R
R
N
N
I
I
L
L
G
G
G
G
T
T
V
V
F
F
R
R
110
|
E
E
A
A
I
I
I
I
C
C
K
K
N
N
I
I
P
P
R
R
120
|
L
L
V
V
S
S
G
G
W
W
V
V
K
K
P
P
I
I
I
I
130
|
I
I
G
G
R
H
H
H
A
A
Y
Y
G
G
D
D
Q
Q
Y
Y
140
|
R
R
A
A
T
T
D
D
F
F
V
V
V
V
P
P
G
G
P
P
150
|
G
G
K
K
V
V
E
E
I
I
T
T
Y
Y
T
T
P
P
S
S
160
|
D
D
G
G
T
T
Q
Q
K
K
V
V
T
T
Y
Y
L
L
V
V
170
|
H
H
N
N
F
F
E
E
E
E
G
G
G
G
G
G
V
V
A
A
180
|
M
M
G
G
M
M
Y
Y
N
N
Q
Q
D
D
K
K
S
S
I
I
190
|
E
E
D
D
F
F
A
A
H
H
S
S
S
S
F
F
Q
Q
M
M
200
|
A
A
L
L
S
S
K
K
G
G
W
W
P
P
L
L
Y
Y
L
L
210
|
S
S
T
T
K
K
N
N
T
T
I
I
L
L
K
K
K
K
Y
Y
220
|
D
D
G
G
R
R
F
F
K
K
D
D
I
I
F
F
Q
Q
E
E
230
|
I
I
Y
Y
D
D
K
K
Q
Q
Y
Y
K
K
S
S
Q
Q
F
F
240
|
E
E
A
A
Q
Q
K
K
I
I
W
W
Y
Y
E
E
H
H
R
R
250
|
L
L
I
I
D
D
D
D
M
M
V
V
A
A
Q
Q
A
A
M
M
260
|
K
K
S
S
E
E
G
G
G
G
F
F
I
I
W
W
A
A
C
C
270
|
K
K
N
N
Y
Y
D
D
G
G
D
D
V
V
Q
Q
S
S
D
D
280
|
S
S
V
V
A
A
Q
Q
G
G
Y
Y
G
G
S
S
L
L
G
G
290
|
M
M
M
M
T
T
S
S
V
V
L
L
V
V
C
C
P
P
D
D
300
|
G
G
K
K
T
T
V
V
E
E
A
A
E
E
A
A
A
A
H
H
310
|
G
G
T
T
V
V
T
T
R
R
H
H
Y
Y
R
R
M
M
Y
Y
320
|
Q
Q
K
K
G
G
Q
Q
E
E
T
T
S
S
T
T
N
N
P
P
330
|
I
I
A
A
S
S
I
I
F
F
A
A
W
W
T
T
R
R
G
G
340
|
L
L
A
A
H
H
R
R
A
A
K
K
L
L
D
D
N
N
N
N
350
|
K
K
E
E
L
L
A
A
F
F
F
F
A
A
N
N
A
A
L
L
360
|
E
E
E
E
V
V
S
S
I
I
E
E
T
T
I
I
E
E
A
A
370
|
G
G
F
F
M
M
T
T
K
K
D
D
L
L
A
A
A
A
C
C
380
|
I
I
K
K
G
G
L
L
P
P
N
N
V
V
Q
Q
R
R
S
S
390
|
D
D
Y
Y
L
L
N
N
T
T
F
F
E
E
F
F
M
M
D
D
400
|
K
K
L
L
G
G
E
E
N
N
L
L
K
K
I
I
K
K
L
L
410
|
A
A
Q
Q
A
A
K
K
L
L
S
S
L
L
E
E
H
H
H
H
420
|
H
H
H
H
H
H
H
H
H
H
H
H
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | IDH2 cells | Ovary | Homo sapiens (Human) | CVCL_D3DY | |||||||||
| IDH1 cells | Ovary | Homo sapiens (Human) | CVCL_D3DY | ||||||||||
| In Vivo Model | Female athymic nu/nu mouse PDX model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
Promega assay | ||||||||||||
| Mechanism Description | The oncometabolite, 2-hydroxyglutarate, renders IDH1/2 mutant cancer cells deficient in homologous recombination and confers vulnerability to synthetic lethal targeting with PARP inhibitors. | ||||||||||||
| Disease Class: Brain glioma [ICD-11: 2A00.0] | [5] | ||||||||||||
| Sensitive Disease | Brain glioma [ICD-11: 2A00.0] | ||||||||||||
| Sensitive Drug | Talazoparib | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132C (c.394C>T) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.93 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
M
-
S
-
K
-
K
-
I
-
S
-
G
-
G
-
S
10
|
-
V
-
V
-
E
-
M
-
Q
-
G
-
D
-
E
-
M
-
T
20
|
-
R
-
I
-
I
-
W
-
E
-
L
-
I
-
K
-
E
-
K
30
|
-
L
-
I
-
F
-
P
-
Y
-
V
-
E
-
L
-
D
-
L
40
|
-
H
-
S
-
Y
-
D
-
L
-
G
-
I
-
E
-
N
-
R
50
|
-
D
-
A
-
T
-
N
-
D
-
Q
-
V
-
T
-
K
-
D
60
|
-
A
-
A
-
E
-
A
-
I
-
K
-
K
-
H
-
N
-
V
70
|
-
G
-
V
-
K
-
C
-
A
-
T
-
I
-
T
-
P
-
D
80
|
-
E
-
K
-
R
-
V
-
E
-
E
-
F
-
K
-
L
-
K
90
|
-
Q
-
M
-
W
-
K
-
S
-
P
-
N
-
G
-
T
-
I
100
|
-
R
-
N
-
I
-
L
-
G
-
G
-
T
-
V
-
F
-
R
110
|
-
E
-
A
-
I
-
I
-
C
-
K
-
N
-
I
-
P
-
R
120
|
-
L
-
V
-
S
-
G
-
W
-
V
K
K
P
P
I
I
I
I
130
|
I
I
G
G
S
C
H
H
A
A
Y
Y
G
G
D
D
-
Q
-
Y
140
|
-
R
-
A
-
T
-
D
-
F
-
V
-
V
-
P
-
G
-
P
150
|
-
G
-
K
-
V
-
E
-
I
-
T
-
Y
-
T
-
P
-
S
160
|
-
D
-
G
-
T
-
Q
-
K
-
V
-
T
-
Y
-
L
-
V
170
|
-
H
-
N
-
F
-
E
-
E
-
G
-
G
-
G
-
V
-
A
180
|
-
M
-
G
-
M
-
Y
-
N
-
Q
-
D
-
K
-
S
-
I
190
|
-
E
-
D
-
F
-
A
-
H
-
S
-
S
-
F
-
Q
-
M
200
|
-
A
-
L
-
S
-
K
-
G
-
W
-
P
-
L
-
Y
-
L
210
|
-
S
-
T
-
K
-
N
-
T
-
I
-
L
-
K
-
K
-
Y
220
|
-
D
-
G
-
R
-
F
-
K
-
D
-
I
-
F
-
Q
-
E
230
|
-
I
-
Y
-
D
-
K
-
Q
-
Y
-
K
-
S
-
Q
-
F
240
|
-
E
-
A
-
Q
-
K
-
I
-
W
-
Y
-
E
-
H
-
R
250
|
-
L
-
I
-
D
-
D
-
M
-
V
-
A
-
Q
-
A
-
M
260
|
-
K
-
S
-
E
-
G
-
G
-
F
-
I
-
W
-
A
-
C
270
|
-
K
-
N
-
Y
-
D
-
G
-
D
-
V
-
Q
-
S
-
D
280
|
-
S
-
V
-
A
-
Q
-
G
-
Y
-
G
-
S
-
L
-
G
290
|
-
M
-
M
-
T
-
S
-
V
-
L
-
V
-
C
-
P
-
D
300
|
-
G
-
K
-
T
-
V
-
E
-
A
-
E
-
A
-
A
-
H
310
|
-
G
-
T
-
V
-
T
-
R
-
H
-
Y
-
R
-
M
-
Y
320
|
-
Q
-
K
-
G
-
Q
-
E
-
T
-
S
-
T
-
N
-
P
330
|
-
I
-
A
-
S
-
I
-
F
-
A
-
W
-
T
-
R
-
G
340
|
-
L
-
A
-
H
-
R
-
A
-
K
-
L
-
D
-
N
-
N
350
|
-
K
-
E
-
L
-
A
-
F
-
F
-
A
-
N
-
A
-
L
360
|
-
E
-
E
-
V
-
S
-
I
-
E
-
T
-
I
-
E
-
A
370
|
-
G
-
F
-
M
-
T
-
K
-
D
-
L
-
A
-
A
-
C
380
|
-
I
-
K
-
G
-
L
-
P
-
N
-
V
-
Q
-
R
-
S
390
|
-
D
-
Y
-
L
-
N
-
T
-
F
-
E
-
F
-
M
-
D
400
|
-
K
-
L
-
G
-
E
-
N
-
L
-
K
-
I
-
K
-
L
410
|
-
A
-
Q
-
A
-
K
-
L
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | IDH2 cells | Ovary | Homo sapiens (Human) | CVCL_D3DY | |||||||||
| IDH1 cells | Ovary | Homo sapiens (Human) | CVCL_D3DY | ||||||||||
| In Vivo Model | Female athymic nu/nu mouse PDX model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
Promega assay | ||||||||||||
| Mechanism Description | The oncometabolite, 2-hydroxyglutarate, renders IDH1/2 mutant cancer cells deficient in homologous recombination and confers vulnerability to synthetic lethal targeting with PARP inhibitors. | ||||||||||||
Clinical Trial Drug(s)
3 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [5] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | Niraparib | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132H (c.395G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.65 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.88 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
M
S
S
K
K
K
K
I
I
S
S
G
G
G
G
S
S
10
|
V
V
V
V
E
E
M
M
Q
Q
G
G
D
D
E
E
M
M
T
T
20
|
R
R
I
I
I
I
W
W
E
E
L
L
I
I
K
K
E
E
K
K
30
|
L
L
I
I
F
F
P
P
Y
Y
V
V
E
E
L
L
D
D
L
L
40
|
H
H
S
S
Y
Y
D
D
L
L
G
G
I
I
E
E
N
N
R
R
50
|
D
D
A
A
T
T
N
N
D
D
Q
Q
V
V
T
T
K
K
D
D
60
|
A
A
A
A
E
E
A
A
I
I
K
K
K
K
H
H
N
N
V
V
70
|
G
G
V
V
K
K
C
C
A
A
T
T
I
I
T
T
P
P
D
D
80
|
E
E
K
K
R
R
V
V
E
E
E
E
F
F
K
K
L
L
K
K
90
|
Q
Q
M
M
W
W
K
K
S
S
P
P
N
N
G
G
T
T
I
I
100
|
R
R
N
N
I
I
L
L
G
G
G
G
T
T
V
V
F
F
R
R
110
|
E
E
A
A
I
I
I
I
C
C
K
K
N
N
I
I
P
P
R
R
120
|
L
L
V
V
S
S
G
G
W
W
V
V
K
K
P
P
I
I
I
I
130
|
I
I
G
G
R
H
H
H
A
A
Y
Y
G
G
D
D
Q
Q
Y
Y
140
|
R
R
A
A
T
T
D
D
F
F
V
V
V
V
P
P
G
G
P
P
150
|
G
G
K
K
V
V
E
E
I
I
T
T
Y
Y
T
T
P
P
S
S
160
|
D
D
G
G
T
T
Q
Q
K
K
V
V
T
T
Y
Y
L
L
V
V
170
|
H
H
N
N
F
F
E
E
E
E
G
G
G
G
G
G
V
V
A
A
180
|
M
M
G
G
M
M
Y
Y
N
N
Q
Q
D
D
K
K
S
S
I
I
190
|
E
E
D
D
F
F
A
A
H
H
S
S
S
S
F
F
Q
Q
M
M
200
|
A
A
L
L
S
S
K
K
G
G
W
W
P
P
L
L
Y
Y
L
L
210
|
S
S
T
T
K
K
N
N
T
T
I
I
L
L
K
K
K
K
Y
Y
220
|
D
D
G
G
R
R
F
F
K
K
D
D
I
I
F
F
Q
Q
E
E
230
|
I
I
Y
Y
D
D
K
K
Q
Q
Y
Y
K
K
S
S
Q
Q
F
F
240
|
E
E
A
A
Q
Q
K
K
I
I
W
W
Y
Y
E
E
H
H
R
R
250
|
L
L
I
I
D
D
D
D
M
M
V
V
A
A
Q
Q
A
A
M
M
260
|
K
K
S
S
E
E
G
G
G
G
F
F
I
I
W
W
A
A
C
C
270
|
K
K
N
N
Y
Y
D
D
G
G
D
D
V
V
Q
Q
S
S
D
D
280
|
S
S
V
V
A
A
Q
Q
G
G
Y
Y
G
G
S
S
L
L
G
G
290
|
M
M
M
M
T
T
S
S
V
V
L
L
V
V
C
C
P
P
D
D
300
|
G
G
K
K
T
T
V
V
E
E
A
A
E
E
A
A
A
A
H
H
310
|
G
G
T
T
V
V
T
T
R
R
H
H
Y
Y
R
R
M
M
Y
Y
320
|
Q
Q
K
K
G
G
Q
Q
E
E
T
T
S
S
T
T
N
N
P
P
330
|
I
I
A
A
S
S
I
I
F
F
A
A
W
W
T
T
R
R
G
G
340
|
L
L
A
A
H
H
R
R
A
A
K
K
L
L
D
D
N
N
N
N
350
|
K
K
E
E
L
L
A
A
F
F
F
F
A
A
N
N
A
A
L
L
360
|
E
E
E
E
V
V
S
S
I
I
E
E
T
T
I
I
E
E
A
A
370
|
G
G
F
F
M
M
T
T
K
K
D
D
L
L
A
A
A
A
C
C
380
|
I
I
K
K
G
G
L
L
P
P
N
N
V
V
Q
Q
R
R
S
S
390
|
D
D
Y
Y
L
L
N
N
T
T
F
F
E
E
F
F
M
M
D
D
400
|
K
K
L
L
G
G
E
E
N
N
L
L
K
K
I
I
K
K
L
L
410
|
A
A
Q
Q
A
A
K
K
L
L
S
S
L
L
E
E
H
H
H
H
420
|
H
H
H
H
H
H
H
H
H
H
H
H
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | IDH2 cells | Ovary | Homo sapiens (Human) | CVCL_D3DY | |||||||||
| IDH1 cells | Ovary | Homo sapiens (Human) | CVCL_D3DY | ||||||||||
| In Vivo Model | Female athymic nu/nu mouse PDX model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
Promega assay | ||||||||||||
| Mechanism Description | The oncometabolite, 2-hydroxyglutarate, renders IDH1/2 mutant cancer cells deficient in homologous recombination and confers vulnerability to synthetic lethal targeting with PARP inhibitors. | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [6] | ||||||||||||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Sensitive Drug | BAY1436032 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132H (c.395G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.65 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.88 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
M
S
S
K
K
K
K
I
I
S
S
G
G
G
G
S
S
10
|
V
V
V
V
E
E
M
M
Q
Q
G
G
D
D
E
E
M
M
T
T
20
|
R
R
I
I
I
I
W
W
E
E
L
L
I
I
K
K
E
E
K
K
30
|
L
L
I
I
F
F
P
P
Y
Y
V
V
E
E
L
L
D
D
L
L
40
|
H
H
S
S
Y
Y
D
D
L
L
G
G
I
I
E
E
N
N
R
R
50
|
D
D
A
A
T
T
N
N
D
D
Q
Q
V
V
T
T
K
K
D
D
60
|
A
A
A
A
E
E
A
A
I
I
K
K
K
K
H
H
N
N
V
V
70
|
G
G
V
V
K
K
C
C
A
A
T
T
I
I
T
T
P
P
D
D
80
|
E
E
K
K
R
R
V
V
E
E
E
E
F
F
K
K
L
L
K
K
90
|
Q
Q
M
M
W
W
K
K
S
S
P
P
N
N
G
G
T
T
I
I
100
|
R
R
N
N
I
I
L
L
G
G
G
G
T
T
V
V
F
F
R
R
110
|
E
E
A
A
I
I
I
I
C
C
K
K
N
N
I
I
P
P
R
R
120
|
L
L
V
V
S
S
G
G
W
W
V
V
K
K
P
P
I
I
I
I
130
|
I
I
G
G
R
H
H
H
A
A
Y
Y
G
G
D
D
Q
Q
Y
Y
140
|
R
R
A
A
T
T
D
D
F
F
V
V
V
V
P
P
G
G
P
P
150
|
G
G
K
K
V
V
E
E
I
I
T
T
Y
Y
T
T
P
P
S
S
160
|
D
D
G
G
T
T
Q
Q
K
K
V
V
T
T
Y
Y
L
L
V
V
170
|
H
H
N
N
F
F
E
E
E
E
G
G
G
G
G
G
V
V
A
A
180
|
M
M
G
G
M
M
Y
Y
N
N
Q
Q
D
D
K
K
S
S
I
I
190
|
E
E
D
D
F
F
A
A
H
H
S
S
S
S
F
F
Q
Q
M
M
200
|
A
A
L
L
S
S
K
K
G
G
W
W
P
P
L
L
Y
Y
L
L
210
|
S
S
T
T
K
K
N
N
T
T
I
I
L
L
K
K
K
K
Y
Y
220
|
D
D
G
G
R
R
F
F
K
K
D
D
I
I
F
F
Q
Q
E
E
230
|
I
I
Y
Y
D
D
K
K
Q
Q
Y
Y
K
K
S
S
Q
Q
F
F
240
|
E
E
A
A
Q
Q
K
K
I
I
W
W
Y
Y
E
E
H
H
R
R
250
|
L
L
I
I
D
D
D
D
M
M
V
V
A
A
Q
Q
A
A
M
M
260
|
K
K
S
S
E
E
G
G
G
G
F
F
I
I
W
W
A
A
C
C
270
|
K
K
N
N
Y
Y
D
D
G
G
D
D
V
V
Q
Q
S
S
D
D
280
|
S
S
V
V
A
A
Q
Q
G
G
Y
Y
G
G
S
S
L
L
G
G
290
|
M
M
M
M
T
T
S
S
V
V
L
L
V
V
C
C
P
P
D
D
300
|
G
G
K
K
T
T
V
V
E
E
A
A
E
E
A
A
A
A
H
H
310
|
G
G
T
T
V
V
T
T
R
R
H
H
Y
Y
R
R
M
M
Y
Y
320
|
Q
Q
K
K
G
G
Q
Q
E
E
T
T
S
S
T
T
N
N
P
P
330
|
I
I
A
A
S
S
I
I
F
F
A
A
W
W
T
T
R
R
G
G
340
|
L
L
A
A
H
H
R
R
A
A
K
K
L
L
D
D
N
N
N
N
350
|
K
K
E
E
L
L
A
A
F
F
F
F
A
A
N
N
A
A
L
L
360
|
E
E
E
E
V
V
S
S
I
I
E
E
T
T
I
I
E
E
A
A
370
|
G
G
F
F
M
M
T
T
K
K
D
D
L
L
A
A
A
A
C
C
380
|
I
I
K
K
G
G
L
L
P
P
N
N
V
V
Q
Q
R
R
S
S
390
|
D
D
Y
Y
L
L
N
N
T
T
F
F
E
E
F
F
M
M
D
D
400
|
K
K
L
L
G
G
E
E
N
N
L
L
K
K
I
I
K
K
L
L
410
|
A
A
Q
Q
A
A
K
K
L
L
S
S
L
L
E
E
H
H
H
H
420
|
H
H
H
H
H
H
H
H
H
H
H
H
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | HoxA9-IDH2R140Q cells | N.A. | N.A. | N.A. | |||||||||
| HoxA9-IDH2172K cells | N.A. | N.A. | N.A. | ||||||||||
| HoxA9-IDH1R132H cells | N.A. | N.A. | N.A. | ||||||||||
| HoxA9-IDH1R132C cells | N.A. | N.A. | N.A. | ||||||||||
| In Vivo Model | Mouse PDX model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
FACS assay | ||||||||||||
| Mechanism Description | BAY1436032 inhibits proliferation and induces differentiation in primary human AML cells. BAY1436032 clears AML blasts in vivo and prolongs survival in PDX models of IDH1 mutant AML. BAY1436032 induces myeloid differentiation in IDH1 mutant AML PDX models in vivo and depletes leukemic stem cells by induction of myeloid differentiation and inhibition of cell cycle progression. | ||||||||||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [6] | ||||||||||||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Sensitive Drug | BAY1436032 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132C (c.394C>T) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.93 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
M
-
S
-
K
-
K
-
I
-
S
-
G
-
G
-
S
10
|
-
V
-
V
-
E
-
M
-
Q
-
G
-
D
-
E
-
M
-
T
20
|
-
R
-
I
-
I
-
W
-
E
-
L
-
I
-
K
-
E
-
K
30
|
-
L
-
I
-
F
-
P
-
Y
-
V
-
E
-
L
-
D
-
L
40
|
-
H
-
S
-
Y
-
D
-
L
-
G
-
I
-
E
-
N
-
R
50
|
-
D
-
A
-
T
-
N
-
D
-
Q
-
V
-
T
-
K
-
D
60
|
-
A
-
A
-
E
-
A
-
I
-
K
-
K
-
H
-
N
-
V
70
|
-
G
-
V
-
K
-
C
-
A
-
T
-
I
-
T
-
P
-
D
80
|
-
E
-
K
-
R
-
V
-
E
-
E
-
F
-
K
-
L
-
K
90
|
-
Q
-
M
-
W
-
K
-
S
-
P
-
N
-
G
-
T
-
I
100
|
-
R
-
N
-
I
-
L
-
G
-
G
-
T
-
V
-
F
-
R
110
|
-
E
-
A
-
I
-
I
-
C
-
K
-
N
-
I
-
P
-
R
120
|
-
L
-
V
-
S
-
G
-
W
-
V
K
K
P
P
I
I
I
I
130
|
I
I
G
G
S
C
H
H
A
A
Y
Y
G
G
D
D
-
Q
-
Y
140
|
-
R
-
A
-
T
-
D
-
F
-
V
-
V
-
P
-
G
-
P
150
|
-
G
-
K
-
V
-
E
-
I
-
T
-
Y
-
T
-
P
-
S
160
|
-
D
-
G
-
T
-
Q
-
K
-
V
-
T
-
Y
-
L
-
V
170
|
-
H
-
N
-
F
-
E
-
E
-
G
-
G
-
G
-
V
-
A
180
|
-
M
-
G
-
M
-
Y
-
N
-
Q
-
D
-
K
-
S
-
I
190
|
-
E
-
D
-
F
-
A
-
H
-
S
-
S
-
F
-
Q
-
M
200
|
-
A
-
L
-
S
-
K
-
G
-
W
-
P
-
L
-
Y
-
L
210
|
-
S
-
T
-
K
-
N
-
T
-
I
-
L
-
K
-
K
-
Y
220
|
-
D
-
G
-
R
-
F
-
K
-
D
-
I
-
F
-
Q
-
E
230
|
-
I
-
Y
-
D
-
K
-
Q
-
Y
-
K
-
S
-
Q
-
F
240
|
-
E
-
A
-
Q
-
K
-
I
-
W
-
Y
-
E
-
H
-
R
250
|
-
L
-
I
-
D
-
D
-
M
-
V
-
A
-
Q
-
A
-
M
260
|
-
K
-
S
-
E
-
G
-
G
-
F
-
I
-
W
-
A
-
C
270
|
-
K
-
N
-
Y
-
D
-
G
-
D
-
V
-
Q
-
S
-
D
280
|
-
S
-
V
-
A
-
Q
-
G
-
Y
-
G
-
S
-
L
-
G
290
|
-
M
-
M
-
T
-
S
-
V
-
L
-
V
-
C
-
P
-
D
300
|
-
G
-
K
-
T
-
V
-
E
-
A
-
E
-
A
-
A
-
H
310
|
-
G
-
T
-
V
-
T
-
R
-
H
-
Y
-
R
-
M
-
Y
320
|
-
Q
-
K
-
G
-
Q
-
E
-
T
-
S
-
T
-
N
-
P
330
|
-
I
-
A
-
S
-
I
-
F
-
A
-
W
-
T
-
R
-
G
340
|
-
L
-
A
-
H
-
R
-
A
-
K
-
L
-
D
-
N
-
N
350
|
-
K
-
E
-
L
-
A
-
F
-
F
-
A
-
N
-
A
-
L
360
|
-
E
-
E
-
V
-
S
-
I
-
E
-
T
-
I
-
E
-
A
370
|
-
G
-
F
-
M
-
T
-
K
-
D
-
L
-
A
-
A
-
C
380
|
-
I
-
K
-
G
-
L
-
P
-
N
-
V
-
Q
-
R
-
S
390
|
-
D
-
Y
-
L
-
N
-
T
-
F
-
E
-
F
-
M
-
D
400
|
-
K
-
L
-
G
-
E
-
N
-
L
-
K
-
I
-
K
-
L
410
|
-
A
-
Q
-
A
-
K
-
L
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | HoxA9-IDH2R140Q cells | N.A. | N.A. | N.A. | |||||||||
| HoxA9-IDH2172K cells | N.A. | N.A. | N.A. | ||||||||||
| HoxA9-IDH1R132H cells | N.A. | N.A. | N.A. | ||||||||||
| HoxA9-IDH1R132C cells | N.A. | N.A. | N.A. | ||||||||||
| In Vivo Model | Mouse PDX model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
FACS assay | ||||||||||||
| Mechanism Description | BAY1436032 inhibits proliferation and induces differentiation in primary human AML cells. BAY1436032 clears AML blasts in vivo and prolongs survival in PDX models of IDH1 mutant AML. BAY1436032 induces myeloid differentiation in IDH1 mutant AML PDX models in vivo and depletes leukemic stem cells by induction of myeloid differentiation and inhibition of cell cycle progression. | ||||||||||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [6] | ||||||||||||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Sensitive Drug | BAY1436032 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132S (c.394C>A) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | HoxA9-IDH2R140Q cells | N.A. | N.A. | N.A. | |||||||||
| HoxA9-IDH2172K cells | N.A. | N.A. | N.A. | ||||||||||
| HoxA9-IDH1R132H cells | N.A. | N.A. | N.A. | ||||||||||
| HoxA9-IDH1R132C cells | N.A. | N.A. | N.A. | ||||||||||
| In Vivo Model | Mouse PDX model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
FACS assay | ||||||||||||
| Mechanism Description | BAY1436032 inhibits proliferation and induces differentiation in primary human AML cells. BAY1436032 clears AML blasts in vivo and prolongs survival in PDX models of IDH1 mutant AML. BAY1436032 induces myeloid differentiation in IDH1 mutant AML PDX models in vivo and depletes leukemic stem cells by induction of myeloid differentiation and inhibition of cell cycle progression. | ||||||||||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [6] | ||||||||||||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Sensitive Drug | BAY1436032 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132G (c.394C>G) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | HoxA9-IDH2R140Q cells | N.A. | N.A. | N.A. | |||||||||
| HoxA9-IDH2172K cells | N.A. | N.A. | N.A. | ||||||||||
| HoxA9-IDH1R132H cells | N.A. | N.A. | N.A. | ||||||||||
| HoxA9-IDH1R132C cells | N.A. | N.A. | N.A. | ||||||||||
| In Vivo Model | Mouse PDX model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
FACS assay | ||||||||||||
| Mechanism Description | BAY1436032 inhibits proliferation and induces differentiation in primary human AML cells. BAY1436032 clears AML blasts in vivo and prolongs survival in PDX models of IDH1 mutant AML. BAY1436032 induces myeloid differentiation in IDH1 mutant AML PDX models in vivo and depletes leukemic stem cells by induction of myeloid differentiation and inhibition of cell cycle progression. | ||||||||||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [6] | ||||||||||||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Sensitive Drug | BAY1436032 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132L (c.395G>T) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | HoxA9-IDH2R140Q cells | N.A. | N.A. | N.A. | |||||||||
| HoxA9-IDH2172K cells | N.A. | N.A. | N.A. | ||||||||||
| HoxA9-IDH1R132H cells | N.A. | N.A. | N.A. | ||||||||||
| HoxA9-IDH1R132C cells | N.A. | N.A. | N.A. | ||||||||||
| In Vivo Model | Mouse PDX model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
FACS assay | ||||||||||||
| Mechanism Description | BAY1436032 inhibits proliferation and induces differentiation in primary human AML cells. BAY1436032 clears AML blasts in vivo and prolongs survival in PDX models of IDH1 mutant AML. BAY1436032 induces myeloid differentiation in IDH1 mutant AML PDX models in vivo and depletes leukemic stem cells by induction of myeloid differentiation and inhibition of cell cycle progression. | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [5] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | Berzosertib | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132H (c.395G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.65 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.88 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
M
S
S
K
K
K
K
I
I
S
S
G
G
G
G
S
S
10
|
V
V
V
V
E
E
M
M
Q
Q
G
G
D
D
E
E
M
M
T
T
20
|
R
R
I
I
I
I
W
W
E
E
L
L
I
I
K
K
E
E
K
K
30
|
L
L
I
I
F
F
P
P
Y
Y
V
V
E
E
L
L
D
D
L
L
40
|
H
H
S
S
Y
Y
D
D
L
L
G
G
I
I
E
E
N
N
R
R
50
|
D
D
A
A
T
T
N
N
D
D
Q
Q
V
V
T
T
K
K
D
D
60
|
A
A
A
A
E
E
A
A
I
I
K
K
K
K
H
H
N
N
V
V
70
|
G
G
V
V
K
K
C
C
A
A
T
T
I
I
T
T
P
P
D
D
80
|
E
E
K
K
R
R
V
V
E
E
E
E
F
F
K
K
L
L
K
K
90
|
Q
Q
M
M
W
W
K
K
S
S
P
P
N
N
G
G
T
T
I
I
100
|
R
R
N
N
I
I
L
L
G
G
G
G
T
T
V
V
F
F
R
R
110
|
E
E
A
A
I
I
I
I
C
C
K
K
N
N
I
I
P
P
R
R
120
|
L
L
V
V
S
S
G
G
W
W
V
V
K
K
P
P
I
I
I
I
130
|
I
I
G
G
R
H
H
H
A
A
Y
Y
G
G
D
D
Q
Q
Y
Y
140
|
R
R
A
A
T
T
D
D
F
F
V
V
V
V
P
P
G
G
P
P
150
|
G
G
K
K
V
V
E
E
I
I
T
T
Y
Y
T
T
P
P
S
S
160
|
D
D
G
G
T
T
Q
Q
K
K
V
V
T
T
Y
Y
L
L
V
V
170
|
H
H
N
N
F
F
E
E
E
E
G
G
G
G
G
G
V
V
A
A
180
|
M
M
G
G
M
M
Y
Y
N
N
Q
Q
D
D
K
K
S
S
I
I
190
|
E
E
D
D
F
F
A
A
H
H
S
S
S
S
F
F
Q
Q
M
M
200
|
A
A
L
L
S
S
K
K
G
G
W
W
P
P
L
L
Y
Y
L
L
210
|
S
S
T
T
K
K
N
N
T
T
I
I
L
L
K
K
K
K
Y
Y
220
|
D
D
G
G
R
R
F
F
K
K
D
D
I
I
F
F
Q
Q
E
E
230
|
I
I
Y
Y
D
D
K
K
Q
Q
Y
Y
K
K
S
S
Q
Q
F
F
240
|
E
E
A
A
Q
Q
K
K
I
I
W
W
Y
Y
E
E
H
H
R
R
250
|
L
L
I
I
D
D
D
D
M
M
V
V
A
A
Q
Q
A
A
M
M
260
|
K
K
S
S
E
E
G
G
G
G
F
F
I
I
W
W
A
A
C
C
270
|
K
K
N
N
Y
Y
D
D
G
G
D
D
V
V
Q
Q
S
S
D
D
280
|
S
S
V
V
A
A
Q
Q
G
G
Y
Y
G
G
S
S
L
L
G
G
290
|
M
M
M
M
T
T
S
S
V
V
L
L
V
V
C
C
P
P
D
D
300
|
G
G
K
K
T
T
V
V
E
E
A
A
E
E
A
A
A
A
H
H
310
|
G
G
T
T
V
V
T
T
R
R
H
H
Y
Y
R
R
M
M
Y
Y
320
|
Q
Q
K
K
G
G
Q
Q
E
E
T
T
S
S
T
T
N
N
P
P
330
|
I
I
A
A
S
S
I
I
F
F
A
A
W
W
T
T
R
R
G
G
340
|
L
L
A
A
H
H
R
R
A
A
K
K
L
L
D
D
N
N
N
N
350
|
K
K
E
E
L
L
A
A
F
F
F
F
A
A
N
N
A
A
L
L
360
|
E
E
E
E
V
V
S
S
I
I
E
E
T
T
I
I
E
E
A
A
370
|
G
G
F
F
M
M
T
T
K
K
D
D
L
L
A
A
A
A
C
C
380
|
I
I
K
K
G
G
L
L
P
P
N
N
V
V
Q
Q
R
R
S
S
390
|
D
D
Y
Y
L
L
N
N
T
T
F
F
E
E
F
F
M
M
D
D
400
|
K
K
L
L
G
G
E
E
N
N
L
L
K
K
I
I
K
K
L
L
410
|
A
A
Q
Q
A
A
K
K
L
L
S
S
L
L
E
E
H
H
H
H
420
|
H
H
H
H
H
H
H
H
H
H
H
H
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | IDH2 cells | Ovary | Homo sapiens (Human) | CVCL_D3DY | |||||||||
| IDH1 cells | Ovary | Homo sapiens (Human) | CVCL_D3DY | ||||||||||
| In Vivo Model | Female athymic nu/nu mouse PDX model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
Promega assay | ||||||||||||
| Mechanism Description | The oncometabolite, 2-hydroxyglutarate, renders IDH1/2 mutant cancer cells deficient in homologous recombination and confers vulnerability to synthetic lethal targeting with PARP inhibitors. | ||||||||||||
Preclinical Drug(s)
9 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: FGFR-tacc positive glioblastoma [ICD-11: 2A00.01] | [7] | ||||||||||||
| Sensitive Disease | FGFR-tacc positive glioblastoma [ICD-11: 2A00.01] | ||||||||||||
| Sensitive Drug | AGI-5198 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132H (c.395G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.65 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.88 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
M
S
S
K
K
K
K
I
I
S
S
G
G
G
G
S
S
10
|
V
V
V
V
E
E
M
M
Q
Q
G
G
D
D
E
E
M
M
T
T
20
|
R
R
I
I
I
I
W
W
E
E
L
L
I
I
K
K
E
E
K
K
30
|
L
L
I
I
F
F
P
P
Y
Y
V
V
E
E
L
L
D
D
L
L
40
|
H
H
S
S
Y
Y
D
D
L
L
G
G
I
I
E
E
N
N
R
R
50
|
D
D
A
A
T
T
N
N
D
D
Q
Q
V
V
T
T
K
K
D
D
60
|
A
A
A
A
E
E
A
A
I
I
K
K
K
K
H
H
N
N
V
V
70
|
G
G
V
V
K
K
C
C
A
A
T
T
I
I
T
T
P
P
D
D
80
|
E
E
K
K
R
R
V
V
E
E
E
E
F
F
K
K
L
L
K
K
90
|
Q
Q
M
M
W
W
K
K
S
S
P
P
N
N
G
G
T
T
I
I
100
|
R
R
N
N
I
I
L
L
G
G
G
G
T
T
V
V
F
F
R
R
110
|
E
E
A
A
I
I
I
I
C
C
K
K
N
N
I
I
P
P
R
R
120
|
L
L
V
V
S
S
G
G
W
W
V
V
K
K
P
P
I
I
I
I
130
|
I
I
G
G
R
H
H
H
A
A
Y
Y
G
G
D
D
Q
Q
Y
Y
140
|
R
R
A
A
T
T
D
D
F
F
V
V
V
V
P
P
G
G
P
P
150
|
G
G
K
K
V
V
E
E
I
I
T
T
Y
Y
T
T
P
P
S
S
160
|
D
D
G
G
T
T
Q
Q
K
K
V
V
T
T
Y
Y
L
L
V
V
170
|
H
H
N
N
F
F
E
E
E
E
G
G
G
G
G
G
V
V
A
A
180
|
M
M
G
G
M
M
Y
Y
N
N
Q
Q
D
D
K
K
S
S
I
I
190
|
E
E
D
D
F
F
A
A
H
H
S
S
S
S
F
F
Q
Q
M
M
200
|
A
A
L
L
S
S
K
K
G
G
W
W
P
P
L
L
Y
Y
L
L
210
|
S
S
T
T
K
K
N
N
T
T
I
I
L
L
K
K
K
K
Y
Y
220
|
D
D
G
G
R
R
F
F
K
K
D
D
I
I
F
F
Q
Q
E
E
230
|
I
I
Y
Y
D
D
K
K
Q
Q
Y
Y
K
K
S
S
Q
Q
F
F
240
|
E
E
A
A
Q
Q
K
K
I
I
W
W
Y
Y
E
E
H
H
R
R
250
|
L
L
I
I
D
D
D
D
M
M
V
V
A
A
Q
Q
A
A
M
M
260
|
K
K
S
S
E
E
G
G
G
G
F
F
I
I
W
W
A
A
C
C
270
|
K
K
N
N
Y
Y
D
D
G
G
D
D
V
V
Q
Q
S
S
D
D
280
|
S
S
V
V
A
A
Q
Q
G
G
Y
Y
G
G
S
S
L
L
G
G
290
|
M
M
M
M
T
T
S
S
V
V
L
L
V
V
C
C
P
P
D
D
300
|
G
G
K
K
T
T
V
V
E
E
A
A
E
E
A
A
A
A
H
H
310
|
G
G
T
T
V
V
T
T
R
R
H
H
Y
Y
R
R
M
M
Y
Y
320
|
Q
Q
K
K
G
G
Q
Q
E
E
T
T
S
S
T
T
N
N
P
P
330
|
I
I
A
A
S
S
I
I
F
F
A
A
W
W
T
T
R
R
G
G
340
|
L
L
A
A
H
H
R
R
A
A
K
K
L
L
D
D
N
N
N
N
350
|
K
K
E
E
L
L
A
A
F
F
F
F
A
A
N
N
A
A
L
L
360
|
E
E
E
E
V
V
S
S
I
I
E
E
T
T
I
I
E
E
A
A
370
|
G
G
F
F
M
M
T
T
K
K
D
D
L
L
A
A
A
A
C
C
380
|
I
I
K
K
G
G
L
L
P
P
N
N
V
V
Q
Q
R
R
S
S
390
|
D
D
Y
Y
L
L
N
N
T
T
F
F
E
E
F
F
M
M
D
D
400
|
K
K
L
L
G
G
E
E
N
N
L
L
K
K
I
I
K
K
L
L
410
|
A
A
Q
Q
A
A
K
K
L
L
S
S
L
L
E
E
H
H
H
H
420
|
H
H
H
H
H
H
H
H
H
H
H
H
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | TS676 cells | Brain | Homo sapiens (Human) | CVCL_A5HX | |||||||||
| TS603 cells | Brain | Homo sapiens (Human) | CVCL_A5HW | ||||||||||
| TS516 cells | Brain | Homo sapiens (Human) | CVCL_A5HY | ||||||||||
| In Vivo Model | SCID mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
Soft agar assay | ||||||||||||
| Mechanism Description | The missense mutation p.R132H (c.395G>A) in gene IDH1 cause the sensitivity of AGI-5198 by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Brain glioma [ICD-11: 2A00.0] | [7] | ||||||||||||
| Sensitive Disease | Brain glioma [ICD-11: 2A00.0] | ||||||||||||
| Sensitive Drug | AGI-5198 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132H (c.395G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.65 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.88 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
M
S
S
K
K
K
K
I
I
S
S
G
G
G
G
S
S
10
|
V
V
V
V
E
E
M
M
Q
Q
G
G
D
D
E
E
M
M
T
T
20
|
R
R
I
I
I
I
W
W
E
E
L
L
I
I
K
K
E
E
K
K
30
|
L
L
I
I
F
F
P
P
Y
Y
V
V
E
E
L
L
D
D
L
L
40
|
H
H
S
S
Y
Y
D
D
L
L
G
G
I
I
E
E
N
N
R
R
50
|
D
D
A
A
T
T
N
N
D
D
Q
Q
V
V
T
T
K
K
D
D
60
|
A
A
A
A
E
E
A
A
I
I
K
K
K
K
H
H
N
N
V
V
70
|
G
G
V
V
K
K
C
C
A
A
T
T
I
I
T
T
P
P
D
D
80
|
E
E
K
K
R
R
V
V
E
E
E
E
F
F
K
K
L
L
K
K
90
|
Q
Q
M
M
W
W
K
K
S
S
P
P
N
N
G
G
T
T
I
I
100
|
R
R
N
N
I
I
L
L
G
G
G
G
T
T
V
V
F
F
R
R
110
|
E
E
A
A
I
I
I
I
C
C
K
K
N
N
I
I
P
P
R
R
120
|
L
L
V
V
S
S
G
G
W
W
V
V
K
K
P
P
I
I
I
I
130
|
I
I
G
G
R
H
H
H
A
A
Y
Y
G
G
D
D
Q
Q
Y
Y
140
|
R
R
A
A
T
T
D
D
F
F
V
V
V
V
P
P
G
G
P
P
150
|
G
G
K
K
V
V
E
E
I
I
T
T
Y
Y
T
T
P
P
S
S
160
|
D
D
G
G
T
T
Q
Q
K
K
V
V
T
T
Y
Y
L
L
V
V
170
|
H
H
N
N
F
F
E
E
E
E
G
G
G
G
G
G
V
V
A
A
180
|
M
M
G
G
M
M
Y
Y
N
N
Q
Q
D
D
K
K
S
S
I
I
190
|
E
E
D
D
F
F
A
A
H
H
S
S
S
S
F
F
Q
Q
M
M
200
|
A
A
L
L
S
S
K
K
G
G
W
W
P
P
L
L
Y
Y
L
L
210
|
S
S
T
T
K
K
N
N
T
T
I
I
L
L
K
K
K
K
Y
Y
220
|
D
D
G
G
R
R
F
F
K
K
D
D
I
I
F
F
Q
Q
E
E
230
|
I
I
Y
Y
D
D
K
K
Q
Q
Y
Y
K
K
S
S
Q
Q
F
F
240
|
E
E
A
A
Q
Q
K
K
I
I
W
W
Y
Y
E
E
H
H
R
R
250
|
L
L
I
I
D
D
D
D
M
M
V
V
A
A
Q
Q
A
A
M
M
260
|
K
K
S
S
E
E
G
G
G
G
F
F
I
I
W
W
A
A
C
C
270
|
K
K
N
N
Y
Y
D
D
G
G
D
D
V
V
Q
Q
S
S
D
D
280
|
S
S
V
V
A
A
Q
Q
G
G
Y
Y
G
G
S
S
L
L
G
G
290
|
M
M
M
M
T
T
S
S
V
V
L
L
V
V
C
C
P
P
D
D
300
|
G
G
K
K
T
T
V
V
E
E
A
A
E
E
A
A
A
A
H
H
310
|
G
G
T
T
V
V
T
T
R
R
H
H
Y
Y
R
R
M
M
Y
Y
320
|
Q
Q
K
K
G
G
Q
Q
E
E
T
T
S
S
T
T
N
N
P
P
330
|
I
I
A
A
S
S
I
I
F
F
A
A
W
W
T
T
R
R
G
G
340
|
L
L
A
A
H
H
R
R
A
A
K
K
L
L
D
D
N
N
N
N
350
|
K
K
E
E
L
L
A
A
F
F
F
F
A
A
N
N
A
A
L
L
360
|
E
E
E
E
V
V
S
S
I
I
E
E
T
T
I
I
E
E
A
A
370
|
G
G
F
F
M
M
T
T
K
K
D
D
L
L
A
A
A
A
C
C
380
|
I
I
K
K
G
G
L
L
P
P
N
N
V
V
Q
Q
R
R
S
S
390
|
D
D
Y
Y
L
L
N
N
T
T
F
F
E
E
F
F
M
M
D
D
400
|
K
K
L
L
G
G
E
E
N
N
L
L
K
K
I
I
K
K
L
L
410
|
A
A
Q
Q
A
A
K
K
L
L
S
S
L
L
E
E
H
H
H
H
420
|
H
H
H
H
H
H
H
H
H
H
H
H
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | TS676 cells | Brain | Homo sapiens (Human) | CVCL_A5HX | |||||||||
| TS603 cells | Brain | Homo sapiens (Human) | CVCL_A5HW | ||||||||||
| TS516 cells | Brain | Homo sapiens (Human) | CVCL_A5HY | ||||||||||
| In Vivo Model | SCID mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
Soft agar assay | ||||||||||||
| Mechanism Description | The missense mutation p.R132H (c.395G>A) in gene IDH1 cause the sensitivity of AGI-5198 by aberration of the drug's therapeutic target | ||||||||||||
| Disease Class: Brain glioma [ICD-11: 2A00.0] | [7] | ||||||||||||
| Sensitive Disease | Brain glioma [ICD-11: 2A00.0] | ||||||||||||
| Sensitive Drug | AGI-5198 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132C (c.394C>T) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.93 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
M
-
S
-
K
-
K
-
I
-
S
-
G
-
G
-
S
10
|
-
V
-
V
-
E
-
M
-
Q
-
G
-
D
-
E
-
M
-
T
20
|
-
R
-
I
-
I
-
W
-
E
-
L
-
I
-
K
-
E
-
K
30
|
-
L
-
I
-
F
-
P
-
Y
-
V
-
E
-
L
-
D
-
L
40
|
-
H
-
S
-
Y
-
D
-
L
-
G
-
I
-
E
-
N
-
R
50
|
-
D
-
A
-
T
-
N
-
D
-
Q
-
V
-
T
-
K
-
D
60
|
-
A
-
A
-
E
-
A
-
I
-
K
-
K
-
H
-
N
-
V
70
|
-
G
-
V
-
K
-
C
-
A
-
T
-
I
-
T
-
P
-
D
80
|
-
E
-
K
-
R
-
V
-
E
-
E
-
F
-
K
-
L
-
K
90
|
-
Q
-
M
-
W
-
K
-
S
-
P
-
N
-
G
-
T
-
I
100
|
-
R
-
N
-
I
-
L
-
G
-
G
-
T
-
V
-
F
-
R
110
|
-
E
-
A
-
I
-
I
-
C
-
K
-
N
-
I
-
P
-
R
120
|
-
L
-
V
-
S
-
G
-
W
-
V
K
K
P
P
I
I
I
I
130
|
I
I
G
G
S
C
H
H
A
A
Y
Y
G
G
D
D
-
Q
-
Y
140
|
-
R
-
A
-
T
-
D
-
F
-
V
-
V
-
P
-
G
-
P
150
|
-
G
-
K
-
V
-
E
-
I
-
T
-
Y
-
T
-
P
-
S
160
|
-
D
-
G
-
T
-
Q
-
K
-
V
-
T
-
Y
-
L
-
V
170
|
-
H
-
N
-
F
-
E
-
E
-
G
-
G
-
G
-
V
-
A
180
|
-
M
-
G
-
M
-
Y
-
N
-
Q
-
D
-
K
-
S
-
I
190
|
-
E
-
D
-
F
-
A
-
H
-
S
-
S
-
F
-
Q
-
M
200
|
-
A
-
L
-
S
-
K
-
G
-
W
-
P
-
L
-
Y
-
L
210
|
-
S
-
T
-
K
-
N
-
T
-
I
-
L
-
K
-
K
-
Y
220
|
-
D
-
G
-
R
-
F
-
K
-
D
-
I
-
F
-
Q
-
E
230
|
-
I
-
Y
-
D
-
K
-
Q
-
Y
-
K
-
S
-
Q
-
F
240
|
-
E
-
A
-
Q
-
K
-
I
-
W
-
Y
-
E
-
H
-
R
250
|
-
L
-
I
-
D
-
D
-
M
-
V
-
A
-
Q
-
A
-
M
260
|
-
K
-
S
-
E
-
G
-
G
-
F
-
I
-
W
-
A
-
C
270
|
-
K
-
N
-
Y
-
D
-
G
-
D
-
V
-
Q
-
S
-
D
280
|
-
S
-
V
-
A
-
Q
-
G
-
Y
-
G
-
S
-
L
-
G
290
|
-
M
-
M
-
T
-
S
-
V
-
L
-
V
-
C
-
P
-
D
300
|
-
G
-
K
-
T
-
V
-
E
-
A
-
E
-
A
-
A
-
H
310
|
-
G
-
T
-
V
-
T
-
R
-
H
-
Y
-
R
-
M
-
Y
320
|
-
Q
-
K
-
G
-
Q
-
E
-
T
-
S
-
T
-
N
-
P
330
|
-
I
-
A
-
S
-
I
-
F
-
A
-
W
-
T
-
R
-
G
340
|
-
L
-
A
-
H
-
R
-
A
-
K
-
L
-
D
-
N
-
N
350
|
-
K
-
E
-
L
-
A
-
F
-
F
-
A
-
N
-
A
-
L
360
|
-
E
-
E
-
V
-
S
-
I
-
E
-
T
-
I
-
E
-
A
370
|
-
G
-
F
-
M
-
T
-
K
-
D
-
L
-
A
-
A
-
C
380
|
-
I
-
K
-
G
-
L
-
P
-
N
-
V
-
Q
-
R
-
S
390
|
-
D
-
Y
-
L
-
N
-
T
-
F
-
E
-
F
-
M
-
D
400
|
-
K
-
L
-
G
-
E
-
N
-
L
-
K
-
I
-
K
-
L
410
|
-
A
-
Q
-
A
-
K
-
L
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | TS676 cells | Brain | Homo sapiens (Human) | CVCL_A5HX | |||||||||
| TS603 cells | Brain | Homo sapiens (Human) | CVCL_A5HW | ||||||||||
| TS516 cells | Brain | Homo sapiens (Human) | CVCL_A5HY | ||||||||||
| In Vivo Model | SCID mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
Soft agar assay | ||||||||||||
| Mechanism Description | The missense mutation p.R132C (c.394C>T) in gene IDH1 cause the sensitivity of AGI-5198 by aberration of the drug's therapeutic target | ||||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [8] | ||||||||||||
| Resistant Disease | Colorectal cancer [ICD-11: 2B91.1] | ||||||||||||
| Resistant Drug | AGI-5198/Metformin | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132H (c.395G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.65 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.88 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
M
S
S
K
K
K
K
I
I
S
S
G
G
G
G
S
S
10
|
V
V
V
V
E
E
M
M
Q
Q
G
G
D
D
E
E
M
M
T
T
20
|
R
R
I
I
I
I
W
W
E
E
L
L
I
I
K
K
E
E
K
K
30
|
L
L
I
I
F
F
P
P
Y
Y
V
V
E
E
L
L
D
D
L
L
40
|
H
H
S
S
Y
Y
D
D
L
L
G
G
I
I
E
E
N
N
R
R
50
|
D
D
A
A
T
T
N
N
D
D
Q
Q
V
V
T
T
K
K
D
D
60
|
A
A
A
A
E
E
A
A
I
I
K
K
K
K
H
H
N
N
V
V
70
|
G
G
V
V
K
K
C
C
A
A
T
T
I
I
T
T
P
P
D
D
80
|
E
E
K
K
R
R
V
V
E
E
E
E
F
F
K
K
L
L
K
K
90
|
Q
Q
M
M
W
W
K
K
S
S
P
P
N
N
G
G
T
T
I
I
100
|
R
R
N
N
I
I
L
L
G
G
G
G
T
T
V
V
F
F
R
R
110
|
E
E
A
A
I
I
I
I
C
C
K
K
N
N
I
I
P
P
R
R
120
|
L
L
V
V
S
S
G
G
W
W
V
V
K
K
P
P
I
I
I
I
130
|
I
I
G
G
R
H
H
H
A
A
Y
Y
G
G
D
D
Q
Q
Y
Y
140
|
R
R
A
A
T
T
D
D
F
F
V
V
V
V
P
P
G
G
P
P
150
|
G
G
K
K
V
V
E
E
I
I
T
T
Y
Y
T
T
P
P
S
S
160
|
D
D
G
G
T
T
Q
Q
K
K
V
V
T
T
Y
Y
L
L
V
V
170
|
H
H
N
N
F
F
E
E
E
E
G
G
G
G
G
G
V
V
A
A
180
|
M
M
G
G
M
M
Y
Y
N
N
Q
Q
D
D
K
K
S
S
I
I
190
|
E
E
D
D
F
F
A
A
H
H
S
S
S
S
F
F
Q
Q
M
M
200
|
A
A
L
L
S
S
K
K
G
G
W
W
P
P
L
L
Y
Y
L
L
210
|
S
S
T
T
K
K
N
N
T
T
I
I
L
L
K
K
K
K
Y
Y
220
|
D
D
G
G
R
R
F
F
K
K
D
D
I
I
F
F
Q
Q
E
E
230
|
I
I
Y
Y
D
D
K
K
Q
Q
Y
Y
K
K
S
S
Q
Q
F
F
240
|
E
E
A
A
Q
Q
K
K
I
I
W
W
Y
Y
E
E
H
H
R
R
250
|
L
L
I
I
D
D
D
D
M
M
V
V
A
A
Q
Q
A
A
M
M
260
|
K
K
S
S
E
E
G
G
G
G
F
F
I
I
W
W
A
A
C
C
270
|
K
K
N
N
Y
Y
D
D
G
G
D
D
V
V
Q
Q
S
S
D
D
280
|
S
S
V
V
A
A
Q
Q
G
G
Y
Y
G
G
S
S
L
L
G
G
290
|
M
M
M
M
T
T
S
S
V
V
L
L
V
V
C
C
P
P
D
D
300
|
G
G
K
K
T
T
V
V
E
E
A
A
E
E
A
A
A
A
H
H
310
|
G
G
T
T
V
V
T
T
R
R
H
H
Y
Y
R
R
M
M
Y
Y
320
|
Q
Q
K
K
G
G
Q
Q
E
E
T
T
S
S
T
T
N
N
P
P
330
|
I
I
A
A
S
S
I
I
F
F
A
A
W
W
T
T
R
R
G
G
340
|
L
L
A
A
H
H
R
R
A
A
K
K
L
L
D
D
N
N
N
N
350
|
K
K
E
E
L
L
A
A
F
F
F
F
A
A
N
N
A
A
L
L
360
|
E
E
E
E
V
V
S
S
I
I
E
E
T
T
I
I
E
E
A
A
370
|
G
G
F
F
M
M
T
T
K
K
D
D
L
L
A
A
A
A
C
C
380
|
I
I
K
K
G
G
L
L
P
P
N
N
V
V
Q
Q
R
R
S
S
390
|
D
D
Y
Y
L
L
N
N
T
T
F
F
E
E
F
F
M
M
D
D
400
|
K
K
L
L
G
G
E
E
N
N
L
L
K
K
I
I
K
K
L
L
410
|
A
A
Q
Q
A
A
K
K
L
L
S
S
L
L
E
E
H
H
H
H
420
|
H
H
H
H
H
H
H
H
H
H
H
H
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| IDH1 cells | Ovary | Homo sapiens (Human) | CVCL_D3DY | ||||||||||
| Experiment for Molecule Alteration |
Western blot analysis; gama-H2AX immunofluorescence staining and measurement | ||||||||||||
| Experiment for Drug Resistance |
Colony formation assay | ||||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Sarcoma [ICD-11: 2C35.0] | [5] | ||||||||||||
| Resistant Disease | Sarcoma [ICD-11: 2C35.0] | ||||||||||||
| Resistant Drug | AGI-5198/Olaparib | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132C (c.394C>T) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.93 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
M
-
S
-
K
-
K
-
I
-
S
-
G
-
G
-
S
10
|
-
V
-
V
-
E
-
M
-
Q
-
G
-
D
-
E
-
M
-
T
20
|
-
R
-
I
-
I
-
W
-
E
-
L
-
I
-
K
-
E
-
K
30
|
-
L
-
I
-
F
-
P
-
Y
-
V
-
E
-
L
-
D
-
L
40
|
-
H
-
S
-
Y
-
D
-
L
-
G
-
I
-
E
-
N
-
R
50
|
-
D
-
A
-
T
-
N
-
D
-
Q
-
V
-
T
-
K
-
D
60
|
-
A
-
A
-
E
-
A
-
I
-
K
-
K
-
H
-
N
-
V
70
|
-
G
-
V
-
K
-
C
-
A
-
T
-
I
-
T
-
P
-
D
80
|
-
E
-
K
-
R
-
V
-
E
-
E
-
F
-
K
-
L
-
K
90
|
-
Q
-
M
-
W
-
K
-
S
-
P
-
N
-
G
-
T
-
I
100
|
-
R
-
N
-
I
-
L
-
G
-
G
-
T
-
V
-
F
-
R
110
|
-
E
-
A
-
I
-
I
-
C
-
K
-
N
-
I
-
P
-
R
120
|
-
L
-
V
-
S
-
G
-
W
-
V
K
K
P
P
I
I
I
I
130
|
I
I
G
G
S
C
H
H
A
A
Y
Y
G
G
D
D
-
Q
-
Y
140
|
-
R
-
A
-
T
-
D
-
F
-
V
-
V
-
P
-
G
-
P
150
|
-
G
-
K
-
V
-
E
-
I
-
T
-
Y
-
T
-
P
-
S
160
|
-
D
-
G
-
T
-
Q
-
K
-
V
-
T
-
Y
-
L
-
V
170
|
-
H
-
N
-
F
-
E
-
E
-
G
-
G
-
G
-
V
-
A
180
|
-
M
-
G
-
M
-
Y
-
N
-
Q
-
D
-
K
-
S
-
I
190
|
-
E
-
D
-
F
-
A
-
H
-
S
-
S
-
F
-
Q
-
M
200
|
-
A
-
L
-
S
-
K
-
G
-
W
-
P
-
L
-
Y
-
L
210
|
-
S
-
T
-
K
-
N
-
T
-
I
-
L
-
K
-
K
-
Y
220
|
-
D
-
G
-
R
-
F
-
K
-
D
-
I
-
F
-
Q
-
E
230
|
-
I
-
Y
-
D
-
K
-
Q
-
Y
-
K
-
S
-
Q
-
F
240
|
-
E
-
A
-
Q
-
K
-
I
-
W
-
Y
-
E
-
H
-
R
250
|
-
L
-
I
-
D
-
D
-
M
-
V
-
A
-
Q
-
A
-
M
260
|
-
K
-
S
-
E
-
G
-
G
-
F
-
I
-
W
-
A
-
C
270
|
-
K
-
N
-
Y
-
D
-
G
-
D
-
V
-
Q
-
S
-
D
280
|
-
S
-
V
-
A
-
Q
-
G
-
Y
-
G
-
S
-
L
-
G
290
|
-
M
-
M
-
T
-
S
-
V
-
L
-
V
-
C
-
P
-
D
300
|
-
G
-
K
-
T
-
V
-
E
-
A
-
E
-
A
-
A
-
H
310
|
-
G
-
T
-
V
-
T
-
R
-
H
-
Y
-
R
-
M
-
Y
320
|
-
Q
-
K
-
G
-
Q
-
E
-
T
-
S
-
T
-
N
-
P
330
|
-
I
-
A
-
S
-
I
-
F
-
A
-
W
-
T
-
R
-
G
340
|
-
L
-
A
-
H
-
R
-
A
-
K
-
L
-
D
-
N
-
N
350
|
-
K
-
E
-
L
-
A
-
F
-
F
-
A
-
N
-
A
-
L
360
|
-
E
-
E
-
V
-
S
-
I
-
E
-
T
-
I
-
E
-
A
370
|
-
G
-
F
-
M
-
T
-
K
-
D
-
L
-
A
-
A
-
C
380
|
-
I
-
K
-
G
-
L
-
P
-
N
-
V
-
Q
-
R
-
S
390
|
-
D
-
Y
-
L
-
N
-
T
-
F
-
E
-
F
-
M
-
D
400
|
-
K
-
L
-
G
-
E
-
N
-
L
-
K
-
I
-
K
-
L
410
|
-
A
-
Q
-
A
-
K
-
L
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | IDH2 cells | Ovary | Homo sapiens (Human) | CVCL_D3DY | |||||||||
| IDH1 cells | Ovary | Homo sapiens (Human) | CVCL_D3DY | ||||||||||
| In Vivo Model | Female athymic nu/nu mouse PDX model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
Promega assay | ||||||||||||
| Mechanism Description | The oncometabolite, 2-hydroxyglutarate, renders IDH1/2 mutant cancer cells deficient in homologous recombination and confers vulnerability to synthetic lethal targeting with PARP inhibitors. | ||||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [5] | ||||||||||||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Resistant Drug | AGI-5198/Talazoparib | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132H (c.395G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.65 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.88 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
M
S
S
K
K
K
K
I
I
S
S
G
G
G
G
S
S
10
|
V
V
V
V
E
E
M
M
Q
Q
G
G
D
D
E
E
M
M
T
T
20
|
R
R
I
I
I
I
W
W
E
E
L
L
I
I
K
K
E
E
K
K
30
|
L
L
I
I
F
F
P
P
Y
Y
V
V
E
E
L
L
D
D
L
L
40
|
H
H
S
S
Y
Y
D
D
L
L
G
G
I
I
E
E
N
N
R
R
50
|
D
D
A
A
T
T
N
N
D
D
Q
Q
V
V
T
T
K
K
D
D
60
|
A
A
A
A
E
E
A
A
I
I
K
K
K
K
H
H
N
N
V
V
70
|
G
G
V
V
K
K
C
C
A
A
T
T
I
I
T
T
P
P
D
D
80
|
E
E
K
K
R
R
V
V
E
E
E
E
F
F
K
K
L
L
K
K
90
|
Q
Q
M
M
W
W
K
K
S
S
P
P
N
N
G
G
T
T
I
I
100
|
R
R
N
N
I
I
L
L
G
G
G
G
T
T
V
V
F
F
R
R
110
|
E
E
A
A
I
I
I
I
C
C
K
K
N
N
I
I
P
P
R
R
120
|
L
L
V
V
S
S
G
G
W
W
V
V
K
K
P
P
I
I
I
I
130
|
I
I
G
G
R
H
H
H
A
A
Y
Y
G
G
D
D
Q
Q
Y
Y
140
|
R
R
A
A
T
T
D
D
F
F
V
V
V
V
P
P
G
G
P
P
150
|
G
G
K
K
V
V
E
E
I
I
T
T
Y
Y
T
T
P
P
S
S
160
|
D
D
G
G
T
T
Q
Q
K
K
V
V
T
T
Y
Y
L
L
V
V
170
|
H
H
N
N
F
F
E
E
E
E
G
G
G
G
G
G
V
V
A
A
180
|
M
M
G
G
M
M
Y
Y
N
N
Q
Q
D
D
K
K
S
S
I
I
190
|
E
E
D
D
F
F
A
A
H
H
S
S
S
S
F
F
Q
Q
M
M
200
|
A
A
L
L
S
S
K
K
G
G
W
W
P
P
L
L
Y
Y
L
L
210
|
S
S
T
T
K
K
N
N
T
T
I
I
L
L
K
K
K
K
Y
Y
220
|
D
D
G
G
R
R
F
F
K
K
D
D
I
I
F
F
Q
Q
E
E
230
|
I
I
Y
Y
D
D
K
K
Q
Q
Y
Y
K
K
S
S
Q
Q
F
F
240
|
E
E
A
A
Q
Q
K
K
I
I
W
W
Y
Y
E
E
H
H
R
R
250
|
L
L
I
I
D
D
D
D
M
M
V
V
A
A
Q
Q
A
A
M
M
260
|
K
K
S
S
E
E
G
G
G
G
F
F
I
I
W
W
A
A
C
C
270
|
K
K
N
N
Y
Y
D
D
G
G
D
D
V
V
Q
Q
S
S
D
D
280
|
S
S
V
V
A
A
Q
Q
G
G
Y
Y
G
G
S
S
L
L
G
G
290
|
M
M
M
M
T
T
S
S
V
V
L
L
V
V
C
C
P
P
D
D
300
|
G
G
K
K
T
T
V
V
E
E
A
A
E
E
A
A
A
A
H
H
310
|
G
G
T
T
V
V
T
T
R
R
H
H
Y
Y
R
R
M
M
Y
Y
320
|
Q
Q
K
K
G
G
Q
Q
E
E
T
T
S
S
T
T
N
N
P
P
330
|
I
I
A
A
S
S
I
I
F
F
A
A
W
W
T
T
R
R
G
G
340
|
L
L
A
A
H
H
R
R
A
A
K
K
L
L
D
D
N
N
N
N
350
|
K
K
E
E
L
L
A
A
F
F
F
F
A
A
N
N
A
A
L
L
360
|
E
E
E
E
V
V
S
S
I
I
E
E
T
T
I
I
E
E
A
A
370
|
G
G
F
F
M
M
T
T
K
K
D
D
L
L
A
A
A
A
C
C
380
|
I
I
K
K
G
G
L
L
P
P
N
N
V
V
Q
Q
R
R
S
S
390
|
D
D
Y
Y
L
L
N
N
T
T
F
F
E
E
F
F
M
M
D
D
400
|
K
K
L
L
G
G
E
E
N
N
L
L
K
K
I
I
K
K
L
L
410
|
A
A
Q
Q
A
A
K
K
L
L
S
S
L
L
E
E
H
H
H
H
420
|
H
H
H
H
H
H
H
H
H
H
H
H
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | IDH2 cells | Ovary | Homo sapiens (Human) | CVCL_D3DY | |||||||||
| IDH1 cells | Ovary | Homo sapiens (Human) | CVCL_D3DY | ||||||||||
| In Vivo Model | Female athymic nu/nu mouse PDX model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
Promega assay | ||||||||||||
| Mechanism Description | The oncometabolite, 2-hydroxyglutarate, renders IDH1/2 mutant cancer cells deficient in homologous recombination and confers vulnerability to synthetic lethal targeting with PARP inhibitors. | ||||||||||||
| Disease Class: Sarcoma [ICD-11: 2C35.0] | [5] | ||||||||||||
| Resistant Disease | Sarcoma [ICD-11: 2C35.0] | ||||||||||||
| Resistant Drug | AGI-5198/Talazoparib | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132C (c.394C>T) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.93 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
M
-
S
-
K
-
K
-
I
-
S
-
G
-
G
-
S
10
|
-
V
-
V
-
E
-
M
-
Q
-
G
-
D
-
E
-
M
-
T
20
|
-
R
-
I
-
I
-
W
-
E
-
L
-
I
-
K
-
E
-
K
30
|
-
L
-
I
-
F
-
P
-
Y
-
V
-
E
-
L
-
D
-
L
40
|
-
H
-
S
-
Y
-
D
-
L
-
G
-
I
-
E
-
N
-
R
50
|
-
D
-
A
-
T
-
N
-
D
-
Q
-
V
-
T
-
K
-
D
60
|
-
A
-
A
-
E
-
A
-
I
-
K
-
K
-
H
-
N
-
V
70
|
-
G
-
V
-
K
-
C
-
A
-
T
-
I
-
T
-
P
-
D
80
|
-
E
-
K
-
R
-
V
-
E
-
E
-
F
-
K
-
L
-
K
90
|
-
Q
-
M
-
W
-
K
-
S
-
P
-
N
-
G
-
T
-
I
100
|
-
R
-
N
-
I
-
L
-
G
-
G
-
T
-
V
-
F
-
R
110
|
-
E
-
A
-
I
-
I
-
C
-
K
-
N
-
I
-
P
-
R
120
|
-
L
-
V
-
S
-
G
-
W
-
V
K
K
P
P
I
I
I
I
130
|
I
I
G
G
S
C
H
H
A
A
Y
Y
G
G
D
D
-
Q
-
Y
140
|
-
R
-
A
-
T
-
D
-
F
-
V
-
V
-
P
-
G
-
P
150
|
-
G
-
K
-
V
-
E
-
I
-
T
-
Y
-
T
-
P
-
S
160
|
-
D
-
G
-
T
-
Q
-
K
-
V
-
T
-
Y
-
L
-
V
170
|
-
H
-
N
-
F
-
E
-
E
-
G
-
G
-
G
-
V
-
A
180
|
-
M
-
G
-
M
-
Y
-
N
-
Q
-
D
-
K
-
S
-
I
190
|
-
E
-
D
-
F
-
A
-
H
-
S
-
S
-
F
-
Q
-
M
200
|
-
A
-
L
-
S
-
K
-
G
-
W
-
P
-
L
-
Y
-
L
210
|
-
S
-
T
-
K
-
N
-
T
-
I
-
L
-
K
-
K
-
Y
220
|
-
D
-
G
-
R
-
F
-
K
-
D
-
I
-
F
-
Q
-
E
230
|
-
I
-
Y
-
D
-
K
-
Q
-
Y
-
K
-
S
-
Q
-
F
240
|
-
E
-
A
-
Q
-
K
-
I
-
W
-
Y
-
E
-
H
-
R
250
|
-
L
-
I
-
D
-
D
-
M
-
V
-
A
-
Q
-
A
-
M
260
|
-
K
-
S
-
E
-
G
-
G
-
F
-
I
-
W
-
A
-
C
270
|
-
K
-
N
-
Y
-
D
-
G
-
D
-
V
-
Q
-
S
-
D
280
|
-
S
-
V
-
A
-
Q
-
G
-
Y
-
G
-
S
-
L
-
G
290
|
-
M
-
M
-
T
-
S
-
V
-
L
-
V
-
C
-
P
-
D
300
|
-
G
-
K
-
T
-
V
-
E
-
A
-
E
-
A
-
A
-
H
310
|
-
G
-
T
-
V
-
T
-
R
-
H
-
Y
-
R
-
M
-
Y
320
|
-
Q
-
K
-
G
-
Q
-
E
-
T
-
S
-
T
-
N
-
P
330
|
-
I
-
A
-
S
-
I
-
F
-
A
-
W
-
T
-
R
-
G
340
|
-
L
-
A
-
H
-
R
-
A
-
K
-
L
-
D
-
N
-
N
350
|
-
K
-
E
-
L
-
A
-
F
-
F
-
A
-
N
-
A
-
L
360
|
-
E
-
E
-
V
-
S
-
I
-
E
-
T
-
I
-
E
-
A
370
|
-
G
-
F
-
M
-
T
-
K
-
D
-
L
-
A
-
A
-
C
380
|
-
I
-
K
-
G
-
L
-
P
-
N
-
V
-
Q
-
R
-
S
390
|
-
D
-
Y
-
L
-
N
-
T
-
F
-
E
-
F
-
M
-
D
400
|
-
K
-
L
-
G
-
E
-
N
-
L
-
K
-
I
-
K
-
L
410
|
-
A
-
Q
-
A
-
K
-
L
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | IDH2 cells | Ovary | Homo sapiens (Human) | CVCL_D3DY | |||||||||
| IDH1 cells | Ovary | Homo sapiens (Human) | CVCL_D3DY | ||||||||||
| In Vivo Model | Female athymic nu/nu mouse PDX model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
Promega assay | ||||||||||||
| Mechanism Description | The oncometabolite, 2-hydroxyglutarate, renders IDH1/2 mutant cancer cells deficient in homologous recombination and confers vulnerability to synthetic lethal targeting with PARP inhibitors. | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Brain glioma [ICD-11: 2A00.0] | [9] | ||||||||||||
| Sensitive Disease | Brain glioma [ICD-11: 2A00.0] | ||||||||||||
| Sensitive Drug | BPTES | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132H (c.395G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.65 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.88 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
M
S
S
K
K
K
K
I
I
S
S
G
G
G
G
S
S
10
|
V
V
V
V
E
E
M
M
Q
Q
G
G
D
D
E
E
M
M
T
T
20
|
R
R
I
I
I
I
W
W
E
E
L
L
I
I
K
K
E
E
K
K
30
|
L
L
I
I
F
F
P
P
Y
Y
V
V
E
E
L
L
D
D
L
L
40
|
H
H
S
S
Y
Y
D
D
L
L
G
G
I
I
E
E
N
N
R
R
50
|
D
D
A
A
T
T
N
N
D
D
Q
Q
V
V
T
T
K
K
D
D
60
|
A
A
A
A
E
E
A
A
I
I
K
K
K
K
H
H
N
N
V
V
70
|
G
G
V
V
K
K
C
C
A
A
T
T
I
I
T
T
P
P
D
D
80
|
E
E
K
K
R
R
V
V
E
E
E
E
F
F
K
K
L
L
K
K
90
|
Q
Q
M
M
W
W
K
K
S
S
P
P
N
N
G
G
T
T
I
I
100
|
R
R
N
N
I
I
L
L
G
G
G
G
T
T
V
V
F
F
R
R
110
|
E
E
A
A
I
I
I
I
C
C
K
K
N
N
I
I
P
P
R
R
120
|
L
L
V
V
S
S
G
G
W
W
V
V
K
K
P
P
I
I
I
I
130
|
I
I
G
G
R
H
H
H
A
A
Y
Y
G
G
D
D
Q
Q
Y
Y
140
|
R
R
A
A
T
T
D
D
F
F
V
V
V
V
P
P
G
G
P
P
150
|
G
G
K
K
V
V
E
E
I
I
T
T
Y
Y
T
T
P
P
S
S
160
|
D
D
G
G
T
T
Q
Q
K
K
V
V
T
T
Y
Y
L
L
V
V
170
|
H
H
N
N
F
F
E
E
E
E
G
G
G
G
G
G
V
V
A
A
180
|
M
M
G
G
M
M
Y
Y
N
N
Q
Q
D
D
K
K
S
S
I
I
190
|
E
E
D
D
F
F
A
A
H
H
S
S
S
S
F
F
Q
Q
M
M
200
|
A
A
L
L
S
S
K
K
G
G
W
W
P
P
L
L
Y
Y
L
L
210
|
S
S
T
T
K
K
N
N
T
T
I
I
L
L
K
K
K
K
Y
Y
220
|
D
D
G
G
R
R
F
F
K
K
D
D
I
I
F
F
Q
Q
E
E
230
|
I
I
Y
Y
D
D
K
K
Q
Q
Y
Y
K
K
S
S
Q
Q
F
F
240
|
E
E
A
A
Q
Q
K
K
I
I
W
W
Y
Y
E
E
H
H
R
R
250
|
L
L
I
I
D
D
D
D
M
M
V
V
A
A
Q
Q
A
A
M
M
260
|
K
K
S
S
E
E
G
G
G
G
F
F
I
I
W
W
A
A
C
C
270
|
K
K
N
N
Y
Y
D
D
G
G
D
D
V
V
Q
Q
S
S
D
D
280
|
S
S
V
V
A
A
Q
Q
G
G
Y
Y
G
G
S
S
L
L
G
G
290
|
M
M
M
M
T
T
S
S
V
V
L
L
V
V
C
C
P
P
D
D
300
|
G
G
K
K
T
T
V
V
E
E
A
A
E
E
A
A
A
A
H
H
310
|
G
G
T
T
V
V
T
T
R
R
H
H
Y
Y
R
R
M
M
Y
Y
320
|
Q
Q
K
K
G
G
Q
Q
E
E
T
T
S
S
T
T
N
N
P
P
330
|
I
I
A
A
S
S
I
I
F
F
A
A
W
W
T
T
R
R
G
G
340
|
L
L
A
A
H
H
R
R
A
A
K
K
L
L
D
D
N
N
N
N
350
|
K
K
E
E
L
L
A
A
F
F
F
F
A
A
N
N
A
A
L
L
360
|
E
E
E
E
V
V
S
S
I
I
E
E
T
T
I
I
E
E
A
A
370
|
G
G
F
F
M
M
T
T
K
K
D
D
L
L
A
A
A
A
C
C
380
|
I
I
K
K
G
G
L
L
P
P
N
N
V
V
Q
Q
R
R
S
S
390
|
D
D
Y
Y
L
L
N
N
T
T
F
F
E
E
F
F
M
M
D
D
400
|
K
K
L
L
G
G
E
E
N
N
L
L
K
K
I
I
K
K
L
L
410
|
A
A
Q
Q
A
A
K
K
L
L
S
S
L
L
E
E
H
H
H
H
420
|
H
H
H
H
H
H
H
H
H
H
H
H
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Experiment for Drug Resistance |
3H-thymidine incorporation assay | ||||||||||||
| Mechanism Description | The missense mutation p.R132H (c.395G>A) in gene IDH1 cause the sensitivity of BPTES by unusual activation of pro-survival pathway | ||||||||||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [10] | ||||||||||||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Sensitive Drug | BPTES | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132C (c.394C>T) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.93 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
M
-
S
-
K
-
K
-
I
-
S
-
G
-
G
-
S
10
|
-
V
-
V
-
E
-
M
-
Q
-
G
-
D
-
E
-
M
-
T
20
|
-
R
-
I
-
I
-
W
-
E
-
L
-
I
-
K
-
E
-
K
30
|
-
L
-
I
-
F
-
P
-
Y
-
V
-
E
-
L
-
D
-
L
40
|
-
H
-
S
-
Y
-
D
-
L
-
G
-
I
-
E
-
N
-
R
50
|
-
D
-
A
-
T
-
N
-
D
-
Q
-
V
-
T
-
K
-
D
60
|
-
A
-
A
-
E
-
A
-
I
-
K
-
K
-
H
-
N
-
V
70
|
-
G
-
V
-
K
-
C
-
A
-
T
-
I
-
T
-
P
-
D
80
|
-
E
-
K
-
R
-
V
-
E
-
E
-
F
-
K
-
L
-
K
90
|
-
Q
-
M
-
W
-
K
-
S
-
P
-
N
-
G
-
T
-
I
100
|
-
R
-
N
-
I
-
L
-
G
-
G
-
T
-
V
-
F
-
R
110
|
-
E
-
A
-
I
-
I
-
C
-
K
-
N
-
I
-
P
-
R
120
|
-
L
-
V
-
S
-
G
-
W
-
V
K
K
P
P
I
I
I
I
130
|
I
I
G
G
S
C
H
H
A
A
Y
Y
G
G
D
D
-
Q
-
Y
140
|
-
R
-
A
-
T
-
D
-
F
-
V
-
V
-
P
-
G
-
P
150
|
-
G
-
K
-
V
-
E
-
I
-
T
-
Y
-
T
-
P
-
S
160
|
-
D
-
G
-
T
-
Q
-
K
-
V
-
T
-
Y
-
L
-
V
170
|
-
H
-
N
-
F
-
E
-
E
-
G
-
G
-
G
-
V
-
A
180
|
-
M
-
G
-
M
-
Y
-
N
-
Q
-
D
-
K
-
S
-
I
190
|
-
E
-
D
-
F
-
A
-
H
-
S
-
S
-
F
-
Q
-
M
200
|
-
A
-
L
-
S
-
K
-
G
-
W
-
P
-
L
-
Y
-
L
210
|
-
S
-
T
-
K
-
N
-
T
-
I
-
L
-
K
-
K
-
Y
220
|
-
D
-
G
-
R
-
F
-
K
-
D
-
I
-
F
-
Q
-
E
230
|
-
I
-
Y
-
D
-
K
-
Q
-
Y
-
K
-
S
-
Q
-
F
240
|
-
E
-
A
-
Q
-
K
-
I
-
W
-
Y
-
E
-
H
-
R
250
|
-
L
-
I
-
D
-
D
-
M
-
V
-
A
-
Q
-
A
-
M
260
|
-
K
-
S
-
E
-
G
-
G
-
F
-
I
-
W
-
A
-
C
270
|
-
K
-
N
-
Y
-
D
-
G
-
D
-
V
-
Q
-
S
-
D
280
|
-
S
-
V
-
A
-
Q
-
G
-
Y
-
G
-
S
-
L
-
G
290
|
-
M
-
M
-
T
-
S
-
V
-
L
-
V
-
C
-
P
-
D
300
|
-
G
-
K
-
T
-
V
-
E
-
A
-
E
-
A
-
A
-
H
310
|
-
G
-
T
-
V
-
T
-
R
-
H
-
Y
-
R
-
M
-
Y
320
|
-
Q
-
K
-
G
-
Q
-
E
-
T
-
S
-
T
-
N
-
P
330
|
-
I
-
A
-
S
-
I
-
F
-
A
-
W
-
T
-
R
-
G
340
|
-
L
-
A
-
H
-
R
-
A
-
K
-
L
-
D
-
N
-
N
350
|
-
K
-
E
-
L
-
A
-
F
-
F
-
A
-
N
-
A
-
L
360
|
-
E
-
E
-
V
-
S
-
I
-
E
-
T
-
I
-
E
-
A
370
|
-
G
-
F
-
M
-
T
-
K
-
D
-
L
-
A
-
A
-
C
380
|
-
I
-
K
-
G
-
L
-
P
-
N
-
V
-
Q
-
R
-
S
390
|
-
D
-
Y
-
L
-
N
-
T
-
F
-
E
-
F
-
M
-
D
400
|
-
K
-
L
-
G
-
E
-
N
-
L
-
K
-
I
-
K
-
L
410
|
-
A
-
Q
-
A
-
K
-
L
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | 5637 cells | Bladder | Homo sapiens (Human) | CVCL_0126 | |||||||||
| AML cells | N.A. | Homo sapiens (Human) | N.A. | ||||||||||
| Experiment for Drug Resistance |
Manually cell counting assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [5] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | Cisplatin/Talazoparib | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132H (c.395G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.65 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.88 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
M
S
S
K
K
K
K
I
I
S
S
G
G
G
G
S
S
10
|
V
V
V
V
E
E
M
M
Q
Q
G
G
D
D
E
E
M
M
T
T
20
|
R
R
I
I
I
I
W
W
E
E
L
L
I
I
K
K
E
E
K
K
30
|
L
L
I
I
F
F
P
P
Y
Y
V
V
E
E
L
L
D
D
L
L
40
|
H
H
S
S
Y
Y
D
D
L
L
G
G
I
I
E
E
N
N
R
R
50
|
D
D
A
A
T
T
N
N
D
D
Q
Q
V
V
T
T
K
K
D
D
60
|
A
A
A
A
E
E
A
A
I
I
K
K
K
K
H
H
N
N
V
V
70
|
G
G
V
V
K
K
C
C
A
A
T
T
I
I
T
T
P
P
D
D
80
|
E
E
K
K
R
R
V
V
E
E
E
E
F
F
K
K
L
L
K
K
90
|
Q
Q
M
M
W
W
K
K
S
S
P
P
N
N
G
G
T
T
I
I
100
|
R
R
N
N
I
I
L
L
G
G
G
G
T
T
V
V
F
F
R
R
110
|
E
E
A
A
I
I
I
I
C
C
K
K
N
N
I
I
P
P
R
R
120
|
L
L
V
V
S
S
G
G
W
W
V
V
K
K
P
P
I
I
I
I
130
|
I
I
G
G
R
H
H
H
A
A
Y
Y
G
G
D
D
Q
Q
Y
Y
140
|
R
R
A
A
T
T
D
D
F
F
V
V
V
V
P
P
G
G
P
P
150
|
G
G
K
K
V
V
E
E
I
I
T
T
Y
Y
T
T
P
P
S
S
160
|
D
D
G
G
T
T
Q
Q
K
K
V
V
T
T
Y
Y
L
L
V
V
170
|
H
H
N
N
F
F
E
E
E
E
G
G
G
G
G
G
V
V
A
A
180
|
M
M
G
G
M
M
Y
Y
N
N
Q
Q
D
D
K
K
S
S
I
I
190
|
E
E
D
D
F
F
A
A
H
H
S
S
S
S
F
F
Q
Q
M
M
200
|
A
A
L
L
S
S
K
K
G
G
W
W
P
P
L
L
Y
Y
L
L
210
|
S
S
T
T
K
K
N
N
T
T
I
I
L
L
K
K
K
K
Y
Y
220
|
D
D
G
G
R
R
F
F
K
K
D
D
I
I
F
F
Q
Q
E
E
230
|
I
I
Y
Y
D
D
K
K
Q
Q
Y
Y
K
K
S
S
Q
Q
F
F
240
|
E
E
A
A
Q
Q
K
K
I
I
W
W
Y
Y
E
E
H
H
R
R
250
|
L
L
I
I
D
D
D
D
M
M
V
V
A
A
Q
Q
A
A
M
M
260
|
K
K
S
S
E
E
G
G
G
G
F
F
I
I
W
W
A
A
C
C
270
|
K
K
N
N
Y
Y
D
D
G
G
D
D
V
V
Q
Q
S
S
D
D
280
|
S
S
V
V
A
A
Q
Q
G
G
Y
Y
G
G
S
S
L
L
G
G
290
|
M
M
M
M
T
T
S
S
V
V
L
L
V
V
C
C
P
P
D
D
300
|
G
G
K
K
T
T
V
V
E
E
A
A
E
E
A
A
A
A
H
H
310
|
G
G
T
T
V
V
T
T
R
R
H
H
Y
Y
R
R
M
M
Y
Y
320
|
Q
Q
K
K
G
G
Q
Q
E
E
T
T
S
S
T
T
N
N
P
P
330
|
I
I
A
A
S
S
I
I
F
F
A
A
W
W
T
T
R
R
G
G
340
|
L
L
A
A
H
H
R
R
A
A
K
K
L
L
D
D
N
N
N
N
350
|
K
K
E
E
L
L
A
A
F
F
F
F
A
A
N
N
A
A
L
L
360
|
E
E
E
E
V
V
S
S
I
I
E
E
T
T
I
I
E
E
A
A
370
|
G
G
F
F
M
M
T
T
K
K
D
D
L
L
A
A
A
A
C
C
380
|
I
I
K
K
G
G
L
L
P
P
N
N
V
V
Q
Q
R
R
S
S
390
|
D
D
Y
Y
L
L
N
N
T
T
F
F
E
E
F
F
M
M
D
D
400
|
K
K
L
L
G
G
E
E
N
N
L
L
K
K
I
I
K
K
L
L
410
|
A
A
Q
Q
A
A
K
K
L
L
S
S
L
L
E
E
H
H
H
H
420
|
H
H
H
H
H
H
H
H
H
H
H
H
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | IDH2 cells | Ovary | Homo sapiens (Human) | CVCL_D3DY | |||||||||
| IDH1 cells | Ovary | Homo sapiens (Human) | CVCL_D3DY | ||||||||||
| In Vivo Model | Female athymic nu/nu mouse PDX model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
Promega assay | ||||||||||||
| Mechanism Description | The oncometabolite, 2-hydroxyglutarate, renders IDH1/2 mutant cancer cells deficient in homologous recombination and confers vulnerability to synthetic lethal targeting with PARP inhibitors. | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [11] | ||||||||||||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Sensitive Drug | GSK321 | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132C (c.394C>T) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.93 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.20 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
M
-
S
-
K
-
K
-
I
-
S
-
G
-
G
-
S
10
|
-
V
-
V
-
E
-
M
-
Q
-
G
-
D
-
E
-
M
-
T
20
|
-
R
-
I
-
I
-
W
-
E
-
L
-
I
-
K
-
E
-
K
30
|
-
L
-
I
-
F
-
P
-
Y
-
V
-
E
-
L
-
D
-
L
40
|
-
H
-
S
-
Y
-
D
-
L
-
G
-
I
-
E
-
N
-
R
50
|
-
D
-
A
-
T
-
N
-
D
-
Q
-
V
-
T
-
K
-
D
60
|
-
A
-
A
-
E
-
A
-
I
-
K
-
K
-
H
-
N
-
V
70
|
-
G
-
V
-
K
-
C
-
A
-
T
-
I
-
T
-
P
-
D
80
|
-
E
-
K
-
R
-
V
-
E
-
E
-
F
-
K
-
L
-
K
90
|
-
Q
-
M
-
W
-
K
-
S
-
P
-
N
-
G
-
T
-
I
100
|
-
R
-
N
-
I
-
L
-
G
-
G
-
T
-
V
-
F
-
R
110
|
-
E
-
A
-
I
-
I
-
C
-
K
-
N
-
I
-
P
-
R
120
|
-
L
-
V
-
S
-
G
-
W
-
V
K
K
P
P
I
I
I
I
130
|
I
I
G
G
S
C
H
H
A
A
Y
Y
G
G
D
D
-
Q
-
Y
140
|
-
R
-
A
-
T
-
D
-
F
-
V
-
V
-
P
-
G
-
P
150
|
-
G
-
K
-
V
-
E
-
I
-
T
-
Y
-
T
-
P
-
S
160
|
-
D
-
G
-
T
-
Q
-
K
-
V
-
T
-
Y
-
L
-
V
170
|
-
H
-
N
-
F
-
E
-
E
-
G
-
G
-
G
-
V
-
A
180
|
-
M
-
G
-
M
-
Y
-
N
-
Q
-
D
-
K
-
S
-
I
190
|
-
E
-
D
-
F
-
A
-
H
-
S
-
S
-
F
-
Q
-
M
200
|
-
A
-
L
-
S
-
K
-
G
-
W
-
P
-
L
-
Y
-
L
210
|
-
S
-
T
-
K
-
N
-
T
-
I
-
L
-
K
-
K
-
Y
220
|
-
D
-
G
-
R
-
F
-
K
-
D
-
I
-
F
-
Q
-
E
230
|
-
I
-
Y
-
D
-
K
-
Q
-
Y
-
K
-
S
-
Q
-
F
240
|
-
E
-
A
-
Q
-
K
-
I
-
W
-
Y
-
E
-
H
-
R
250
|
-
L
-
I
-
D
-
D
-
M
-
V
-
A
-
Q
-
A
-
M
260
|
-
K
-
S
-
E
-
G
-
G
-
F
-
I
-
W
-
A
-
C
270
|
-
K
-
N
-
Y
-
D
-
G
-
D
-
V
-
Q
-
S
-
D
280
|
-
S
-
V
-
A
-
Q
-
G
-
Y
-
G
-
S
-
L
-
G
290
|
-
M
-
M
-
T
-
S
-
V
-
L
-
V
-
C
-
P
-
D
300
|
-
G
-
K
-
T
-
V
-
E
-
A
-
E
-
A
-
A
-
H
310
|
-
G
-
T
-
V
-
T
-
R
-
H
-
Y
-
R
-
M
-
Y
320
|
-
Q
-
K
-
G
-
Q
-
E
-
T
-
S
-
T
-
N
-
P
330
|
-
I
-
A
-
S
-
I
-
F
-
A
-
W
-
T
-
R
-
G
340
|
-
L
-
A
-
H
-
R
-
A
-
K
-
L
-
D
-
N
-
N
350
|
-
K
-
E
-
L
-
A
-
F
-
F
-
A
-
N
-
A
-
L
360
|
-
E
-
E
-
V
-
S
-
I
-
E
-
T
-
I
-
E
-
A
370
|
-
G
-
F
-
M
-
T
-
K
-
D
-
L
-
A
-
A
-
C
380
|
-
I
-
K
-
G
-
L
-
P
-
N
-
V
-
Q
-
R
-
S
390
|
-
D
-
Y
-
L
-
N
-
T
-
F
-
E
-
F
-
M
-
D
400
|
-
K
-
L
-
G
-
E
-
N
-
L
-
K
-
I
-
K
-
L
410
|
-
A
-
Q
-
A
-
K
-
L
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | HT-1080 cells | Acetabulum | Homo sapiens (Human) | CVCL_0317 | |||||||||
| In Vivo Model | Male CD-1 xenograft mouse model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Diaphorase/resazurin coupled assay; Colony formation assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Brain glioma [ICD-11: 2A00.0] | [7] | ||||||||||||
| Sensitive Disease | Brain glioma [ICD-11: 2A00.0] | ||||||||||||
| Sensitive Drug | IDH1 inhibitors | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132H (c.395G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.65 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.88 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
M
S
S
K
K
K
K
I
I
S
S
G
G
G
G
S
S
10
|
V
V
V
V
E
E
M
M
Q
Q
G
G
D
D
E
E
M
M
T
T
20
|
R
R
I
I
I
I
W
W
E
E
L
L
I
I
K
K
E
E
K
K
30
|
L
L
I
I
F
F
P
P
Y
Y
V
V
E
E
L
L
D
D
L
L
40
|
H
H
S
S
Y
Y
D
D
L
L
G
G
I
I
E
E
N
N
R
R
50
|
D
D
A
A
T
T
N
N
D
D
Q
Q
V
V
T
T
K
K
D
D
60
|
A
A
A
A
E
E
A
A
I
I
K
K
K
K
H
H
N
N
V
V
70
|
G
G
V
V
K
K
C
C
A
A
T
T
I
I
T
T
P
P
D
D
80
|
E
E
K
K
R
R
V
V
E
E
E
E
F
F
K
K
L
L
K
K
90
|
Q
Q
M
M
W
W
K
K
S
S
P
P
N
N
G
G
T
T
I
I
100
|
R
R
N
N
I
I
L
L
G
G
G
G
T
T
V
V
F
F
R
R
110
|
E
E
A
A
I
I
I
I
C
C
K
K
N
N
I
I
P
P
R
R
120
|
L
L
V
V
S
S
G
G
W
W
V
V
K
K
P
P
I
I
I
I
130
|
I
I
G
G
R
H
H
H
A
A
Y
Y
G
G
D
D
Q
Q
Y
Y
140
|
R
R
A
A
T
T
D
D
F
F
V
V
V
V
P
P
G
G
P
P
150
|
G
G
K
K
V
V
E
E
I
I
T
T
Y
Y
T
T
P
P
S
S
160
|
D
D
G
G
T
T
Q
Q
K
K
V
V
T
T
Y
Y
L
L
V
V
170
|
H
H
N
N
F
F
E
E
E
E
G
G
G
G
G
G
V
V
A
A
180
|
M
M
G
G
M
M
Y
Y
N
N
Q
Q
D
D
K
K
S
S
I
I
190
|
E
E
D
D
F
F
A
A
H
H
S
S
S
S
F
F
Q
Q
M
M
200
|
A
A
L
L
S
S
K
K
G
G
W
W
P
P
L
L
Y
Y
L
L
210
|
S
S
T
T
K
K
N
N
T
T
I
I
L
L
K
K
K
K
Y
Y
220
|
D
D
G
G
R
R
F
F
K
K
D
D
I
I
F
F
Q
Q
E
E
230
|
I
I
Y
Y
D
D
K
K
Q
Q
Y
Y
K
K
S
S
Q
Q
F
F
240
|
E
E
A
A
Q
Q
K
K
I
I
W
W
Y
Y
E
E
H
H
R
R
250
|
L
L
I
I
D
D
D
D
M
M
V
V
A
A
Q
Q
A
A
M
M
260
|
K
K
S
S
E
E
G
G
G
G
F
F
I
I
W
W
A
A
C
C
270
|
K
K
N
N
Y
Y
D
D
G
G
D
D
V
V
Q
Q
S
S
D
D
280
|
S
S
V
V
A
A
Q
Q
G
G
Y
Y
G
G
S
S
L
L
G
G
290
|
M
M
M
M
T
T
S
S
V
V
L
L
V
V
C
C
P
P
D
D
300
|
G
G
K
K
T
T
V
V
E
E
A
A
E
E
A
A
A
A
H
H
310
|
G
G
T
T
V
V
T
T
R
R
H
H
Y
Y
R
R
M
M
Y
Y
320
|
Q
Q
K
K
G
G
Q
Q
E
E
T
T
S
S
T
T
N
N
P
P
330
|
I
I
A
A
S
S
I
I
F
F
A
A
W
W
T
T
R
R
G
G
340
|
L
L
A
A
H
H
R
R
A
A
K
K
L
L
D
D
N
N
N
N
350
|
K
K
E
E
L
L
A
A
F
F
F
F
A
A
N
N
A
A
L
L
360
|
E
E
E
E
V
V
S
S
I
I
E
E
T
T
I
I
E
E
A
A
370
|
G
G
F
F
M
M
T
T
K
K
D
D
L
L
A
A
A
A
C
C
380
|
I
I
K
K
G
G
L
L
P
P
N
N
V
V
Q
Q
R
R
S
S
390
|
D
D
Y
Y
L
L
N
N
T
T
F
F
E
E
F
F
M
M
D
D
400
|
K
K
L
L
G
G
E
E
N
N
L
L
K
K
I
I
K
K
L
L
410
|
A
A
Q
Q
A
A
K
K
L
L
S
S
L
L
E
E
H
H
H
H
420
|
H
H
H
H
H
H
H
H
H
H
H
H
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | TS676 cells | Brain | Homo sapiens (Human) | CVCL_A5HX | |||||||||
| TS603 cells | Brain | Homo sapiens (Human) | CVCL_A5HW | ||||||||||
| TS516 cells | Brain | Homo sapiens (Human) | CVCL_A5HY | ||||||||||
| In Vivo Model | SCID mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
Soft agar assay | ||||||||||||
| Mechanism Description | The missense mutation p.R132H (c.395G>A) in gene IDH1 cause the sensitivity of IDH1 inhibitors by aberration of the drug's therapeutic target | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [5] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | PARP inhibitors | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132H (c.395G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.65 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.88 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
M
S
S
K
K
K
K
I
I
S
S
G
G
G
G
S
S
10
|
V
V
V
V
E
E
M
M
Q
Q
G
G
D
D
E
E
M
M
T
T
20
|
R
R
I
I
I
I
W
W
E
E
L
L
I
I
K
K
E
E
K
K
30
|
L
L
I
I
F
F
P
P
Y
Y
V
V
E
E
L
L
D
D
L
L
40
|
H
H
S
S
Y
Y
D
D
L
L
G
G
I
I
E
E
N
N
R
R
50
|
D
D
A
A
T
T
N
N
D
D
Q
Q
V
V
T
T
K
K
D
D
60
|
A
A
A
A
E
E
A
A
I
I
K
K
K
K
H
H
N
N
V
V
70
|
G
G
V
V
K
K
C
C
A
A
T
T
I
I
T
T
P
P
D
D
80
|
E
E
K
K
R
R
V
V
E
E
E
E
F
F
K
K
L
L
K
K
90
|
Q
Q
M
M
W
W
K
K
S
S
P
P
N
N
G
G
T
T
I
I
100
|
R
R
N
N
I
I
L
L
G
G
G
G
T
T
V
V
F
F
R
R
110
|
E
E
A
A
I
I
I
I
C
C
K
K
N
N
I
I
P
P
R
R
120
|
L
L
V
V
S
S
G
G
W
W
V
V
K
K
P
P
I
I
I
I
130
|
I
I
G
G
R
H
H
H
A
A
Y
Y
G
G
D
D
Q
Q
Y
Y
140
|
R
R
A
A
T
T
D
D
F
F
V
V
V
V
P
P
G
G
P
P
150
|
G
G
K
K
V
V
E
E
I
I
T
T
Y
Y
T
T
P
P
S
S
160
|
D
D
G
G
T
T
Q
Q
K
K
V
V
T
T
Y
Y
L
L
V
V
170
|
H
H
N
N
F
F
E
E
E
E
G
G
G
G
G
G
V
V
A
A
180
|
M
M
G
G
M
M
Y
Y
N
N
Q
Q
D
D
K
K
S
S
I
I
190
|
E
E
D
D
F
F
A
A
H
H
S
S
S
S
F
F
Q
Q
M
M
200
|
A
A
L
L
S
S
K
K
G
G
W
W
P
P
L
L
Y
Y
L
L
210
|
S
S
T
T
K
K
N
N
T
T
I
I
L
L
K
K
K
K
Y
Y
220
|
D
D
G
G
R
R
F
F
K
K
D
D
I
I
F
F
Q
Q
E
E
230
|
I
I
Y
Y
D
D
K
K
Q
Q
Y
Y
K
K
S
S
Q
Q
F
F
240
|
E
E
A
A
Q
Q
K
K
I
I
W
W
Y
Y
E
E
H
H
R
R
250
|
L
L
I
I
D
D
D
D
M
M
V
V
A
A
Q
Q
A
A
M
M
260
|
K
K
S
S
E
E
G
G
G
G
F
F
I
I
W
W
A
A
C
C
270
|
K
K
N
N
Y
Y
D
D
G
G
D
D
V
V
Q
Q
S
S
D
D
280
|
S
S
V
V
A
A
Q
Q
G
G
Y
Y
G
G
S
S
L
L
G
G
290
|
M
M
M
M
T
T
S
S
V
V
L
L
V
V
C
C
P
P
D
D
300
|
G
G
K
K
T
T
V
V
E
E
A
A
E
E
A
A
A
A
H
H
310
|
G
G
T
T
V
V
T
T
R
R
H
H
Y
Y
R
R
M
M
Y
Y
320
|
Q
Q
K
K
G
G
Q
Q
E
E
T
T
S
S
T
T
N
N
P
P
330
|
I
I
A
A
S
S
I
I
F
F
A
A
W
W
T
T
R
R
G
G
340
|
L
L
A
A
H
H
R
R
A
A
K
K
L
L
D
D
N
N
N
N
350
|
K
K
E
E
L
L
A
A
F
F
F
F
A
A
N
N
A
A
L
L
360
|
E
E
E
E
V
V
S
S
I
I
E
E
T
T
I
I
E
E
A
A
370
|
G
G
F
F
M
M
T
T
K
K
D
D
L
L
A
A
A
A
C
C
380
|
I
I
K
K
G
G
L
L
P
P
N
N
V
V
Q
Q
R
R
S
S
390
|
D
D
Y
Y
L
L
N
N
T
T
F
F
E
E
F
F
M
M
D
D
400
|
K
K
L
L
G
G
E
E
N
N
L
L
K
K
I
I
K
K
L
L
410
|
A
A
Q
Q
A
A
K
K
L
L
S
S
L
L
E
E
H
H
H
H
420
|
H
H
H
H
H
H
H
H
H
H
H
H
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | IDH2 cells | Ovary | Homo sapiens (Human) | CVCL_D3DY | |||||||||
| IDH1 cells | Ovary | Homo sapiens (Human) | CVCL_D3DY | ||||||||||
| In Vivo Model | Female athymic nu/nu mouse PDX model | Mus musculus | |||||||||||
| Experiment for Drug Resistance |
Promega assay | ||||||||||||
| Mechanism Description | The oncometabolite, 2-hydroxyglutarate, renders IDH1/2 mutant cancer cells deficient in homologous recombination and confers vulnerability to synthetic lethal targeting with PARP inhibitors. | ||||||||||||
Investigative Drug(s)
1 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: FGFR-tacc positive glioblastoma [ICD-11: 2A00.01] | [12] | ||||||||||||
| Sensitive Disease | FGFR-tacc positive glioblastoma [ICD-11: 2A00.01] | ||||||||||||
| Sensitive Drug | Temozolomide/Vandetanib | ||||||||||||
| Molecule Alteration | Missense mutation | p.R132H (c.395G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.65 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.88 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
M
S
S
K
K
K
K
I
I
S
S
G
G
G
G
S
S
10
|
V
V
V
V
E
E
M
M
Q
Q
G
G
D
D
E
E
M
M
T
T
20
|
R
R
I
I
I
I
W
W
E
E
L
L
I
I
K
K
E
E
K
K
30
|
L
L
I
I
F
F
P
P
Y
Y
V
V
E
E
L
L
D
D
L
L
40
|
H
H
S
S
Y
Y
D
D
L
L
G
G
I
I
E
E
N
N
R
R
50
|
D
D
A
A
T
T
N
N
D
D
Q
Q
V
V
T
T
K
K
D
D
60
|
A
A
A
A
E
E
A
A
I
I
K
K
K
K
H
H
N
N
V
V
70
|
G
G
V
V
K
K
C
C
A
A
T
T
I
I
T
T
P
P
D
D
80
|
E
E
K
K
R
R
V
V
E
E
E
E
F
F
K
K
L
L
K
K
90
|
Q
Q
M
M
W
W
K
K
S
S
P
P
N
N
G
G
T
T
I
I
100
|
R
R
N
N
I
I
L
L
G
G
G
G
T
T
V
V
F
F
R
R
110
|
E
E
A
A
I
I
I
I
C
C
K
K
N
N
I
I
P
P
R
R
120
|
L
L
V
V
S
S
G
G
W
W
V
V
K
K
P
P
I
I
I
I
130
|
I
I
G
G
R
H
H
H
A
A
Y
Y
G
G
D
D
Q
Q
Y
Y
140
|
R
R
A
A
T
T
D
D
F
F
V
V
V
V
P
P
G
G
P
P
150
|
G
G
K
K
V
V
E
E
I
I
T
T
Y
Y
T
T
P
P
S
S
160
|
D
D
G
G
T
T
Q
Q
K
K
V
V
T
T
Y
Y
L
L
V
V
170
|
H
H
N
N
F
F
E
E
E
E
G
G
G
G
G
G
V
V
A
A
180
|
M
M
G
G
M
M
Y
Y
N
N
Q
Q
D
D
K
K
S
S
I
I
190
|
E
E
D
D
F
F
A
A
H
H
S
S
S
S
F
F
Q
Q
M
M
200
|
A
A
L
L
S
S
K
K
G
G
W
W
P
P
L
L
Y
Y
L
L
210
|
S
S
T
T
K
K
N
N
T
T
I
I
L
L
K
K
K
K
Y
Y
220
|
D
D
G
G
R
R
F
F
K
K
D
D
I
I
F
F
Q
Q
E
E
230
|
I
I
Y
Y
D
D
K
K
Q
Q
Y
Y
K
K
S
S
Q
Q
F
F
240
|
E
E
A
A
Q
Q
K
K
I
I
W
W
Y
Y
E
E
H
H
R
R
250
|
L
L
I
I
D
D
D
D
M
M
V
V
A
A
Q
Q
A
A
M
M
260
|
K
K
S
S
E
E
G
G
G
G
F
F
I
I
W
W
A
A
C
C
270
|
K
K
N
N
Y
Y
D
D
G
G
D
D
V
V
Q
Q
S
S
D
D
280
|
S
S
V
V
A
A
Q
Q
G
G
Y
Y
G
G
S
S
L
L
G
G
290
|
M
M
M
M
T
T
S
S
V
V
L
L
V
V
C
C
P
P
D
D
300
|
G
G
K
K
T
T
V
V
E
E
A
A
E
E
A
A
A
A
H
H
310
|
G
G
T
T
V
V
T
T
R
R
H
H
Y
Y
R
R
M
M
Y
Y
320
|
Q
Q
K
K
G
G
Q
Q
E
E
T
T
S
S
T
T
N
N
P
P
330
|
I
I
A
A
S
S
I
I
F
F
A
A
W
W
T
T
R
R
G
G
340
|
L
L
A
A
H
H
R
R
A
A
K
K
L
L
D
D
N
N
N
N
350
|
K
K
E
E
L
L
A
A
F
F
F
F
A
A
N
N
A
A
L
L
360
|
E
E
E
E
V
V
S
S
I
I
E
E
T
T
I
I
E
E
A
A
370
|
G
G
F
F
M
M
T
T
K
K
D
D
L
L
A
A
A
A
C
C
380
|
I
I
K
K
G
G
L
L
P
P
N
N
V
V
Q
Q
R
R
S
S
390
|
D
D
Y
Y
L
L
N
N
T
T
F
F
E
E
F
F
M
M
D
D
400
|
K
K
L
L
G
G
E
E
N
N
L
L
K
K
I
I
K
K
L
L
410
|
A
A
Q
Q
A
A
K
K
L
L
S
S
L
L
E
E
H
H
H
H
420
|
H
H
H
H
H
H
H
H
H
H
H
H
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Brain | N.A. | |||||||||||
| Experiment for Molecule Alteration |
Multiplex array; Standard ELISA assay | ||||||||||||
| Experiment for Drug Resistance |
Pharmacokinetics analysis | ||||||||||||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Nervous tissue | |
| The Specified Disease | Brain cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.42E-176; Fold-change: 1.87E+00; Z-score: 2.54E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.08E-01; Fold-change: -1.90E-01; Z-score: -2.22E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
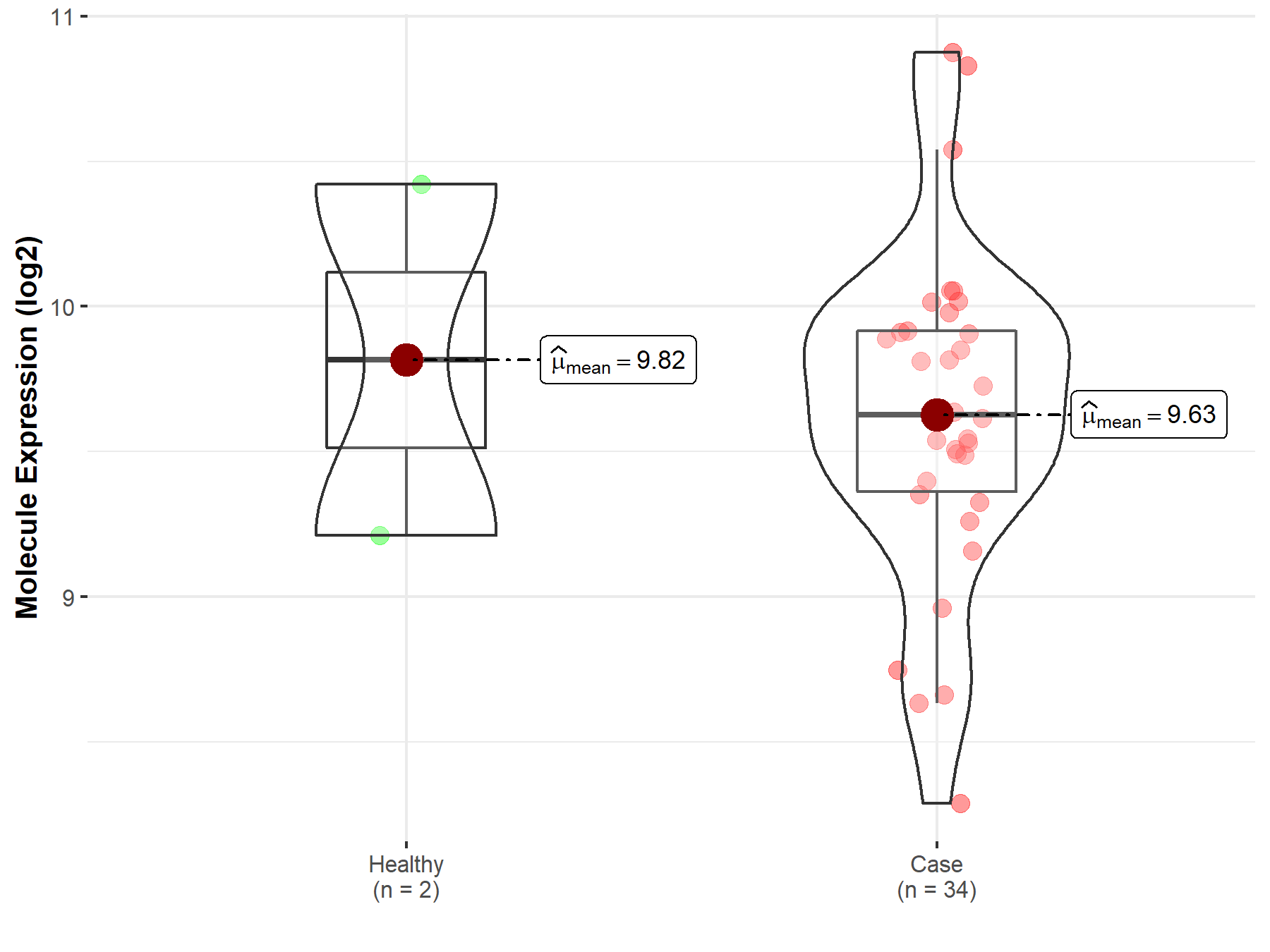
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | White matter | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.47E-01; Fold-change: 2.98E-01; Z-score: 6.75E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
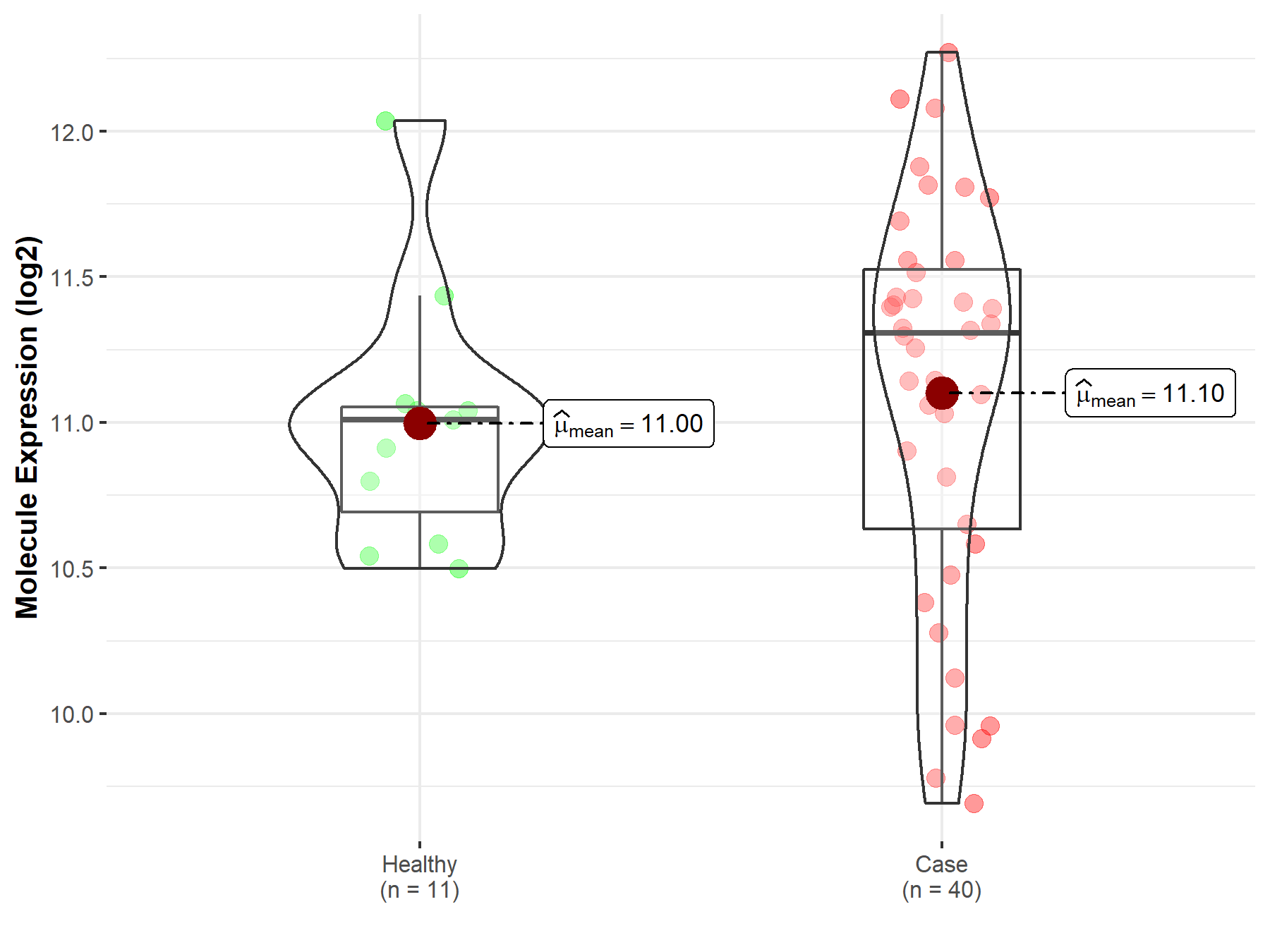
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Neuroectodermal tumor | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.39E-07; Fold-change: 2.16E+00; Z-score: 3.41E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
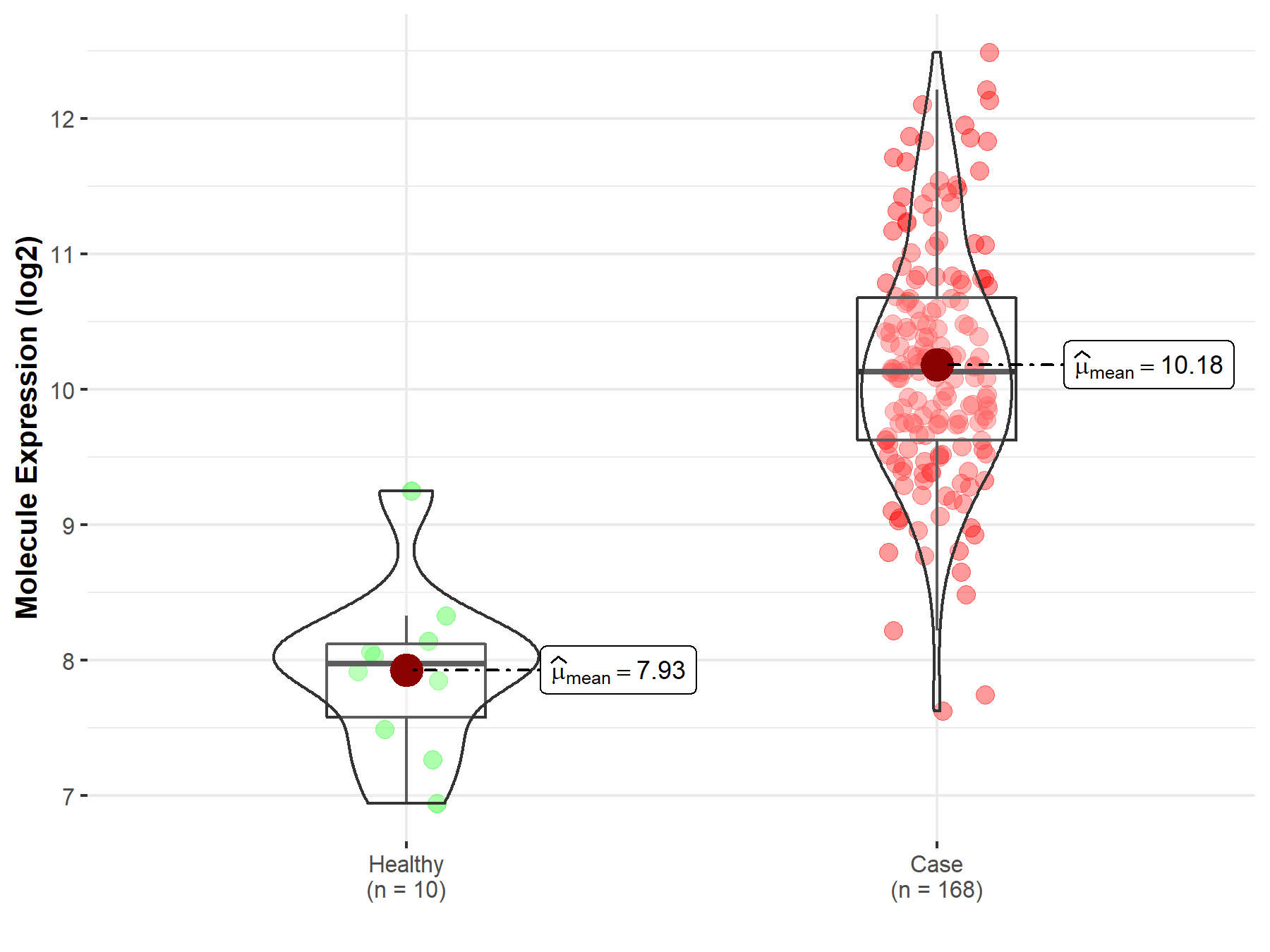
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Bone marrow | |
| The Specified Disease | Acute myeloid leukemia | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.34E-03; Fold-change: -5.35E-02; Z-score: -4.78E-02 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
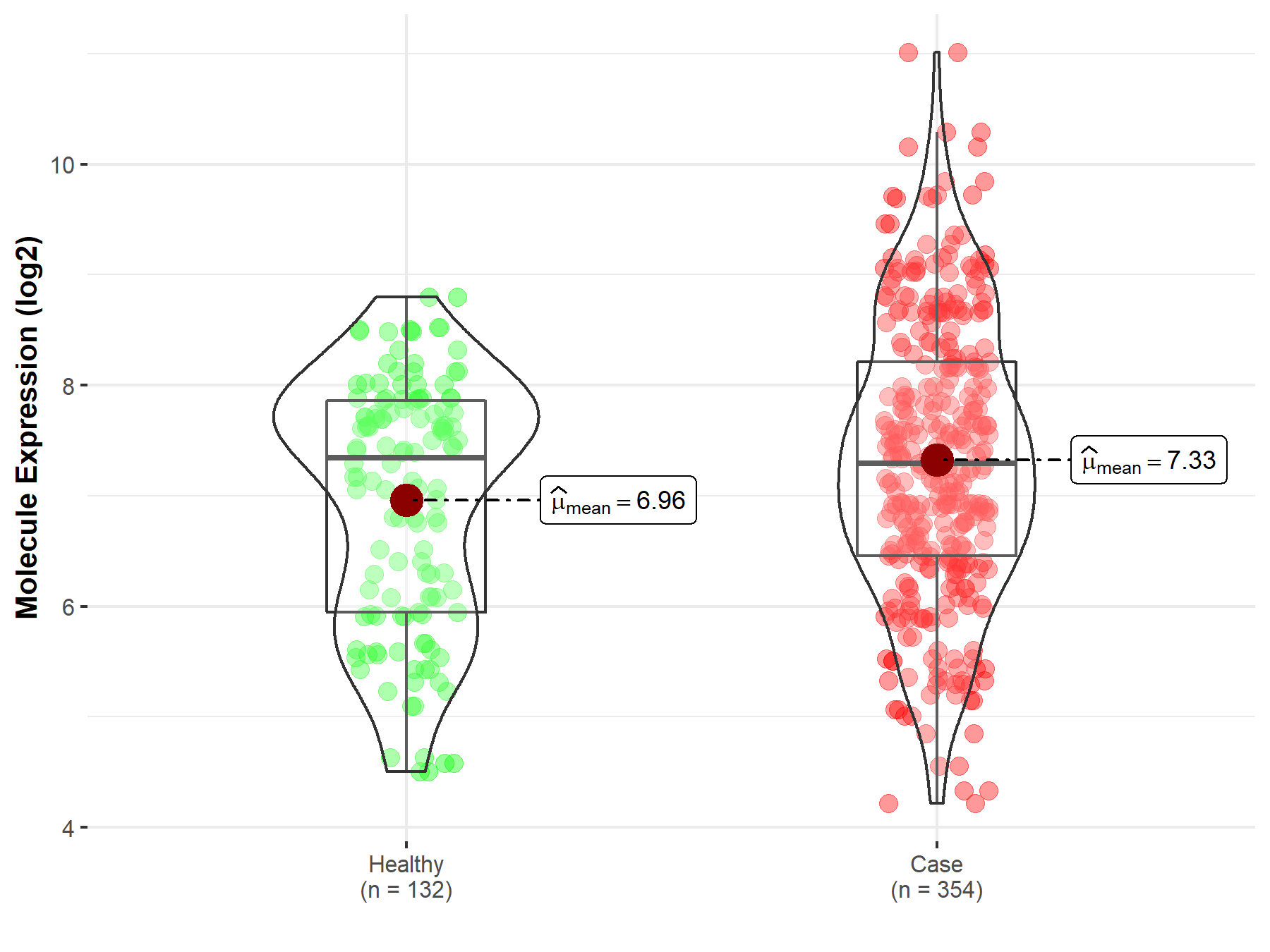
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Colon | |
| The Specified Disease | Colon cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.24E-32; Fold-change: -4.05E-01; Z-score: -1.35E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 4.33E-01; Fold-change: -1.34E-01; Z-score: -2.49E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
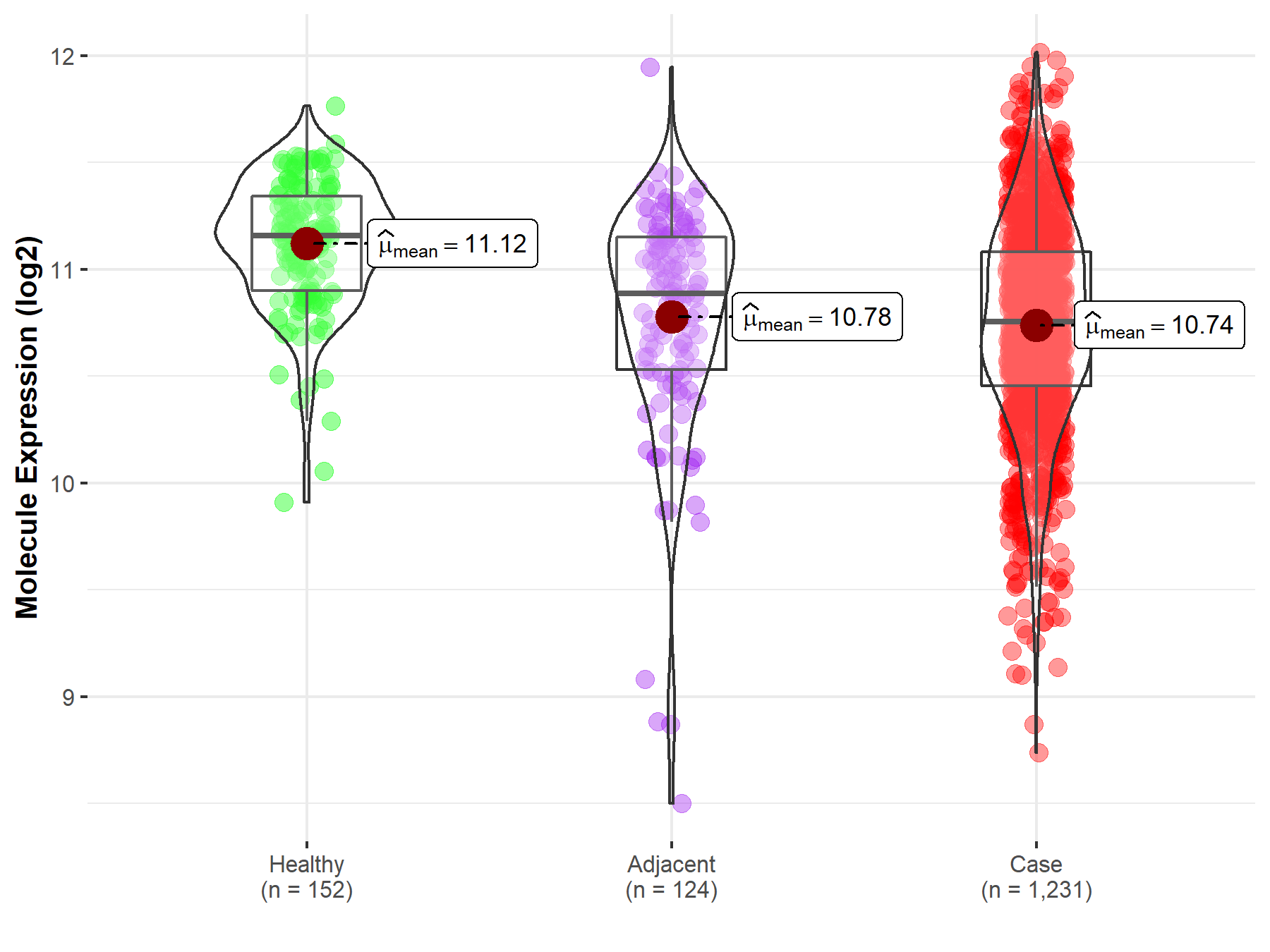
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Muscle | |
| The Specified Disease | Sarcoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.89E-33; Fold-change: 4.54E-01; Z-score: 9.43E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.10E-06; Fold-change: 6.55E-01; Z-score: 8.02E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
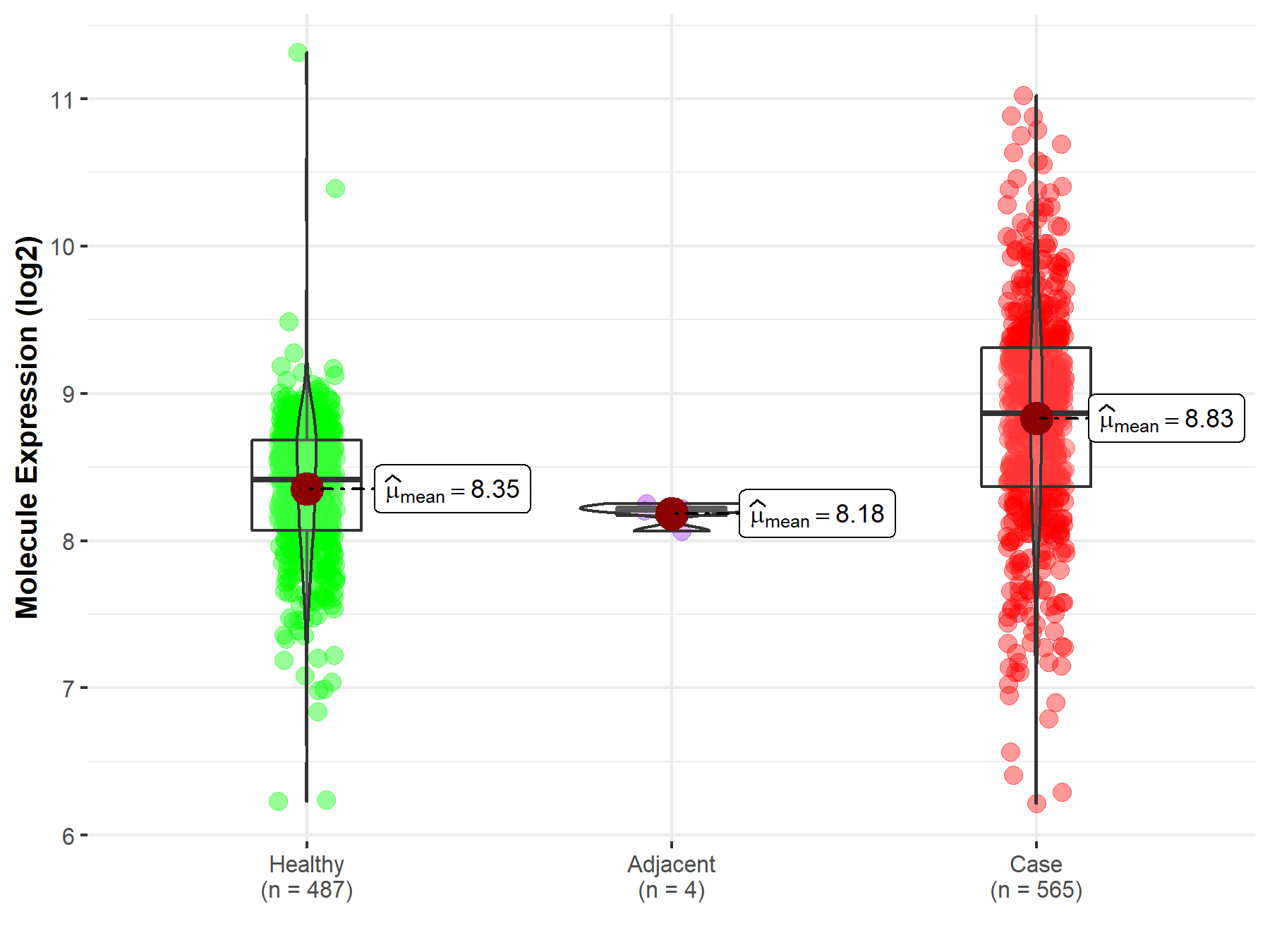
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

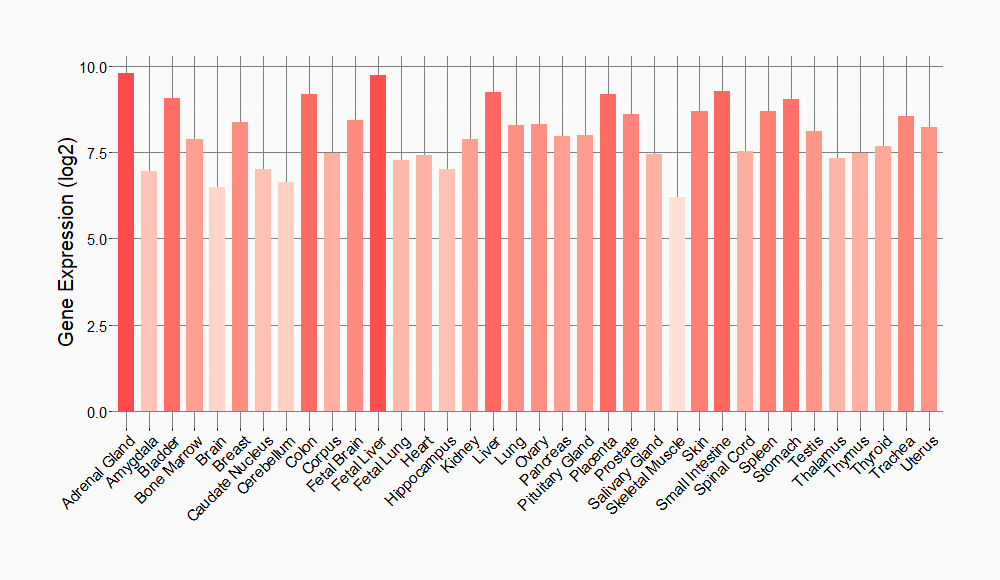
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
