Drug Information
Drug (ID: DG00001) and It's Reported Resistant Information
| Name |
Triclosan
|
||||
|---|---|---|---|---|---|
| Synonyms |
Triclosan; 3380-34-5; 5-CHLORO-2-(2,4-DICHLOROPHENOXY)PHENOL; 2,4,4'-Trichloro-2'-hydroxydiphenyl ether; Irgasan; Cloxifenolum; Triclosanum; Irgasan DP300; Stri-Dex Face Wash; Phenol, 5-chloro-2-(2,4-dichlorophenoxy)-; Lexol 300; Stri-Dex Cleansing Bar; 5-Chloro-2-(2,4-dichloro-phenoxy)-phenol; Triclosanum [INN-Latin]; CH 3565; DP-300; Caswell No 186A; C12H7Cl3O2; UNII-4NM5039Y5X; HSDB 7194; CHEBI:164200; Ether, 2'-hydroxy-2,4,4'-trichlorodiphenyl; Phenyl ether, 2'-hydroxy-2,4,4'-trichloro-; EINECS 222-182-2; CHEMBL849; EPA; Aquasept; Cliniclean; Cloxifenol; Manusept; Sapoderm; TCL; Trisan; Clearasil DailyFace Wash; Dermtek Brand of Triclosan; GlaxoSmithKline Brand of Triclosan; Microshield T; Oxy Skin Wash; Pharmachem Brand of Triclosan; Reckitt Brand of Triclosan; SSL Brand of Triclosan; Ster Zac Bath Concentrate; SterZac Bath Concentrate; Trans Canaderm Brand of Triclosan; Triclosan Pharmachem Brand; Triclosan Reckitt Brand; IN1424; Irgasan DP 300; CH-3565; Johnson & Johnson Brand of Triclosan; Procter & Gamble Brand of Triclosan; Ster-Zac Bath Concentrate; Stri-Dex cleansing bar (TN); Triclosan (USP/INN); Triclosan [USAN:BAN:INN]; 2,4,4'-Trichloro-2'-hydroxy diphenyl ether; 2-Hydroxy-2',4,4'-trichlorodiphenyl Ether; WL-1001
Click to Show/Hide
|
||||
| Indication |
In total 2 Indication(s)
|
||||
| Structure |
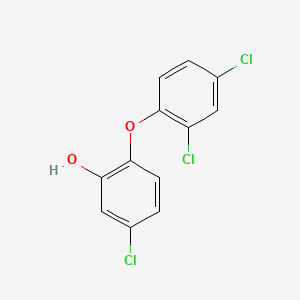
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Clinically Reported Resistance for This Drug
(1 diseases)
[2]
Disease(s) with Resistance Information Validated by in-vivo Model for This Drug
(1 diseases)
[1]
|
||||
| Target | Plasmodium Enoyl-ACP reductase (Malaria fabI) | Q965D5_PLAFA | [1] | ||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C12H7Cl3O2
|
||||
| IsoSMILES |
C1=CC(=C(C=C1Cl)O)OC2=C(C=C(C=C2)Cl)Cl
|
||||
| InChI |
1S/C12H7Cl3O2/c13-7-1-3-11(9(15)5-7)17-12-4-2-8(14)6-10(12)16/h1-6,16H
|
||||
| InChIKey |
XEFQLINVKFYRCS-UHFFFAOYSA-N
|
||||
| PubChem CID | |||||
| ChEBI ID | |||||
| TTD Drug ID | |||||
| DrugBank ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-01: Infectious/parasitic diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Key Molecule: NADH-dependent enoyl-ACP reductase (FABL) | [1] | ||||||||||||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | ||||||||||||
| Molecule Alteration | Missense mutation | p.G93V |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.54 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.90 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
G
-
D
-
H
-
0
|
G
-
M
M
G
G
F
F
L
L
S
S
G
G
K
K
R
R
I
I
10
|
L
L
V
V
T
T
G
G
V
V
A
A
S
S
K
K
L
L
S
S
20
|
I
I
A
A
Y
Y
G
G
I
I
A
A
Q
Q
A
A
M
M
H
H
30
|
R
R
E
E
G
G
A
A
E
E
L
L
A
A
F
F
T
T
Y
Y
40
|
Q
Q
N
N
D
D
K
K
L
L
K
K
G
G
R
R
V
V
E
E
50
|
E
E
F
F
A
A
A
A
Q
Q
L
L
G
G
S
S
D
D
I
I
60
|
V
V
L
L
Q
Q
C
C
D
D
V
V
A
A
E
E
D
D
A
A
70
|
S
S
I
I
D
D
T
T
M
M
F
F
A
A
E
E
L
L
G
G
80
|
K
K
V
V
W
W
P
P
K
K
F
F
D
D
G
G
F
F
V
V
90
|
H
H
S
S
I
I
G
V
F
F
A
A
P
P
G
G
D
D
Q
Q
100
|
L
L
D
D
G
G
D
D
Y
Y
V
V
N
N
A
A
V
V
T
T
110
|
R
R
E
E
G
G
F
F
K
K
I
I
A
A
H
H
D
D
I
I
120
|
S
S
S
S
Y
Y
S
S
F
F
V
V
A
A
M
M
A
A
K
K
130
|
A
A
C
C
R
R
S
S
M
M
L
L
N
N
P
P
G
G
S
S
140
|
A
A
L
L
L
L
T
T
L
L
S
S
Y
Y
L
L
G
G
A
A
150
|
E
E
R
R
A
A
I
I
P
P
N
N
Y
Y
N
N
V
V
M
M
160
|
G
G
L
L
A
A
K
K
A
A
S
S
L
L
E
E
A
A
N
N
170
|
V
V
R
R
Y
Y
M
M
A
A
N
N
A
A
M
M
G
G
P
P
180
|
E
E
G
G
V
V
R
R
V
V
N
N
A
A
I
I
S
S
A
A
190
|
G
G
P
P
I
I
R
R
T
T
L
L
A
A
A
A
S
S
G
G
200
|
I
I
K
K
D
D
F
F
R
R
K
K
M
M
L
L
A
A
H
H
210
|
C
C
E
E
A
A
V
V
T
T
P
P
I
I
R
R
R
R
T
T
220
|
V
V
T
T
I
I
E
E
D
D
V
V
G
G
N
N
S
S
A
A
230
|
A
A
F
F
L
L
C
C
S
S
D
D
L
L
S
S
A
A
G
G
240
|
I
I
S
S
G
G
E
E
V
V
V
V
H
H
V
V
D
D
G
G
250
|
G
G
F
F
S
S
I
I
A
A
A
A
M
M
N
N
E
E
L
L
260
|
E
E
L
L
K
K
-
L
-
E
-
H
-
H
-
H
-
H
-
H
270
|
-
H
|
|||||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | E. coli IFN4 | 562 | |||||||||||
| E. coli imp4231 FabI(G93V) strain | 562 | ||||||||||||
| Experiment for Drug Resistance |
MIC assay | ||||||||||||
| Mechanism Description | Among the triclosan-resistant mutants, E. coli AGT11, which had a Gly93 Val mutation in fabI, FabI(G93V), showed a 95-fold higher MIC than the wild-type. On the basis of these results, Levy et al.7 carried out structural analysis and inhibition experiments on a complex of E. coli FabI with triclosan and NAD+ and found that a FabI NAD+ triclosan ternary complex was formed by face-to-face interaction between the phenol ring of triclosan and the nicotinamide ring of NAD+ in the active site of FabI. In contrast to intact FabI, overexpression of FabI(G93V) conferred greater triclosan resistance by preventing formation of the FabI NAD+ triclosan ternary complex. | ||||||||||||
|
|
|||||||||||||
| Key Molecule: Uncharacterized oxidoreductase YeiQ (YEIQ) | [1] | ||||||||||||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | E. coli IFN4 | 562 | |||||||||||
| Experiment for Molecule Alteration |
Microarray hybridization assay | ||||||||||||
| Experiment for Drug Resistance |
MIC assay | ||||||||||||
| Mechanism Description | A FabI NAD+ triclosan ternary complex is formed by face-to-face interaction between the phenol ring of triclosan and the nicotinamide ring of NAD+ in the active site of FabI (enoyl-acyl carrier protein reductase) and therefore highly expressed reductases (narGHJI, garR, hcp and yeaA) and dehydrogenases (fdnGHI, ykgEF, garD, gldA and yeiQ) could bind triclosan which were as the NAD(P) cofactor, thus lowering the effective triclosan concentration. | ||||||||||||
|
|
|||||||||||||
| Key Molecule: Tartronate semialdehyde reductase (TSAR) | [1] | ||||||||||||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | E. coli IFN4 | 562 | |||||||||||
| Experiment for Molecule Alteration |
Microarray hybridization assay | ||||||||||||
| Experiment for Drug Resistance |
MIC assay | ||||||||||||
| Mechanism Description | A FabI NAD+ triclosan ternary complex is formed by face-to-face interaction between the phenol ring of triclosan and the nicotinamide ring of NAD+ in the active site of FabI (enoyl-acyl carrier protein reductase) and therefore highly expressed reductases (narGHJI, garR, hcp and yeaA) and dehydrogenases (fdnGHI, ykgEF, garD, gldA and yeiQ) could bind triclosan which were as the NAD(P) cofactor, thus lowering the effective triclosan concentration. | ||||||||||||
| Key Molecule: Glycerol dehydrogenase (GLDA) | [1] | ||||||||||||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | E. coli IFN4 | 562 | |||||||||||
| Experiment for Molecule Alteration |
Microarray hybridization assay | ||||||||||||
| Experiment for Drug Resistance |
MIC assay | ||||||||||||
| Mechanism Description | A FabI NAD+ triclosan ternary complex is formed by face-to-face interaction between the phenol ring of triclosan and the nicotinamide ring of NAD+ in the active site of FabI (enoyl-acyl carrier protein reductase) and therefore highly expressed reductases (narGHJI, garR, hcp and yeaA) and dehydrogenases (fdnGHI, ykgEF, garD, gldA and yeiQ) could bind triclosan which were as the NAD(P) cofactor, thus lowering the effective triclosan concentration. | ||||||||||||
| Key Molecule: Nitrate/nitrite transporter NarU (NARU) | [1] | ||||||||||||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | E. coli IFN4 | 562 | |||||||||||
| Experiment for Molecule Alteration |
Microarray hybridization assay | ||||||||||||
| Experiment for Drug Resistance |
MIC assay | ||||||||||||
| Mechanism Description | Concerning up-regulated porins and transporters (ompF, ompC, ykgG, ydjXYZ), they can either provide an efflux mechanism to export triclosan from the cells or accelerate the import of triclosan into the cytoplasm before the cell membrane is destabilized, thereby contributing to increasing the MICs of triclosan. | ||||||||||||
| Key Molecule: Outer membrane porin C (OMPC) | [1] | ||||||||||||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | E. coli IFN4 | 562 | |||||||||||
| Experiment for Molecule Alteration |
Microarray hybridization assay | ||||||||||||
| Experiment for Drug Resistance |
MIC assay | ||||||||||||
| Mechanism Description | Concerning up-regulated porins and transporters (ompF, ompC, ykgG, ydjXYZ), they can either provide an efflux mechanism to export triclosan from the cells or accelerate the import of triclosan into the cytoplasm before the cell membrane is destabilized, thereby contributing to increasing the MICs of triclosan. | ||||||||||||
| Key Molecule: Outer membrane porin F (OMPF) | [1] | ||||||||||||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | E. coli IFN4 | 562 | |||||||||||
| Experiment for Molecule Alteration |
Microarray hybridization assay | ||||||||||||
| Experiment for Drug Resistance |
MIC assay | ||||||||||||
| Mechanism Description | Concerning up-regulated porins and transporters (ompF, ompC, ykgG, ydjXYZ), they can either provide an efflux mechanism to export triclosan from the cells or accelerate the import of triclosan into the cytoplasm before the cell membrane is destabilized, thereby contributing to increasing the MICs of triclosan. | ||||||||||||
| Key Molecule: Unclear drug transporter ydjXYZ (DT-unclear) | [1] | ||||||||||||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | E. coli IFN4 | 562 | |||||||||||
| Experiment for Molecule Alteration |
Microarray hybridization assay | ||||||||||||
| Experiment for Drug Resistance |
MIC assay | ||||||||||||
| Mechanism Description | Concerning up-regulated porins and transporters (ompF, ompC, ykgG, ydjXYZ), they can either provide an efflux mechanism to export triclosan from the cells or accelerate the import of triclosan into the cytoplasm before the cell membrane is destabilized, thereby contributing to increasing the MICs of triclosan. | ||||||||||||
| Key Molecule: Unclear drug transporter ykgEF (DT-unclear) | [1] | ||||||||||||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | E. coli IFN4 | 562 | |||||||||||
| Experiment for Molecule Alteration |
Microarray hybridization assay | ||||||||||||
| Experiment for Drug Resistance |
MIC assay | ||||||||||||
| Mechanism Description | A FabI NAD+ triclosan ternary complex is formed by face-to-face interaction between the phenol ring of triclosan and the nicotinamide ring of NAD+ in the active site of FabI (enoyl-acyl carrier protein reductase) and therefore highly expressed reductases (narGHJI, garR, hcp and yeaA) and dehydrogenases (fdnGHI, ykgEF, garD, gldA and yeiQ) could bind triclosan which were as the NAD(P) cofactor, thus lowering the effective triclosan concentration. | ||||||||||||
| Key Molecule: Uncharacterized protein YkgG (YKGG) | [1] | ||||||||||||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | E. coli IFN4 | 562 | |||||||||||
| Experiment for Molecule Alteration |
Microarray hybridization assay | ||||||||||||
| Experiment for Drug Resistance |
MIC assay | ||||||||||||
| Mechanism Description | Concerning up-regulated porins and transporters (ompF, ompC, ykgG, ydjXYZ), they can either provide an efflux mechanism to export triclosan from the cells or accelerate the import of triclosan into the cytoplasm before the cell membrane is destabilized, thereby contributing to increasing the MICs of triclosan. | ||||||||||||
|
|
|||||||||||||
| Key Molecule: FDH-N subunit gamma (FDHNI) | [1] | ||||||||||||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | E. coli IFN4 | 562 | |||||||||||
| Experiment for Molecule Alteration |
Microarray hybridization assay | ||||||||||||
| Experiment for Drug Resistance |
MIC assay | ||||||||||||
| Mechanism Description | A FabI NAD+ triclosan ternary complex is formed by face-to-face interaction between the phenol ring of triclosan and the nicotinamide ring of NAD+ in the active site of FabI (enoyl-acyl carrier protein reductase) and therefore highly expressed reductases (narGHJI, garR, hcp and yeaA) and dehydrogenases (fdnGHI, ykgEF, garD, gldA and yeiQ) could bind triclosan which were as the NAD(P) cofactor, thus lowering the effective triclosan concentration. | ||||||||||||
| Key Molecule: Galactarate dehydratase (L-threo-forming) (GARD) | [1] | ||||||||||||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | E. coli IFN4 | 562 | |||||||||||
| Experiment for Molecule Alteration |
Microarray hybridization assay | ||||||||||||
| Experiment for Drug Resistance |
MIC assay | ||||||||||||
| Mechanism Description | A FabI NAD+ triclosan ternary complex is formed by face-to-face interaction between the phenol ring of triclosan and the nicotinamide ring of NAD+ in the active site of FabI (enoyl-acyl carrier protein reductase) and therefore highly expressed reductases (narGHJI, garR, hcp and yeaA) and dehydrogenases (fdnGHI, ykgEF, garD, gldA and yeiQ) could bind triclosan which were as the NAD(P) cofactor, thus lowering the effective triclosan concentration. | ||||||||||||
| Key Molecule: Flavohemoprotein (HCP) | [1] | ||||||||||||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | E. coli IFN4 | 562 | |||||||||||
| Experiment for Molecule Alteration |
Microarray hybridization assay | ||||||||||||
| Experiment for Drug Resistance |
MIC assay | ||||||||||||
| Mechanism Description | A FabI NAD+ triclosan ternary complex is formed by face-to-face interaction between the phenol ring of triclosan and the nicotinamide ring of NAD+ in the active site of FabI (enoyl-acyl carrier protein reductase) and therefore highly expressed reductases (narGHJI, garR, hcp and yeaA) and dehydrogenases (fdnGHI, ykgEF, garD, gldA and yeiQ) could bind triclosan which were as the NAD(P) cofactor, thus lowering the effective triclosan concentration. | ||||||||||||
| Key Molecule: Nitrate reductase 1 (narGHJI) | [1] | ||||||||||||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | E. coli IFN4 | 562 | |||||||||||
| Experiment for Molecule Alteration |
Microarray hybridization assay | ||||||||||||
| Experiment for Drug Resistance |
MIC assay | ||||||||||||
| Mechanism Description | A FabI NAD+ triclosan ternary complex is formed by face-to-face interaction between the phenol ring of triclosan and the nicotinamide ring of NAD+ in the active site of FabI (enoyl-acyl carrier protein reductase) and therefore highly expressed reductases (narGHJI, garR, hcp and yeaA) and dehydrogenases (fdnGHI, ykgEF, garD, gldA and yeiQ) could bind triclosan which were as the NAD(P) cofactor, thus lowering the effective triclosan concentration. | ||||||||||||
| Key Molecule: Peptide methionine sulfoxide reductase MsrB (MSRB) | [1] | ||||||||||||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | ||||||||||||
| Molecule Alteration | Expression | Up-regulation |
|||||||||||
| Experimental Note | Discovered Using In-vivo Testing Model | ||||||||||||
| In Vitro Model | E. coli IFN4 | 562 | |||||||||||
| Experiment for Molecule Alteration |
Microarray hybridization assay | ||||||||||||
| Experiment for Drug Resistance |
MIC assay | ||||||||||||
| Mechanism Description | A FabI NAD+ triclosan ternary complex is formed by face-to-face interaction between the phenol ring of triclosan and the nicotinamide ring of NAD+ in the active site of FabI (enoyl-acyl carrier protein reductase) and therefore highly expressed reductases (narGHJI, garR, hcp and yeaA) and dehydrogenases (fdnGHI, ykgEF, garD, gldA and yeiQ) could bind triclosan which were as the NAD(P) cofactor, thus lowering the effective triclosan concentration. | ||||||||||||
ICD-12: Respiratory system diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: MATE family efflux transporter (ABEM) | [2] | |||
| Resistant Disease | Acinetobacter baumannii infection [ICD-11: CA40.4] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli kAM32 | 562 | ||
| Experiment for Drug Resistance |
MIC assay | |||
| Mechanism Description | AbeM was found to be an H+-coupled multidrug efflux pump and a unique member of the MATE family which lead to drug resistance. | |||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
