Molecule Information
General Information of the Molecule (ID: Mol01917)
| Name |
Squalene epoxidase (SQLE)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
SQLE; ERG1
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
SQLE
|
||||
| Gene ID | |||||
| Location |
chr8:124,998,497-125,022,283[+]
|
||||
| Sequence |
MWTFLGIATFTYFYKKFGDFITLANREVLLCVLVFLSLGLVLSYRCRHRNGGLLGRQQSG
SQFALFSDILSGLPFIGFFWAKSPPESENKEQLEARRRRKGTNISETSLIGTAACTSTSS QNDPEVIIVGAGVLGSALAAVLSRDGRKVTVIERDLKEPDRIVGEFLQPGGYHVLKDLGL GDTVEGLDAQVVNGYMIHDQESKSEVQIPYPLSENNQVQSGRAFHHGRFIMSLRKAAMAE PNAKFIEGVVLQLLEEDDVVMGVQYKDKETGDIKELHAPLTVVADGLFSKFRKSLVSNKV SVSSHFVGFLMKNAPQFKANHAELILANPSPVLIYQISSSETRVLVDIRGEMPRNLREYM VEKIYPQIPDHLKEPFLEATDNSHLRSMPASFLPPSSVKKRGVLLLGDAYNMRHPLTGGG MTVAFKDIKLWRKLLKGIPDLYDDAAIFEAKKSFYWARKTSHSFVVNILAQALYELFSAT DDSLHQLRKACFLYFKLGGECVAGPVGLLSVLSPNPLVLIGHFFAVAIYAVYFCFKSEPW ITKPRALLSSGAVLYKACSVIFPLIYSEMKYMVH Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Catalyzes the stereospecific oxidation of squalene to (S)-2,3-epoxysqualene, and is considered to be a rate-limiting enzyme in steroid biosynthesis.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
3 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [1] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Fluvastatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Breast cancer [ICD-11: 2C60] | |||
| The Specified Disease | Breast cancer | |||
| The Studied Tissue | Breast tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.09E-160 Fold-change: 3.12E-01 Z-score: 3.56E+01 |
|||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| Cell Pathway Regulation | Steroid biosynthesis signaling pathway | Activation | hsa00100 | |
| Terpenoid bacKbone biosynthesis signaling pathway | Activation | hsa00900 | ||
| Steroid hormone biosynthesis signaling pathway | Activation | hsa00140 | ||
| In Vitro Model | MCF-10A-neoT cells | Breast | Homo sapiens (Human) | CVCL_5554 |
| In Vivo Model | SV40 C3TAg transgenic mouse model | Mus musculus | ||
| Experiment for Molecule Alteration |
Clariom D RNA profiling assay | |||
| Experiment for Drug Resistance |
MTT assay; Colony formation assay | |||
| Mechanism Description | Acquired resistance to fluvastatin is mediated by restorative upregulation of cholesterol biosynthesis pathway genes. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Prostate cancer [ICD-11: 2C82.0] | [2] | |||
| Metabolic Type | Lipid metabolism | |||
| Resistant Disease | Prostate cancer [ICD-11: 2C82.0] | |||
| Resistant Drug | Bicalutamide | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vivo Model | Male NOD/SCID nude mice, With LNCaP, C4-2B, and C4-2B_shSQLE cells | Mice | ||
| Experiment for Molecule Alteration |
qRT-PCR; Western blot analysis | |||
| Experiment for Drug Resistance |
Tumor volume assay | |||
| Mechanism Description | In our study, we found that the expression level of SQLE was significantly increased in bicalutamide-resistant-C4-2B cells compared to LNCaP cells. SQLE knockdown partly restored the sensitivity of drug-resistant cells to bicalutamide and reduced lymph node metastasis by inhibiting fatty acid oxidation in mitochondria. We also found that terbinafine, the specific inhibitor of SQLE, can enhance the sensitivity of prostate cancer cells to bicalutamide. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: t. indotineae infection [ICD-11: 1F28] | [3] | |||
| Resistant Disease | t. indotineae infection [ICD-11: 1F28] | |||
| Resistant Drug | Terbinafine | |||
| Molecule Alteration | Missense mutation | L38HL |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | T. indotineae | 5475 | ||
| Experiment for Molecule Alteration |
PCR; PCR-ELISA assay; GeneSeq assay | |||
| Experiment for Drug Resistance |
In vitro drug susceptibility testing; Resistance testing | |||
| Mechanism Description | Mutations L393S, L393F, F397L, and F397I of the SQLE gene were associated with terbinafine resistance. Resistance to itraconazole could not be explained by mutations in the ERG11B gene. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.09E-160; Fold-change: 1.33E+00; Z-score: 2.70E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 9.79E-30; Fold-change: 1.20E+00; Z-score: 2.73E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
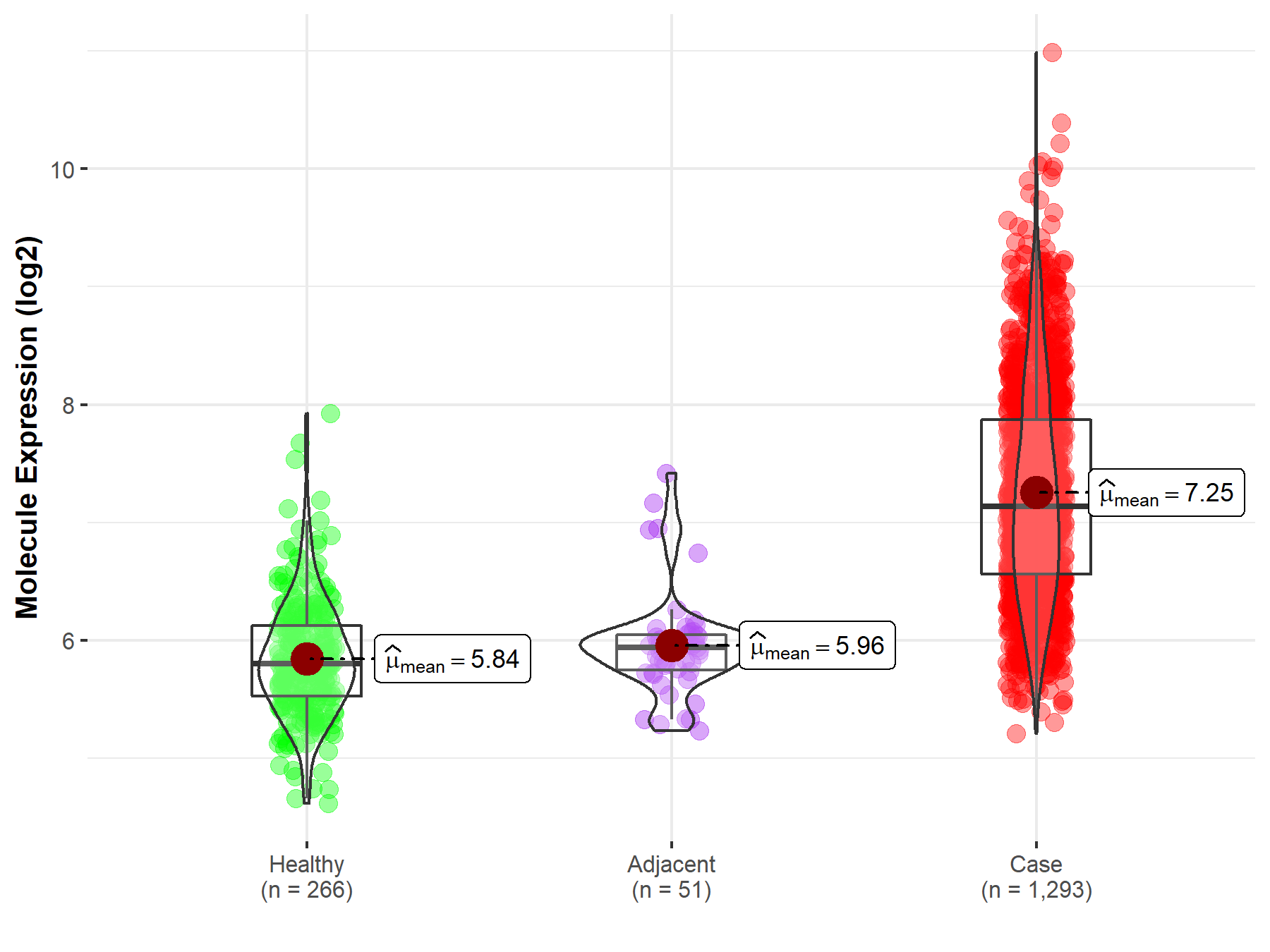
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

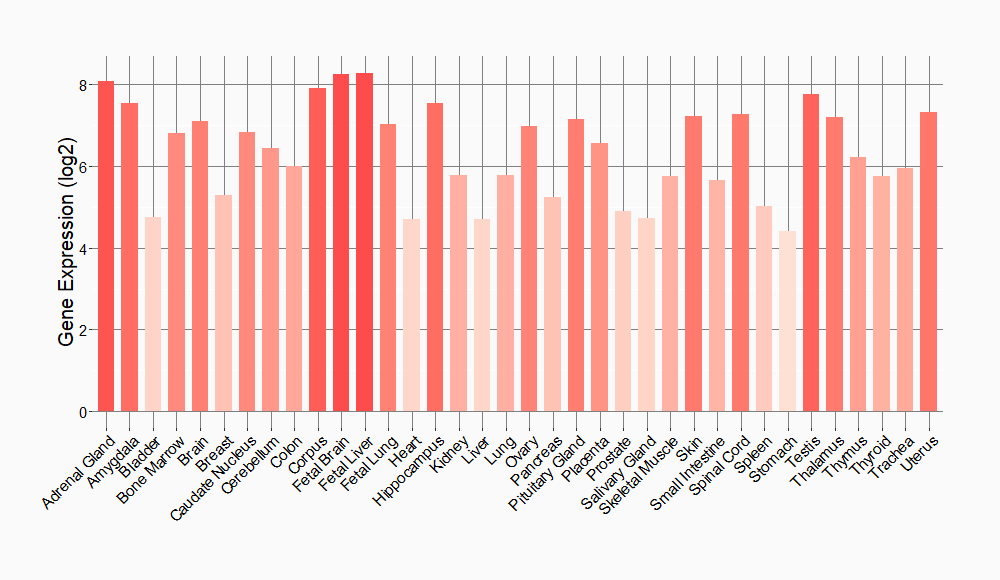
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
