Drug Information
Drug (ID: DG00125) and It's Reported Resistant Information
| Name |
Chloroquine
|
||||
|---|---|---|---|---|---|
| Synonyms |
Amokin; Aralen; Aralen HCl; Arechin; Arechine; Arequin; Arolen; Arthrochin; Artrichin; Avlochlor; Avloclor; Bemaco; Bemaphate; Bemasulph; Benaquin; Bipiquin; CU-01000012392-2; Capquin; Chemochin; Chingamin; Chloraquine; Chlorochin; Chlorochine; Chlorochinum; Chloroin; Chloroquin; Chloroquina; Chloroquine (USP/INN); Chloroquine (VAN); Chloroquine Bis-Phosphoric Acid; Chloroquine FNA (TN); Chloroquine [USAN:INN:BAN]; Chloroquine phosphate; Chloroquinium; Chloroquinum; Chloroquinum [INN-Latin]; Chlorquin; Cidanchin; Clorochina; Clorochina [DCIT]; Cloroquina; Cloroquina [INN-Spanish]; Cocartrit; Dawaquin (TN); Delagil; Dichinalex; Elestol; Gontochin; Gontochin phosphate; Heliopar; Imagon; Ipsen 225; Iroquine; Khingamin; Klorokin; Lapaquin; Malaquin; Malaquin (*Diphosphate*); Malaren; Malarex; Mesylith; Miniquine; Neochin; Nivachine; Nivaquine B; Pfizerquine; Quinachlor; Quinagamin; Quinagamine; Quinercyl; Quingamine; Quinilon; Quinoscan; RP 3377; RP-3377; Resochen; Resochin; Resochin (TN); Resoquina; Resoquine; Reumachlor; Reumaquin; Rivoquine; Ro 01-6014/N2; Ronaquine; Roquine; SN 6718; SN-7618; ST 21; ST 21 (pharmaceutical); Sanoquin; Silbesan;Siragan; Solprina; Sopaquin; Tanakan; Tanakene; Tresochin; Trochin; W 7618;WIN 244
Click to Show/Hide
|
||||
| Indication |
In total 1 Indication(s)
|
||||
| Structure |
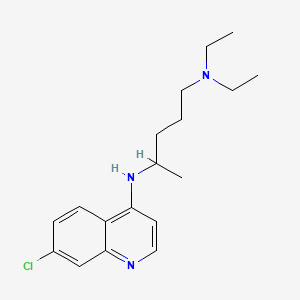
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Resistance Information Discovered by Cell Line Test for This Drug
(1 diseases)
[5]
Disease(s) with Clinically Reported Resistance for This Drug
(2 diseases)
[6]
[7]
Disease(s) with Resistance Information Validated by in-vivo Model for This Drug
(1 diseases)
|
||||
| Target | Duffy antigen chemokine receptor (ACKR1) | ACKR1_HUMAN | [4] | ||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C18H26ClN3
|
||||
| IsoSMILES |
CCN(CC)CCCC(C)NC1=C2C=CC(=CC2=NC=C1)Cl
|
||||
| InChI |
1S/C18H26ClN3/c1-4-22(5-2)12-6-7-14(3)21-17-10-11-20-18-13-15(19)8-9-16(17)18/h8-11,13-14H,4-7,12H2,1-3H3,(H,20,21)
|
||||
| InChIKey |
WHTVZRBIWZFKQO-UHFFFAOYSA-N
|
||||
| PubChem CID | |||||
| ChEBI ID | |||||
| TTD Drug ID | |||||
| VARIDT ID | |||||
| INTEDE ID | |||||
| DrugBank ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-01: Infectious/parasitic diseases
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Chloroquine resistance transporter (CRT) | [11] | |||
| Sensitive Disease | Fungal infection [ICD-11: 1F29-1F2F] | |||
| Molecule Alteration | Missense mutation | p.A144T |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Saccharomyces cerevisiae BY4741 (MATa his3deta1 leu2deta met15deta ura3deta) | 1247190 | ||
| Saccharomyces cerevisiae CH1305 (MAT a ade2 ade3 ura3 - 52 leu2 lys2 - 801 ) | 4932 | |||
| Saccharomyces cerevisiae detaVma (MATa leu2deta met15deta ura3deta) | 4932 | |||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Colony formation assay | |||
| Mechanism Description | The sequences of PfCRT isoforms 'PH1' and 'PH2', which harbour novel mutations A144T and L160Y. Two isoforms (PH1 and PH2 PfCRT) were found to be intrinsically toxic to yeast, even in the absence. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [11] | |||
| Sensitive Disease | Fungal infection [ICD-11: 1F29-1F2F] | |||
| Molecule Alteration | Missense mutation | p.L160Y |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Saccharomyces cerevisiae BY4741 (MATa his3deta1 leu2deta met15deta ura3deta) | 1247190 | ||
| Saccharomyces cerevisiae CH1305 (MAT a ade2 ade3 ura3 - 52 leu2 lys2 - 801 ) | 4932 | |||
| Saccharomyces cerevisiae detaVma (MATa leu2deta met15deta ura3deta) | 4932 | |||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Colony formation assay | |||
| Mechanism Description | The sequences of PfCRT isoforms 'PH1' and 'PH2', which harbour novel mutations A144T and L160Y. Two isoforms (PH1 and PH2 PfCRT) were found to be intrinsically toxic to yeast, even in the absence. | |||
ICD-02: Benign/in-situ/malignant neoplasm
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: . | [5] | |||
| Resistant Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Molecule Alteration | . | . |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Wnt/beta-catenin Signalling Pathway | Regulation | N.A. | |
| In Vitro Model | HCT8 cells | Colon | Homo sapiens (Human) | CVCL_2478 |
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | The results showed that Huaier can regulate autophagy, inhibit the Wnt/-catenin signalling pathway and reverse the drug resistance of OXA-resistant CRC cells. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Chloroquine resistance transporter (CRT) | [1], [2], [3] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.H97Y |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
PacBio amplicon sequencing assay; Whole genome sequencing assay | |||
| Experiment for Drug Resistance |
Piperaquine susceptibility testing assay | |||
| Mechanism Description | In parasites with single-copy pfpm2, those with the PfCRT F145I, G353V, or I218F mutations had a significantly greater log10-transformed piperaquine IC90 compared to Dd2 (linear regression; P <.0001, P =.0022, and P =.019, respectively), while other mutations did not show a significant difference in piperaquine IC90 compared to Dd2 (perhaps owing to smaller sample. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [8], [9], [10] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.K76T |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
MIP probes and PCR sequencing assay | |||
| Experiment for Drug Resistance |
SYBR Green I detection assay; [3H]-hypoxanthine assay | |||
| Mechanism Description | Notably, the PfCRT Lys76Thr substitution was associated with significantly decreased susceptibility to chloroquine, monodesethylamodiaquine, and AQ-13; associations with other aminoquinolines were not conclusive. | |||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [4], [12], [13] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.N86Y |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
MIP probes and PCR sequencing assay | |||
| Experiment for Drug Resistance |
SYBR Green I detection assay | |||
| Mechanism Description | Increasingly, molecular genetic markers for antimalarial drug resistance have been identified, an advance that facilitates the monitoring of the emergence and spread of resistance. Currently, reliable molecular markers are available for P. falciparum resistance to artemisinins (mutations in the propeller region of Pfkelch), sulfadoxine-pyrimethamine (mutations in the dihydrofolate reductase [PfDHFR] and dihydropteroate synthase [PfDHPS] genes), mefloquine (MQ) (amplification of the multidrug resistance-1 gene [PfMDR1]), and piperaquine (amplification of PfPlasmepsin2/3 and specific mutations in the P. falciparum chloroquine resistance transporter gene. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [2], [14], [15] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.C350R |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
Amino acid sequence alignments assay | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | Functional studies on the newly emerging PfCRT F145I and C350R mutations, associated with decreased PPQ susceptibility in Asia and South America respectively reveal their ability to mediate PPQ transport in 7G8 variant proteins and to confer resistance in gene-edited parasites. The apparent dichotomy observed between CQ and PPQ for the F145I and C350R mutants, which evolved on CQ-R isoforms and caused CQ resensitization along with a gain of PPQ resistance, highlights the value of extending this research to rapidly emerging PfCRT mutations. | |||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [16], [17], [18] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.Y184F |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
Nested PCR; Sequence assay | |||
| Experiment for Drug Resistance |
[3H]-hypoxanthine assay | |||
| Mechanism Description | Parasites with a chloroquine IC50 > 100 nM were significantly associated with PfCRT 97L and pfmdr1 184F, and a pfmdr1 copy number >= 4 was more common in those with a chloroquine IC50 <=100 nM. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [19], [20] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.M74I |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
Direct DNA sequencing method assay | |||
| Mechanism Description | High prevalence of mutant Pfcrt genotypes associated with chloroquine resistance in Assam and Arunachal Pradesh, India. The k76T mutation was observed in 77.78% cases followed by M74I (61.11%), N75E (61.11%) and C72S (16.67%). Triple mutant allele M74I+N75E+k76T was found in 61.11% P. falciparum field isolates. Double mutant allele C72S+k76T was seen among 16.67% samples. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [19], [20] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.N75E |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
Direct DNA sequencing method assay | |||
| Mechanism Description | High prevalence of mutant Pfcrt genotypes associated with chloroquine resistance in Assam and Arunachal Pradesh, India. The k76T mutation was observed in 77.78% cases followed by M74I (61.11%), N75E (61.11%) and C72S (16.67%). Triple mutant allele M74I+N75E+k76T was found in 61.11% P. falciparum field isolates. Double mutant allele C72S+k76T was seen among 16.67% samples. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [9], [20] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.A220S |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
[3H]-hypoxanthine assay | |||
| Mechanism Description | Plasmodium falciparum chloroquine resistance (CQR) transporter protein (PfCRT) is the important key of CQR. There is a link between mutations in the gene pfcrt and resistance to chloroquine in P. falciparum. Almost all of the parasites characterized carried the previously reported mutations k76T, A220S, Q271E, N326S, I356T and R371I. On complete sequencing, isolates were identified with novel mutations at k76A and E198K. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [9], [20] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.I356T |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
[3H]-hypoxanthine assay | |||
| Mechanism Description | Plasmodium falciparum chloroquine resistance (CQR) transporter protein (PfCRT) is the important key of CQR. There is a link between mutations in the gene pfcrt and resistance to chloroquine in P. falciparum. Almost all of the parasites characterized carried the previously reported mutations k76T, A220S, Q271E, N326S, I356T and R371I. On complete sequencing, isolates were identified with novel mutations at k76A and E198K. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [9], [20] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.N326S |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
[3H]-hypoxanthine assay | |||
| Mechanism Description | Plasmodium falciparum chloroquine resistance (CQR) transporter protein (PfCRT) is the important key of CQR. There is a link between mutations in the gene pfcrt and resistance to chloroquine in P. falciparum. Almost all of the parasites characterized carried the previously reported mutations k76T, A220S, Q271E, N326S, I356T and R371I. On complete sequencing, isolates were identified with novel mutations at k76A and E198K. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [9], [20] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.R371I |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
[3H]-hypoxanthine assay | |||
| Mechanism Description | Plasmodium falciparum chloroquine resistance (CQR) transporter protein (PfCRT) is the important key of CQR. There is a link between mutations in the gene pfcrt and resistance to chloroquine in P. falciparum. Almost all of the parasites characterized carried the previously reported mutations k76T, A220S, Q271E, N326S, I356T and R371I. On complete sequencing, isolates were identified with novel mutations at k76A and E198K. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [21] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.C101F+p.F145I+p.M343L+p.G353V+T93S+I218F |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum D10 | 5833 | ||
| Plasmodium falciparum Dd2 | 5833 | |||
| Plasmodium falciparum PfCRT | 5833 | |||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Mechanism Description | Plasmodium falciparum parasites resistant to chloroquine, amodiaquine, or piperaquine harbor mutations in the P. falciparum chloroquine resistance transporter (PfCRT), a transporter resident on the digestive vacuole membrane that in its variant forms can transport these weak-base 4-aminoquinoline drugs out of this acidic organelle, thus preventing these drugs from binding heme and inhibiting its detoxification. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [22] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.C101 |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum isolate 3D7 | 5833 | ||
| Plasmodium falciparum isolate 7G8 | 5833 | |||
| Plasmodium falciparum isolate Dd2 | 5833 | |||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
P. falciparum proliferation assay | |||
| Mechanism Description | Drug-resistance-conferring mutations reduce both the peptide transport capacity and substrate range of PfCRT, explaining the impaired fitness of drug-resistant parasites. Two PfCRT mutations that arose separately under in vitro drug pressure (C101F and L272F) incur both a fitness cost and a monstrously swollen DV. Three laboratory-derived isoforms that cause a gross enlargement of the DV-L272F-PfCRT3D7, L272F-PfCRTDd2 and C101F-PfCRTDd2-displayed unusually low capacities for VF-6 tra. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [22] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.L272F |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum isolate 3D7 | 5833 | ||
| Plasmodium falciparum isolate 7G8 | 5833 | |||
| Plasmodium falciparum isolate Dd2 | 5833 | |||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
P. falciparum proliferation assay | |||
| Mechanism Description | Drug-resistance-conferring mutations reduce both the peptide transport capacity and substrate range of PfCRT, explaining the impaired fitness of drug-resistant parasites. Two PfCRT mutations that arose separately under in vitro drug pressure (C101F and L272F) incur both a fitness cost and a monstrously swollen DV. Three laboratory-derived isoforms that cause a gross enlargement of the DV-L272F-PfCRT3D7, L272F-PfCRTDd2 and C101F-PfCRTDd2-displayed unusually low capacities for VF-6 tra. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [2] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.C101F |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum W2 | 5833 | ||
| Experiment for Molecule Alteration |
Pfcrt genotyping assay | |||
| Experiment for Drug Resistance |
HRP2 ELISA-based assay Malaria Ag Celisa kit assay | |||
| Mechanism Description | In a context of dihydroartemisinin-piperaquine resistance in Cambodia and high prevalence of k13 C580Y mutation associated with artemisinin resistance, new pfcrt mutations (H97Y, M343L, and G353V) were revealed to induce in vitro piperaquine resistance. Treatment failures with dihydroartemisinin-piperaquine were associated with T93S, H97Y, F145I and I218F mutations in PfCRT and with plasmepsin 2/3 amplification in Cambodia, Thailand and Vietnam. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [2] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.F145I |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum W2 | 5833 | ||
| Experiment for Molecule Alteration |
Pfcrt genotyping assay | |||
| Experiment for Drug Resistance |
HRP2 ELISA-based assay Malaria Ag Celisa kit assay | |||
| Mechanism Description | In a context of dihydroartemisinin-piperaquine resistance in Cambodia and high prevalence of k13 C580Y mutation associated with artemisinin resistance, new pfcrt mutations (H97Y, M343L, and G353V) were revealed to induce in vitro piperaquine resistance. Treatment failures with dihydroartemisinin-piperaquine were associated with T93S, H97Y, F145I and I218F mutations in PfCRT and with plasmepsin 2/3 amplification in Cambodia, Thailand and Vietnam. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [2] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.G353V |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum W2 | 5833 | ||
| Experiment for Molecule Alteration |
Pfcrt genotyping assay | |||
| Experiment for Drug Resistance |
HRP2 ELISA-based assay Malaria Ag Celisa kit assay | |||
| Mechanism Description | In a context of dihydroartemisinin-piperaquine resistance in Cambodia and high prevalence of k13 C580Y mutation associated with artemisinin resistance, new pfcrt mutations (H97Y, M343L, and G353V) were revealed to induce in vitro piperaquine resistance. Treatment failures with dihydroartemisinin-piperaquine were associated with T93S, H97Y, F145I and I218F mutations in PfCRT and with plasmepsin 2/3 amplification in Cambodia, Thailand and Vietnam. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [2] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.M343L |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum W2 | 5833 | ||
| Experiment for Molecule Alteration |
Pfcrt genotyping assay | |||
| Experiment for Drug Resistance |
HRP2 ELISA-based assay Malaria Ag Celisa kit assay | |||
| Mechanism Description | In a context of dihydroartemisinin-piperaquine resistance in Cambodia and high prevalence of k13 C580Y mutation associated with artemisinin resistance, new pfcrt mutations (H97Y, M343L, and G353V) were revealed to induce in vitro piperaquine resistance. Treatment failures with dihydroartemisinin-piperaquine were associated with T93S, H97Y, F145I and I218F mutations in PfCRT and with plasmepsin 2/3 amplification in Cambodia, Thailand and Vietnam. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [2] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.T93S |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum W2 | 5833 | ||
| Experiment for Molecule Alteration |
Pfcrt genotyping assay | |||
| Experiment for Drug Resistance |
HRP2 ELISA-based assay Malaria Ag Celisa kit assay | |||
| Mechanism Description | In a context of dihydroartemisinin-piperaquine resistance in Cambodia and high prevalence of k13 C580Y mutation associated with artemisinin resistance, new pfcrt mutations (H97Y, M343L, and G353V) were revealed to induce in vitro piperaquine resistance. Treatment failures with dihydroartemisinin-piperaquine were associated with T93S, H97Y, F145I and I218F mutations in PfCRT and with plasmepsin 2/3 amplification in Cambodia, Thailand and Vietnam. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [20] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.C72S |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| Mechanism Description | Mutant PfCRT molecules have acquired the ability to expel CQ out of the DV. All CQR haplotypes carry the key k76T mutation that removes a positive charge in TM 1, suggesting a charge-dependent transport mechanism as CQ is di-protonated in the acidic DV10,13. The k76T mutation is always accompanied by additional mutations which may increase the CQR level and/or attenuate the fitness cost of resistance. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [20] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.I356L |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| Mechanism Description | Mutant PfCRT molecules have acquired the ability to expel CQ out of the DV. All CQR haplotypes carry the key k76T mutation that removes a positive charge in TM 1, suggesting a charge-dependent transport mechanism as CQ is di-protonated in the acidic DV10,13. The k76T mutation is always accompanied by additional mutations which may increase the CQR level and/or attenuate the fitness cost of resistance. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [20] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.N326D |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| Mechanism Description | Mutant PfCRT molecules have acquired the ability to expel CQ out of the DV. All CQR haplotypes carry the key k76T mutation that removes a positive charge in TM 1, suggesting a charge-dependent transport mechanism as CQ is di-protonated in the acidic DV10,13. The k76T mutation is always accompanied by additional mutations which may increase the CQR level and/or attenuate the fitness cost of resistance. | |||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [12] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.N86F |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Mechanism Description | Resistance to chloroquine (CQ) in P. falciparum parasites is predominantly linked to a single mutation in the P. falciparum transporter gene (Pfcrt) on chromosome 7, which encodes a protein localized on the parasite digestive vacuole (DV) membrane. The replacement of lysine (k) at position 76 to a threonine (T), i.e. the k76T mutation, has been established as the most important prognostic marker of treatment failure. Another point mutation N86Y in P. falciparum multidrug resistance gene 1 (Pfmdr1), on chromosome 5, that encodes a P-glycoprotein homologue and is located on the parasite DV membrane has also been implicated in CQ resistance. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [14] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.F145I; |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
Amino acid sequence alignments assay | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | Functional studies on the newly emerging PfCRT F145I and C350R mutations, associated with decreased PPQ susceptibility in Asia and South America respectively reveal their ability to mediate PPQ transport in 7G8 variant proteins and to confer resistance in gene-edited parasites. The apparent dichotomy observed between CQ and PPQ for the F145I and C350R mutants, which evolved on CQ-R isoforms and caused CQ resensitization along with a gain of PPQ resistance, highlights the value of extending this research to rapidly emerging PfCRT mutations. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [23] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Phosphorylation | Up-regulation |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
SYBR Green I detection assay | |||
| Mechanism Description | Phosphorylation at Ser-33 and Ser-411 of PfCRT of the chloroquine-resistant P. falciparum strain Dd2 and kinase inhibitors can sensitize drug responsiveness. | |||
| Key Molecule: Putative chloroquine resistance transporter (PVCRT) | [24] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.K10 insertion (c.AAG) |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| Mechanism Description | Mutations in k10 insertion in the Pvcrt-o gene have been identified as a possible molecular marker of CQ resistance in P.vivax. | |||
| Key Molecule: Putative chloroquine resistance transporter (PVCRT) | [24] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.K10 insertion (c.AAG) |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium vivax isolates | 5855 | ||
| Experiment for Molecule Alteration |
Nested PCR | |||
| Mechanism Description | Mutations in k10 insertion in the Pvcrt-o gene have been identified as a possible molecular marker of CQ resistance in P.vivax. | |||
| Key Molecule: Putative chloroquine resistance transporter (PVCRT) | [25] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.S249P |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Yeast CH1305 | 4932 | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Quantitative Growth Rate assay | |||
| Mechanism Description | It is surprising then that the data presented here suggests a single mutation (S249P in PvCRT isoform CQR3) can increase CQ transport by 32% relative to wild type. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [19] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.C72S |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
Direct DNA sequencing method assay | |||
| Mechanism Description | High prevalence of mutant Pfcrt genotypes associated with chloroquine resistance in Assam and Arunachal Pradesh, India. The k76T mutation was observed in 77.78% cases followed by M74I (61.11%), N75E (61.11%) and C72S (16.67%). Triple mutant allele M74I+N75E+k76T was found in 61.11% P. falciparum field isolates. Double mutant allele C72S+k76T was seen among 16.67% samples. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [19] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.C72S+p.K76T |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
Direct DNA sequencing method assay | |||
| Mechanism Description | High prevalence of mutant Pfcrt genotypes associated with chloroquine resistance in Assam and Arunachal Pradesh, India. The k76T mutation was observed in 77.78% cases followed by M74I (61.11%), N75E (61.11%) and C72S (16.67%). Triple mutant allele M74I+N75E+k76T was found in 61.11% P. falciparum field isolates. Double mutant allele C72S+k76T was seen among 16.67% samples. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [19] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.M74I+p.N75E+p.K76T |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
Direct DNA sequencing method assay | |||
| Mechanism Description | High prevalence of mutant Pfcrt genotypes associated with chloroquine resistance in Assam and Arunachal Pradesh, India. The k76T mutation was observed in 77.78% cases followed by M74I (61.11%), N75E (61.11%) and C72S (16.67%). Triple mutant allele M74I+N75E+k76T was found in 61.11% P. falciparum field isolates. Double mutant allele C72S+k76T was seen among 16.67% samples. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [26] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Chromosome variation | Dd2 genotype |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Drug Resistance |
lactate dehydrogenase (pLDH) assay | |||
| Mechanism Description | Chloroquine resistance-conferring isoforms of PfCRT reduced the susceptibility of the parasite to QC, MB, and AO. In chloroquine-resistant (but not chloroquine-sensitive) parasites, AO and QC increased the parasite's accumulation of, and susceptibility to, chloroquine. All 3 compounds were shown to bind to Pf. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [26] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Chromosome variation | K1 genotype |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Drug Resistance |
lactate dehydrogenase (pLDH) assay | |||
| Mechanism Description | Chloroquine resistance-conferring isoforms of PfCRT reduced the susceptibility of the parasite to QC, MB, and AO. In chloroquine-resistant (but not chloroquine-sensitive) parasites, AO and QC increased the parasite's accumulation of, and susceptibility to, chloroquine. All 3 compounds were shown to bind to Pf. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [27] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | Mutant pfcrt alleles PH1 and PH2 |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Drug Resistance |
[3H]-hypoxanthine assay | |||
| Mechanism Description | Variant alleles from the Philippines (PH1 and PH2, which differ solely by the C72S mutation) both conferred a moderate gain of chloroquine resistance and a reduction in growth rates in vitro. Of the two, PH2 showed higher IC50 values, contrasting with reduced. | |||
| Key Molecule: Putative chloroquine resistance transporter (PVCRT) | [28] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium vivax strains | 5855 | ||
| Mechanism Description | Patients with CQ-resistant P. vivax parasites presented a higher gene expression of pvcrt-o and pvmdr-1 at D0 and DR when compared to the susceptible group. For the CQR patients, median gene expression values at D0 and DR, presented 2.4 fold (95% CI: 0.96-7.1) and 6.1 fold (95% CI: 3.8-14.3) increase in pvcrt-o levels compared to the susceptible patients at D0 with 0.12 fold (95% CI: 0.034-0.324). Median gene expression for pvmdr-1 presented 2.0 fold (95% CI: 0.95-3.8) and 2.4 fold (95% CI: 0.53-9.1) increase levels at D0 and DR, for the CQR patients versus 0.288 fold (95% CI: 0.068-0.497) for the susceptible patients. | |||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [28] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium vivax strains | 5855 | ||
| Mechanism Description | Patients with CQ-resistant P. vivax parasites presented a higher gene expression of pvcrt-o and pvmdr-1 at D0 and DR when compared to the susceptible group. For the CQR patients, median gene expression values at D0 and DR, presented 2.4 fold (95% CI: 0.96-7.1) and 6.1 fold (95% CI: 3.8-14.3) increase in pvcrt-o levels compared to the susceptible patients at D0 with 0.12 fold (95% CI: 0.034-0.324). Median gene expression for pvmdr-1 presented 2.0 fold (95% CI: 0.95-3.8) and 2.4 fold (95% CI: 0.53-9.1) increase levels at D0 and DR, for the CQR patients versus 0.288 fold (95% CI: 0.068-0.497) for the susceptible patients. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [10] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.K76T |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum C-1Dd2 | 5833 | ||
| Plasmodium falciparum C2GC03 | 5833 | |||
| Plasmodium falciparum C3Dd2 | 5833 | |||
| Plasmodium falciparum C67G8 | 5833 | |||
| Plasmodium falciparum GC03 | 5833 | |||
| Plasmodium falciparum T76k-1Dd2 | 5833 | |||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
[3H]-hypoxanthine assay | |||
| Mechanism Description | The k76T mutation in PfCRT generates structural changes that are sufficient to allow GSH transport, but not CQ transport. mutant pfcrt allows enhanced transport of GSH into the parasite's DV. The elevated levels of GSH in the DV reduce the level of free heme available for CQ binding, which mediates the lower susceptibility to CQ in the PfCRT mutant parasites. | |||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [29] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.976F |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium vivax isolates | 5855 | ||
| Mechanism Description | In Southeast Asia the pvmdr1 976 F allele has been associated with reduced susceptibility to CQ. Finding the pvmdr1 976 F allele in 7/41 (17%) P. vivax might thus indicate a degree of CQ tolerance but probably not resistance in Honduras. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [16] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.H97L |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
Nested PCR | |||
| Experiment for Drug Resistance |
[3H]-hypoxanthine assay | |||
| Mechanism Description | Parasites with a chloroquine IC50 > 100 nM were significantly associated with PfCRT 97L and pfmdr1 184F, and a pfmdr1 copy number >= 4 was more common in those with a chloroquine IC50 <=100 nM. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [9] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.E198K |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
[3H]-hypoxanthine assay | |||
| Mechanism Description | Plasmodium falciparum chloroquine resistance (CQR) transporter protein (PfCRT) is the important key of CQR. There is a link between mutations in the gene pfcrt and resistance to chloroquine in P. falciparum. Almost all of the parasites characterized carried the previously reported mutations k76T, A220S, Q271E, N326S, I356T and R371I. On complete sequencing, isolates were identified with novel mutations at k76A and E198K. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [9] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.K76A |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
[3H]-hypoxanthine assay | |||
| Mechanism Description | Plasmodium falciparum chloroquine resistance (CQR) transporter protein (PfCRT) is the important key of CQR. There is a link between mutations in the gene pfcrt and resistance to chloroquine in P. falciparum. Almost all of the parasites characterized carried the previously reported mutations k76T, A220S, Q271E, N326S, I356T and R371I. On complete sequencing, isolates were identified with novel mutations at k76A and E198K. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [9] | |||
| Resistant Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.Q271E |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
[3H]-hypoxanthine assay | |||
| Mechanism Description | Plasmodium falciparum chloroquine resistance (CQR) transporter protein (PfCRT) is the important key of CQR. There is a link between mutations in the gene pfcrt and resistance to chloroquine in P. falciparum. Almost all of the parasites characterized carried the previously reported mutations k76T, A220S, Q271E, N326S, I356T and R371I. On complete sequencing, isolates were identified with novel mutations at k76A and E198K. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Chloroquine resistance transporter (CRT) | [30], [31] | |||
| Sensitive Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | Human foreskin fibroblasts | N.A. | Saccharomyces cerevisiae | N.A. |
| Toxoplasma gondii Prudetaku80S/Luc | 1080348 | |||
| In Vivo Model | C57BL/6J mice xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Tachyzoite plaque assay | |||
| Mechanism Description | Rrecombinant PfCRT transports amino acids, small peptides, and chloroquine, suggests that PfCRT functions to transport products of hemoglobin digestion out of the digestive vacuole. TgCRT is also able to transport amino acids and small peptides out of the VAC. TgCRT-deficient tachyzoites also grow more slowly in vitro and are compromised in their ability to cause mortality in mice during acute infection, suggesting that an inability to transport digested material out of the VAC and into the parasite cytosol has a moderate effect on T. gondii tachyzoites. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [32], [33], [34] | |||
| Sensitive Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
SYBR Green I detection assay; [3H]-hypoxanthine assay | |||
| Mechanism Description | Phosphorylation at Ser-33 and Ser-411 of PfCRT of the chloroquine-resistant P. falciparum strain Dd2 and kinase inhibitors can sensitize drug responsiveness. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [35] | |||
| Sensitive Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.C101F |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum asexual blood-stage parasites | 5833 | ||
| Experiment for Molecule Alteration |
DNA clones asssay | |||
| Experiment for Drug Resistance |
SYBR Green I detection assay | |||
| Mechanism Description | This mutation (C101F) also reversed Dd2-mediated CQ resistance, sensitized parasites to amodiaquine, quinine, and artemisinin, and conferred amantadine and blasticidin resistance. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [36] | |||
| Sensitive Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.C101F |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum 3D7 | 36329 | ||
| Plasmodium falciparum 3D7L272F mutant | 36329 | |||
| Experiment for Drug Resistance |
SYBR Green I detection assay | |||
| Mechanism Description | Carry PfCRT mutations (C101F or L272F), causing the development of enlarged food vacuoles. These parasites also have increased sensitivity to chloroquine and some other quinoline antimalarials. Furthermore, the introduction of the C101F or L272F mutation into a chloroquine-resistant variant of PfCRT reduced the ability of this protein to transport chloroquine by approximately 93 and 82% when expressed in Xenopus oocytes. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [36] | |||
| Sensitive Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.L272F |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum 3D7 | 36329 | ||
| Plasmodium falciparum 3D7L272F mutant | 36329 | |||
| Experiment for Drug Resistance |
SYBR Green I detection assay | |||
| Mechanism Description | Carry PfCRT mutations (C101F or L272F), causing the development of enlarged food vacuoles. These parasites also have increased sensitivity to chloroquine and some other quinoline antimalarials. Furthermore, the introduction of the C101F or L272F mutation into a chloroquine-resistant variant of PfCRT reduced the ability of this protein to transport chloroquine by approximately 93 and 82% when expressed in Xenopus oocytes. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [33] | |||
| Sensitive Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.S163R |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Plasmodium falciparum D10 | 5833 | ||
| Plasmodium falciparum Dd2 | 5833 | |||
| Plasmodium falciparum PfCRT | 5833 | |||
| Mechanism Description | T76k and S163R mutations in PfCRTCQR restore CQ sensitivity to CQR parasites. The introduction of either one of these changes to PfCRTCQR, each of which entailed the addition of a positive charge to the putative substrate-binding site of the protein, resulted in the loss of CQ transport activity. | |||
| Key Molecule: Chloroquine resistance transporter (CRT) | [33] | |||
| Sensitive Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Missense mutation | p.T76K |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Plasmodium falciparum D10 | 5833 | ||
| Plasmodium falciparum Dd2 | 5833 | |||
| Plasmodium falciparum PfCRT | 5833 | |||
| Mechanism Description | T76k and S163R mutations in PfCRTCQR restore CQ sensitivity to CQR parasites. The introduction of either one of these changes to PfCRTCQR, each of which entailed the addition of a positive charge to the putative substrate-binding site of the protein, resulted in the loss of CQ transport activity. | |||
| Key Molecule: ABC transporter (ABCT) | [32] | |||
| Sensitive Disease | Malaria [ICD-11: 1F45.0] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Plasmodium falciparum strains | 5833 | ||
| Mechanism Description | Phenothiazine drugs (chlor-promazine, trifluoperazine, prochlorperazine, methotrime-prazin or fluphenazin) can enhance in vitro the potency of CQ against P. falciparum CQ-resistant strains. phenothiazines may exert their CQ resistance reversal activity by interacting with Pgh1. Combinations of chlorpromazine or prochlorperazine with CQ confirm the reversal effect of these drugs on the CQ resistance as showed by cures obtained in Aotus monkeys infected with CQ-resistant P. falciparum. | |||
ICD-15: Musculoskeletal/connective-tissue diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [7] | |||
| Resistant Disease | Rheumatoid arthritis [ICD-11: FA20.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Mechanism Description | MTX is a substrate for eight ABC transporters. In vitro studies demonstrated that RAFLS treated with MTX had higher ABCB1 expression levels than controls, with a positive correlation between ABCB1 expression levels and RA treatment duration. In addition to MTX, other DMARDs (e.g. sulfasalazine, leflunomide, bucillamine, azathioprine), glucocorticoids (e.g. betamethasone, dexamethasone), and NSAIDs (e.g. celecoxib and indomethacin) are also substrates of ABC transporters. | |||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
