Molecule Information
General Information of the Molecule (ID: Mol00713)
| Name |
Y-box-binding protein 1 (YBX1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
YB-1; CCAAT-binding transcription factor I subunit A; CBF-A; DNA-binding protein B; DBPB; Enhancer factor I subunit A; EFI-A; Nuclease-sensitive element-binding protein 1; Y-box transcription factor; NSEP1; YB1
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
YBX1
|
||||
| Gene ID | |||||
| Location |
chr1:42682418-42703805[+]
|
||||
| Sequence |
MSSEAETQQPPAAPPAAPALSAADTKPGTTGSGAGSGGPGGLTSAAPAGGDKKVIATKVL
GTVKWFNVRNGYGFINRNDTKEDVFVHQTAIKKNNPRKYLRSVGDGETVEFDVVEGEKGA EAANVTGPGGVPVQGSKYAADRNHYRRYPRRRGPPRNYQQNYQNSESGEKNEGSESAPEG QAQQRRPYRRRRFPPYYMRRPYGRRPQYSNPPVQGEVMEGADNQGAGEQGRPVRQNMYRG YRPRFRRGPPRQRQPREDGNEEDKENQGDETQGQQPPQRRYRRNFNYRRRRPENPKPQDG KETKAADPPAENSSAPEAEQGGAE Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
DNA- and RNA-binding protein involved in various processes, such as translational repression, RNA stabilization, mRNA splicing, DNA repair and transcription regulation. Predominantly acts as a RNA-binding protein: binds preferentially to the 5'-[CU]CUGCG-3' RNA motif and specifically recognizes mRNA transcripts modified by C5-methylcytosine (m5C). Promotes mRNA stabilization: acts by binding to m5C-containing mRNAs and recruiting the mRNA stability maintainer ELAVL1, thereby preventing mRNA decay. Component of the CRD-mediated complex that promotes MYC mRNA stability. Contributes to the regulation of translation by modulating the interaction between the mRNA and eukaryotic initiation factors. Plays a key role in RNA composition of extracellular exosomes by defining the sorting of small non-coding RNAs, such as tRNAs, Y RNAs, Vault RNAs and miRNAs. Probably sorts RNAs in exosomes by recognizing and binding C5-methylcytosine (m5C)-containing RNAs. Acts as a key effector of epidermal progenitors by preventing epidermal progenitor senescence: acts by regulating the translation of a senescence-associated subset of cytokine mRNAs, possibly by binding to m5C-containing mRNAs. Also involved in pre-mRNA alternative splicing regulation: binds to splice sites in pre-mRNA and regulates splice site selection. Also able to bind DNA: regulates transcription of the multidrug resistance gene MDR1 is enhanced in presence of the APEX1 acetylated form at 'Lys-6' and 'Lys-7'. Binds to promoters that contain a Y-box (5'-CTGATTGGCCAA-3'), such as MDR1 and HLA class II genes. Promotes separation of DNA strands that contain mismatches or are modified by cisplatin. Has endonucleolytic activity and can introduce nicks or breaks into double-stranded DNA, suggesting a role in DNA repair. The secreted form acts as an extracellular mitogen and stimulates cell migration and proliferation.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
10 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hepatocellular carcinoma [ICD-11: 2C12.2] | [1] | |||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Liver cancer [ICD-11: 2C12] | |||
| The Specified Disease | Liver cancer | |||
| The Studied Tissue | Liver tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.97E-03 Fold-change: 7.34E-02 Z-score: 3.27E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Activation | hsa05200 | |
| Tumorigenesis | Activation | hsa05206 | ||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| SMMC7721 cells | Uterus | Homo sapiens (Human) | CVCL_0534 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
xCELLigence assay | |||
| Mechanism Description | HULC promotes the phosphorylation of YB-1 through the extracellular signal-regulated kinase pathway, in turn leads to the release of YB-1 from its bound mRNA. | |||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [4] | |||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | p53 signaling pathway | Inhibition | hsa04115 | |
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| MkN-45 cells | Gastric | Homo sapiens (Human) | CVCL_0434 | |
| GCIY cells | Gastric | Homo sapiens (Human) | CVCL_1228 | |
| KATO-3 cells | Gastric | Homo sapiens (Human) | CVCL_0371 | |
| MkN-7 cells | Gastric | Homo sapiens (Human) | CVCL_1417 | |
| SNU-1 cells | Gastric | Homo sapiens (Human) | CVCL_0099 | |
| TGBC11TkB cells | Gastric | Homo sapiens (Human) | CVCL_1768 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; TUNEL assay; xCELLigence Real-Time invasion and migration assays | |||
| Mechanism Description | TP53TG1, a p53-induced LncRNA, binds to the multifaceted RNA/RNA binding protein YBX1 to prevent its nuclear localization and thus the YBX1-mediated activation of oncogenes. The epigenetic silencing of TP53TG1 in cancer cells promotes the YBX1-mediated activation of the PI3k/AkT pathway, which then creates further resistance not only to common chemotherapy RNA-damaging agents but also to small drug-targeted inhibitors. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [2] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Breast cancer [ICD-11: 2C60] | |||
| The Specified Disease | Breast cancer | |||
| The Studied Tissue | Breast tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.77E-05 Fold-change: 2.43E-02 Z-score: 4.02E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Elevated miR-137 expression could sensitize breast cancer cells to chemotherapeutic agents (like Vincristine) through modulating the expression of P-gp by targeting YB-1. | |||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [4] | |||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Resistant Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | p53 signaling pathway | Inhibition | hsa04115 | |
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| MkN-45 cells | Gastric | Homo sapiens (Human) | CVCL_0434 | |
| GCIY cells | Gastric | Homo sapiens (Human) | CVCL_1228 | |
| KATO-3 cells | Gastric | Homo sapiens (Human) | CVCL_0371 | |
| MkN-7 cells | Gastric | Homo sapiens (Human) | CVCL_1417 | |
| SNU-1 cells | Gastric | Homo sapiens (Human) | CVCL_0099 | |
| TGBC11TkB cells | Gastric | Homo sapiens (Human) | CVCL_1768 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; TUNEL assay; xCELLigence Real-Time invasion and migration assays | |||
| Mechanism Description | TP53TG1, a p53-induced LncRNA, binds to the multifaceted RNA/RNA binding protein YBX1 to prevent its nuclear localization and thus the YBX1-mediated activation of oncogenes. The epigenetic silencing of TP53TG1 in cancer cells promotes the YBX1-mediated activation of the PI3k/AkT pathway, which then creates further resistance not only to common chemotherapy RNA-damaging agents but also to small drug-targeted inhibitors. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [5] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | MCF-7/PTX cells | Breast | Homo sapiens (Human) | CVCL_4V97 |
| Experiment for Molecule Alteration |
Dual luciferase assay; Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-375 is downregulated in MCF-7/ADM and MCF-7/PTX cells, and its downregulation is a result of promoter methylation. miR-375 can directly target 3'UTR of YBX1 and, thereby, decrease its expression, which might be an important mechanism of MDR in breast cancer cells. miR-375 is downregulated in MCF-7/ADM and MCF-7/PTX cells, and its downregulation is a result of promoter methylation. miR-375 can directly target 3'UTR of YBX1 and thereby decrease its expression, which might be an important mechanism of MDR in breast cancer cells. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Chordoma [ICD-11: 2B5J.0] | [3] | |||
| Resistant Disease | Chordoma [ICD-11: 2B5J.0] | |||
| Resistant Drug | Afatinib | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | EGFR/AKT signaling pathway | Regulation | N.A. | |
| Cell invasion | Activation | hsa05200 | ||
| In Vitro Model | Chordoma tissue | N.A. | ||
| In Vivo Model | NOD/SCID/IL2Rgamma null (NOG) mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Mechanism Description | YBX1 regulated protein expression of pEGFR, pAKT and its downstream target genes that influenced cell apoptosis, cell cycle transition and cell invasion. YBX1 activated the EGFR/AKT pathway in chordoma and YBX1-induced elevated expression of key molecules in the EGFR/AKT pathway were downregulated by EGFR and AKT pathway inhibitors. These in vitro results were further confirmed by in vivo data. These data showed that YBX1 promoted tumorigenesis and progression in spinal chordoma via the EGFR/AKT pathway. YBX1 might serve as a prognostic and predictive biomarker, as well as a rational therapeutic target, for chordoma. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [4] | |||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Resistant Drug | Carboplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | p53 signaling pathway | Inhibition | hsa04115 | |
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| MkN-45 cells | Gastric | Homo sapiens (Human) | CVCL_0434 | |
| GCIY cells | Gastric | Homo sapiens (Human) | CVCL_1228 | |
| KATO-3 cells | Gastric | Homo sapiens (Human) | CVCL_0371 | |
| MkN-7 cells | Gastric | Homo sapiens (Human) | CVCL_1417 | |
| SNU-1 cells | Gastric | Homo sapiens (Human) | CVCL_0099 | |
| TGBC11TkB cells | Gastric | Homo sapiens (Human) | CVCL_1768 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; TUNEL assay; xCELLigence Real-Time invasion and migration assays | |||
| Mechanism Description | TP53TG1, a p53-induced LncRNA, binds to the multifaceted RNA/RNA binding protein YBX1 to prevent its nuclear localization and thus the YBX1-mediated activation of oncogenes. The epigenetic silencing of TP53TG1 in cancer cells promotes the YBX1-mediated activation of the PI3k/AkT pathway, which then creates further resistance not only to common chemotherapy RNA-damaging agents but also to small drug-targeted inhibitors. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Chordoma [ICD-11: 2B5J.0] | [3] | |||
| Resistant Disease | Chordoma [ICD-11: 2B5J.0] | |||
| Resistant Drug | Erlotinib HCI | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | EGFR/AKT signaling pathway | Regulation | N.A. | |
| Cell invasion | Activation | hsa05200 | ||
| In Vitro Model | Chordoma tissue | N.A. | ||
| In Vivo Model | NOD/SCID/IL2Rgamma null (NOG) mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Mechanism Description | YBX1 regulated protein expression of pEGFR, pAKT and its downstream target genes that influenced cell apoptosis, cell cycle transition and cell invasion. YBX1 activated the EGFR/AKT pathway in chordoma and YBX1-induced elevated expression of key molecules in the EGFR/AKT pathway were downregulated by EGFR and AKT pathway inhibitors. These in vitro results were further confirmed by in vivo data. These data showed that YBX1 promoted tumorigenesis and progression in spinal chordoma via the EGFR/AKT pathway. YBX1 might serve as a prognostic and predictive biomarker, as well as a rational therapeutic target, for chordoma. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [4] | |||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Resistant Drug | Irinotecan | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | p53 signaling pathway | Inhibition | hsa04115 | |
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| MkN-45 cells | Gastric | Homo sapiens (Human) | CVCL_0434 | |
| GCIY cells | Gastric | Homo sapiens (Human) | CVCL_1228 | |
| KATO-3 cells | Gastric | Homo sapiens (Human) | CVCL_0371 | |
| MkN-7 cells | Gastric | Homo sapiens (Human) | CVCL_1417 | |
| SNU-1 cells | Gastric | Homo sapiens (Human) | CVCL_0099 | |
| TGBC11TkB cells | Gastric | Homo sapiens (Human) | CVCL_1768 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; TUNEL assay; xCELLigence Real-Time invasion and migration assays | |||
| Mechanism Description | TP53TG1, a p53-induced LncRNA, binds to the multifaceted RNA/RNA binding protein YBX1 to prevent its nuclear localization and thus the YBX1-mediated activation of oncogenes. The epigenetic silencing of TP53TG1 in cancer cells promotes the YBX1-mediated activation of the PI3k/AkT pathway, which then creates further resistance not only to common chemotherapy RNA-damaging agents but also to small drug-targeted inhibitors. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [4] | |||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Resistant Drug | Oxaliplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | p53 signaling pathway | Inhibition | hsa04115 | |
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| MkN-45 cells | Gastric | Homo sapiens (Human) | CVCL_0434 | |
| GCIY cells | Gastric | Homo sapiens (Human) | CVCL_1228 | |
| KATO-3 cells | Gastric | Homo sapiens (Human) | CVCL_0371 | |
| MkN-7 cells | Gastric | Homo sapiens (Human) | CVCL_1417 | |
| SNU-1 cells | Gastric | Homo sapiens (Human) | CVCL_0099 | |
| TGBC11TkB cells | Gastric | Homo sapiens (Human) | CVCL_1768 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; TUNEL assay; xCELLigence Real-Time invasion and migration assays | |||
| Mechanism Description | TP53TG1, a p53-induced LncRNA, binds to the multifaceted RNA/RNA binding protein YBX1 to prevent its nuclear localization and thus the YBX1-mediated activation of oncogenes. The epigenetic silencing of TP53TG1 in cancer cells promotes the YBX1-mediated activation of the PI3k/AkT pathway, which then creates further resistance not only to common chemotherapy RNA-damaging agents but also to small drug-targeted inhibitors. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colon cancer [ICD-11: 2B90.1] | [6] | |||
| Sensitive Disease | Colon cancer [ICD-11: 2B90.1] | |||
| Sensitive Drug | Oxaliplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SW480 cells | Colon | Homo sapiens (Human) | CVCL_0546 |
| HCT15 cells | Colon | Homo sapiens (Human) | CVCL_0292 | |
| Experiment for Molecule Alteration |
Dual luciferase assay; Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | microRNA-137 chemosensitizes colon cancer cells to the chemotherapeutic drug OXA by targeting YBX1, miR137 was involved in repression of YBX1 expression through targeting its 3'-untranslated region. Down-regulation of miR137 conferred OXA resistance in parental cells, while over-expression of miR137 sensitized resistant cells to OXA, which was partly rescued by YBX1 siRNA. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [2] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Elevated miR-137 expression could sensitize breast cancer cells to chemotherapeutic agents (like Vincristine) through modulating the expression of P-gp by targeting YB-1. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [5] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | MCF-7/PTX cells | Breast | Homo sapiens (Human) | CVCL_4V97 |
| Experiment for Molecule Alteration |
Dual luciferase assay; Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-375 is downregulated in MCF-7/ADM and MCF-7/PTX cells, and its downregulation is a result of promoter methylation. miR-375 can directly target 3'UTR of YBX1 and, thereby, decrease its expression, which might be an important mechanism of MDR in breast cancer cells. miR-375 is downregulated in MCF-7/ADM and MCF-7/PTX cells, and its downregulation is a result of promoter methylation. miR-375 can directly target 3'UTR of YBX1 and thereby decrease its expression, which might be an important mechanism of MDR in breast cancer cells. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Chordoma [ICD-11: 2B5J.0] | [3] | |||
| Resistant Disease | Chordoma [ICD-11: 2B5J.0] | |||
| Resistant Drug | Sirolimus | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | EGFR/AKT signaling pathway | Regulation | N.A. | |
| Cell invasion | Activation | hsa05200 | ||
| In Vitro Model | Chordoma tissue | N.A. | ||
| In Vivo Model | NOD/SCID/IL2Rgamma null (NOG) mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Mechanism Description | YBX1 regulated protein expression of pEGFR, pAKT and its downstream target genes that influenced cell apoptosis, cell cycle transition and cell invasion. YBX1 activated the EGFR/AKT pathway in chordoma and YBX1-induced elevated expression of key molecules in the EGFR/AKT pathway were downregulated by EGFR and AKT pathway inhibitors. These in vitro results were further confirmed by in vivo data. These data showed that YBX1 promoted tumorigenesis and progression in spinal chordoma via the EGFR/AKT pathway. YBX1 might serve as a prognostic and predictive biomarker, as well as a rational therapeutic target, for chordoma. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [2] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Vincristine | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Elevated miR-137 expression could sensitize breast cancer cells to chemotherapeutic agents (like Vincristine) through modulating the expression of P-gp by targeting YB-1. | |||
Investigative Drug(s)
1 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Chordoma [ICD-11: 2B5J.0] | [3] | |||
| Resistant Disease | Chordoma [ICD-11: 2B5J.0] | |||
| Resistant Drug | PKI-587 | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | EGFR/AKT signaling pathway | Regulation | N.A. | |
| Cell invasion | Activation | hsa05200 | ||
| In Vitro Model | Chordoma tissue | N.A. | ||
| In Vivo Model | NOD/SCID/IL2Rgamma null (NOG) mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Mechanism Description | YBX1 regulated protein expression of pEGFR, pAKT and its downstream target genes that influenced cell apoptosis, cell cycle transition and cell invasion. YBX1 activated the EGFR/AKT pathway in chordoma and YBX1-induced elevated expression of key molecules in the EGFR/AKT pathway were downregulated by EGFR and AKT pathway inhibitors. These in vitro results were further confirmed by in vivo data. These data showed that YBX1 promoted tumorigenesis and progression in spinal chordoma via the EGFR/AKT pathway. YBX1 might serve as a prognostic and predictive biomarker, as well as a rational therapeutic target, for chordoma. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Colon | |
| The Specified Disease | Colon cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.71E-01; Fold-change: 7.94E-02; Z-score: 2.76E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 3.10E-15; Fold-change: 5.07E-01; Z-score: 5.77E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
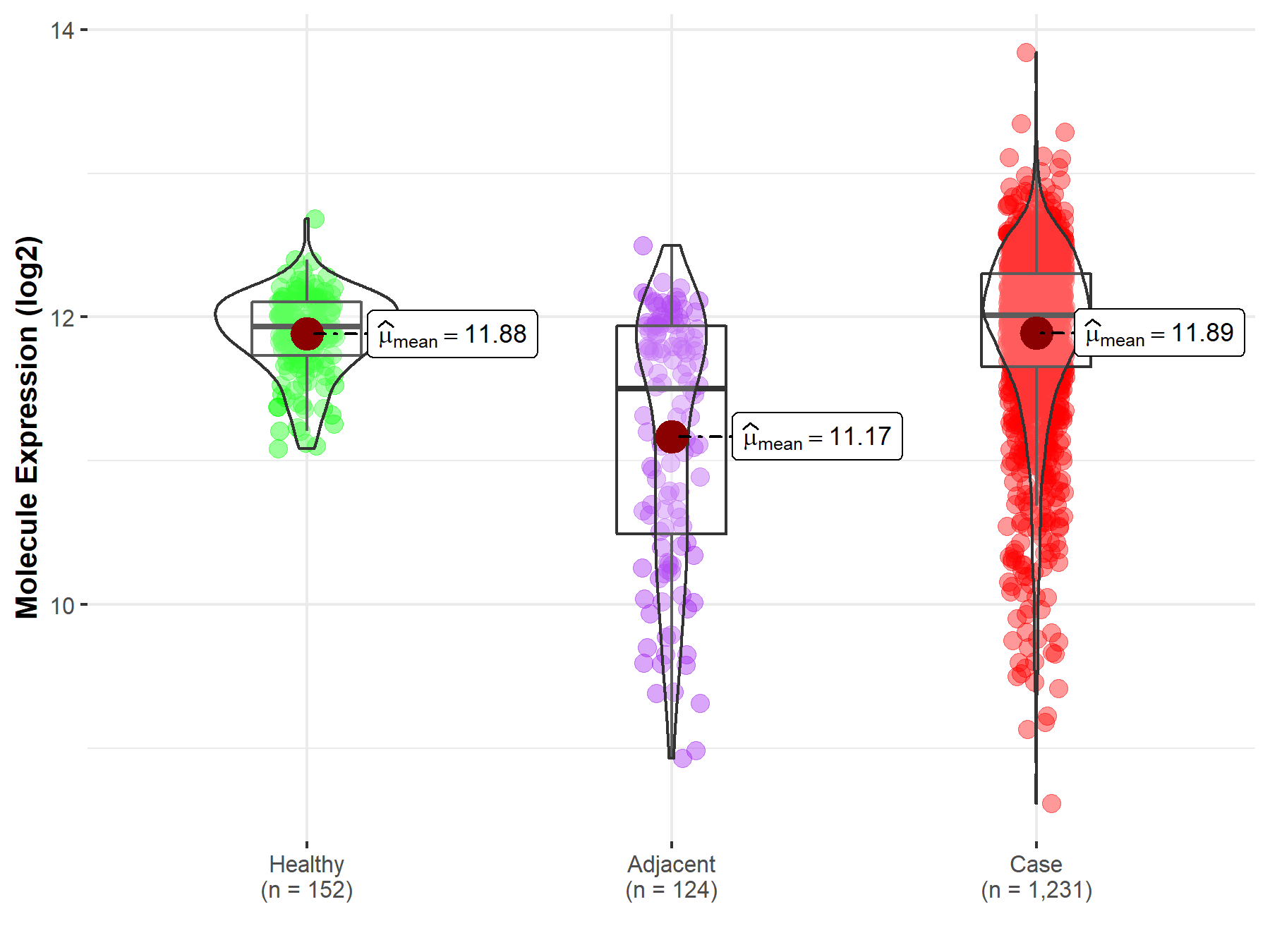
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Liver | |
| The Specified Disease | Liver cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.97E-03; Fold-change: 4.36E-01; Z-score: 4.53E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 3.44E-25; Fold-change: 8.13E-01; Z-score: 9.49E-01 | |
| The Expression Level of Disease Section Compare with the Other Disease Section | p-value: 8.64E-01; Fold-change: 1.61E-01; Z-score: 6.99E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
Molecule expression in tissue other than the diseased tissue of patients
|
||
| Disease-specific Molecule Abundances |
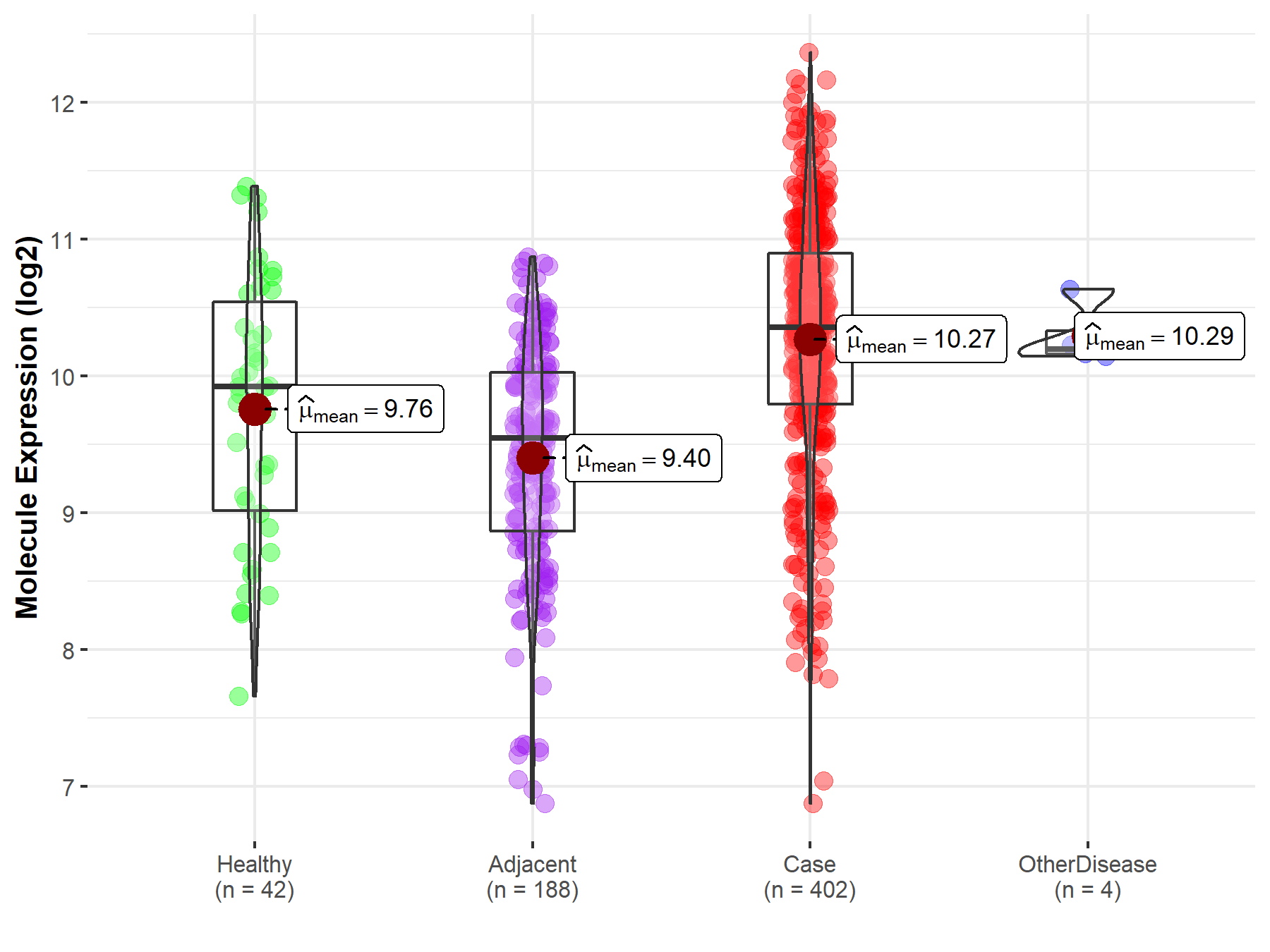
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.77E-05; Fold-change: 2.67E-01; Z-score: 4.22E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.18E-01; Fold-change: 8.16E-02; Z-score: 7.43E-02 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

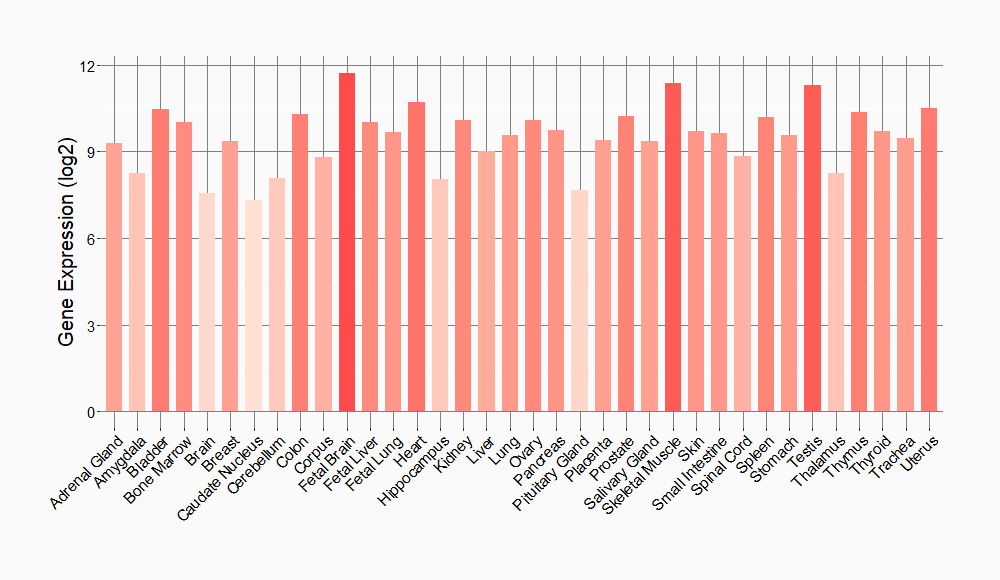
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
