Drug Information
Drug (ID: DG00074) and It's Reported Resistant Information
| Name |
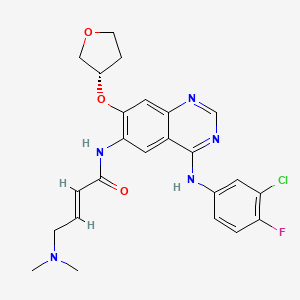
Afatinib
|
||||
|---|---|---|---|---|---|
| Synonyms |
Afatinib; Tomtovok; Tovok; BIBW-2992; Tovok (TN); Tovok, BIBW2992; (2E)-N-{4-[(3-chloro-4-fluorophenyl)amino]-7-[(3S)-tetrahydrofuran-3-yloxy]quinazolin-6-yl}-4-(dimethylamino)but-2-enamide; EGFR inhibitor 2nd gens
Click to Show/Hide
|
||||
| Indication |
In total 1 Indication(s)
|
||||
| Structure |
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Clinically Reported Resistance for This Drug
(3 diseases)
[2]
[8]
Disease(s) with Resistance Information Discovered by Cell Line Test for This Drug
(1 diseases)
[6]
|
||||
| Target | Epidermal growth factor receptor (EGFR) | EGFR_HUMAN | [1] | ||
| Erbb2 tyrosine kinase receptor (HER2) | ERBB2_HUMAN | [1] | |||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C24H25ClFN5O3
|
||||
| IsoSMILES |
CN(C)C/C=C/C(=O)NC1=C(C=C2C(=C1)C(=NC=N2)NC3=CC(=C(C=C3)F)Cl)O[C@H]4CCOC4
|
||||
| InChI |
1S/C24H25ClFN5O3/c1-31(2)8-3-4-23(32)30-21-11-17-20(12-22(21)34-16-7-9-33-13-16)27-14-28-24(17)29-15-5-6-19(26)18(25)10-15/h3-6,10-12,14,16H,7-9,13H2,1-2H3,(H,30,32)(H,27,28,29)/b4-3+/t16-/m0/s1
|
||||
| InChIKey |
ULXXDDBFHOBEHA-CWDCEQMOSA-N
|
||||
| PubChem CID | |||||
| ChEBI ID | |||||
| TTD Drug ID | |||||
| VARIDT ID | |||||
| DrugBank ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-02: Benign/in-situ/malignant neoplasm
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Y-box-binding protein 1 (YBX1) | [2] | |||
| Resistant Disease | Chordoma [ICD-11: 2B5J.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | EGFR/AKT signaling pathway | Regulation | N.A. | |
| Cell invasion | Activation | hsa05200 | ||
| In Vitro Model | Chordoma tissue | N.A. | ||
| In Vivo Model | NOD/SCID/IL2Rgamma null (NOG) mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Mechanism Description | YBX1 regulated protein expression of pEGFR, pAKT and its downstream target genes that influenced cell apoptosis, cell cycle transition and cell invasion. YBX1 activated the EGFR/AKT pathway in chordoma and YBX1-induced elevated expression of key molecules in the EGFR/AKT pathway were downregulated by EGFR and AKT pathway inhibitors. These in vitro results were further confirmed by in vivo data. These data showed that YBX1 promoted tumorigenesis and progression in spinal chordoma via the EGFR/AKT pathway. YBX1 might serve as a prognostic and predictive biomarker, as well as a rational therapeutic target, for chordoma. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Hepatocyte growth factor receptor (MET) | [8] | |||
| Resistant Disease | Esophagogastric cancer [ICD-11: 2B71.1] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Mechanism Description | We find that concurrent amplification of EGFR and ERBB2 is associated with response to the HER kinase inhibitor afatinib in patients with trastuzumab-refractory EG cancer. Heterogeneous uptake of 89Zr-trastuzumab measured noninvasively by PET was associated with disease progression. Analyses of multiple disease sites sampled at the time of disease progression indicated several potential mediators of afatinib resistance, including loss of EGFR amplification and gain of MET amplification. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [8] | |||
| Sensitive Disease | Esophagogastric cancer [ICD-11: 2B71.1] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Mechanism Description | In summary, we find that concurrent amplification of EGFR and ERBB2 is associated with response to the HER kinase inhibitor afatinib in patients with trastuzumab-refractory EG cancer. Heterogeneous uptake of 89Zr-trastuzumab measured noninvasively by PET was associated with disease progression. Analyses of multiple disease sites sampled at the time of disease progression indicated several potential mediators of afatinib resistance, including loss of EGFR amplification and gain of MET amplification. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [3], [4], [5] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Missense mutation | p.T790M |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 3.10 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.05 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
S
G
G
E
E
A
A
P
P
700
|
N
N
Q
Q
A
A
L
L
L
L
R
R
I
I
L
L
K
K
E
E
710
|
T
T
E
E
F
F
K
K
K
K
I
I
K
K
V
V
L
L
G
G
720
|
S
S
G
G
A
A
F
F
G
G
T
T
V
V
Y
Y
K
K
G
G
730
|
L
L
W
W
I
I
P
P
E
E
G
G
E
E
K
K
V
V
K
K
740
|
I
I
P
P
V
V
A
A
I
I
K
K
E
E
L
L
R
R
E
E
750
|
A
A
T
T
S
S
P
P
K
K
A
A
N
N
K
K
E
E
I
I
760
|
L
L
D
D
E
E
A
A
Y
Y
V
V
M
M
A
A
S
S
V
V
770
|
D
D
N
N
P
P
H
H
V
V
C
C
R
R
L
L
L
L
G
G
780
|
I
I
C
C
L
L
T
T
S
S
T
T
V
V
Q
Q
L
L
I
I
790
|
T
M
Q
Q
L
L
M
M
P
P
F
F
G
G
C
C
L
L
L
L
800
|
D
D
Y
Y
V
V
R
R
E
E
H
H
K
K
D
D
N
N
I
I
810
|
G
G
S
S
Q
Q
Y
Y
L
L
L
L
N
N
W
W
C
C
V
V
820
|
Q
Q
I
I
A
A
K
K
G
G
M
M
N
N
Y
Y
L
L
E
E
830
|
D
D
R
R
R
R
L
L
V
V
H
H
R
R
D
D
L
L
A
A
840
|
A
A
R
R
N
N
V
V
L
L
V
V
K
K
T
T
P
P
Q
Q
850
|
H
H
V
V
K
K
I
I
T
T
D
D
F
F
G
G
L
L
A
A
860
|
K
K
L
L
L
L
G
G
A
A
E
E
E
E
K
K
E
E
Y
Y
870
|
H
H
A
A
E
E
G
G
G
G
K
K
V
V
P
P
I
I
K
K
880
|
W
W
M
M
A
A
L
L
E
E
S
S
I
I
L
L
H
H
R
R
890
|
I
I
Y
Y
T
T
H
H
Q
Q
S
S
D
D
V
V
W
W
S
S
900
|
Y
Y
G
G
V
V
T
T
V
V
W
W
E
E
L
L
M
M
T
T
910
|
F
F
G
G
S
S
K
K
P
P
Y
Y
D
D
G
G
I
I
P
P
920
|
A
A
S
S
E
E
I
I
S
S
S
S
I
I
L
L
E
E
K
K
930
|
G
G
E
E
R
R
L
L
P
P
Q
Q
P
P
P
P
I
I
C
C
940
|
T
T
I
I
D
D
V
V
Y
Y
M
M
I
I
M
M
V
V
K
K
950
|
C
C
W
W
M
M
I
I
D
D
A
A
D
D
S
S
R
R
P
P
960
|
K
K
F
F
R
R
E
E
L
L
I
I
I
I
E
E
F
F
S
S
970
|
K
K
M
M
A
A
R
R
D
D
P
P
Q
Q
R
R
Y
Y
L
L
980
|
V
V
I
I
Q
Q
G
G
D
D
E
E
R
R
M
M
H
H
L
L
990
|
P
P
S
S
P
P
T
T
D
D
S
S
N
N
F
F
Y
Y
R
R
1000
|
A
A
L
L
M
M
D
D
E
E
E
E
D
D
M
M
D
D
D
D
1010
|
V
V
V
V
D
D
A
A
D
D
E
E
Y
Y
L
L
I
I
P
P
1020
|
Q
Q
Q
Q
G
G
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
Exon sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Progression-free and post-progression survival asaay | ||||||||||||
| Mechanism Description | T790M was detected in half of the lung adenocarcinoma after acquiring resistance to afatinib. T790M is still the major acquired resistance mechanism. First-generation EGFR TkI exposure did not influence the prevalence of T790M in lung cancer acquired resistance to afatinib. | ||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [9], [10], [11] | ||||||||||||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | ||||||||||||
| Molecule Alteration | Missense mutation | p.T790M |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 3.10 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 3.05 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
S
G
G
E
E
A
A
P
P
700
|
N
N
Q
Q
A
A
L
L
L
L
R
R
I
I
L
L
K
K
E
E
710
|
T
T
E
E
F
F
K
K
K
K
I
I
K
K
V
V
L
L
G
G
720
|
S
S
G
G
A
A
F
F
G
G
T
T
V
V
Y
Y
K
K
G
G
730
|
L
L
W
W
I
I
P
P
E
E
G
G
E
E
K
K
V
V
K
K
740
|
I
I
P
P
V
V
A
A
I
I
K
K
E
E
L
L
R
R
E
E
750
|
A
A
T
T
S
S
P
P
K
K
A
A
N
N
K
K
E
E
I
I
760
|
L
L
D
D
E
E
A
A
Y
Y
V
V
M
M
A
A
S
S
V
V
770
|
D
D
N
N
P
P
H
H
V
V
C
C
R
R
L
L
L
L
G
G
780
|
I
I
C
C
L
L
T
T
S
S
T
T
V
V
Q
Q
L
L
I
I
790
|
T
M
Q
Q
L
L
M
M
P
P
F
F
G
G
C
C
L
L
L
L
800
|
D
D
Y
Y
V
V
R
R
E
E
H
H
K
K
D
D
N
N
I
I
810
|
G
G
S
S
Q
Q
Y
Y
L
L
L
L
N
N
W
W
C
C
V
V
820
|
Q
Q
I
I
A
A
K
K
G
G
M
M
N
N
Y
Y
L
L
E
E
830
|
D
D
R
R
R
R
L
L
V
V
H
H
R
R
D
D
L
L
A
A
840
|
A
A
R
R
N
N
V
V
L
L
V
V
K
K
T
T
P
P
Q
Q
850
|
H
H
V
V
K
K
I
I
T
T
D
D
F
F
G
G
L
L
A
A
860
|
K
K
L
L
L
L
G
G
A
A
E
E
E
E
K
K
E
E
Y
Y
870
|
H
H
A
A
E
E
G
G
G
G
K
K
V
V
P
P
I
I
K
K
880
|
W
W
M
M
A
A
L
L
E
E
S
S
I
I
L
L
H
H
R
R
890
|
I
I
Y
Y
T
T
H
H
Q
Q
S
S
D
D
V
V
W
W
S
S
900
|
Y
Y
G
G
V
V
T
T
V
V
W
W
E
E
L
L
M
M
T
T
910
|
F
F
G
G
S
S
K
K
P
P
Y
Y
D
D
G
G
I
I
P
P
920
|
A
A
S
S
E
E
I
I
S
S
S
S
I
I
L
L
E
E
K
K
930
|
G
G
E
E
R
R
L
L
P
P
Q
Q
P
P
P
P
I
I
C
C
940
|
T
T
I
I
D
D
V
V
Y
Y
M
M
I
I
M
M
V
V
K
K
950
|
C
C
W
W
M
M
I
I
D
D
A
A
D
D
S
S
R
R
P
P
960
|
K
K
F
F
R
R
E
E
L
L
I
I
I
I
E
E
F
F
S
S
970
|
K
K
M
M
A
A
R
R
D
D
P
P
Q
Q
R
R
Y
Y
L
L
980
|
V
V
I
I
Q
Q
G
G
D
D
E
E
R
R
M
M
H
H
L
L
990
|
P
P
S
S
P
P
T
T
D
D
S
S
N
N
F
F
Y
Y
R
R
1000
|
A
A
L
L
M
M
D
D
E
E
E
E
D
D
M
M
D
D
D
D
1010
|
V
V
V
V
D
D
A
A
D
D
E
E
Y
Y
L
L
I
I
P
P
1020
|
Q
Q
Q
Q
G
G
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | |||||||||||
| Experiment for Molecule Alteration |
Directional sequencing assay; Direct sequencing assay; Sanger sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Progression-free and post-progression survival asaay; Analysis of progression-free survival (PFS) assay; Overall survival assay | ||||||||||||
| Mechanism Description | The identification of T790M as acquired resistance mechanism was clinically feasible. Although T790M had no prognostic or predictive role in the present study, further research is necessary to identify patients with T790M-mutant tumors who might benefit from newly developed T790M-specific TkIs.T790M is likely a common resistance mechanism in patients treated with first-line afatinib. Although repeat biopsies at progression are crucial in elucidating resistance mechanisms, this study suggests that clinical and technical issues often limit their feasibility, highlighting the importance of developing. | ||||||||||||
| Key Molecule: Oncogenic epidermal growth factor receptor (EGFR) | [6] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Missense mutation | Exon 20 insertion mutations |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | PI3K-Akt signaling pathway | Inhibition | hsa04151 | ||||||||||
| In Vitro Model | Ba/F3 murine cells | Bone marrow | Homo sapiens (Human) | N.A. | |||||||||
| Bosc23 cells | Fetal kidney | Homo sapiens (Human) | CVCL_4401 | ||||||||||
| Experiment for Molecule Alteration |
GeneSeq assay | ||||||||||||
| Experiment for Drug Resistance |
Cell proliferation assay; Immunoblotting assay | ||||||||||||
| Mechanism Description | Mechanisms of acquired EGFR TKI resistance of this mutant remained underreported. | ||||||||||||
| Key Molecule: Epidermal growth factor receptor (EGFR) | [7] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Mutation | . |
|||||||||||
| Experiment for Drug Resistance |
Statistical analysis | ||||||||||||
| Mechanism Description | EGFR-TKI Rechallenge With Another TKI may be a useful treatment option after first-line osimertinib. | ||||||||||||
|
|
|||||||||||||
| Key Molecule: Hepatocyte growth factor receptor (MET) | [12] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Missense mutation | p.D1228V |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.71 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.67 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
G
-
D
1040
|
-
S
-
D
-
I
-
S
-
S
-
P
-
L
-
L
-
Q
N
N
1050
|
T
T
V
V
H
H
I
I
D
D
L
L
S
S
A
A
L
L
N
N
1060
|
P
P
E
E
L
L
V
V
Q
Q
A
A
V
V
Q
Q
H
H
V
V
1070
|
V
V
I
I
G
G
P
P
S
S
S
S
L
L
I
I
V
V
H
H
1080
|
F
F
N
N
E
E
V
V
I
I
G
G
R
R
G
G
H
H
F
F
1090
|
G
G
C
C
V
V
Y
Y
H
H
G
G
T
T
L
L
L
L
D
D
1100
|
N
N
D
D
G
G
K
K
K
K
I
I
H
H
C
C
A
A
V
V
1110
|
K
K
S
S
L
L
N
N
R
R
I
I
T
T
D
D
I
I
G
G
1120
|
E
E
V
V
S
S
Q
Q
F
F
L
L
T
T
E
E
G
G
I
I
1130
|
I
I
M
M
K
K
D
D
F
F
S
S
H
H
P
P
N
N
V
V
1140
|
L
L
S
S
L
L
L
L
G
G
I
I
C
C
L
L
R
R
S
S
1150
|
E
E
G
G
S
S
P
P
L
L
V
V
V
V
L
L
P
P
Y
Y
1160
|
M
M
K
K
H
H
G
G
D
D
L
L
R
R
N
N
F
F
I
I
1170
|
R
R
N
N
E
E
T
T
H
H
N
N
P
P
T
T
V
V
K
K
1180
|
D
D
L
L
I
I
G
G
F
F
G
G
L
L
Q
Q
V
V
A
A
1190
|
K
K
G
G
M
M
K
K
F
Y
L
L
A
A
S
S
K
K
K
K
1200
|
F
F
V
V
H
H
R
R
D
D
L
L
A
A
A
A
R
R
N
N
1210
|
C
C
M
M
L
L
D
D
E
E
K
K
F
F
T
T
V
V
K
K
1220
|
V
V
A
A
D
D
F
F
G
G
L
L
A
A
R
R
D
V
M
M
1230
|
Y
Y
D
D
K
K
E
E
F
Y
D
Y
S
S
V
V
H
H
N
N
1240
|
K
K
T
T
G
G
A
A
K
K
L
L
P
P
V
V
K
K
W
W
1250
|
M
M
A
A
L
L
E
E
S
S
L
L
Q
Q
T
T
Q
Q
K
K
1260
|
F
F
T
T
T
T
K
K
S
S
D
D
V
V
W
W
S
S
F
F
1270
|
G
G
V
V
L
L
L
L
W
W
E
E
L
L
M
M
T
T
R
R
1280
|
G
G
A
A
P
P
P
P
Y
Y
P
P
D
D
V
V
N
N
T
T
1290
|
F
F
D
D
I
I
T
T
V
V
Y
Y
L
L
L
L
Q
Q
G
G
1300
|
R
R
R
R
L
L
L
L
Q
Q
P
P
E
E
Y
Y
C
C
P
P
1310
|
D
D
P
P
L
L
Y
Y
E
E
V
V
M
M
L
L
K
K
C
C
1320
|
W
W
H
H
P
P
K
K
A
A
E
E
M
M
R
R
P
P
S
S
1330
|
F
F
S
S
E
E
L
L
V
V
S
S
R
R
I
I
S
S
A
A
1340
|
I
I
F
F
S
S
T
T
F
F
I
I
G
G
E
-
H
-
Y
-
1350
|
V
-
H
-
V
-
N
-
A
-
T
-
Y
-
V
-
N
-
V
-
1360
|
K
-
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
MTS assay | ||||||||||||
| Mechanism Description | Our in vitro findings demonstrate that MET D1228V induces resistance to type I MET TkIs through impaired drug binding while sensitivity to type II MET TkIs is maintained. | ||||||||||||
| Key Molecule: Merlin (NF2) | [5] | ||||||||||||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Molecule Alteration | Missense mutation | p.R198* |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Cell Pathway Regulation | mTOR signaling pathway | Activation | hsa04150 | ||||||||||
| Experiment for Molecule Alteration |
Deep amplicon-based resequencing assay; Whole exome sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Magnetic resonance imaging assay; Computerized tomography assay | ||||||||||||
| Mechanism Description | Whole exome sequencing (WES) of the A+C-resistant #16 and #24 tumors did not detect mutations in 23 mTOR-pathway related genes, strongly suggesting that non-mutational processes account for sustained activation of this pathway in these tumors. | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Oncogenic epidermal growth factor receptor (EGFR) | [6] | |||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Molecule Alteration | Missense mutation | Exon 20 insertion mutations |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K-Akt signaling pathway | Inhibition | hsa04151 | |
| In Vitro Model | Ba/F3 murine cells | Bone marrow | Homo sapiens (Human) | N.A. |
| Bosc23 cells | Fetal kidney | Homo sapiens (Human) | CVCL_4401 | |
| Experiment for Molecule Alteration |
GeneSeq assay | |||
| Experiment for Drug Resistance |
Cell proliferation assay; Immunoblotting assay | |||
| Mechanism Description | Mechanisms of acquired EGFR TKI resistance of this mutant remained underreported. | |||
|
|
||||
| Key Molecule: BLACAT1 overlapping LEMD1 locus (BLACAT1) | [1] | |||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| STAT3 signaling pathway | Activation | hsa04550 | ||
| In Vitro Model | Calu3 cells | Lung | Homo sapiens (Human) | CVCL_0609 |
| H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | |
| In Vivo Model | BALB/c nude xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR; Western blot analysis; Immunohistochemistry | |||
| Experiment for Drug Resistance |
MTT assay; Colony formation assay; Flow cytometry assay | |||
| Mechanism Description | BLACAT1 was up-regulated in afatinib-resistant NSCLC cells and knockdown of BLACAT1 reversed the resistance of afatinib to NSCLC cells by modulating STAT3 signalling. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Transmembrane protease serine 2 (TMPRSS2) | [13] | |||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SK-BR-3 cells | Pleural effusion | Homo sapiens (Human) | CVCL_0033 |
| Experiment for Molecule Alteration |
Western blot assay | |||
| Experiment for Drug Resistance |
MTT assay; Clonogenic assay | |||
| Mechanism Description | Analysis of the Cancer Genome Atlas (TCGA) revealed diminished expression of transmembrane serine protease 2 (TMPRSS2), a subfamily of membrane proteolytic enzymes, in breast cancer patients, correlating with unfavorable outcomes. Intriguingly, lapatinib-responsive patients exhibited higher TMPRSS2 expression. Our study unveiled that the compounds from?Artemisia argyi, eriodictyol, and umbelliferone could inhibit the growth of lapatinib-resistant HER2-positive breast cancer cells. Mechanistically, they suppressed HER2 kinase activation by enhancing TMPRSS2 activity. Our findings propose TMPRSS2 as a critical determinant in lapatinib sensitivity, and?Artemisia argyi?emerges as a potential agent to overcome lapatinib via activating TMPRSS2 in HER2-positive breast cancer.? | |||
|
|
||||
| Key Molecule: hsa-miR-630 | [14] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| In Vitro Model | SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 |
| HCC1954 cells | Breast | Homo sapiens (Human) | CVCL_1259 | |
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Introducing miR-630 into cells with innate- or acquired- resistance to HER-drugs significantly restored the efficacy of lapatinib, neratinib and afatinib; through a mechanism that at least partly, involve miR-630's regulation of IGF1R. Blocking miR-630 induced resistance/insensitivity to these drugs. Cellular motility, invasion, and anoikis were also observed as significantly altered by miR-630 manipulation, whereby introducing miR-630 into cells reduced cellular aggression while inhibition of miR-630 induced a more aggressive cellular phenotype. | |||
|
|
||||
| Key Molecule: Insulin-like growth factor 1 receptor (IGF1R) | [14] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| In Vitro Model | SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 |
| HCC1954 cells | Breast | Homo sapiens (Human) | CVCL_1259 | |
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Introducing miR-630 into cells with innate- or acquired- resistance to HER-drugs significantly restored the efficacy of lapatinib, neratinib and afatinib; through a mechanism that at least partly, involve miR-630's regulation of IGF1R. Blocking miR-630 induced resistance/insensitivity to these drugs. Cellular motility, invasion, and anoikis were also observed as significantly altered by miR-630 manipulation, whereby introducing miR-630 into cells reduced cellular aggression while inhibition of miR-630 induced a more aggressive cellular phenotype. | |||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
