Molecule Information
General Information of the Molecule (ID: Mol00591)
| Name |
GTPase Hras (HRAS)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
H-Ras-1; Ha-Ras; Transforming protein p21; c-H-ras; p21ras; HRAS1
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
HRAS
|
||||
| Gene ID | |||||
| Location |
chr11:532242-537321[-]
|
||||
| Sequence |
MTEYKLVVVGAGGVGKSALTIQLIQNHFVDEYDPTIEDSYRKQVVIDGETCLLDILDTAG
QEEYSAMRDQYMRTGEGFLCVFAINNTKSFEDIHQYREQIKRVKDSDDVPMVLVGNKCDL AARTVESRQAQDLARSYGIPYIETSAKTRQGVEDAFYTLVREIRQHKLRKLNPPDESGPG CMSCKCVLS Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Involved in the activation of Ras protein signal transduction. Ras proteins bind GDP/GTP and possess intrinsic GTPase activity.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
6 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [1] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | Binimetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.G12V (c.35G>T) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.98 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.96 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
G
-
0
|
S
-
M
M
T
T
E
E
Y
Y
K
K
L
L
V
V
V
V
V
V
10
|
G
G
A
A
G
V
G
G
V
V
G
G
K
K
S
S
A
A
L
L
20
|
T
T
I
I
Q
Q
L
L
I
I
Q
Q
N
N
H
H
F
F
V
V
30
|
D
D
E
E
Y
Y
D
D
P
P
T
T
I
I
E
E
D
D
S
S
40
|
Y
Y
R
R
K
K
Q
Q
V
V
V
V
I
I
D
D
G
G
E
E
50
|
T
T
C
C
L
L
L
L
D
D
I
I
L
L
D
D
T
T
A
A
60
|
G
G
Q
Q
E
E
E
E
Y
Y
S
S
A
A
M
M
R
R
D
D
70
|
Q
Q
Y
Y
M
M
R
R
T
T
G
G
E
E
G
G
F
F
L
L
80
|
C
C
V
V
F
F
A
A
I
I
N
N
N
N
T
T
K
K
S
S
90
|
F
F
E
E
D
D
I
I
H
H
H
H
Y
Y
R
R
E
E
Q
Q
100
|
I
I
K
K
R
R
V
V
K
K
D
D
S
S
E
E
D
D
V
V
110
|
P
P
M
M
V
V
L
L
V
V
G
G
N
N
K
K
S
S
D
D
120
|
L
L
P
P
S
S
R
R
T
T
V
V
D
D
T
T
K
K
Q
Q
130
|
A
A
Q
Q
D
D
L
L
A
A
R
R
S
S
Y
Y
G
G
I
I
140
|
P
P
F
F
I
I
E
E
T
T
S
S
A
A
K
K
T
T
R
R
150
|
Q
Q
G
G
V
V
D
D
D
D
A
A
F
F
Y
Y
T
T
L
L
160
|
V
V
R
R
E
E
I
I
R
R
K
K
H
H
K
K
E
E
K
K
170
|
M
M
S
S
K
K
D
D
G
G
K
K
K
K
K
K
K
K
K
K
180
|
K
K
S
S
K
K
T
T
K
K
C
C
V
V
I
I
M
M
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| RL952 cells | Endometrium | Homo sapiens (Human) | CVCL_0505 | ||||||||||
| NCI-H1915 cells | Lung | Homo sapiens (Human) | CVCL_1505 | ||||||||||
| KYSE-30 cells | Esophagus | Homo sapiens (Human) | CVCL_1351 | ||||||||||
| KNS62 cells | Brain | Homo sapiens (Human) | CVCL_1335 | ||||||||||
| HCC78 cells | Pleural effusion | Homo sapiens (Human) | CVCL_2061 | ||||||||||
| HCC44 cells | Lung | Homo sapiens (Human) | CVCL_2060 | ||||||||||
| CAL-12T cells | Lung | Homo sapiens (Human) | CVCL_1105 | ||||||||||
| In Vivo Model | CB17 SCID-/- mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [1] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | Binimetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q61R (c.182A>G) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.59 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.24 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
0
|
S
-
M
M
T
T
E
E
Y
Y
K
K
L
L
V
V
V
V
V
V
10
|
G
G
A
A
G
G
G
G
V
V
G
G
K
K
S
S
A
A
L
L
20
|
T
T
I
I
Q
Q
L
L
I
I
Q
Q
N
N
H
H
F
F
V
V
30
|
D
D
E
E
Y
Y
D
D
P
P
T
T
I
I
E
E
D
D
S
S
40
|
Y
Y
R
R
K
K
Q
Q
V
V
V
V
I
I
D
D
G
G
E
E
50
|
T
T
C
C
L
L
L
L
D
D
I
I
L
L
D
D
T
T
A
A
60
|
G
G
Q
R
E
E
E
E
Y
Y
S
S
A
A
M
M
R
R
D
D
70
|
Q
Q
Y
Y
M
M
R
R
T
T
G
G
E
E
G
G
F
F
L
L
80
|
C
C
V
V
F
F
A
A
I
I
N
N
N
N
S
S
K
K
S
D
90
|
F
F
A
A
D
D
I
I
N
N
L
L
Y
Y
R
R
E
E
Q
Q
100
|
I
I
K
K
R
R
V
V
K
K
D
D
S
S
D
D
D
D
V
V
110
|
P
P
M
M
V
V
L
L
V
V
G
G
N
N
K
K
C
C
D
D
120
|
L
L
P
P
T
T
R
R
T
T
V
V
D
D
T
T
K
K
Q
Q
130
|
A
A
H
H
E
E
L
L
A
A
K
K
S
S
Y
Y
G
G
I
I
140
|
P
P
F
F
I
I
E
E
T
T
S
S
A
A
K
K
T
T
R
R
150
|
Q
Q
G
G
V
V
E
E
D
D
A
A
F
F
Y
Y
T
T
L
L
160
|
V
V
R
R
E
E
I
I
R
R
Q
Q
Y
Y
R
R
M
M
K
K
170
|
K
-
L
-
N
-
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| RL952 cells | Endometrium | Homo sapiens (Human) | CVCL_0505 | ||||||||||
| NCI-H1915 cells | Lung | Homo sapiens (Human) | CVCL_1505 | ||||||||||
| KYSE-30 cells | Esophagus | Homo sapiens (Human) | CVCL_1351 | ||||||||||
| KNS62 cells | Brain | Homo sapiens (Human) | CVCL_1335 | ||||||||||
| HCC78 cells | Pleural effusion | Homo sapiens (Human) | CVCL_2061 | ||||||||||
| HCC44 cells | Lung | Homo sapiens (Human) | CVCL_2060 | ||||||||||
| CAL-12T cells | Lung | Homo sapiens (Human) | CVCL_1105 | ||||||||||
| In Vivo Model | CB17 SCID-/- mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay | ||||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Lapatinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.G12S |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.24 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.71 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
M
T
T
E
E
Y
Y
K
K
L
L
V
V
V
V
V
V
10
|
G
G
A
A
G
S
G
G
V
V
G
G
K
K
S
S
A
A
L
L
20
|
T
T
I
I
Q
Q
L
L
I
I
Q
Q
N
N
H
H
F
F
V
V
30
|
D
D
E
E
Y
Y
D
D
P
P
T
T
I
I
E
E
D
D
S
S
40
|
Y
Y
R
R
K
K
Q
Q
V
V
V
V
I
I
D
D
G
G
E
E
50
|
T
T
C
C
L
L
L
L
D
D
I
I
L
L
D
D
T
T
A
A
60
|
G
G
Q
Q
E
E
E
E
Y
Y
S
S
A
A
M
M
R
R
D
D
70
|
Q
Q
Y
Y
M
M
R
R
T
T
G
G
E
E
G
G
F
F
L
L
80
|
C
C
V
V
F
F
A
A
I
I
N
N
N
N
T
T
K
K
S
S
90
|
F
F
E
E
D
D
I
I
H
H
H
H
Y
Y
R
R
E
E
Q
Q
100
|
I
I
K
K
R
R
V
V
K
K
D
D
S
S
E
E
D
D
V
V
110
|
P
P
M
M
V
V
L
L
V
V
G
G
N
N
K
K
C
C
D
D
120
|
L
L
P
P
S
S
R
R
T
T
V
V
D
D
T
T
K
K
Q
Q
130
|
A
A
Q
Q
D
D
L
L
A
A
R
R
S
S
Y
Y
G
G
I
I
140
|
P
P
F
F
I
I
E
E
T
T
S
S
A
A
K
K
T
T
R
R
150
|
Q
Q
G
G
V
V
D
D
D
D
A
A
F
F
Y
Y
T
T
L
L
160
|
V
V
R
R
E
E
I
I
R
R
K
K
H
H
K
K
E
E
K
K
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Lapatinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.V9A |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Lapatinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.T2A |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Lapatinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.S17N |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Lapatinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q61X |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Lapatinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.N26S |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Lapatinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.D54N |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [3] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Pamidronate | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | U266 cells | Bone marrow | Homo sapiens (Human) | CVCL_0566 |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| Pseudomonas aeruginosa strain B-730P/17 | 287 | |||
| Experiment for Drug Resistance |
CellTiter 96 one solution proliferation assay | |||
| Mechanism Description | In response to pamidronate, there was significant inhibition of cell proliferation in MDA-231 and SKBR-3 cells, compared to MDA-175 cells. This correlated with their respective basal levels of N-ras and H-ras. N-ras and H-ras protein levels were both reduced in MDA-231 cells, and to lesser extent in SKBR-3 cells, following exposure to pamidronate, whereas these markers were not altered in MDA-175 cells, resistance to pamidronate may result from low levels of GTPase-activating proteins, such as N-ras and H-ras, in tumor cells. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Pertuzumab | ||||||||||||
| Molecule Alteration | Missense mutation | p.G12S |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.24 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.71 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
M
T
T
E
E
Y
Y
K
K
L
L
V
V
V
V
V
V
10
|
G
G
A
A
G
S
G
G
V
V
G
G
K
K
S
S
A
A
L
L
20
|
T
T
I
I
Q
Q
L
L
I
I
Q
Q
N
N
H
H
F
F
V
V
30
|
D
D
E
E
Y
Y
D
D
P
P
T
T
I
I
E
E
D
D
S
S
40
|
Y
Y
R
R
K
K
Q
Q
V
V
V
V
I
I
D
D
G
G
E
E
50
|
T
T
C
C
L
L
L
L
D
D
I
I
L
L
D
D
T
T
A
A
60
|
G
G
Q
Q
E
E
E
E
Y
Y
S
S
A
A
M
M
R
R
D
D
70
|
Q
Q
Y
Y
M
M
R
R
T
T
G
G
E
E
G
G
F
F
L
L
80
|
C
C
V
V
F
F
A
A
I
I
N
N
N
N
T
T
K
K
S
S
90
|
F
F
E
E
D
D
I
I
H
H
H
H
Y
Y
R
R
E
E
Q
Q
100
|
I
I
K
K
R
R
V
V
K
K
D
D
S
S
E
E
D
D
V
V
110
|
P
P
M
M
V
V
L
L
V
V
G
G
N
N
K
K
C
C
D
D
120
|
L
L
P
P
S
S
R
R
T
T
V
V
D
D
T
T
K
K
Q
Q
130
|
A
A
Q
Q
D
D
L
L
A
A
R
R
S
S
Y
Y
G
G
I
I
140
|
P
P
F
F
I
I
E
E
T
T
S
S
A
A
K
K
T
T
R
R
150
|
Q
Q
G
G
V
V
D
D
D
D
A
A
F
F
Y
Y
T
T
L
L
160
|
V
V
R
R
E
E
I
I
R
R
K
K
H
H
K
K
E
E
K
K
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Pertuzumab | ||||||||||||
| Molecule Alteration | Missense mutation | p.V9A |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Pertuzumab | ||||||||||||
| Molecule Alteration | Missense mutation | p.T2A |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Pertuzumab | ||||||||||||
| Molecule Alteration | Missense mutation | p.S17N |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Pertuzumab | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q61X |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Pertuzumab | ||||||||||||
| Molecule Alteration | Missense mutation | p.N26S |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Pertuzumab | ||||||||||||
| Molecule Alteration | Missense mutation | p.D54N |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Trastuzumab | ||||||||||||
| Molecule Alteration | Missense mutation | p.G12S |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.24 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.71 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
M
T
T
E
E
Y
Y
K
K
L
L
V
V
V
V
V
V
10
|
G
G
A
A
G
S
G
G
V
V
G
G
K
K
S
S
A
A
L
L
20
|
T
T
I
I
Q
Q
L
L
I
I
Q
Q
N
N
H
H
F
F
V
V
30
|
D
D
E
E
Y
Y
D
D
P
P
T
T
I
I
E
E
D
D
S
S
40
|
Y
Y
R
R
K
K
Q
Q
V
V
V
V
I
I
D
D
G
G
E
E
50
|
T
T
C
C
L
L
L
L
D
D
I
I
L
L
D
D
T
T
A
A
60
|
G
G
Q
Q
E
E
E
E
Y
Y
S
S
A
A
M
M
R
R
D
D
70
|
Q
Q
Y
Y
M
M
R
R
T
T
G
G
E
E
G
G
F
F
L
L
80
|
C
C
V
V
F
F
A
A
I
I
N
N
N
N
T
T
K
K
S
S
90
|
F
F
E
E
D
D
I
I
H
H
H
H
Y
Y
R
R
E
E
Q
Q
100
|
I
I
K
K
R
R
V
V
K
K
D
D
S
S
E
E
D
D
V
V
110
|
P
P
M
M
V
V
L
L
V
V
G
G
N
N
K
K
C
C
D
D
120
|
L
L
P
P
S
S
R
R
T
T
V
V
D
D
T
T
K
K
Q
Q
130
|
A
A
Q
Q
D
D
L
L
A
A
R
R
S
S
Y
Y
G
G
I
I
140
|
P
P
F
F
I
I
E
E
T
T
S
S
A
A
K
K
T
T
R
R
150
|
Q
Q
G
G
V
V
D
D
D
D
A
A
F
F
Y
Y
T
T
L
L
160
|
V
V
R
R
E
E
I
I
R
R
K
K
H
H
K
K
E
E
K
K
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Trastuzumab | ||||||||||||
| Molecule Alteration | Missense mutation | p.V9A |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Trastuzumab | ||||||||||||
| Molecule Alteration | Missense mutation | p.T2A |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Trastuzumab | ||||||||||||
| Molecule Alteration | Missense mutation | p.S17N |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Trastuzumab | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q61X |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Trastuzumab | ||||||||||||
| Molecule Alteration | Missense mutation | p.N26S |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Trastuzumab | ||||||||||||
| Molecule Alteration | Missense mutation | p.D54N |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Trastuzumab emtansine | ||||||||||||
| Molecule Alteration | Missense mutation | p.G12S |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.24 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.71 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
M
M
T
T
E
E
Y
Y
K
K
L
L
V
V
V
V
V
V
10
|
G
G
A
A
G
S
G
G
V
V
G
G
K
K
S
S
A
A
L
L
20
|
T
T
I
I
Q
Q
L
L
I
I
Q
Q
N
N
H
H
F
F
V
V
30
|
D
D
E
E
Y
Y
D
D
P
P
T
T
I
I
E
E
D
D
S
S
40
|
Y
Y
R
R
K
K
Q
Q
V
V
V
V
I
I
D
D
G
G
E
E
50
|
T
T
C
C
L
L
L
L
D
D
I
I
L
L
D
D
T
T
A
A
60
|
G
G
Q
Q
E
E
E
E
Y
Y
S
S
A
A
M
M
R
R
D
D
70
|
Q
Q
Y
Y
M
M
R
R
T
T
G
G
E
E
G
G
F
F
L
L
80
|
C
C
V
V
F
F
A
A
I
I
N
N
N
N
T
T
K
K
S
S
90
|
F
F
E
E
D
D
I
I
H
H
H
H
Y
Y
R
R
E
E
Q
Q
100
|
I
I
K
K
R
R
V
V
K
K
D
D
S
S
E
E
D
D
V
V
110
|
P
P
M
M
V
V
L
L
V
V
G
G
N
N
K
K
C
C
D
D
120
|
L
L
P
P
S
S
R
R
T
T
V
V
D
D
T
T
K
K
Q
Q
130
|
A
A
Q
Q
D
D
L
L
A
A
R
R
S
S
Y
Y
G
G
I
I
140
|
P
P
F
F
I
I
E
E
T
T
S
S
A
A
K
K
T
T
R
R
150
|
Q
Q
G
G
V
V
D
D
D
D
A
A
F
F
Y
Y
T
T
L
L
160
|
V
V
R
R
E
E
I
I
R
R
K
K
H
H
K
K
E
E
K
K
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Trastuzumab emtansine | ||||||||||||
| Molecule Alteration | Missense mutation | p.V9A |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Trastuzumab emtansine | ||||||||||||
| Molecule Alteration | Missense mutation | p.T2A |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Trastuzumab emtansine | ||||||||||||
| Molecule Alteration | Missense mutation | p.S17N |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Trastuzumab emtansine | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q61X |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Trastuzumab emtansine | ||||||||||||
| Molecule Alteration | Missense mutation | p.N26S |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Trastuzumab emtansine | ||||||||||||
| Molecule Alteration | Missense mutation | p.D54N |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [2] | ||||||||||||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | ||||||||||||
| Resistant Drug | Trastuzumab emtansine | ||||||||||||
| Molecule Alteration | Mutation | . |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. | |||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | ||||||||||||
| Experiment for Drug Resistance |
Positron emission tomography/Computed tomography assay | ||||||||||||
| Mechanism Description | Seven genes, including epidermal growth factor receptor (EGFR), G protein subunit alpha S (GNAS), HRas proto-oncogene (HRAS), mutL homolog 1 (MLH1), cadherin 1 (CDH1), neuroblastoma RAS viral oncogene homolog (NRAS), and NOTCH1, that only occurred mutations in the resistant group were associated with the resistance of targeted therapy. | ||||||||||||
Clinical Trial Drug(s)
1 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [4] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | Rigosertib | ||||||||||||
| Molecule Alteration | Missense mutation | p.G12V (c.35G>T) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.98 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.96 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
G
-
0
|
S
-
M
M
T
T
E
E
Y
Y
K
K
L
L
V
V
V
V
V
V
10
|
G
G
A
A
G
V
G
G
V
V
G
G
K
K
S
S
A
A
L
L
20
|
T
T
I
I
Q
Q
L
L
I
I
Q
Q
N
N
H
H
F
F
V
V
30
|
D
D
E
E
Y
Y
D
D
P
P
T
T
I
I
E
E
D
D
S
S
40
|
Y
Y
R
R
K
K
Q
Q
V
V
V
V
I
I
D
D
G
G
E
E
50
|
T
T
C
C
L
L
L
L
D
D
I
I
L
L
D
D
T
T
A
A
60
|
G
G
Q
Q
E
E
E
E
Y
Y
S
S
A
A
M
M
R
R
D
D
70
|
Q
Q
Y
Y
M
M
R
R
T
T
G
G
E
E
G
G
F
F
L
L
80
|
C
C
V
V
F
F
A
A
I
I
N
N
N
N
T
T
K
K
S
S
90
|
F
F
E
E
D
D
I
I
H
H
H
H
Y
Y
R
R
E
E
Q
Q
100
|
I
I
K
K
R
R
V
V
K
K
D
D
S
S
E
E
D
D
V
V
110
|
P
P
M
M
V
V
L
L
V
V
G
G
N
N
K
K
S
S
D
D
120
|
L
L
P
P
S
S
R
R
T
T
V
V
D
D
T
T
K
K
Q
Q
130
|
A
A
Q
Q
D
D
L
L
A
A
R
R
S
S
Y
Y
G
G
I
I
140
|
P
P
F
F
I
I
E
E
T
T
S
S
A
A
K
K
T
T
R
R
150
|
Q
Q
G
G
V
V
D
D
D
D
A
A
F
F
Y
Y
T
T
L
L
160
|
V
V
R
R
E
E
I
I
R
R
K
K
H
H
K
K
E
E
K
K
170
|
M
M
S
S
K
K
D
D
G
G
K
K
K
K
K
K
K
K
K
K
180
|
K
K
S
S
K
K
T
T
K
K
C
C
V
V
I
I
M
M
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Cell Pathway Regulation | RAS/RAF/MEK/ERK signaling pathway | Inhibition | hsa01521 | ||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | ||||||||||
| Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 | ||||||||||
| A431 cells | Skin | Homo sapiens (Human) | CVCL_0037 | ||||||||||
| HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 | ||||||||||
| MIA PaCa-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0428 | ||||||||||
| WM1617 cells | Lymph node | Homo sapiens (Human) | CVCL_6791 | ||||||||||
| In Vivo Model | Female nu/nu mouse PDX model | Mus musculus | |||||||||||
| Mechanism Description | Rigosertib, a styryl-benzyl sulfone, acts as a RAS-mimetic and interacts with the RBDs of RAF kinases, resulting in their inability to bind to RAS, disruption of RAF activation, and inhibition of the RAS-RAF-MEK pathway. We also find that rigosertib binds to the RBDs of Ral-GDS and PI3Ks. | ||||||||||||
Preclinical Drug(s)
9 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [1] | |||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Sensitive Drug | AZD-8055/Binimetinib | |||
| Molecule Alteration | Missense mutation | p.Q61L (c.182A>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| RL952 cells | Endometrium | Homo sapiens (Human) | CVCL_0505 | |
| NCI-H1915 cells | Lung | Homo sapiens (Human) | CVCL_1505 | |
| KYSE-30 cells | Esophagus | Homo sapiens (Human) | CVCL_1351 | |
| KNS62 cells | Brain | Homo sapiens (Human) | CVCL_1335 | |
| HCC78 cells | Pleural effusion | Homo sapiens (Human) | CVCL_2061 | |
| HCC44 cells | Lung | Homo sapiens (Human) | CVCL_2060 | |
| CAL-12T cells | Lung | Homo sapiens (Human) | CVCL_1105 | |
| In Vivo Model | CB17 SCID-/- mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Bladder cancer [ICD-11: 2C94.0] | [1] | ||||||||||||
| Sensitive Disease | Bladder cancer [ICD-11: 2C94.0] | ||||||||||||
| Sensitive Drug | Binimetinib/Everolimus | ||||||||||||
| Molecule Alteration | Missense mutation | p.G12V (c.35G>T) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.98 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.96 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
G
-
0
|
S
-
M
M
T
T
E
E
Y
Y
K
K
L
L
V
V
V
V
V
V
10
|
G
G
A
A
G
V
G
G
V
V
G
G
K
K
S
S
A
A
L
L
20
|
T
T
I
I
Q
Q
L
L
I
I
Q
Q
N
N
H
H
F
F
V
V
30
|
D
D
E
E
Y
Y
D
D
P
P
T
T
I
I
E
E
D
D
S
S
40
|
Y
Y
R
R
K
K
Q
Q
V
V
V
V
I
I
D
D
G
G
E
E
50
|
T
T
C
C
L
L
L
L
D
D
I
I
L
L
D
D
T
T
A
A
60
|
G
G
Q
Q
E
E
E
E
Y
Y
S
S
A
A
M
M
R
R
D
D
70
|
Q
Q
Y
Y
M
M
R
R
T
T
G
G
E
E
G
G
F
F
L
L
80
|
C
C
V
V
F
F
A
A
I
I
N
N
N
N
T
T
K
K
S
S
90
|
F
F
E
E
D
D
I
I
H
H
H
H
Y
Y
R
R
E
E
Q
Q
100
|
I
I
K
K
R
R
V
V
K
K
D
D
S
S
E
E
D
D
V
V
110
|
P
P
M
M
V
V
L
L
V
V
G
G
N
N
K
K
S
S
D
D
120
|
L
L
P
P
S
S
R
R
T
T
V
V
D
D
T
T
K
K
Q
Q
130
|
A
A
Q
Q
D
D
L
L
A
A
R
R
S
S
Y
Y
G
G
I
I
140
|
P
P
F
F
I
I
E
E
T
T
S
S
A
A
K
K
T
T
R
R
150
|
Q
Q
G
G
V
V
D
D
D
D
A
A
F
F
Y
Y
T
T
L
L
160
|
V
V
R
R
E
E
I
I
R
R
K
K
H
H
K
K
E
E
K
K
170
|
M
M
S
S
K
K
D
D
G
G
K
K
K
K
K
K
K
K
K
K
180
|
K
K
S
S
K
K
T
T
K
K
C
C
V
V
I
I
M
M
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| RL952 cells | Endometrium | Homo sapiens (Human) | CVCL_0505 | ||||||||||
| NCI-H1915 cells | Lung | Homo sapiens (Human) | CVCL_1505 | ||||||||||
| KYSE-30 cells | Esophagus | Homo sapiens (Human) | CVCL_1351 | ||||||||||
| KNS62 cells | Brain | Homo sapiens (Human) | CVCL_1335 | ||||||||||
| HCC78 cells | Pleural effusion | Homo sapiens (Human) | CVCL_2061 | ||||||||||
| HCC44 cells | Lung | Homo sapiens (Human) | CVCL_2060 | ||||||||||
| CAL-12T cells | Lung | Homo sapiens (Human) | CVCL_1105 | ||||||||||
| In Vivo Model | CB17 SCID-/- mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay | ||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [1] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | Binimetinib/Everolimus | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q61L (c.182A>T) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| RL952 cells | Endometrium | Homo sapiens (Human) | CVCL_0505 | ||||||||||
| NCI-H1915 cells | Lung | Homo sapiens (Human) | CVCL_1505 | ||||||||||
| KYSE-30 cells | Esophagus | Homo sapiens (Human) | CVCL_1351 | ||||||||||
| KNS62 cells | Brain | Homo sapiens (Human) | CVCL_1335 | ||||||||||
| HCC78 cells | Pleural effusion | Homo sapiens (Human) | CVCL_2061 | ||||||||||
| HCC44 cells | Lung | Homo sapiens (Human) | CVCL_2060 | ||||||||||
| CAL-12T cells | Lung | Homo sapiens (Human) | CVCL_1105 | ||||||||||
| In Vivo Model | CB17 SCID-/- mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Bladder cancer [ICD-11: 2C94.0] | [1] | ||||||||||||
| Sensitive Disease | Bladder cancer [ICD-11: 2C94.0] | ||||||||||||
| Sensitive Drug | Everolimus/Selumetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.G12V (c.35G>T) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.98 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.96 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
G
-
0
|
S
-
M
M
T
T
E
E
Y
Y
K
K
L
L
V
V
V
V
V
V
10
|
G
G
A
A
G
V
G
G
V
V
G
G
K
K
S
S
A
A
L
L
20
|
T
T
I
I
Q
Q
L
L
I
I
Q
Q
N
N
H
H
F
F
V
V
30
|
D
D
E
E
Y
Y
D
D
P
P
T
T
I
I
E
E
D
D
S
S
40
|
Y
Y
R
R
K
K
Q
Q
V
V
V
V
I
I
D
D
G
G
E
E
50
|
T
T
C
C
L
L
L
L
D
D
I
I
L
L
D
D
T
T
A
A
60
|
G
G
Q
Q
E
E
E
E
Y
Y
S
S
A
A
M
M
R
R
D
D
70
|
Q
Q
Y
Y
M
M
R
R
T
T
G
G
E
E
G
G
F
F
L
L
80
|
C
C
V
V
F
F
A
A
I
I
N
N
N
N
T
T
K
K
S
S
90
|
F
F
E
E
D
D
I
I
H
H
H
H
Y
Y
R
R
E
E
Q
Q
100
|
I
I
K
K
R
R
V
V
K
K
D
D
S
S
E
E
D
D
V
V
110
|
P
P
M
M
V
V
L
L
V
V
G
G
N
N
K
K
S
S
D
D
120
|
L
L
P
P
S
S
R
R
T
T
V
V
D
D
T
T
K
K
Q
Q
130
|
A
A
Q
Q
D
D
L
L
A
A
R
R
S
S
Y
Y
G
G
I
I
140
|
P
P
F
F
I
I
E
E
T
T
S
S
A
A
K
K
T
T
R
R
150
|
Q
Q
G
G
V
V
D
D
D
D
A
A
F
F
Y
Y
T
T
L
L
160
|
V
V
R
R
E
E
I
I
R
R
K
K
H
H
K
K
E
E
K
K
170
|
M
M
S
S
K
K
D
D
G
G
K
K
K
K
K
K
K
K
K
K
180
|
K
K
S
S
K
K
T
T
K
K
C
C
V
V
I
I
M
M
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| RL952 cells | Endometrium | Homo sapiens (Human) | CVCL_0505 | ||||||||||
| NCI-H1915 cells | Lung | Homo sapiens (Human) | CVCL_1505 | ||||||||||
| KYSE-30 cells | Esophagus | Homo sapiens (Human) | CVCL_1351 | ||||||||||
| KNS62 cells | Brain | Homo sapiens (Human) | CVCL_1335 | ||||||||||
| HCC78 cells | Pleural effusion | Homo sapiens (Human) | CVCL_2061 | ||||||||||
| HCC44 cells | Lung | Homo sapiens (Human) | CVCL_2060 | ||||||||||
| CAL-12T cells | Lung | Homo sapiens (Human) | CVCL_1105 | ||||||||||
| In Vivo Model | CB17 SCID-/- mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay | ||||||||||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [1] | ||||||||||||
| Sensitive Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | ||||||||||||
| Sensitive Drug | Everolimus/Selumetinib | ||||||||||||
| Molecule Alteration | Missense mutation | p.Q61L (c.182A>T) |
|||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |||||||||
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | ||||||||||
| RL952 cells | Endometrium | Homo sapiens (Human) | CVCL_0505 | ||||||||||
| NCI-H1915 cells | Lung | Homo sapiens (Human) | CVCL_1505 | ||||||||||
| KYSE-30 cells | Esophagus | Homo sapiens (Human) | CVCL_1351 | ||||||||||
| KNS62 cells | Brain | Homo sapiens (Human) | CVCL_1335 | ||||||||||
| HCC78 cells | Pleural effusion | Homo sapiens (Human) | CVCL_2061 | ||||||||||
| HCC44 cells | Lung | Homo sapiens (Human) | CVCL_2060 | ||||||||||
| CAL-12T cells | Lung | Homo sapiens (Human) | CVCL_1105 | ||||||||||
| In Vivo Model | CB17 SCID-/- mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter-Glo assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Thyroid gland cancer [ICD-11: 2D10.0] | [5] | |||
| Sensitive Disease | Thyroid gland cancer [ICD-11: 2D10.0] | |||
| Sensitive Drug | MK2206 | |||
| Molecule Alteration | Missense mutation | p.G13R (c.37G>C) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SW1736 cells | Thyroid | Homo sapiens (Human) | CVCL_3883 |
| C643 cells | Thyroid gland | Homo sapiens (Human) | CVCL_5969 | |
| HTH7 cells | Thyroid gland | Homo sapiens (Human) | CVCL_6289 | |
| Hth74 cells | Thyroid gland | Homo sapiens (Human) | CVCL_6288 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The missense mutation p.G13R (c.37G>C) in gene HRAS cause the sensitivity of MK2206 by unusual activation of pro-survival pathway | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Bladder cancer [ICD-11: 2C94.0] | [6] | |||
| Sensitive Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Sensitive Drug | NS1 | |||
| Molecule Alteration | Missense mutation | p.Q61L (c.182A>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Ras signaling pathway | Inhibition | hsa04014 | |
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 |
| HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 | |
| T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |
| HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |
| NIH3T3 cells | Embryo | Homo sapiens (Human) | CVCL_0594 | |
| COS cells | N.A. | N.A. | N.A. | |
| Experiment for Drug Resistance |
Promega assay | |||
| Mechanism Description | NS1, that bound with high affinity to both GTP- and GDP-bound states of H- and K-RAS but not N-RAS. NS1 potently inhibited growth factor signaling and oncogenic H- and K-RAS-mediated signaling and transformation but did not block oncogenic N-RAS, BRAF or MEK1. NS1 bound the alpha4-beta6-alpha5 region of RAS disrupting RAS dimerization/nanoclustering, which in turn blocked CRAF:BRAF heterodimerization and activation. These results establish the importance of the alpha4-beta6-alpha5 interface in RAS-mediated signaling and define a previously unrecognized site in RAS for inhibiting RAS function. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [7] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | PZ-1 | ||||||||||||
| Molecule Alteration | Missense mutation | p.G12V (c.35G>T) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.98 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.96 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
G
-
0
|
S
-
M
M
T
T
E
E
Y
Y
K
K
L
L
V
V
V
V
V
V
10
|
G
G
A
A
G
V
G
G
V
V
G
G
K
K
S
S
A
A
L
L
20
|
T
T
I
I
Q
Q
L
L
I
I
Q
Q
N
N
H
H
F
F
V
V
30
|
D
D
E
E
Y
Y
D
D
P
P
T
T
I
I
E
E
D
D
S
S
40
|
Y
Y
R
R
K
K
Q
Q
V
V
V
V
I
I
D
D
G
G
E
E
50
|
T
T
C
C
L
L
L
L
D
D
I
I
L
L
D
D
T
T
A
A
60
|
G
G
Q
Q
E
E
E
E
Y
Y
S
S
A
A
M
M
R
R
D
D
70
|
Q
Q
Y
Y
M
M
R
R
T
T
G
G
E
E
G
G
F
F
L
L
80
|
C
C
V
V
F
F
A
A
I
I
N
N
N
N
T
T
K
K
S
S
90
|
F
F
E
E
D
D
I
I
H
H
H
H
Y
Y
R
R
E
E
Q
Q
100
|
I
I
K
K
R
R
V
V
K
K
D
D
S
S
E
E
D
D
V
V
110
|
P
P
M
M
V
V
L
L
V
V
G
G
N
N
K
K
S
S
D
D
120
|
L
L
P
P
S
S
R
R
T
T
V
V
D
D
T
T
K
K
Q
Q
130
|
A
A
Q
Q
D
D
L
L
A
A
R
R
S
S
Y
Y
G
G
I
I
140
|
P
P
F
F
I
I
E
E
T
T
S
S
A
A
K
K
T
T
R
R
150
|
Q
Q
G
G
V
V
D
D
D
D
A
A
F
F
Y
Y
T
T
L
L
160
|
V
V
R
R
E
E
I
I
R
R
K
K
H
H
K
K
E
E
K
K
170
|
M
M
S
S
K
K
D
D
G
G
K
K
K
K
K
K
K
K
K
K
180
|
K
K
S
S
K
K
T
T
K
K
C
C
V
V
I
I
M
M
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | NIH3T3 cells | Embryo | Homo sapiens (Human) | CVCL_0594 | |||||||||
| In Vivo Model | Nu/Nu mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Microfluidic separation based assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Brain glioma [ICD-11: 2A00.0] | [8] | ||||||||||||
| Sensitive Disease | Brain glioma [ICD-11: 2A00.0] | ||||||||||||
| Sensitive Drug | SF1126 | ||||||||||||
| Molecule Alteration | Missense mutation | p.G12V (c.35G>T) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.98 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.96 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
G
-
0
|
S
-
M
M
T
T
E
E
Y
Y
K
K
L
L
V
V
V
V
V
V
10
|
G
G
A
A
G
V
G
G
V
V
G
G
K
K
S
S
A
A
L
L
20
|
T
T
I
I
Q
Q
L
L
I
I
Q
Q
N
N
H
H
F
F
V
V
30
|
D
D
E
E
Y
Y
D
D
P
P
T
T
I
I
E
E
D
D
S
S
40
|
Y
Y
R
R
K
K
Q
Q
V
V
V
V
I
I
D
D
G
G
E
E
50
|
T
T
C
C
L
L
L
L
D
D
I
I
L
L
D
D
T
T
A
A
60
|
G
G
Q
Q
E
E
E
E
Y
Y
S
S
A
A
M
M
R
R
D
D
70
|
Q
Q
Y
Y
M
M
R
R
T
T
G
G
E
E
G
G
F
F
L
L
80
|
C
C
V
V
F
F
A
A
I
I
N
N
N
N
T
T
K
K
S
S
90
|
F
F
E
E
D
D
I
I
H
H
H
H
Y
Y
R
R
E
E
Q
Q
100
|
I
I
K
K
R
R
V
V
K
K
D
D
S
S
E
E
D
D
V
V
110
|
P
P
M
M
V
V
L
L
V
V
G
G
N
N
K
K
S
S
D
D
120
|
L
L
P
P
S
S
R
R
T
T
V
V
D
D
T
T
K
K
Q
Q
130
|
A
A
Q
Q
D
D
L
L
A
A
R
R
S
S
Y
Y
G
G
I
I
140
|
P
P
F
F
I
I
E
E
T
T
S
S
A
A
K
K
T
T
R
R
150
|
Q
Q
G
G
V
V
D
D
D
D
A
A
F
F
Y
Y
T
T
L
L
160
|
V
V
R
R
E
E
I
I
R
R
K
K
H
H
K
K
E
E
K
K
170
|
M
M
S
S
K
K
D
D
G
G
K
K
K
K
K
K
K
K
K
K
180
|
K
K
S
S
K
K
T
T
K
K
C
C
V
V
I
I
M
M
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | U87MG glioma cells | Brain | Homo sapiens (Human) | CVCL_0022 | |||||||||
| In Vivo Model | Athymic female (CD-1 Nu/Nu) mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Kinase-Glo luminescent assay | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Head and neck squamous cell carcinoma [ICD-11: 2D42.1] | [9] | |||
| Sensitive Disease | Head and neck squamous cell carcinoma [ICD-11: 2D42.1] | |||
| Sensitive Drug | Sirolimus/Trametinib | |||
| Molecule Alteration | Missense mutation | p.Q61L (c.182A>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K/mTOR signaling pathway | Inhibition | hsa04151 | |
| In Vitro Model | CAL27 cells | Oral | Homo sapiens (Human) | CVCL_1107 |
| UM-SCC-17B cells | Cervical lymph node | Homo sapiens (Human) | CVCL_7725 | |
| Detroit 562 cells | Pleural effusion | Homo sapiens (Human) | CVCL_1171 | |
| In Vivo Model | Athymic nude mouse tumor xenografts model | Mus musculus | ||
| Experiment for Molecule Alteration |
Immunoblotting analysis | |||
| Experiment for Drug Resistance |
Alamar blue cell viability reagent assay | |||
| Mechanism Description | mTOR and MEK inhibition display a synergistic growth inhibitory activity in HNSCC cells genetically engineered to express activating KRAS and PIK3CA mutations. Antitumoral activity of the rapamycin and trametinib combination therapy increase in genetically engineered HNSCC cells expressing activating RAS or PIK3CA mutations | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [10] | ||||||||||||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | ||||||||||||
| Sensitive Drug | SR-9009 | ||||||||||||
| Molecule Alteration | Missense mutation | p.G12V (c.35G>T) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.98 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.96 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
G
-
0
|
S
-
M
M
T
T
E
E
Y
Y
K
K
L
L
V
V
V
V
V
V
10
|
G
G
A
A
G
V
G
G
V
V
G
G
K
K
S
S
A
A
L
L
20
|
T
T
I
I
Q
Q
L
L
I
I
Q
Q
N
N
H
H
F
F
V
V
30
|
D
D
E
E
Y
Y
D
D
P
P
T
T
I
I
E
E
D
D
S
S
40
|
Y
Y
R
R
K
K
Q
Q
V
V
V
V
I
I
D
D
G
G
E
E
50
|
T
T
C
C
L
L
L
L
D
D
I
I
L
L
D
D
T
T
A
A
60
|
G
G
Q
Q
E
E
E
E
Y
Y
S
S
A
A
M
M
R
R
D
D
70
|
Q
Q
Y
Y
M
M
R
R
T
T
G
G
E
E
G
G
F
F
L
L
80
|
C
C
V
V
F
F
A
A
I
I
N
N
N
N
T
T
K
K
S
S
90
|
F
F
E
E
D
D
I
I
H
H
H
H
Y
Y
R
R
E
E
Q
Q
100
|
I
I
K
K
R
R
V
V
K
K
D
D
S
S
E
E
D
D
V
V
110
|
P
P
M
M
V
V
L
L
V
V
G
G
N
N
K
K
S
S
D
D
120
|
L
L
P
P
S
S
R
R
T
T
V
V
D
D
T
T
K
K
Q
Q
130
|
A
A
Q
Q
D
D
L
L
A
A
R
R
S
S
Y
Y
G
G
I
I
140
|
P
P
F
F
I
I
E
E
T
T
S
S
A
A
K
K
T
T
R
R
150
|
Q
Q
G
G
V
V
D
D
D
D
A
A
F
F
Y
Y
T
T
L
L
160
|
V
V
R
R
E
E
I
I
R
R
K
K
H
H
K
K
E
E
K
K
170
|
M
M
S
S
K
K
D
D
G
G
K
K
K
K
K
K
K
K
K
K
180
|
K
K
S
S
K
K
T
T
K
K
C
C
V
V
I
I
M
M
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | ||||||||||
| Sk-Mel28 cells | Skin | Homo sapiens (Human) | CVCL_0526 | ||||||||||
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | ||||||||||
| PANC-1 cells | Pancreas | Homo sapiens (Human) | CVCL_0480 | ||||||||||
| Jurkat cells | Pleural effusion | Homo sapiens (Human) | CVCL_0065 | ||||||||||
| MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 | ||||||||||
| WI38 cells | Fetal lung | Homo sapiens (Human) | CVCL_0579 | ||||||||||
| BJ-ELR cells | N.A. | Homo sapiens (Human) | N.A. | ||||||||||
| BJ cells | Peripheral blood | Homo sapiens (Human) | CVCL_E483 | ||||||||||
| Becker cells | N.A. | Homo sapiens (Human) | CVCL_1093 | ||||||||||
| In Vivo Model | NOD mouse PDX model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Immunofluorescence microscopy assay; qRT-PCR | ||||||||||||
| Mechanism Description | Pharmacological modulation of circadian regulators is an effective novel antitumor strategy, identifying the existence of a previously unknown class of anticancer agents with a wide therapeutic window. REV-ERB agonists are novel autophagy and de novo lipogenesis inhibitors with selective activity towards malignant and benign neoplasms. | ||||||||||||
Investigative Drug(s)
2 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [11] | ||||||||||||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | ||||||||||||
| Sensitive Drug | CI-1040 | ||||||||||||
| Molecule Alteration | Missense mutation | p.G12V (c.35G>T) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.98 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.96 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
G
-
0
|
S
-
M
M
T
T
E
E
Y
Y
K
K
L
L
V
V
V
V
V
V
10
|
G
G
A
A
G
V
G
G
V
V
G
G
K
K
S
S
A
A
L
L
20
|
T
T
I
I
Q
Q
L
L
I
I
Q
Q
N
N
H
H
F
F
V
V
30
|
D
D
E
E
Y
Y
D
D
P
P
T
T
I
I
E
E
D
D
S
S
40
|
Y
Y
R
R
K
K
Q
Q
V
V
V
V
I
I
D
D
G
G
E
E
50
|
T
T
C
C
L
L
L
L
D
D
I
I
L
L
D
D
T
T
A
A
60
|
G
G
Q
Q
E
E
E
E
Y
Y
S
S
A
A
M
M
R
R
D
D
70
|
Q
Q
Y
Y
M
M
R
R
T
T
G
G
E
E
G
G
F
F
L
L
80
|
C
C
V
V
F
F
A
A
I
I
N
N
N
N
T
T
K
K
S
S
90
|
F
F
E
E
D
D
I
I
H
H
H
H
Y
Y
R
R
E
E
Q
Q
100
|
I
I
K
K
R
R
V
V
K
K
D
D
S
S
E
E
D
D
V
V
110
|
P
P
M
M
V
V
L
L
V
V
G
G
N
N
K
K
S
S
D
D
120
|
L
L
P
P
S
S
R
R
T
T
V
V
D
D
T
T
K
K
Q
Q
130
|
A
A
Q
Q
D
D
L
L
A
A
R
R
S
S
Y
Y
G
G
I
I
140
|
P
P
F
F
I
I
E
E
T
T
S
S
A
A
K
K
T
T
R
R
150
|
Q
Q
G
G
V
V
D
D
D
D
A
A
F
F
Y
Y
T
T
L
L
160
|
V
V
R
R
E
E
I
I
R
R
K
K
H
H
K
K
E
E
K
K
170
|
M
M
S
S
K
K
D
D
G
G
K
K
K
K
K
K
K
K
K
K
180
|
K
K
S
S
K
K
T
T
K
K
C
C
V
V
I
I
M
M
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |||||||||
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | ||||||||||
| In Vivo Model | Female athymic nu/nu mouse xenograft model | Mus musculus | |||||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
Caspase-Glo 3/7 luminogenic assay | ||||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [12] | ||||||||||||
| Resistant Disease | Colorectal cancer [ICD-11: 2B91.1] | ||||||||||||
| Resistant Drug | EGFR inhibitors | ||||||||||||
| Molecule Alteration | Missense mutation | p.G13D (c.38G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.01 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.40 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
0
|
-
G
-
G
T
T
E
E
Y
Y
K
K
L
L
V
V
V
V
V
V
10
|
G
G
A
A
G
G
G
D
V
V
G
G
K
K
S
S
A
A
L
L
20
|
T
T
I
I
Q
Q
L
L
I
I
Q
Q
N
N
H
H
F
F
V
V
30
|
D
D
E
E
Y
Y
D
D
P
P
T
T
I
I
E
E
D
D
S
S
40
|
Y
Y
R
R
K
K
Q
Q
V
V
V
V
I
I
D
D
G
G
E
E
50
|
T
T
C
C
L
L
L
L
D
D
I
I
L
L
D
D
T
T
A
A
60
|
G
G
Q
Q
E
E
E
E
Y
Y
S
S
A
A
M
M
R
R
D
D
70
|
Q
Q
Y
Y
M
M
R
R
T
T
G
G
E
E
G
G
F
F
L
L
80
|
C
C
V
V
F
F
A
A
I
I
N
N
N
N
T
T
K
K
S
S
90
|
F
F
E
E
D
D
I
I
H
H
H
H
Y
Y
R
R
E
E
Q
Q
100
|
I
I
K
K
R
R
V
V
K
K
D
D
S
S
E
E
D
D
V
V
110
|
P
P
M
M
V
V
L
L
V
V
G
G
N
N
K
K
C
C
D
D
120
|
L
L
P
P
S
S
R
R
T
T
V
V
D
D
T
T
K
K
Q
Q
130
|
A
A
Q
Q
D
D
L
L
A
A
R
R
S
S
Y
Y
G
G
I
I
140
|
P
P
F
F
I
I
E
E
T
T
S
S
A
A
K
K
T
T
R
R
150
|
Q
Q
G
R
V
V
D
E
D
D
A
A
F
F
Y
Y
T
T
L
L
160
|
V
V
R
R
E
E
I
I
R
R
K
Q
H
Y
K
R
E
L
K
K
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | Colorectum | N.A. | |||||||||||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Nervous tissue | |
| The Specified Disease | Brain cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.33E-103; Fold-change: -7.23E-01; Z-score: -1.65E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
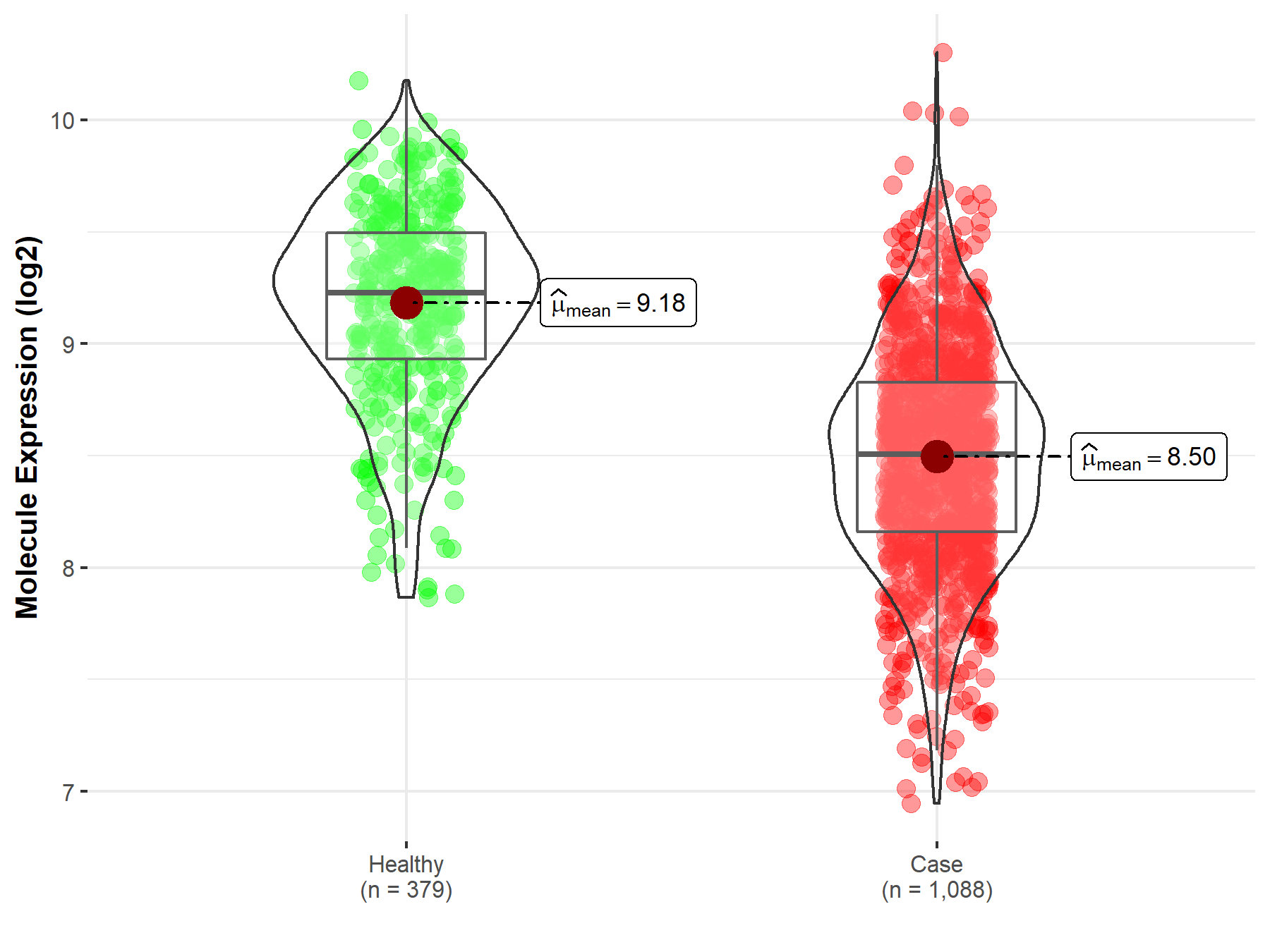
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.81E-02; Fold-change: -1.11E+00; Z-score: -5.27E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
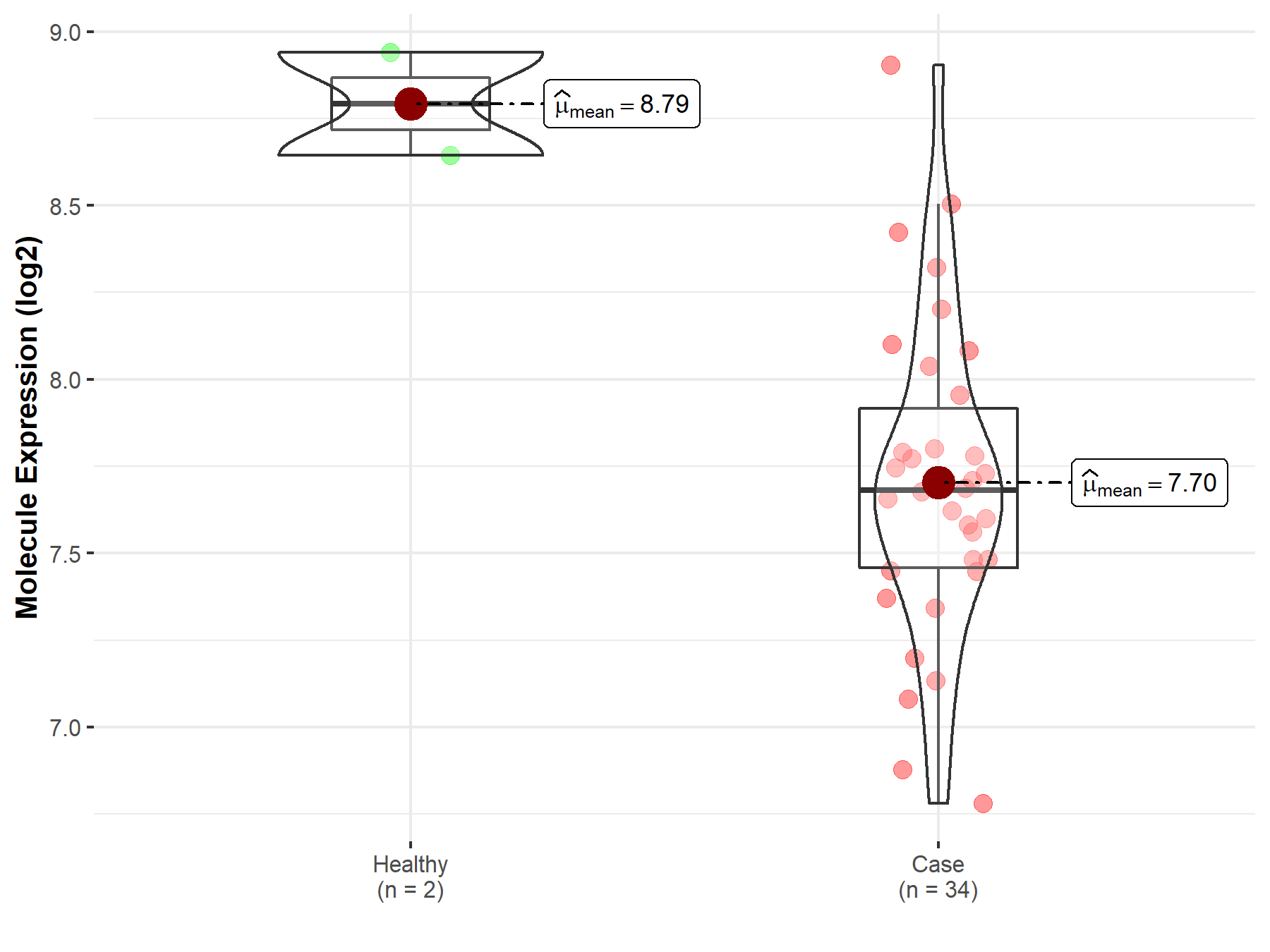
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | White matter | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.23E-03; Fold-change: 5.57E-01; Z-score: 1.68E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Neuroectodermal tumor | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.17E-01; Fold-change: 1.23E-01; Z-score: 2.34E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Lung | |
| The Specified Disease | Lung cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.09E-08; Fold-change: 1.71E-01; Z-score: 4.21E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 8.74E-01; Fold-change: -6.30E-02; Z-score: -1.61E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
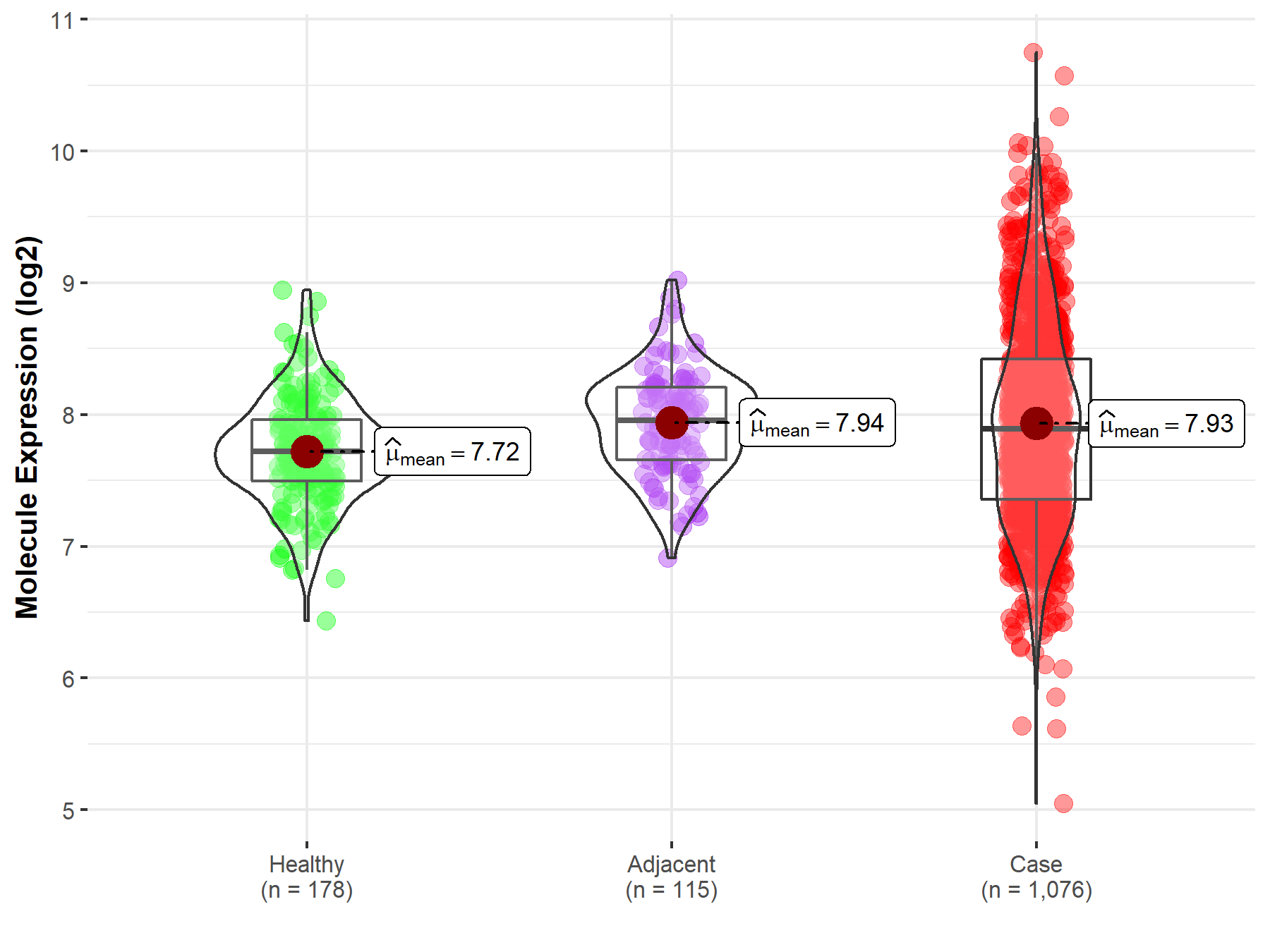
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Skin | |
| The Specified Disease | Melanoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.93E-04; Fold-change: -6.56E-01; Z-score: -1.20E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.17E-25; Fold-change: 2.99E-01; Z-score: 6.35E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.47E-02; Fold-change: 3.15E-01; Z-score: 4.47E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Bladder tissue | |
| The Specified Disease | Bladder cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.45E-03; Fold-change: 6.56E-01; Z-score: 1.56E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
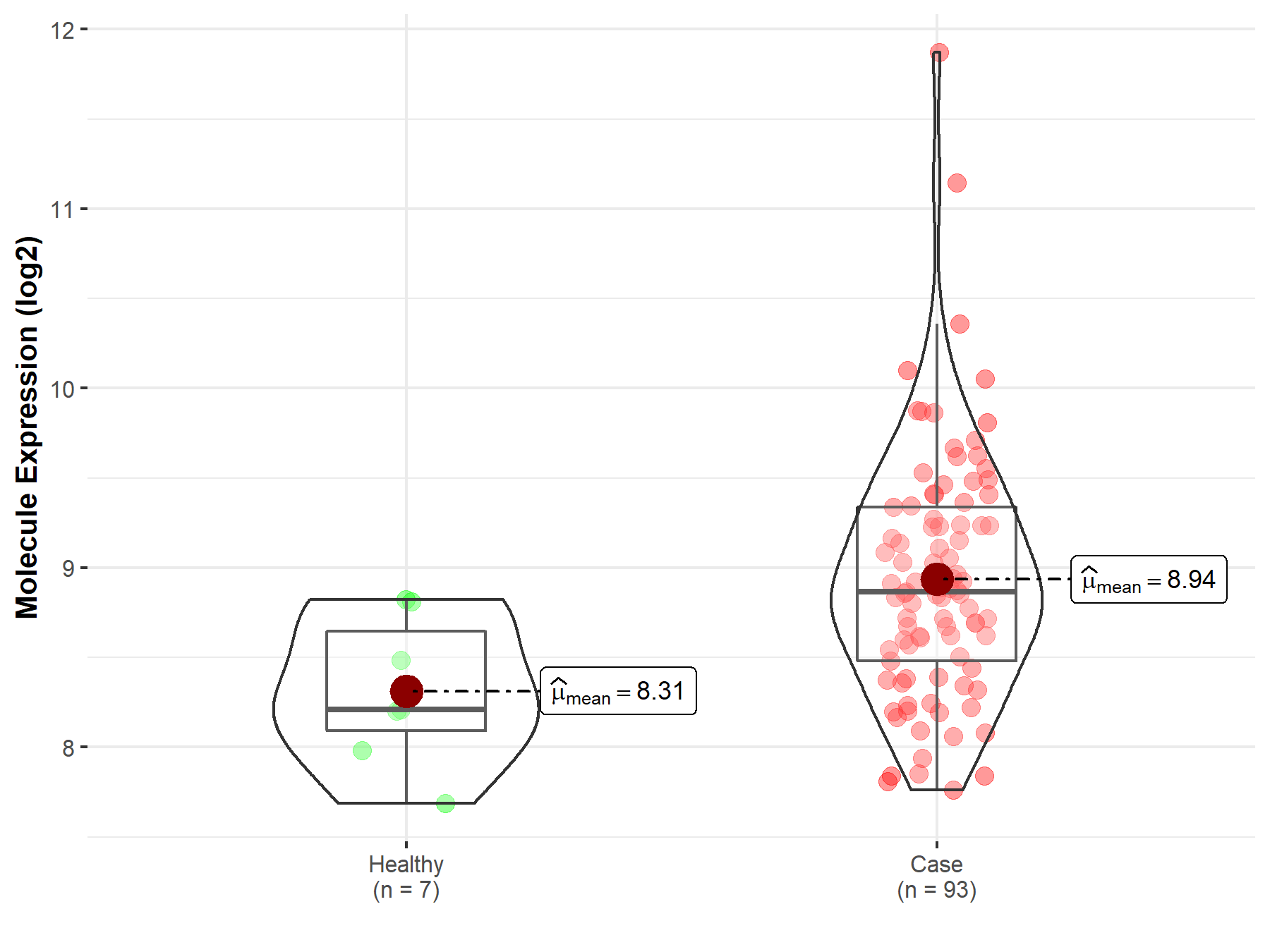
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Thyroid | |
| The Specified Disease | Thyroid cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.30E-11; Fold-change: 3.63E-01; Z-score: 1.03E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.63E-16; Fold-change: 7.02E-01; Z-score: 2.10E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals


|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
