Molecule Information
General Information of the Molecule (ID: Mol00554)
| Name |
Programmed cell death protein 4 (PDCD4)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Neoplastic transformation inhibitor protein; Nuclear antigen H731-like; Protein 197/15a; H731
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
PDCD4
|
||||
| Gene ID | |||||
| Location |
chr10:110871795-110900006[+]
|
||||
| Sequence |
MDVENEQILNVNPADPDNLSDSLFSGDEENAGTEEIKNEINGNWISASSINEARINAKAK
RRLRKNSSRDSGRGDSVSDSGSDALRSGLTVPTSPKGRLLDRRSRSGKGRGLPKKGGAGG KGVWGTPGQVYDVEEVDVKDPNYDDDQENCVYETVVLPLDERAFEKTLTPIIQEYFEHGD TNEVAEMLRDLNLGEMKSGVPVLAVSLALEGKASHREMTSKLLSDLCGTVMSTTDVEKSF DKLLKDLPELALDTPRAPQLVGQFIARAVGDGILCNTYIDSYKGTVDCVQARAALDKATV LLSMSKGGKRKDSVWGSGGGQQSVNHLVKEIDMLLKEYLLSGDISEAEHCLKELEVPHFH HELVYEAIIMVLESTGESTFKMILDLLKSLWKSSTITVDQMKRGYERIYNEIPDINLDVP HSYSVLERFVEECFQAGIISKQLRDLCPSRGRKRFVSEGDGGRLKPESY Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Inhibits translation initiation and cap-dependent translation. May excert its function by hindering the interaction between EIF4A1 and EIF4G. Inhibits the helicase activity of EIF4A. Modulates the activation of JUN kinase. Down-regulates the expression of MAP4K1, thus inhibiting events important in driving invasion, namely, MAPK85 activation and consequent JUN-dependent transcription. May play a role in apoptosis. Tumor suppressor. Inhibits tumor promoter-induced neoplastic transformation. Binds RNA (By similarity).
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
18 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [1] | |||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Resistant Drug | Gefitinib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | |||
| The Specified Disease | Non-small cell lung cancer | |||
| The Studied Tissue | Lung tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.36E-04 Fold-change: -1.45E-01 Z-score: -3.53E+00 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| PI3K/AKT signaling pathway | Activation | hsa04151 | ||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 |
| PC9R cells | Lung | Homo sapiens (Human) | CVCL_D778 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-21 overexpression is associated with the acquired resistance of EGFR-TkI in NSCLC, which might be caused by miR-21's function of activating PI3k/AkT pathway through inhibiting PTEN and PDCD4. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colon cancer [ICD-11: 2B90.1] | [2] | |||
| Resistant Disease | Colon cancer [ICD-11: 2B90.1] | |||
| Resistant Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Colon cancer [ICD-11: 2B90] | |||
| The Specified Disease | Colon cancer | |||
| The Studied Tissue | Colon tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.69E-35 Fold-change: -7.24E-01 Z-score: -1.54E+01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |
| U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 | |
| HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 | |
| PARP cells | Skin | Homo sapiens (Human) | N.A. | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Hypoxia induces miR-424 expression and that miR-424 in turn suppresses the level of PDCD4 protein, a tumor suppressor that is involved in apoptosis, by targeting its 3' untranslated region. Functionally, miR-424 overexpression decreases the sensitivity of cancer cells (HCT116 and A375) to doxorubicin (Dox) and etoposide. In contrast, the inhibition of miR-424 (+) apoptosis and increased the sensitivity of cancer cells to Dox. In a xenograft tumor model, miR-424 overexpression promoted tumor growth following Dox treatment, suggesting that miR-424 promotes tumor cell resistance to Dox. Furthermore, miR-424 levels are inversely correlated with PDCD4 expression in clinical breast cancer samples. | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [2] | |||
| Resistant Disease | Melanoma [ICD-11: 2C30.0] | |||
| Resistant Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |
| U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 | |
| HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 | |
| PARP cells | Skin | Homo sapiens (Human) | N.A. | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Hypoxia induces miR-424 expression and that miR-424 in turn suppresses the level of PDCD4 protein, a tumor suppressor that is involved in apoptosis, by targeting its 3' untranslated region. Functionally, miR-424 overexpression decreases the sensitivity of cancer cells (HCT116 and A375) to doxorubicin (Dox) and etoposide. In contrast, the inhibition of miR-424 (+) apoptosis and increased the sensitivity of cancer cells to Dox. In a xenograft tumor model, miR-424 overexpression promoted tumor growth following Dox treatment, suggesting that miR-424 promotes tumor cell resistance to Dox. Furthermore, miR-424 levels are inversely correlated with PDCD4 expression in clinical breast cancer samples. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colon cancer [ICD-11: 2B90.1] | [3] | |||
| Resistant Disease | Colon cancer [ICD-11: 2B90.1] | |||
| Resistant Drug | Fluorouracil | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Colon cancer [ICD-11: 2B90] | |||
| The Specified Disease | Colon cancer | |||
| The Studied Tissue | Colon tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.69E-35 Fold-change: -7.24E-01 Z-score: -1.54E+01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| PI3K/AKT signaling pathway | Regulation | N.A. | ||
| In Vitro Model | RkO cells | Colon | Homo sapiens (Human) | CVCL_0504 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-21 can mediate the drug resistance to 5-FU by inhibiting its target PDCD4, which can regulate the expression of ABCC5 and CD44 genes. | |||
| Disease Class: Pancreatic cancer [ICD-11: 2C10.3] | [14], [15] | |||
| Resistant Disease | Pancreatic cancer [ICD-11: 2C10.3] | |||
| Resistant Drug | Fluorouracil | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| The Specified Disease | Pancreatic cancer | |||
| The Studied Tissue | Pancreas | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.56E-02 Fold-change: -2.17E-01 Z-score: -2.68E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell invasion | Activation | hsa05200 | ||
| Cell migration | Activation | hsa04670 | ||
| Cell proliferation | Activation | hsa05200 | ||
| PI3K/AKT/mTOR signaling pathway | Regulation | N.A. | ||
| In Vitro Model | PANC-1 cells | Pancreas | Homo sapiens (Human) | CVCL_0480 |
| PATU8988 cells | Pancreas | Homo sapiens (Human) | CVCL_1846 | |
| 293TN cells | Pancreas | Homo sapiens (Human) | CVCL_UL49 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Wound Healing assay; Matrigel transmembrane invasion assay | |||
| Mechanism Description | miR-21 regulates 5-FU drug resistance in pancreatic cancer by reducing the expression of its targets, PTEN and PDCD4. And PTEN and PDCD4, as tumor suppressors, not only can inhibit tumor growth and invasion, but also can downregulate the 5-FU resistance induced by miR-21 in pancreatic cancer cells. | |||
| Disease Class: Hepatocellular carcinoma [ICD-11: 2C12.2] | [24] | |||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Resistant Drug | Fluorouracil | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell viability | Activation | hsa05200 | |
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| PLC/PRF/5 cells | Liver | Homo sapiens (Human) | CVCL_0485 | |
| HLE cells | Liver | Homo sapiens (Human) | CVCL_1281 | |
| HLF cells | Liver | Homo sapiens (Human) | CVCL_2947 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Hepatocellular carcinoma cells transfected with pre-miR-21 were significantly resistant to IFN-alpha/5-FU. Transfection of anti-miR-21 rendered HCC cells sensitive to IFN-alpha/5-FU, and such sensitivity was weakened by transfection of siRNAs of target molecules, PETN and PDCD4, miR-21 induces chemoresistance to IFN-alpha and 5-FU, mediated through PETN and PDCD4. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Renal carcinoma [ICD-11: 2C90.2] | [7] | |||
| Sensitive Disease | Renal carcinoma [ICD-11: 2C90.2] | |||
| Sensitive Drug | Fluorouracil | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Kidney cancer [ICD-11: 2C90] | |||
| The Specified Disease | Renal carcinoma | |||
| The Studied Tissue | Kidney | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.62E-01 Fold-change: 6.66E-03 Z-score: 1.74E-01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K/AKT signaling pathway | Inhibition | hsa04151 | |
| In Vitro Model | 786-O cells | Kidney | Homo sapiens (Human) | CVCL_1051 |
| ACHN cells | Pleural effusion | Homo sapiens (Human) | CVCL_1067 | |
| HK-2 cells | Kidney | Homo sapiens (Human) | CVCL_0302 | |
| RCC10 cells | Kidney | Homo sapiens (Human) | CVCL_6265 | |
| RCC4 cells | Kidney | Homo sapiens (Human) | CVCL_0498 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Celltiter96 Aqueous Non Radioactive Cell Proliferation Assay | |||
| Mechanism Description | Tumor suppressor genes like PTEN, PDCD4 and TIMP3, are target genes of miR21. PTEN is a potent inhibitor of PI3k/Akt pathway, as well as a direct target of miR21. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [4], [5] | |||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | |||
| The Specified Disease | Non-small cell lung cancer | |||
| The Studied Tissue | Lung tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.36E-04 Fold-change: -1.45E-01 Z-score: -3.53E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Activation | hsa05200 | |
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay and TUNEL assay | |||
| Mechanism Description | The expression level of miR-182 in A549 cell line was significantly higher than that in NHBE cell line. Transfection of miR-182 inhibitor induced sensitivity of A549 cells to cisplatin. A549 cells transfected with PDCD4 siRNA became more resistant to cisplatin therapy. We found an increase PDCD4 protein level following the transfection of miR-182 inhibitor using Western blot analysis. In addition, the (+) growth-inhibitory effect by miR-182 inhibitor was weakened after the addition of PDCD4 siRNA. And miR-141 expression was significantly up-regulated in cisplatin-resistant A549/DDP cells compared with the parental cell line A549; and PDCD4, an important apoptosis regulator, was found to be down-regulated. Luciferase activity assay and Western blot analysis confirmed that PDCD4 is a direct target of miR-141. Inhibition of miR-141 in A549/DDP cells markedly increased cisplatin sensitivity and apoptosis, which was partially abrogated by PDCD4 inhibition, indicating that PDCD4 is a functional target of miR-141 in of the regulation of cisplatin sensitivity. | |||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [12], [13] | |||
| Resistant Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Ovarian cancer [ICD-11: 2C73] | |||
| The Specified Disease | Ovarian cancer | |||
| The Studied Tissue | Ovarian tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.37E-01 Fold-change: -1.28E-01 Z-score: -1.28E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | SkOV3 cells | Ovary | Homo sapiens (Human) | CVCL_0532 |
| A2780 cells | Ovary | Homo sapiens (Human) | CVCL_0134 | |
| OVCAR3 cells | Ovary | Homo sapiens (Human) | CVCL_0465 | |
| A2780-CP cells | Ovary | Homo sapiens (Human) | CVCL_H745 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | The inhibition of miR-21 enhanced the sensitivity of ovarian cancer cells to cisplatin, miR-21 knockdown enhanced the expression of tumor suppressor PDCD4, downregulation of PDCD4 results in drug resistance via enhanced expression of c-IAP2 and MDR1. And the enhancement of miR-106a expression contributes to the generation of CDDP-resistant ovarian cancer cells, partly by targeting PDCD4. PDCD4 promoted CDDP-induced apoptosis mainly through the death receptor-mediated pathway. | |||
| Disease Class: Nasopharyngeal carcinoma [ICD-11: 2B6B.0] | [18] | |||
| Resistant Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B.0] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | HNE1 cells | Nasopharynx | Homo sapiens (Human) | CVCL_0308 |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Long non-coding RNA XIST modulates cisplatin resistance by altering PDCD4 and Fas-Lexpressions in human nasopharyngeal carcinoma HNE1 cells in vitro. XIST is up-regulated in HNE1/DDP cells, and down-regulation and up-regulation of XIST expression reduce and increase DDP resistance of the cells, respectively, possibly as a result of changes in the expressions of PDCD4 and Fas-L. | |||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [19] | |||
| Resistant Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Activation | hsa05200 | |
| Cell migration | Activation | hsa04670 | ||
| Cell proliferation | Activation | hsa05200 | ||
| JNk1/c-Jun pathway | Activation | hsa04010 | ||
| In Vitro Model | Hey A8 cells | Ovary | Homo sapiens (Human) | CVCL_8878 |
| SkVO3ip1 cells | Ovary | Homo sapiens (Human) | CVCL_0C84 | |
| A2780CP20 cells | Ovary | Homo sapiens (Human) | CVCL_A5PS | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Alamar blue dye assay | |||
| Mechanism Description | Blocking the JNk-1, the major activator of c-Jun phosphorylation, reduced the expression of pre-mir-21 and increased the expression of its well-known target gene, PDCD4. Overexpression of miR-21 in cisplatin sensitive cells decreased PDCD4 levels and increased cell proliferation. Finally, targeting miR-21 reduced cell growth, proliferation and invasion of cisplatin resistant ovarian cancer cells. These results suggest that the JNk-1/c-Jun/miR-21 pathway contributes to the cisplatin resistance of ovarian cancer cells and demonstrated that miR-21 is a plausible target to overcome cisplatin resistance. | |||
| Disease Class: Tongue squamous cell carcinoma [ICD-11: 2B62.1] | [20] | |||
| Resistant Disease | Tongue squamous cell carcinoma [ICD-11: 2B62.1] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | Tca8113 cells | Tongue | Homo sapiens (Human) | CVCL_6851 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Programmed cell death 4 (PDCD4) is a tumor suppressor gene and loss of PDCD4 expression was found in multiple human cancers. PDCD4 is an important functional target of miR-21 and related to tumor invasion and transformation. miR-21 could modulate chemosensitivity of TSCC cells to cisplatin by targeting PDCD4. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Cervical cancer [ICD-11: 2C77.0] | [21] | |||
| Sensitive Disease | Cervical cancer [ICD-11: 2C77.0] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell migration | Activation | hsa04670 | |
| Cell proliferation | Activation | hsa05200 | ||
| STAT3 signaling pathway | Activation | hsa04550 | ||
| In Vitro Model | Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 |
| Siha cells | Cervix uteri | Homo sapiens (Human) | CVCL_0032 | |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay; Colony formation assay | |||
| Mechanism Description | Down-regulation of LncRNA GAS5 can suppress TIMP3 and PDCD4 expression by enhancing miR-21 expression to suppress apoptosis and promote migration, invasion and cisplatin resistance in cervical cancer through the STAT3 signaling pathway. | |||
| Disease Class: Bladder cancer [ICD-11: 2C94.0] | [22] | |||
| Sensitive Disease | Bladder cancer [ICD-11: 2C94.0] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | 5637 cells | Bladder | Homo sapiens (Human) | CVCL_0126 |
| T24 cells | Bladder | Homo sapiens (Human) | CVCL_0554 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | miR-150 functions as a tumor promoter in reducing chemosensitivity and promoting invasiveness of MIBC cells via downretulating PDCD4. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | [6] | |||
| Sensitive Disease | Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | |||
| Sensitive Drug | Gemcitabine | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Pancreatic cancer [ICD-11: 2C10] | |||
| The Specified Disease | Pancreatic ductal adenocarcinoma | |||
| The Studied Tissue | Pancreas | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.18E-33 Fold-change: -1.18E+00 Z-score: -1.75E+01 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | MIA PaCa-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0428 |
| Panc02 cells | Pancreas | Homo sapiens (Human) | CVCL_D627 | |
| In Vivo Model | Mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR; Immunofluorescence (IF) staining | |||
| Experiment for Drug Resistance |
Costar Transwell Invasion Assay; | |||
| Mechanism Description | Upregulating miR21 in CAFs promoted PDAC desmoplasia and increased its drug resistance to gemcitabine treatment by promoting the activation of cancer-associated fibroblasts (CAFs). miR21 mediates activation of CAFs via down-regulating PDCD4. | |||
|
|
||||
| Disease Class: Pancreatic cancer [ICD-11: 2C10.3] | [25] | |||
| Sensitive Disease | Pancreatic cancer [ICD-11: 2C10.3] | |||
| Sensitive Drug | Gemcitabine | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SW1990 cells | Pancreas | Homo sapiens (Human) | CVCL_1723 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR429 sensitized gemcitabine response in GZ-resistant pancreatic cancer cells via its direct upregulation of PDCD4 expression. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Renal carcinoma [ICD-11: 2C90.2] | [7] | |||
| Sensitive Disease | Renal carcinoma [ICD-11: 2C90.2] | |||
| Sensitive Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Kidney cancer [ICD-11: 2C90] | |||
| The Specified Disease | Renal carcinoma | |||
| The Studied Tissue | Kidney | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.62E-01 Fold-change: 6.66E-03 Z-score: 1.74E-01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K/AKT signaling pathway | Inhibition | hsa04151 | |
| In Vitro Model | 786-O cells | Kidney | Homo sapiens (Human) | CVCL_1051 |
| ACHN cells | Pleural effusion | Homo sapiens (Human) | CVCL_1067 | |
| HK-2 cells | Kidney | Homo sapiens (Human) | CVCL_0302 | |
| RCC10 cells | Kidney | Homo sapiens (Human) | CVCL_6265 | |
| RCC4 cells | Kidney | Homo sapiens (Human) | CVCL_0498 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Celltiter96 Aqueous Non Radioactive Cell Proliferation Assay | |||
| Mechanism Description | Tumor suppressor genes like PTEN, PDCD4 and TIMP3, are target genes of miR21. PTEN is a potent inhibitor of PI3k/Akt pathway, as well as a direct target of miR21. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Renal carcinoma [ICD-11: 2C90.2] | [7] | |||
| Sensitive Disease | Renal carcinoma [ICD-11: 2C90.2] | |||
| Sensitive Drug | Oxaliplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Kidney cancer [ICD-11: 2C90] | |||
| The Specified Disease | Renal carcinoma | |||
| The Studied Tissue | Kidney | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.62E-01 Fold-change: 6.66E-03 Z-score: 1.74E-01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K/AKT signaling pathway | Inhibition | hsa04151 | |
| In Vitro Model | 786-O cells | Kidney | Homo sapiens (Human) | CVCL_1051 |
| ACHN cells | Pleural effusion | Homo sapiens (Human) | CVCL_1067 | |
| HK-2 cells | Kidney | Homo sapiens (Human) | CVCL_0302 | |
| RCC10 cells | Kidney | Homo sapiens (Human) | CVCL_6265 | |
| RCC4 cells | Kidney | Homo sapiens (Human) | CVCL_0498 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Celltiter96 Aqueous Non Radioactive Cell Proliferation Assay | |||
| Mechanism Description | Tumor suppressor genes like PTEN, PDCD4 and TIMP3, are target genes of miR21. PTEN is a potent inhibitor of PI3k/Akt pathway, as well as a direct target of miR21. | |||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [26] | |||
| Sensitive Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Sensitive Drug | Oxaliplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | HT29 Cells | Colon | Homo sapiens (Human) | CVCL_A8EZ |
| SW480 cells | Colon | Homo sapiens (Human) | CVCL_0546 | |
| HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Overexpression of MEG3 improved oxaliplatin sensitivity of HT29/OXA and HCT116/OXA cells via suppressing miR-141 expression and upregulating PDCD4. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [26] | |||
| Resistant Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Resistant Drug | Oxaliplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell viability | Activation | hsa05200 | ||
| In Vitro Model | HT29 Cells | Colon | Homo sapiens (Human) | CVCL_A8EZ |
| SW480 cells | Colon | Homo sapiens (Human) | CVCL_0546 | |
| HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase reporter assay; RNA pull-down assay; RIP assay | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Overexpression of MEG3 improved oxaliplatin sensitivity of HT29/OXA and HCT116/OXA cells via suppressing miR-141 expression and upregulating PDCD4. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Renal carcinoma [ICD-11: 2C90.2] | [7] | |||
| Sensitive Disease | Renal carcinoma [ICD-11: 2C90.2] | |||
| Sensitive Drug | Dovitinib lactate | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Kidney cancer [ICD-11: 2C90] | |||
| The Specified Disease | Renal carcinoma | |||
| The Studied Tissue | Kidney | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.62E-01 Fold-change: 6.66E-03 Z-score: 1.74E-01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PI3K/AKT signaling pathway | Inhibition | hsa04151 | |
| In Vitro Model | 786-O cells | Kidney | Homo sapiens (Human) | CVCL_1051 |
| ACHN cells | Pleural effusion | Homo sapiens (Human) | CVCL_1067 | |
| HK-2 cells | Kidney | Homo sapiens (Human) | CVCL_0302 | |
| RCC10 cells | Kidney | Homo sapiens (Human) | CVCL_6265 | |
| RCC4 cells | Kidney | Homo sapiens (Human) | CVCL_0498 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Celltiter96 Aqueous Non Radioactive Cell Proliferation Assay | |||
| Mechanism Description | Tumor suppressor genes like PTEN, PDCD4 and TIMP3, are target genes of miR21. PTEN is a potent inhibitor of PI3k/Akt pathway, as well as a direct target of miR21. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [8] | |||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Sensitive Drug | Cytarabine | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Acute myeloid leukemia [ICD-11: 2A60] | |||
| The Specified Disease | Acute myelocytic leukemia | |||
| The Studied Tissue | Bone marrow | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.63E-03 Fold-change: 7.01E-02 Z-score: 2.85E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | AMO-miR-21 significantly sensitizes HL60 cells to Ara-C byinducing apoptosis and these effects of AMO-miR-21 may be partially due to its up-regulation ofPDCD4. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [9] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Fulvestrant | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Breast cancer [ICD-11: 2C60] | |||
| The Specified Disease | Breast cancer | |||
| The Studied Tissue | Breast tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.12E-04 Fold-change: 5.26E-02 Z-score: 3.37E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| PI3K/AKT/mTOR signaling pathway | Regulation | N.A. | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-21 is a miRNA that is overexpressed in most tumor types, and acts as an oncogene by targeting many suppressor genes related to proliferation, apoptosis, and invasion. miR-21 facilitates tumor growth and invasion by targeting programmed cell death 4 (PDCD4), PTEN, and Bcl-2. silencing of miR-21 sensitized ER+ breast cancer cells to TAM and FUL induced cell apoptosis. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Renal cell carcinoma [ICD-11: 2C90.0] | [10] | |||
| Sensitive Disease | Renal cell carcinoma [ICD-11: 2C90.0] | |||
| Sensitive Drug | Topotecan | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Kidney cancer [ICD-11: 2C90] | |||
| The Specified Disease | Renal cancer | |||
| The Studied Tissue | Kidney | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.92E-07 Fold-change: 2.01E-01 Z-score: 8.08E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell colony | Inhibition | hsa05200 | ||
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | A498 cells | Kidney | Homo sapiens (Human) | CVCL_1056 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
XTT assay | |||
| Mechanism Description | Inhibition of miR-21 rescues PDCD4 and PTEN protein levels and improves chemosensitivity and therapeutic response. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Glioma [ICD-11: 2A00.1] | [11] | |||
| Resistant Disease | Glioma [ICD-11: 2A00.1] | |||
| Resistant Drug | Temozolomide | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Brain cancer [ICD-11: 2A00] | |||
| The Specified Disease | Brain cancer | |||
| The Studied Tissue | Nervous tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.63E-10 Fold-change: -6.84E-02 Z-score: -6.22E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| U87 cells | Brain | Homo sapiens (Human) | CVCL_0022 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | Ectopic overexpression of miR-497 promotes chemotherapy resistance in glioma cells by targeting PDCD4, a tumor suppressor that is involved in apoptosis. In contrast, the inhibition of miR-497 enhances apoptosis and increases the sensitivity of glioma cells to TMZ. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [2] | |||
| Resistant Disease | Melanoma [ICD-11: 2C30.0] | |||
| Resistant Drug | Etoposide | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Melanoma [ICD-11: 2C30] | |||
| The Specified Disease | Melanoma | |||
| The Studied Tissue | Skin | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.26E-02 Fold-change: -1.06E-01 Z-score: -1.95E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |
| U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 | |
| HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 | |
| PARP cells | Skin | Homo sapiens (Human) | N.A. | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Hypoxia induces miR-424 expression and that miR-424 in turn suppresses the level of PDCD4 protein, a tumor suppressor that is involved in apoptosis, by targeting its 3' untranslated region. Functionally, miR-424 overexpression decreases the sensitivity of cancer cells (HCT116 and A375) to doxorubicin (Dox) and etoposide. In contrast, the inhibition of miR-424 (+) apoptosis and increased the sensitivity of cancer cells to Dox. In a xenograft tumor model, miR-424 overexpression promoted tumor growth following Dox treatment, suggesting that miR-424 promotes tumor cell resistance to Dox. Furthermore, miR-424 levels are inversely correlated with PDCD4 expression in clinical breast cancer samples. | |||
| Disease Class: Colon cancer [ICD-11: 2B90.1] | [2] | |||
| Resistant Disease | Colon cancer [ICD-11: 2B90.1] | |||
| Resistant Drug | Etoposide | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Colon cancer [ICD-11: 2B90] | |||
| The Specified Disease | Colon cancer | |||
| The Studied Tissue | Colon tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.93E-12 Fold-change: -1.62E-01 Z-score: -7.40E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 |
| A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 | |
| U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 | |
| HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 | |
| PARP cells | Skin | Homo sapiens (Human) | N.A. | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Hypoxia induces miR-424 expression and that miR-424 in turn suppresses the level of PDCD4 protein, a tumor suppressor that is involved in apoptosis, by targeting its 3' untranslated region. Functionally, miR-424 overexpression decreases the sensitivity of cancer cells (HCT116 and A375) to doxorubicin (Dox) and etoposide. In contrast, the inhibition of miR-424 (+) apoptosis and increased the sensitivity of cancer cells to Dox. In a xenograft tumor model, miR-424 overexpression promoted tumor growth following Dox treatment, suggesting that miR-424 promotes tumor cell resistance to Dox. Furthermore, miR-424 levels are inversely correlated with PDCD4 expression in clinical breast cancer samples. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Leukemia [ICD-11: 2B33.6] | [16] | |||
| Resistant Disease | Leukemia [ICD-11: 2B33.6] | |||
| Resistant Drug | Arsenic trioxide | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 |
| K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | PDCD4 has been reported to be involved in growth, apoptosis, invasion and cell cycle etc. AMO-miR-21 significantly sensitizes HL60 and k562 cells to ATO by inducing apoptosis, and these effects of AMO-miR-21 may be partially due to its up-regulation of PDCD4. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Chronic myelogenous leukemia [ICD-11: 2A20.3] | [17] | |||
| Sensitive Disease | Chronic myelogenous leukemia [ICD-11: 2A20.3] | |||
| Sensitive Drug | Arsenic trioxide | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell growth | Inhibition | hsa05200 | ||
| In Vitro Model | K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-21 post-transcriptionally down-regulates tumor suppressor PDCD4. AMO-miR-21 sensitized leukemic k562 cells to ATO by inducing apoptosis partially due to its up-regulation of PDCD4 protein level. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Prostate cancer [ICD-11: 2C82.0] | [23] | |||
| Resistant Disease | Prostate cancer [ICD-11: 2C82.0] | |||
| Resistant Drug | Docetaxel | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Activation | hsa05200 | |
| In Vitro Model | PC3 cells | Prostate | Homo sapiens (Human) | CVCL_0035 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Programmed cell death 4 (PDCD4), is a novel suppressor of tumorigenesis, tumor progression and invasion. miR-21 can directly down-regulate the expression of PDCD4 by targeting its 3'UTR in PC3 cells. PDCD4, a direct target gene of miR-21, could mediate chemoresistance to docetaxel in PC3 cells. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hepatocellular carcinoma [ICD-11: 2C12.2] | [24] | |||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Resistant Drug | IFN-alpha | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell viability | Activation | hsa05200 | |
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| PLC/PRF/5 cells | Liver | Homo sapiens (Human) | CVCL_0485 | |
| HLE cells | Liver | Homo sapiens (Human) | CVCL_1281 | |
| HLF cells | Liver | Homo sapiens (Human) | CVCL_2947 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Hepatocellular carcinoma cells transfected with pre-miR-21 were significantly resistant to IFN-alpha/5-FU. Transfection of anti-miR-21 rendered HCC cells sensitive to IFN-alpha/5-FU, and such sensitivity was weakened by transfection of siRNAs of target molecules, PETN and PDCD4, miR-21 induces chemoresistance to IFN-alpha and 5-FU, mediated through PETN and PDCD4. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [9] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Tamoxifen | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| PI3K/AKT/mTOR signaling pathway | Regulation | N.A. | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-21 is a miRNA that is overexpressed in most tumor types, and acts as an oncogene by targeting many suppressor genes related to proliferation, apoptosis, and invasion. miR-21 facilitates tumor growth and invasion by targeting programmed cell death 4 (PDCD4), PTEN, and Bcl-2. silencing of miR-21 sensitized ER+ breast cancer cells to TAM and FUL induced cell apoptosis. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [27] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Trastuzumab | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| PI3K signaling pathway | Activation | hsa04151 | ||
| In Vitro Model | SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | A target prediction analysis coupled with in vitro and in vivo validations revealed that miR-21 levels inversely correlated with the expression of PTEN and PDCD4, which differentially influenced the drug sensitivity of HER2-positive breast cancer cells.miR-21 was able to affect the response to both trastuzumab and chemotherapy, triggering an IL-6/STAT3/NF-kB-mediated signaling loop and activating the PI3k pathway. These findings support the ability of miR-21 signaling to sustain EMT and shape the tumor immune microenvironment in HER2-positive breast cancer. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Nervous tissue | |
| The Specified Disease | Brain cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.63E-10; Fold-change: -1.89E-01; Z-score: -3.42E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
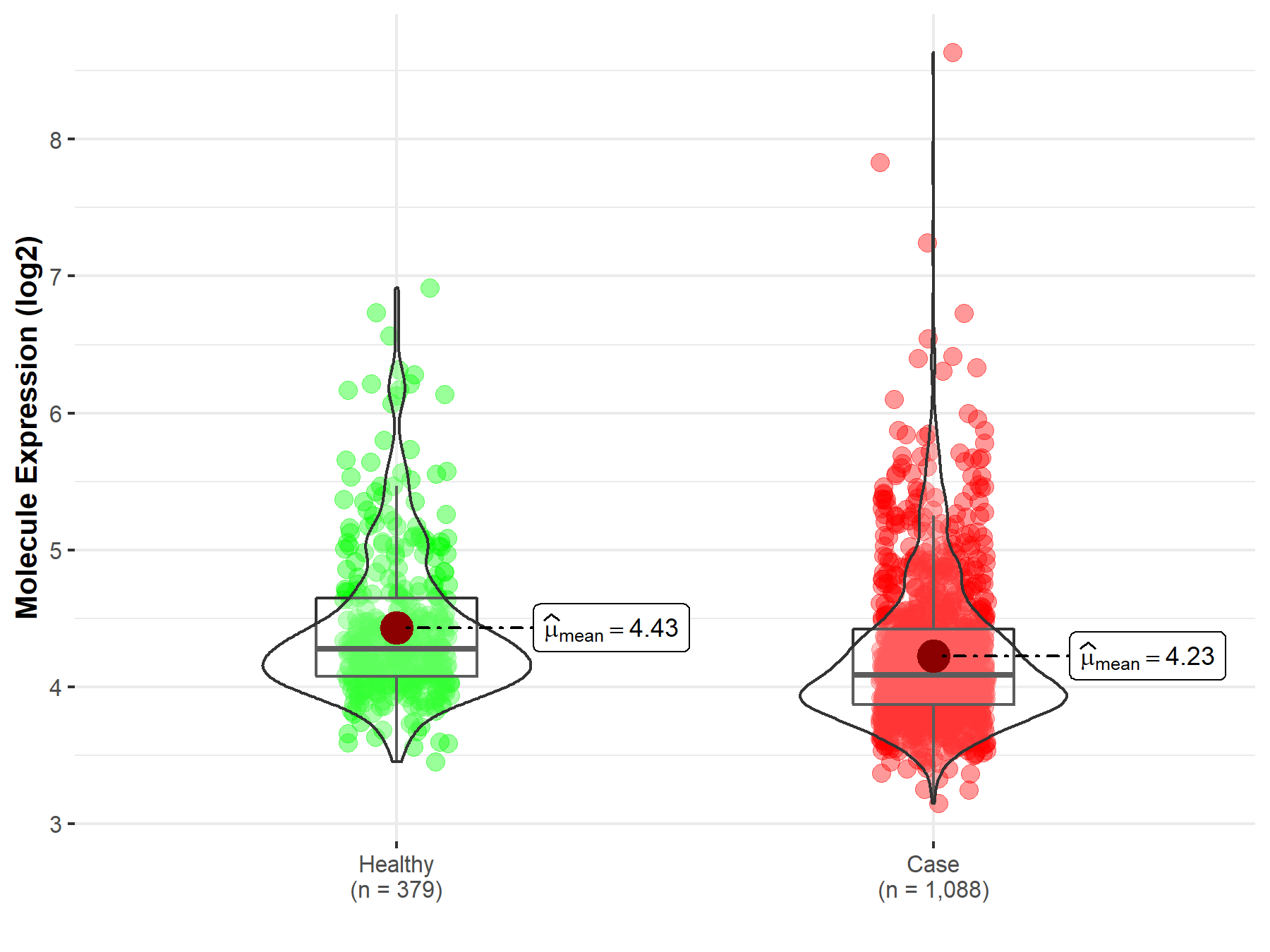
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.61E-01; Fold-change: 6.07E-02; Z-score: 1.96E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
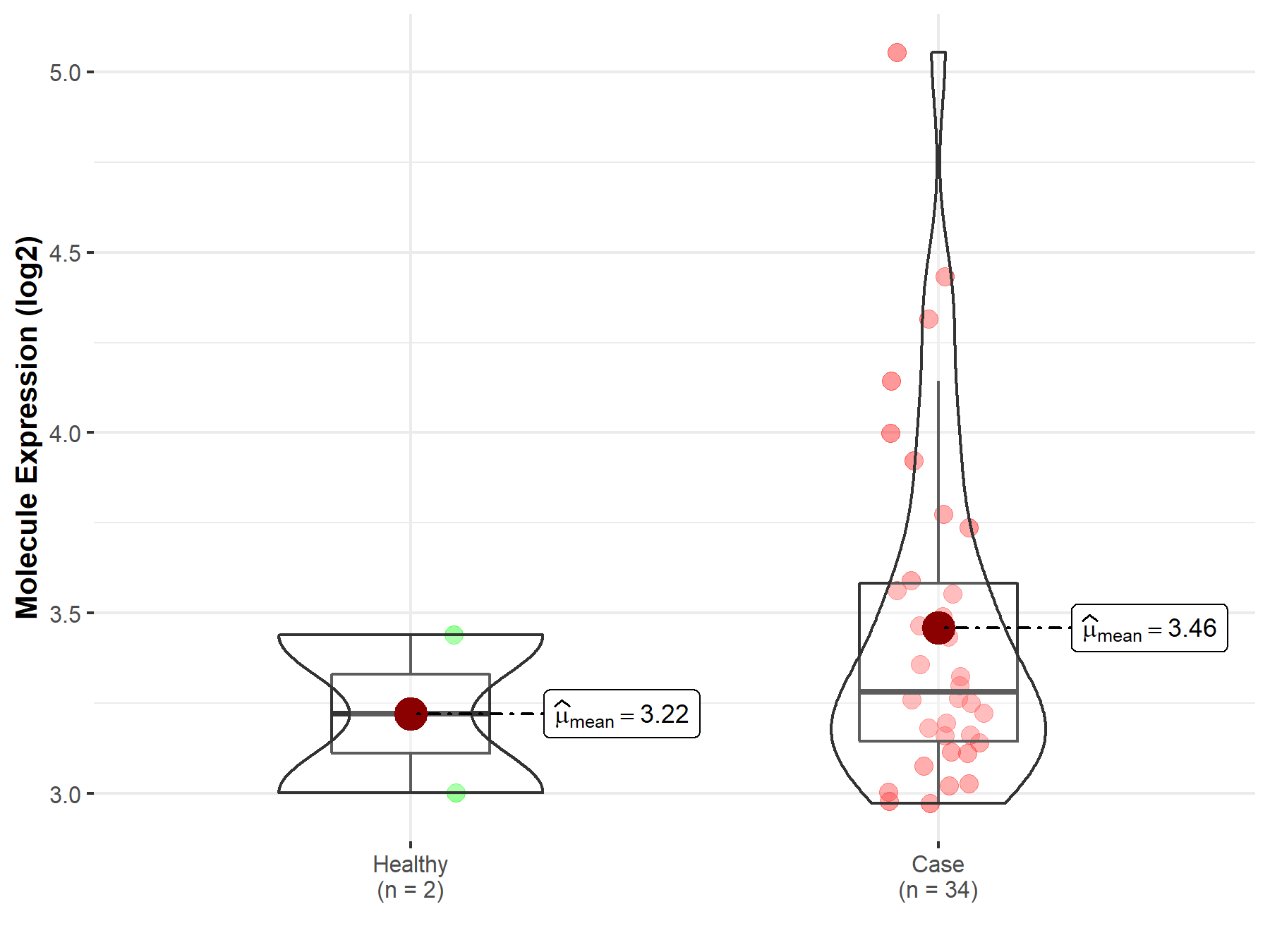
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | White matter | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.81E-02; Fold-change: -5.08E-01; Z-score: -6.17E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
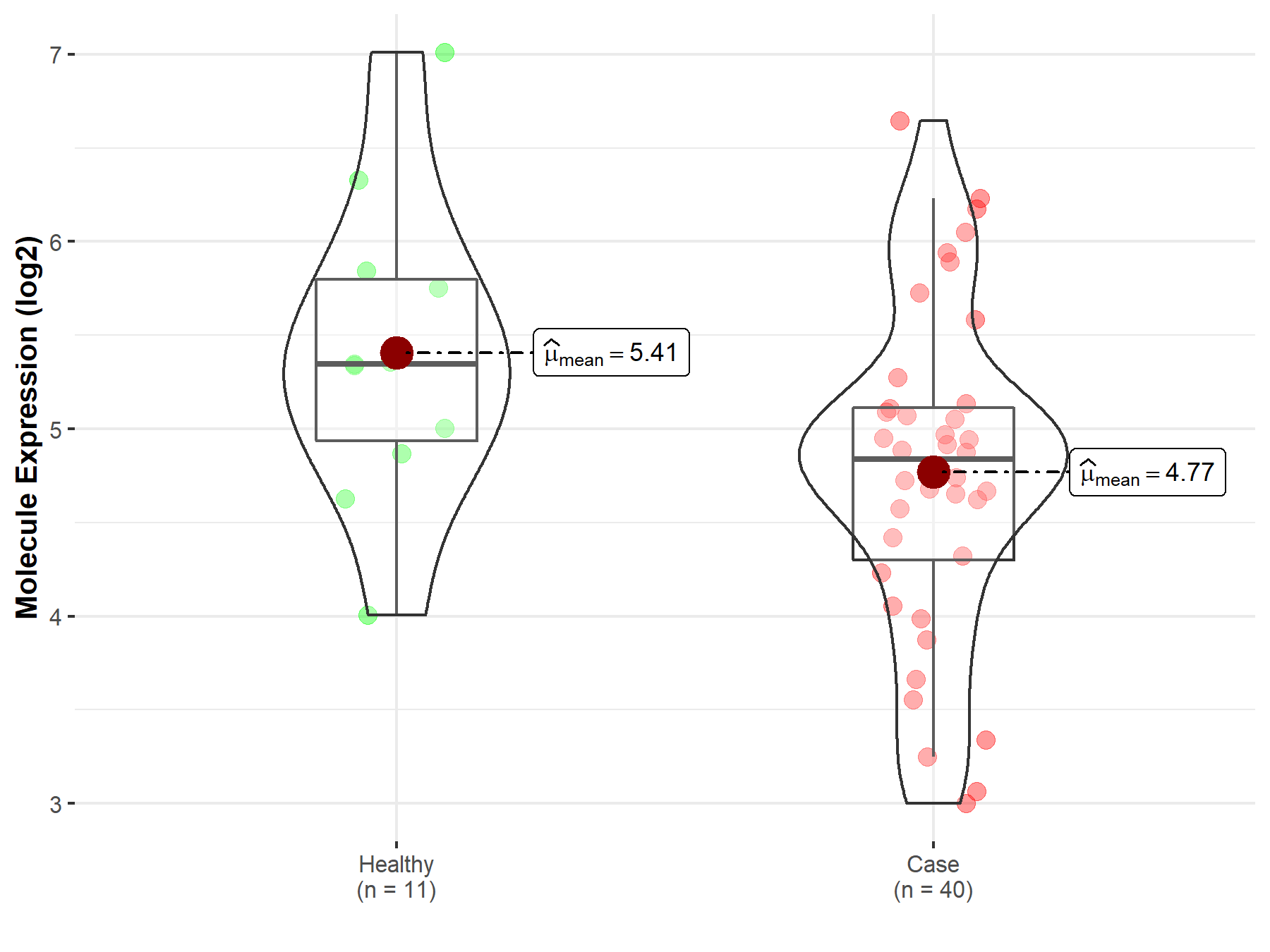
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Neuroectodermal tumor | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.05E-04; Fold-change: 2.39E-01; Z-score: 1.05E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
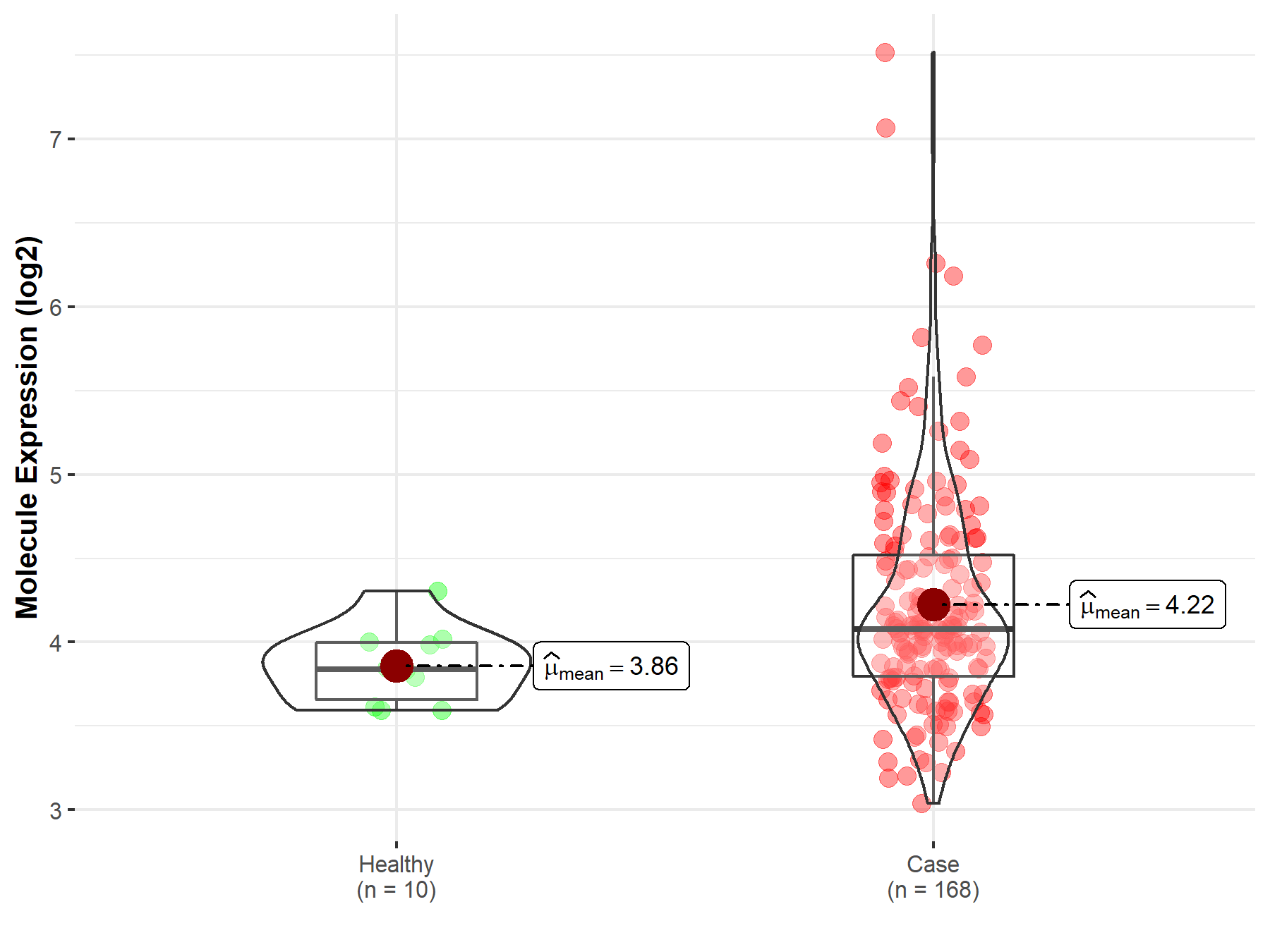
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Whole blood | |
| The Specified Disease | Myelofibrosis | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.34E-01; Fold-change: -6.50E-02; Z-score: -3.54E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
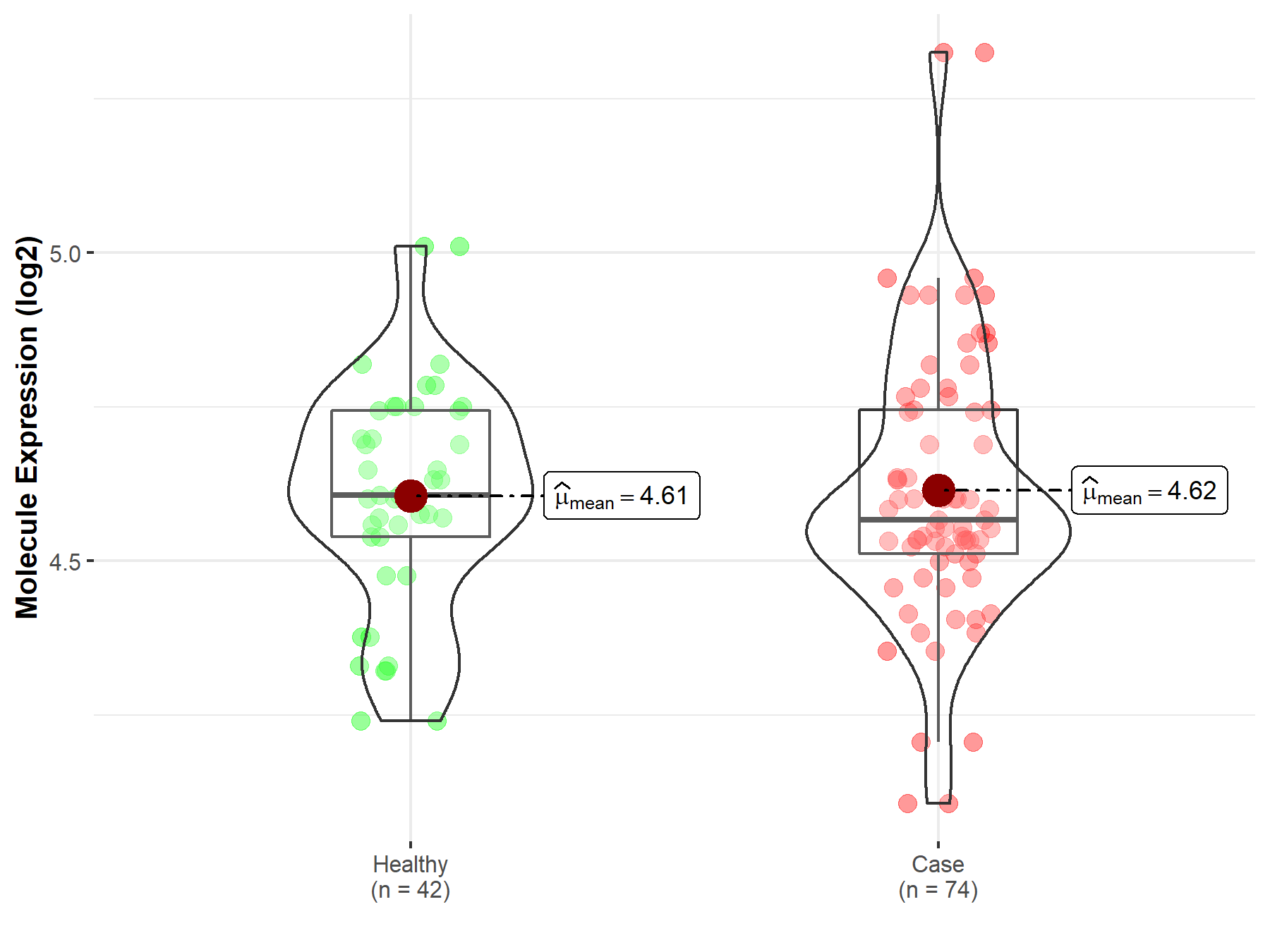
| The Studied Tissue | Whole blood | |
| The Specified Disease | Polycythemia vera | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.18E-01; Fold-change: -4.06E-02; Z-score: -2.21E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
|
Click to View the Clearer Original Diagram |
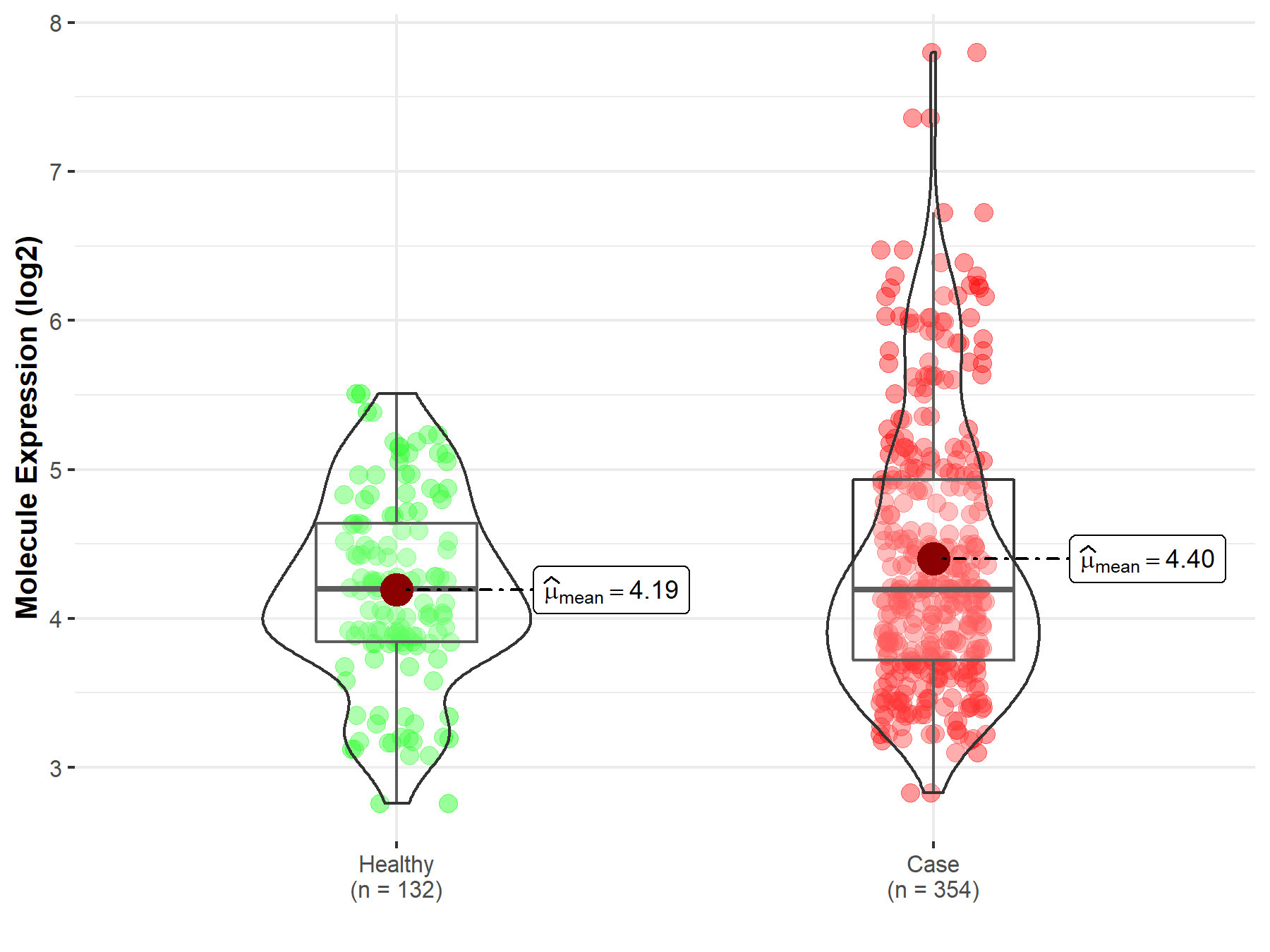
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Bone marrow | |
| The Specified Disease | Acute myeloid leukemia | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.63E-03; Fold-change: -1.26E-03; Z-score: -1.99E-03 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
|
Click to View the Clearer Original Diagram |
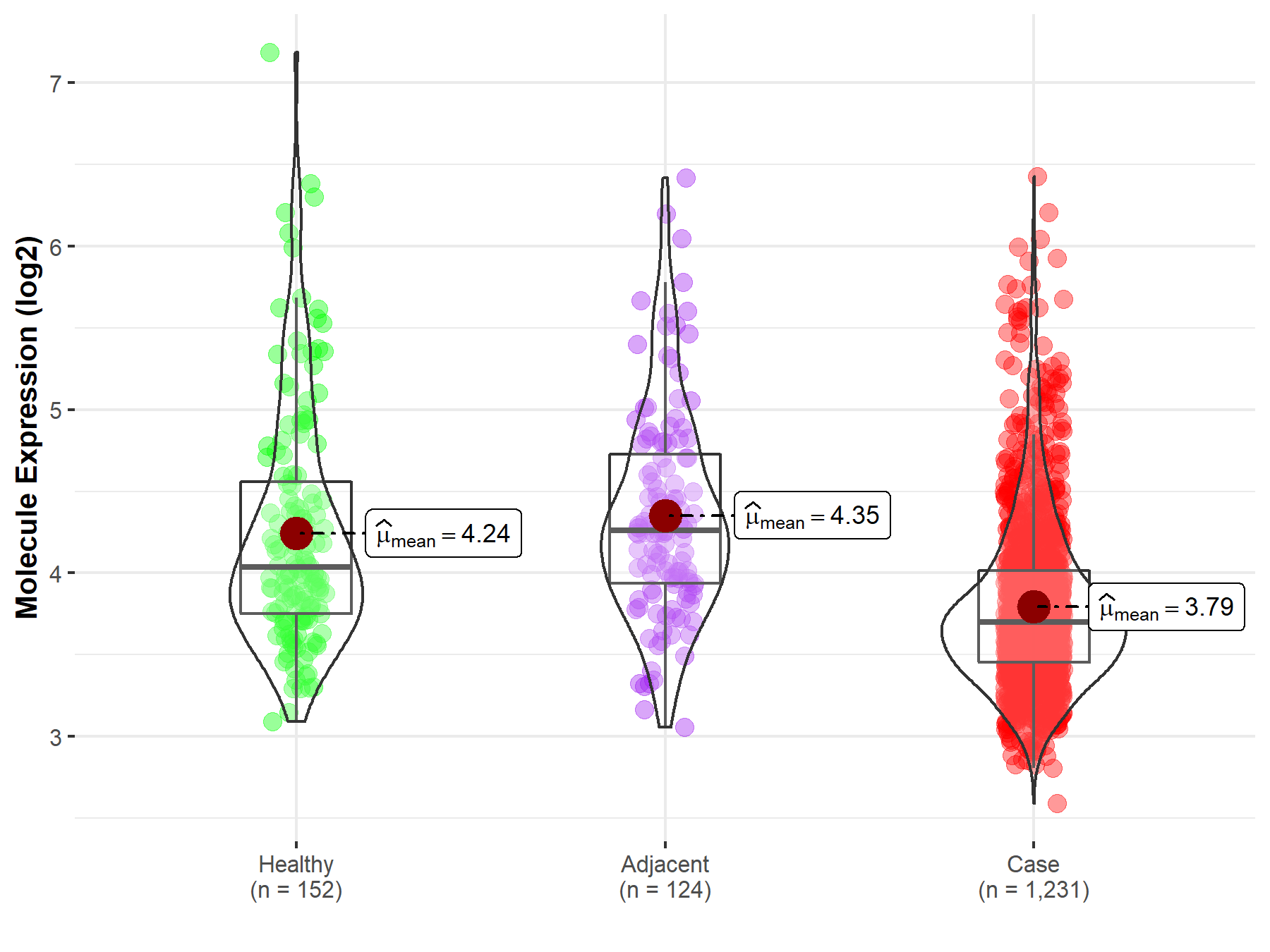
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Colon | |
| The Specified Disease | Colon cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.93E-12; Fold-change: -3.35E-01; Z-score: -4.61E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.29E-16; Fold-change: -5.63E-01; Z-score: -8.77E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
|
Click to View the Clearer Original Diagram |
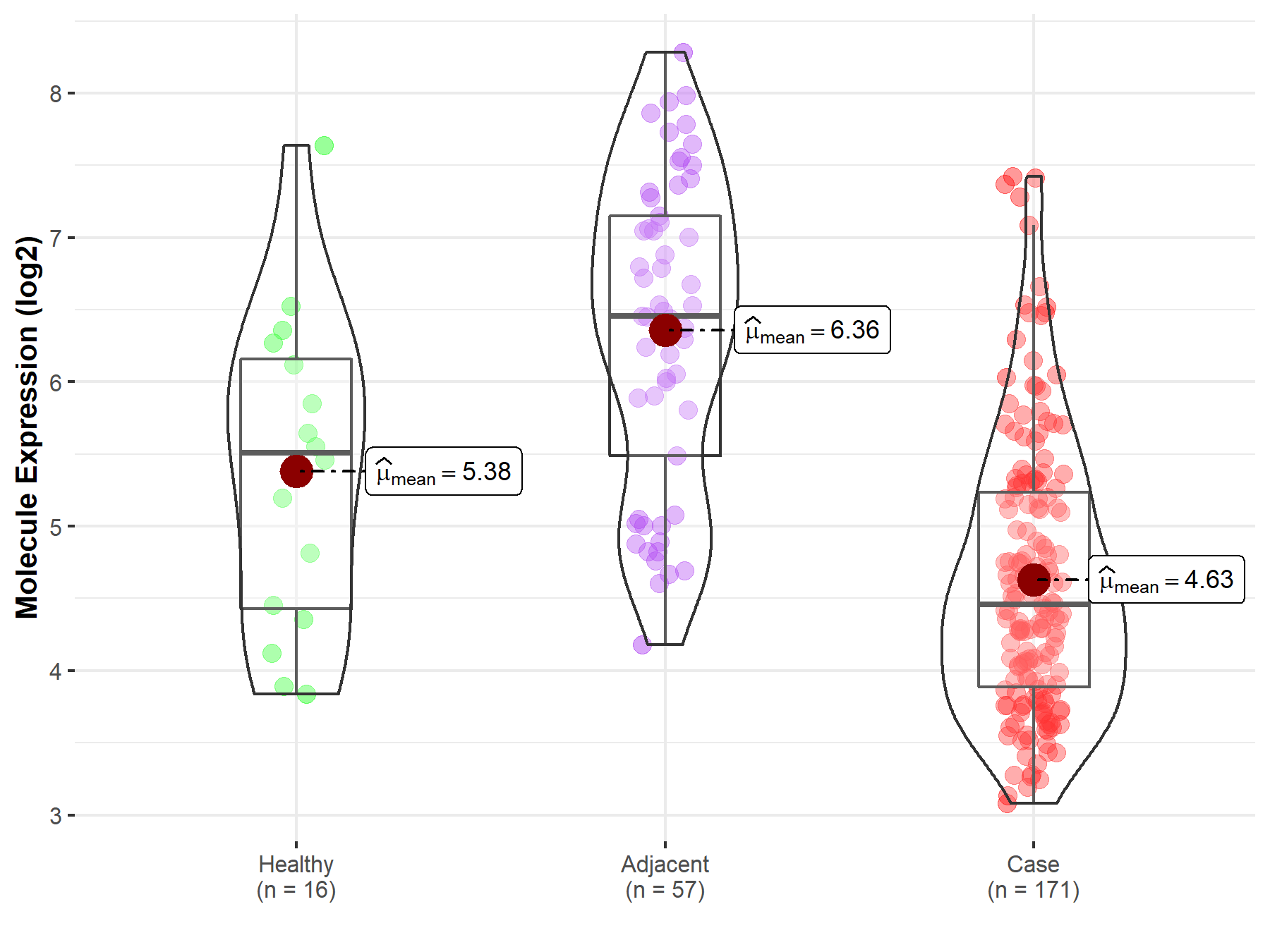
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Pancreas | |
| The Specified Disease | Pancreatic cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.56E-02; Fold-change: -1.05E+00; Z-score: -9.72E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 8.08E-18; Fold-change: -2.00E+00; Z-score: -1.86E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Liver | |
| The Specified Disease | Liver cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.85E-01; Fold-change: -4.51E-02; Z-score: -1.31E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.29E-01; Fold-change: -1.03E-02; Z-score: -2.74E-02 | |
| The Expression Level of Disease Section Compare with the Other Disease Section | p-value: 4.04E-01; Fold-change: -8.09E-02; Z-score: -4.50E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
Molecule expression in tissue other than the diseased tissue of patients
|
||
| Disease-specific Molecule Abundances |
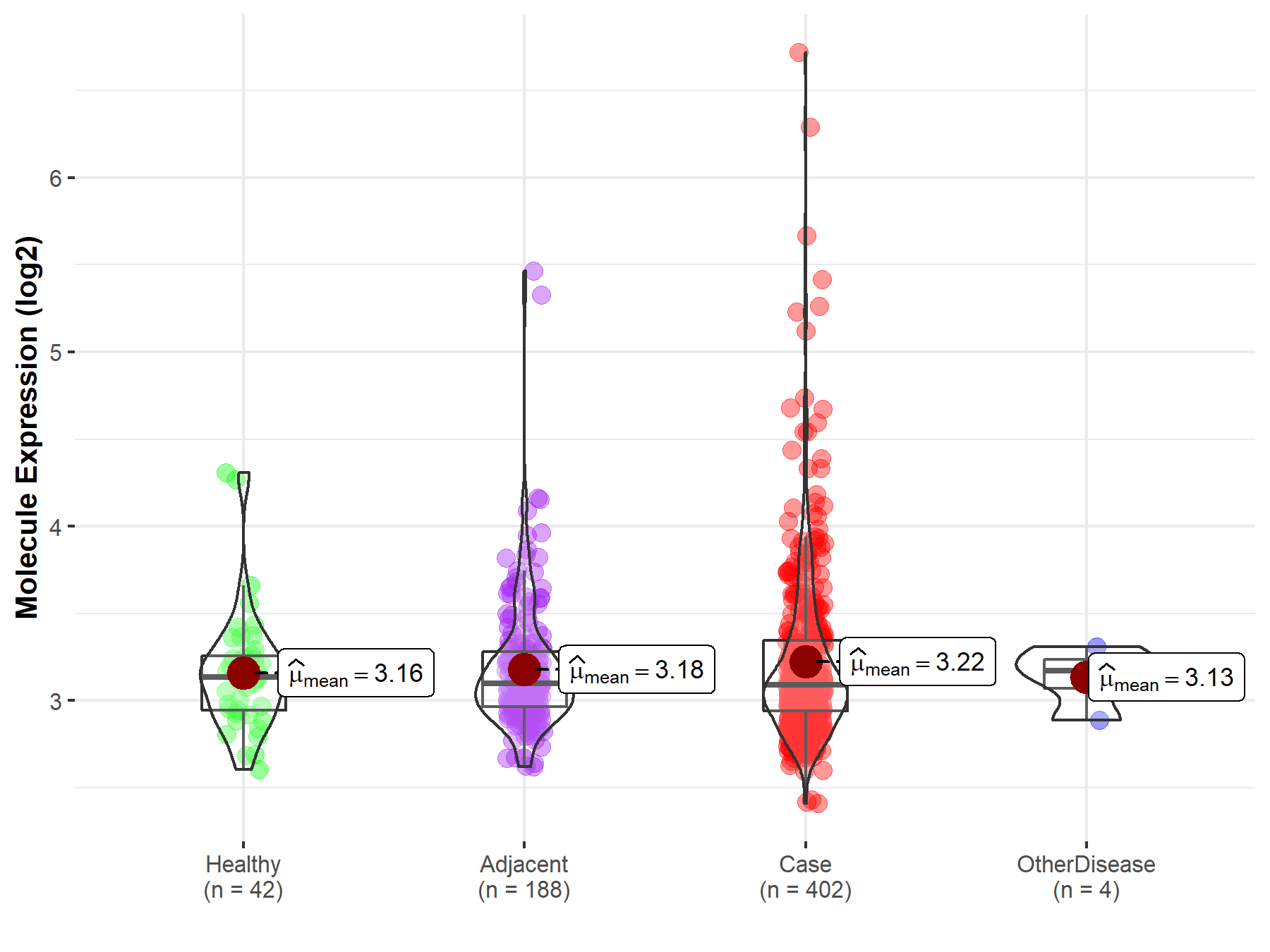
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Lung | |
| The Specified Disease | Lung cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.05E-02; Fold-change: 1.97E-02; Z-score: 5.52E-02 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 6.74E-07; Fold-change: -2.19E-01; Z-score: -5.71E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
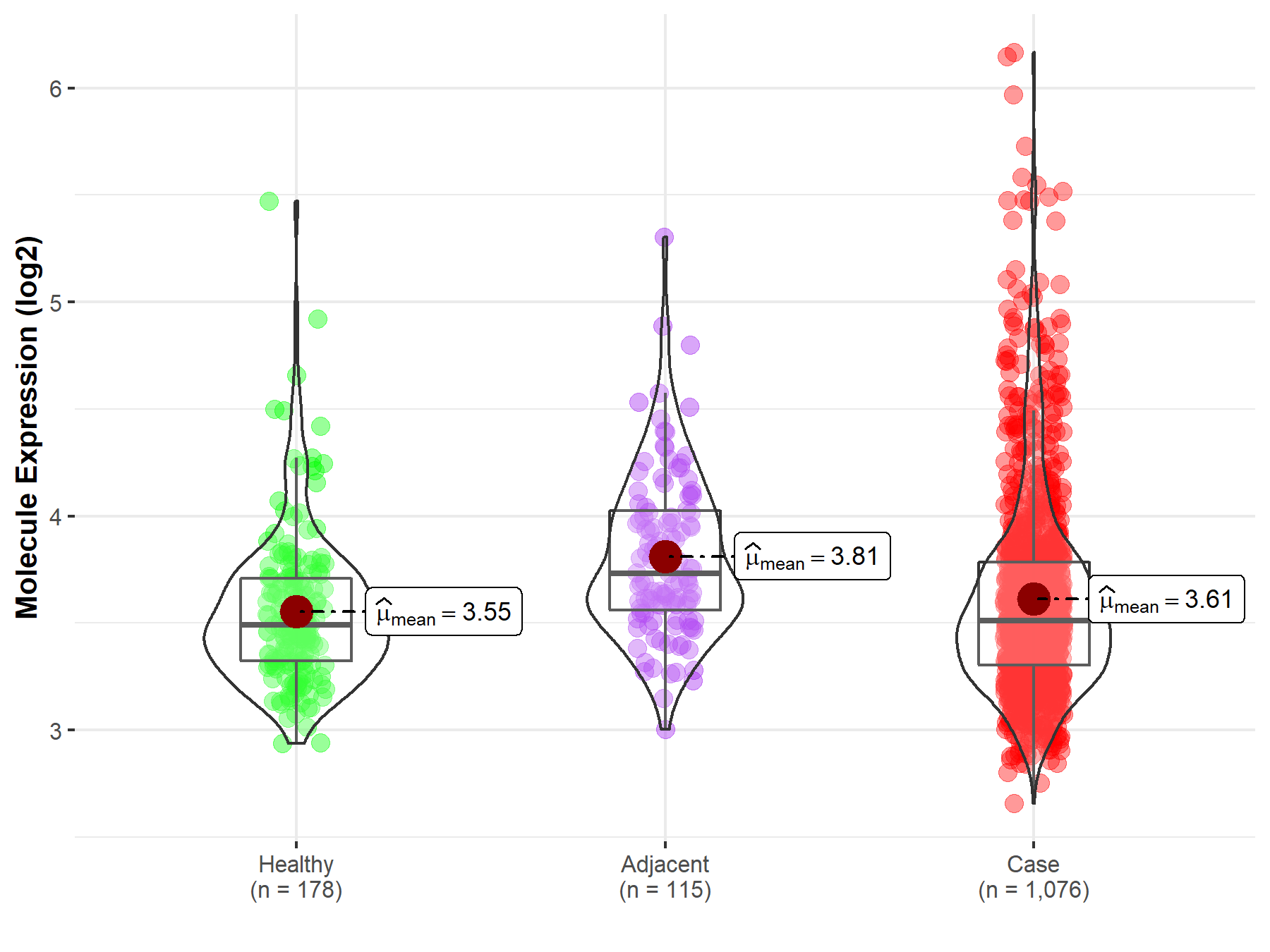
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Skin | |
| The Specified Disease | Melanoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.26E-02; Fold-change: -3.09E-01; Z-score: -3.80E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
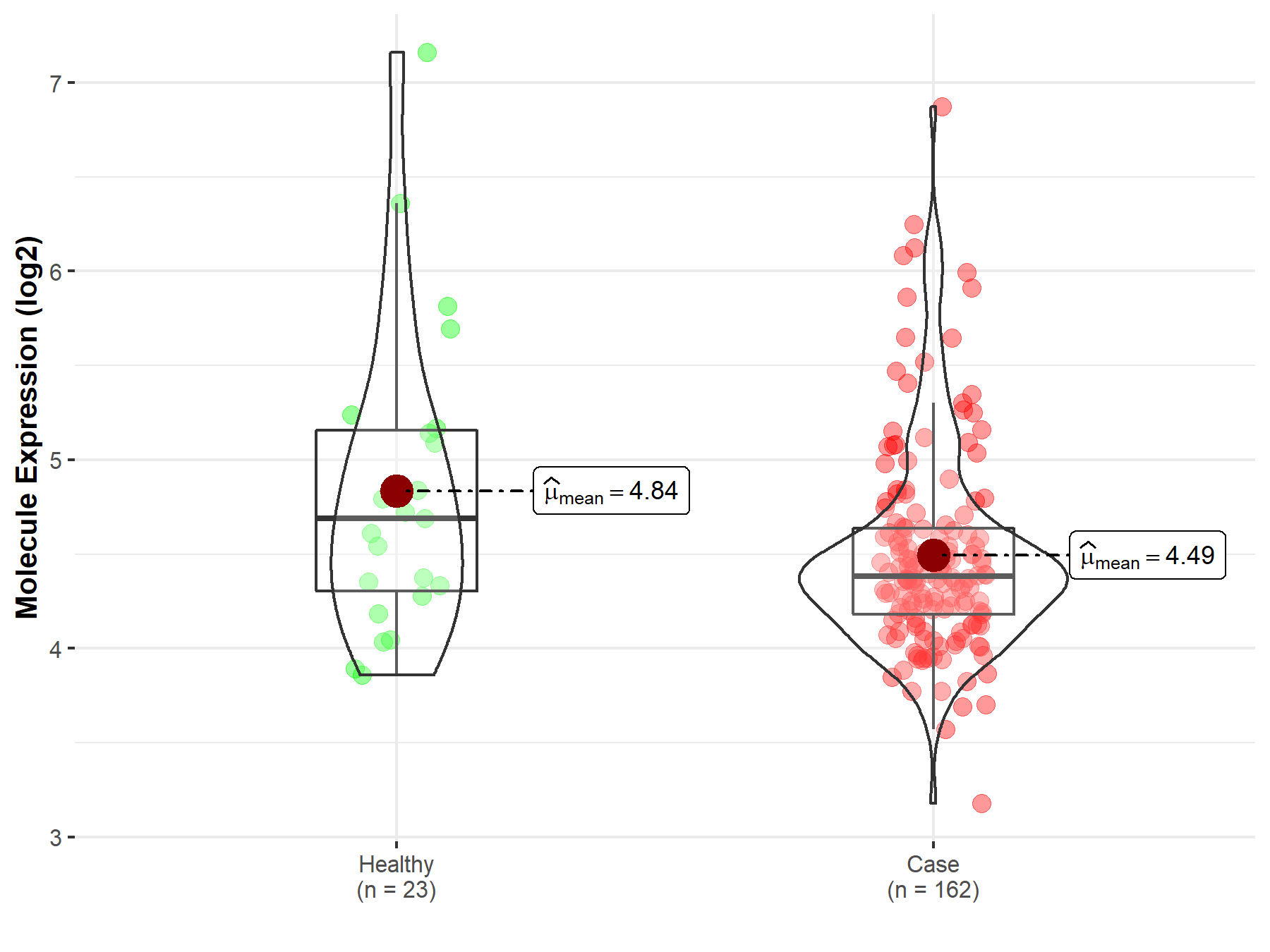
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.12E-04; Fold-change: 1.75E-02; Z-score: 2.67E-02 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.76E-03; Fold-change: -6.83E-01; Z-score: -5.50E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
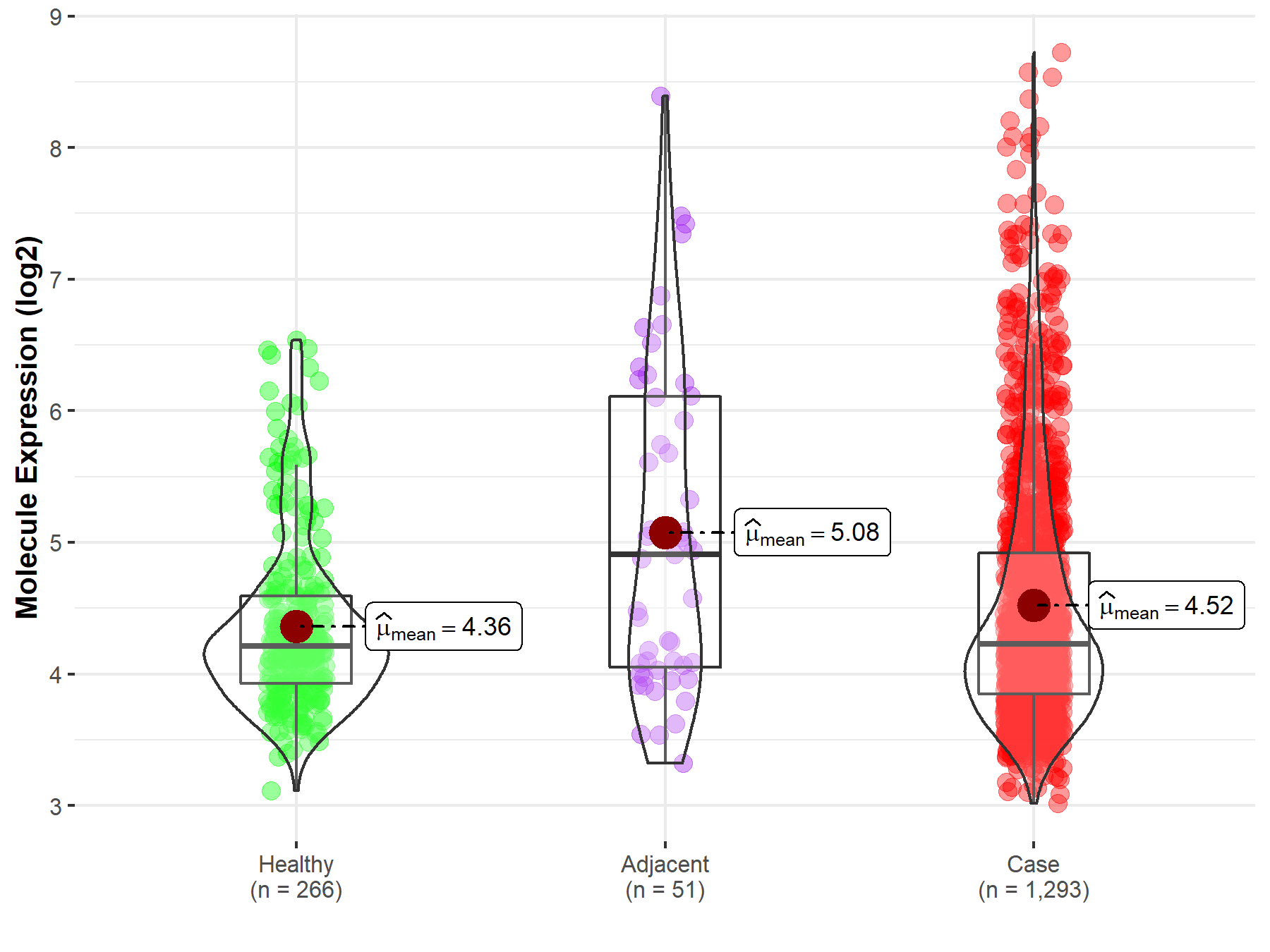
|
Click to View the Clearer Original Diagram |
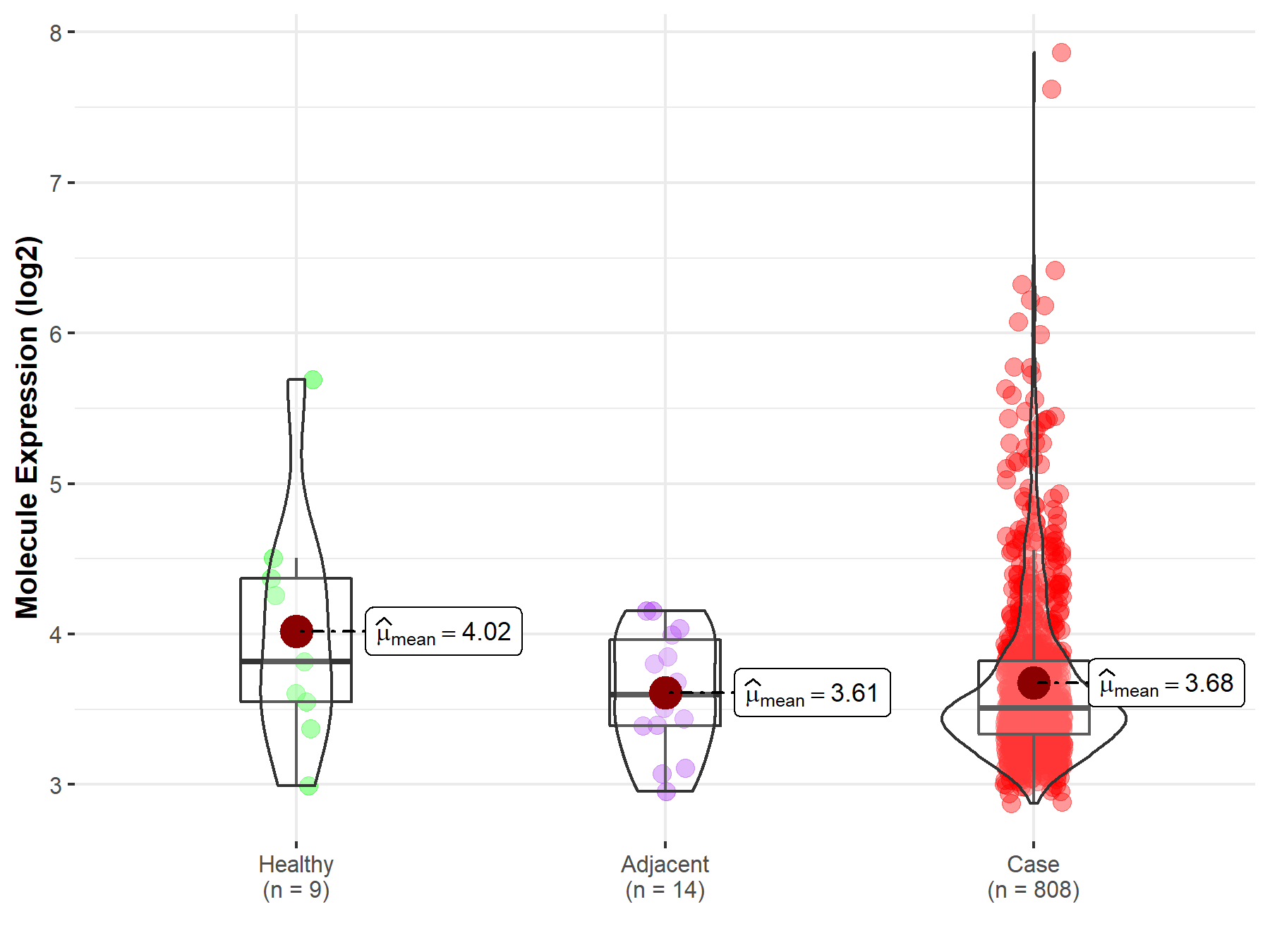
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Ovary | |
| The Specified Disease | Ovarian cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.37E-01; Fold-change: -3.09E-01; Z-score: -3.87E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 5.64E-01; Fold-change: -8.59E-02; Z-score: -2.12E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Cervix uteri | |
| The Specified Disease | Cervical cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.53E-01; Fold-change: 1.16E-01; Z-score: 3.54E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
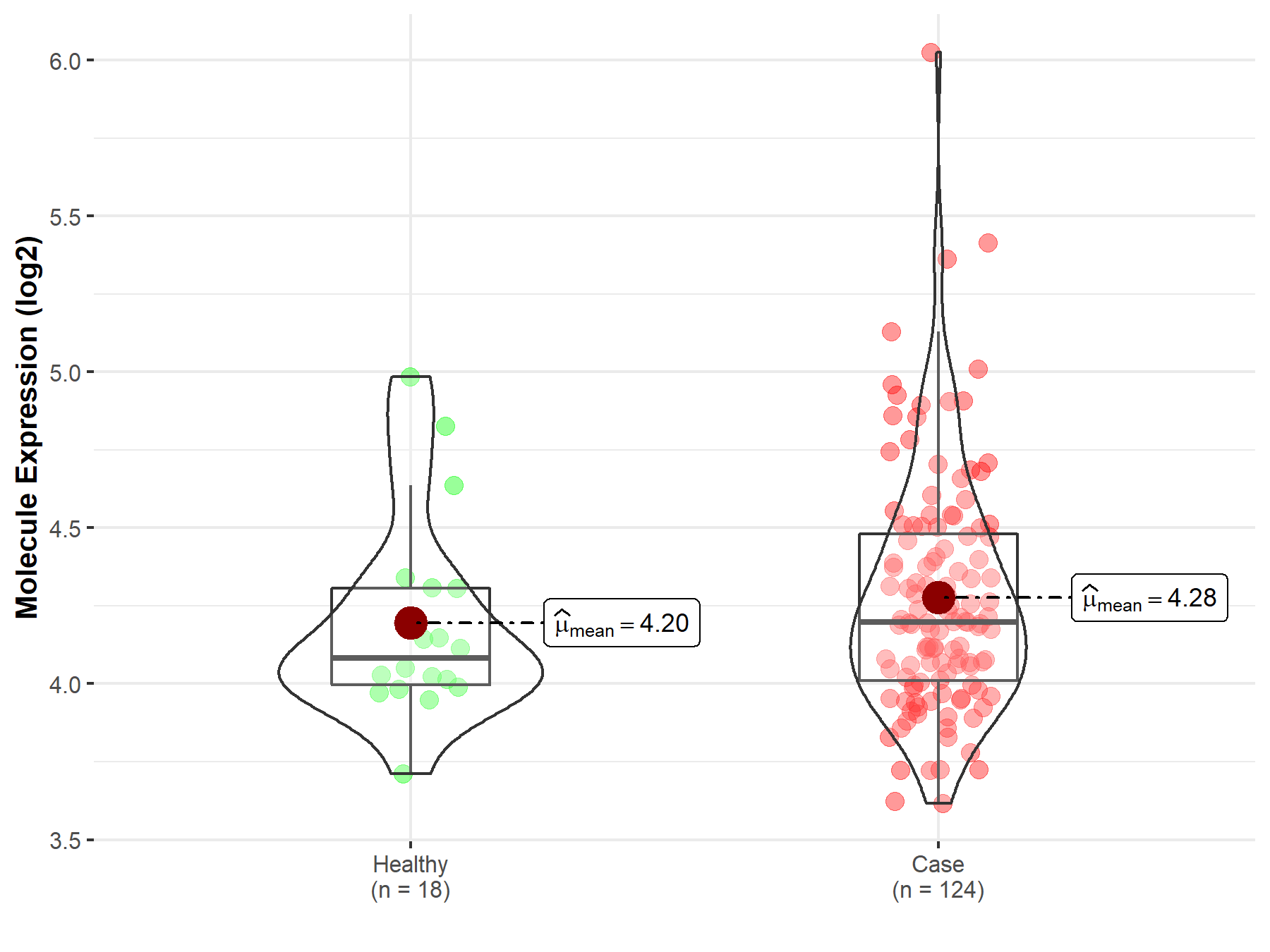
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Prostate | |
| The Specified Disease | Prostate cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.13E-02; Fold-change: 3.88E-01; Z-score: 5.18E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
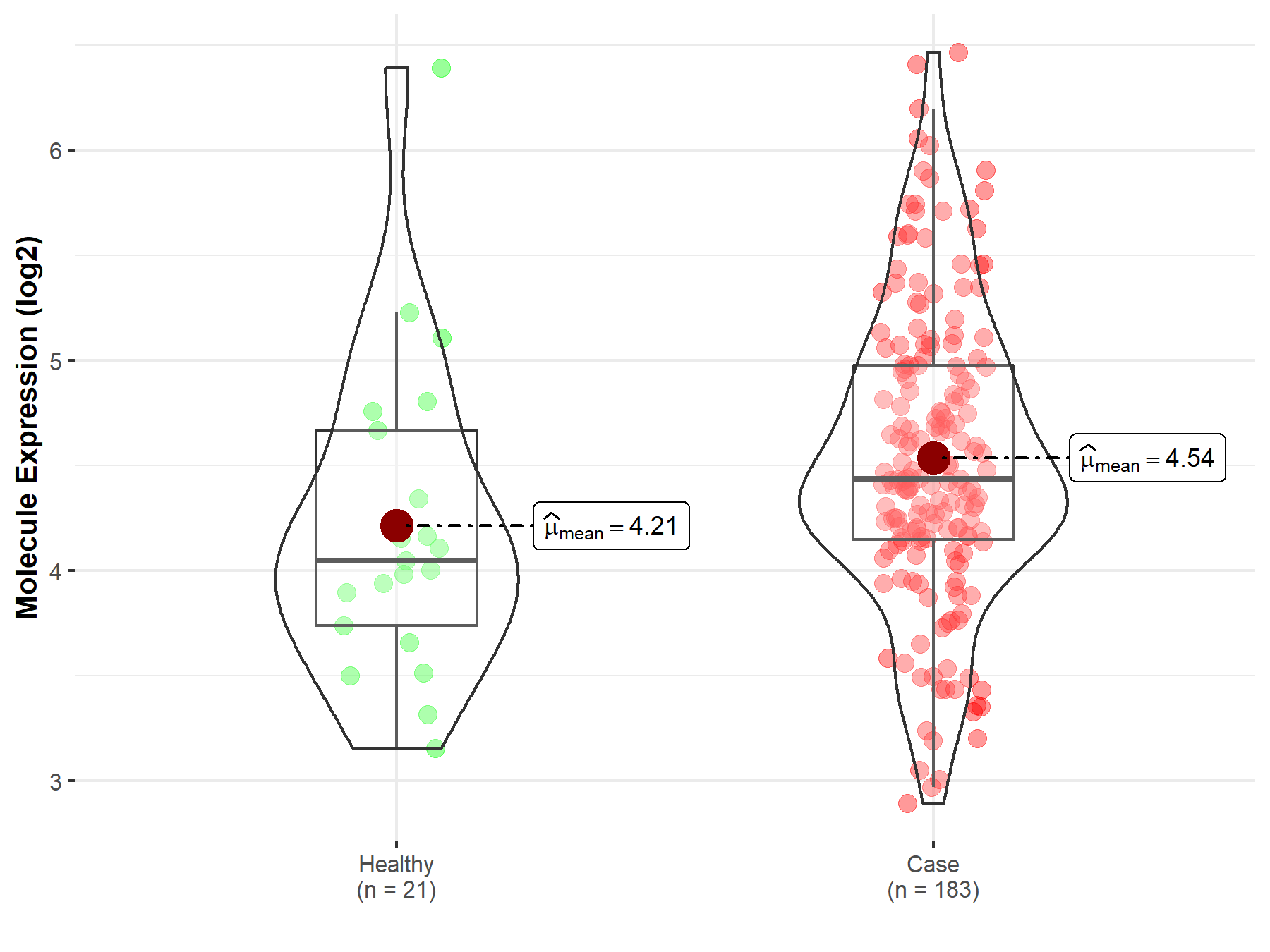
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Kidney | |
| The Specified Disease | Kidney cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.92E-07; Fold-change: 3.74E-01; Z-score: 2.03E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.04E-27; Fold-change: 3.18E-01; Z-score: 1.64E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
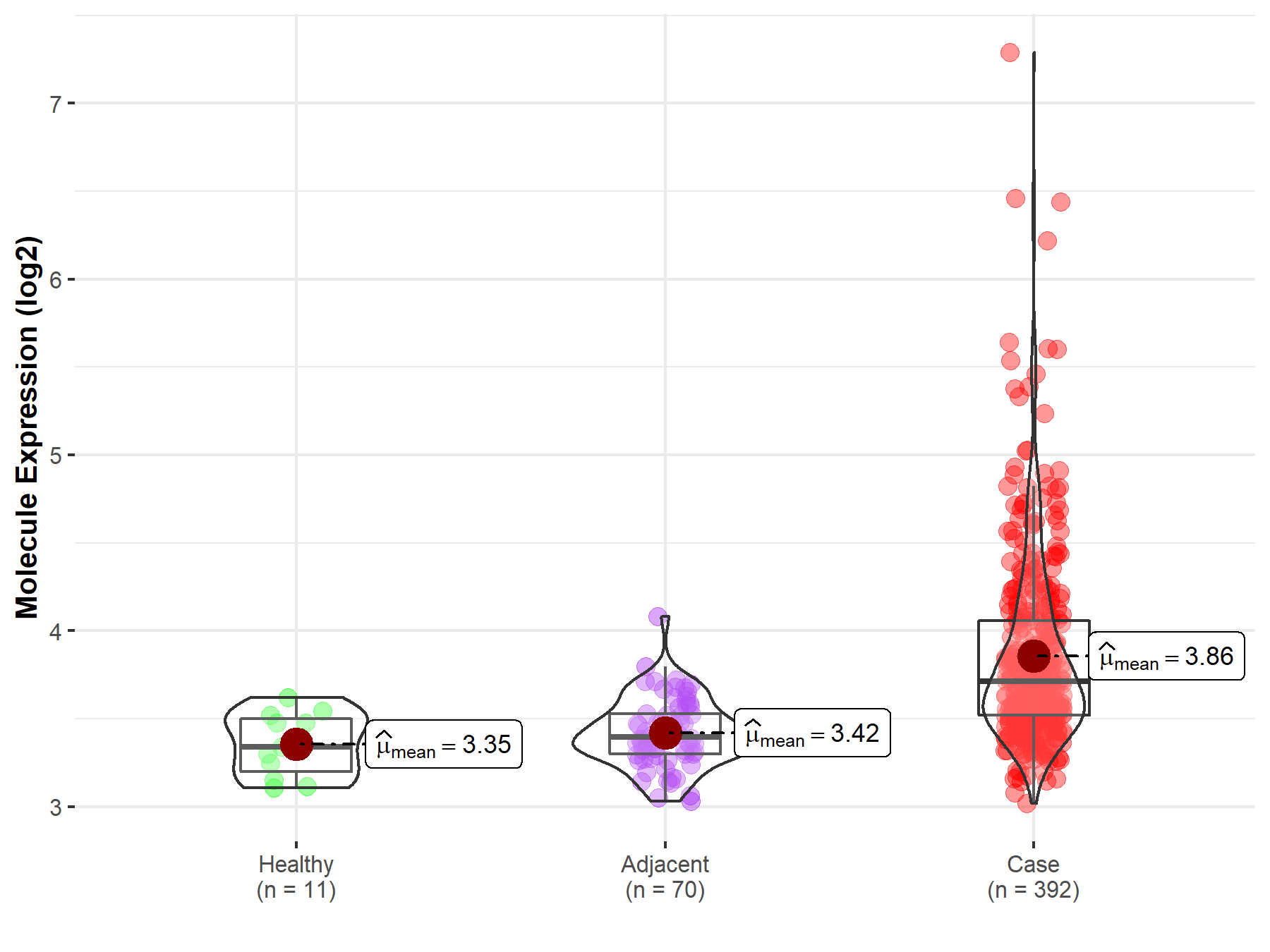
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Bladder tissue | |
| The Specified Disease | Bladder cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.81E-02; Fold-change: -5.74E-01; Z-score: -1.21E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
