Molecule Information
General Information of the Molecule (ID: Mol00110)
| Name |
Induced myeloid leukemia cell differentiation protein Mcl-1 (MCL1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Bcl-2-like protein 3; Bcl2-L-3; Bcl-2-related protein EAT/mcl1; mcl1/EAT; BCL2L3
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
MCL1
|
||||
| Gene ID | |||||
| Location |
chr1:150560895-150579738[-]
|
||||
| Sequence |
MFGLKRNAVIGLNLYCGGAGLGAGSGGATRPGGRLLATEKEASARREIGGGEAGAVIGGS
AGASPPSTLTPDSRRVARPPPIGAEVPDVTATPARLLFFAPTRRAAPLEEMEAPAADAIM SPEEELDGYEPEPLGKRPAVLPLLELVGESGNNTSTDGSLPSTPPPAEEEEDELYRQSLE IISRYLREQATGAKDTKPMGRSGATSRKALETLRRVGDGVQRNHETAFQGMLRKLDIKNE DDVKSLSRVMIHVFSDGVTNWGRIVTLISFGAFVAKHLKTINQESCIEPLAESITDVLVR TKRDWLVKQRGWDGFVEFFHVEDLEGGIRNVLLAFAGVAGVGAGLAYLIR Click to Show/Hide
|
||||
| Function |
Involved in the regulation of apoptosis versus cell survival, and in the maintenance of viability but not of proliferation. Mediates its effects by interactions with a number of other regulators of apoptosis. Isoform 1 inhibits apoptosis. Isoform 2 promotes apoptosis.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
13 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hepatocellular carcinoma [ICD-11: 2C12.2] | [1] | |||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Liver cancer [ICD-11: 2C12] | |||
| The Specified Disease | Hepatocellular carcinoma | |||
| The Studied Tissue | Liver tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.38E-03 Fold-change: 1.01E-01 Z-score: 3.23E+00 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| miR363/Mcl-1 signaling pathway | Regulation | N.A. | ||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Cisplatin-based chemotherapy decreased miR-363 expression in HCC patients. miR-363 expression was also lower in HepG2-R cells than in HepG2 cells, which indicated that the downregulation of miR-363 may be related to cisplatin resistance. overexpression of miR-363 by its mimics can effectively increase the sensitivity of cisplatin-resistant HepG2 cells to cisplatin-induced apoptosis. overexpression of miR-363 could inhibit the expression of Mcl-1 in HepG2-R cells, which implied the inverse correlation between the expression of miR-363 and Mcl-1. More importantly, enforced exogenous Mcl-1 significantly attenuated apoptosis induced by cisplatin. All these results support that Mcl-1 is the target of miR-363 which can enhance sensitivity of human cisplatin-resistant HCC cell cisplatin at least partially. | |||
| Disease Class: Lung cancer [ICD-11: 2C25.5] | [7] | |||
| Resistant Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | miR451/Mcl1/DPP signaling pathway | Inhibition | hsa05206 | |
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| A549/DPP cells | Lung | Homo sapiens (Human) | CVCL_0023 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis; qRT-PCR | |||
| Experiment for Drug Resistance |
MTT and cytotoxicity (IC50) assays | |||
| Mechanism Description | miR451 enhanced DPP chemosensitivity of lung cancer cells by negatively regulating Mcl-1 in vitro and in vivo. | |||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [8] | |||
| Resistant Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| In Vitro Model | A2780 cells | Ovary | Homo sapiens (Human) | CVCL_0134 |
| A2780/DDP cells | Ovary | Homo sapiens (Human) | CVCL_D619 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Knockdown of miR-106a dramatically decreased antiproliferative effects and apoptosis in-duced by cisplatin in A2780 cells, while overexpression of miR-106a significantly increased antiprolif-erative effects and apoptosis induced by cisplatin in A2780/DDP cells. Furthermore, miR-106a inhibited cell survival and cisplatin resistance through downregulating the expression of Mcl-1. Mcl-1 was a di-rect target of miR-106a. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lung cancer [ICD-11: 2C25.5] | [5] | |||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | |||
| The Specified Disease | Lung cancer | |||
| The Studied Tissue | Lung tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.52E-41 Fold-change: -1.91E-01 Z-score: -1.61E+01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| A549/CDDP cells | Lung | Homo sapiens (Human) | CVCL_0023 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Hsa-miR-135a/b could play a role in the development of CDDP resistance in lung cancer cell line at least in partby modulation of apoptosis via targeting MCL1. | |||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [9] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| MCL-1 dependent mitochondria signaling pathway | Activation | hsa04210 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | miR519d impedes cisplatin-resistance in breast cancer stem cells by down-regulating the expression of MCL-1. | |||
| Disease Class: Nasopharyngeal carcinoma [ICD-11: 2B6B.0] | [10] | |||
| Sensitive Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B.0] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | 5-8F cells | Nasopharynx | Homo sapiens (Human) | CVCL_C528 |
| CNE2 cells | Nasopharynx | Homo sapiens (Human) | CVCL_6889 | |
| C666-1 cells | Throat | Homo sapiens (Human) | CVCL_7949 | |
| CNE1 cells | Throat | Homo sapiens (Human) | CVCL_6888 | |
| HONE1 cells | Throat | Homo sapiens (Human) | CVCL_8706 | |
| 6-10B cells | Nasopharynx | Homo sapiens (Human) | CVCL_C529 | |
| SUNE-1 cells | Nasopharynx | Homo sapiens (Human) | CVCL_6946 | |
| HNE-2 cells | Nasopharynx | Homo sapiens (Human) | CVCL_FA07 | |
| In Vivo Model | SCID-Beige nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-29c repressed expression of anti-apoptotic factors, Mcl-1 and Bcl-2 in NPC tissues and cell lines, cause the resstance to Cisplatin. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Multiple myeloma [ICD-11: 2A83.0] | [2] | |||
| Resistant Disease | Multiple myeloma [ICD-11: 2A83.0] | |||
| Resistant Drug | Dexamethasone | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Multiple myeloma [ICD-11: 2A83] | |||
| The Specified Disease | Myeloma | |||
| The Studied Tissue | Bone marrow | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.65E-02 Fold-change: 8.45E-02 Z-score: 2.76E+00 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | U266 cells | Bone marrow | Homo sapiens (Human) | CVCL_0566 |
| ANBL6 cells | Peripheral blood | Homo sapiens (Human) | CVCL_5425 | |
| JJN-3 cells | Bone marrow | Homo sapiens (Human) | CVCL_2078 | |
| MM1R cells | Peripheral blood | Homo sapiens (Human) | CVCL_8794 | |
| MM1S cells | Peripheral blood | Homo sapiens (Human) | CVCL_8792 | |
| OPM-2 cells | Peripheral blood | Homo sapiens (Human) | CVCL_1625 | |
| RPMI-8226 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0014 | |
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometric analysis | |||
| Mechanism Description | LncRNA NEAT1 promotes dexamethasone resistance in multiple myeloma by targeting miR193a/MCL1 pathway. NEAT1 promotes MM cell DEX resistance by competitively binding miR193a. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colon cancer [ICD-11: 2B90.1] | [3] | |||
| Sensitive Disease | Colon cancer [ICD-11: 2B90.1] | |||
| Sensitive Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Colon cancer [ICD-11: 2B90] | |||
| The Specified Disease | Colon cancer | |||
| The Studied Tissue | Colon tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.73E-07 Fold-change: -7.16E-02 Z-score: -5.12E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | HT29 Cells | Colon | Homo sapiens (Human) | CVCL_A8EZ |
| HCT116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |
| In Vivo Model | BALB/C nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Overexpression of miR-125a/b significantly inhibited ALDH1A3 and Mcl1 expression, reduced cell survival, and increased cell apoptosis in HT29-taxol cells. Chemoresistance to paclitaxel is initiated by the downregulation of miR-125a/b expression, which subsequently upregulates ALDH1A3 and Mcl1 expression to promote survival of CSCs. | |||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [20] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| Tumorigenesis | Inhibition | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |
| MDA-MB-435 cells | Breast | Homo sapiens (Human) | CVCL_0417 | |
| 184A1 cells | Breast | Homo sapiens (Human) | CVCL_3040 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | MCL-1(myeloid cell leukemia 1), a pro-survival member of the Bcl-2(B-cell CLL/lymphoma 2) family, several miRNAs induces apoptosis by targeting MCL-1, miR-26a Inhibits MCL-1 expression, increased sensitivity of breast cancer cells to paclitaxel. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hepatitis B virus-associated hepatocellular carcinoma [ICD-11: 2C12.7] | [4] | |||
| Sensitive Disease | Hepatitis B virus-associated hepatocellular carcinoma [ICD-11: 2C12.7] | |||
| Sensitive Drug | Sorafenib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Liver cancer [ICD-11: 2C12] | |||
| The Specified Disease | Liver cancer | |||
| The Studied Tissue | Liver tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.17E-05 Fold-change: -1.54E-01 Z-score: -4.39E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| L02 cells | Liver | Homo sapiens (Human) | CVCL_6926 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | HBV infection in HCC cell lines enhances sorafenib resistance. HBV infection in HCC reduces miR-193b expression and increases Mcl-1 expression. miR-193b directly suppresses the expression of Mcl-1 through its 3'-UTRs. miR-193b facilitates sorafenib-induced apoptosis. miR-193b sensitizes HBV-associated HCC cell lines to sorafenib. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Multiple myeloma [ICD-11: 2A83.0] | [6] | |||
| Resistant Disease | Multiple myeloma [ICD-11: 2A83.0] | |||
| Resistant Drug | Bortezomib | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | 8226 cells | Bone marrow | Homo sapiens (Human) | CVCL_0014 |
| NCI-H929 cells | Bone marrow | Homo sapiens (Human) | CVCL_1600 | |
| U266 cells | Bone marrow | Homo sapiens (Human) | CVCL_0566 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | LncRNA H19 overexpression induces bortezomib resistance in multiple myeloma by targeting MCL-1 via downregulating miR-29b-3p. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [11] | |||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Sensitive Drug | Cytarabine | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell migration | Inhibition | hsa04670 | ||
| In Vitro Model | HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 |
| K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | The ectopic expression of miR-181b in k562/A02 and HL-60/ADM cells robustly suppressed endogenous HMGB1 and Mcl-1 expression both at mRNA and protein levels. Conversely, knockdown of miR-181b by miR-181b inhibitor markedly increased the expression of both HMGB1 and Mcl-1. Restoration of miR-181b increased the drug sensitivity of AML MDR cells by targeting HMGB1 and Mcl-1. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [12] | |||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Sensitive Drug | Daunorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | KG-1 cells | Bone marrow | Homo sapiens (Human) | CVCL_0374 |
| THP-1 cells | Blood | Homo sapiens (Human) | CVCL_0006 | |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | MCL-1 participates in the regulation of DNR sensitivity mediated by miR-33b and overexpression of miR-33b enhances DNR sensitivity by downregulating MCL-1 in AML cells. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Osteosarcoma [ICD-11: 2B51.0] | [13] | |||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Resistant Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell viability | Activation | hsa05200 | ||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| SAOS-2 cells | Bone marrow | Homo sapiens (Human) | CVCL_0548 | |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| HOS cells | Bone | Homo sapiens (Human) | CVCL_0312 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | The up-regulation of MCL1 reversed the sensitivity of doxorubicin induced by miR-320a mimics and knockdown of SNHG12. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hepatocellular carcinoma [ICD-11: 2C12.2] | [14] | |||
| Sensitive Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| PLC cells | Liver | Homo sapiens (Human) | CVCL_0485 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-101 was downregulated in HCC cell lines, while its overexpression (+) the sensitivity of HepG2 cells to the chemotherapeutic agent DOX by facilitating apoptosis. Of note, Mcl-1 was confirmed as a functional target of miR-101 in HCC, demonstrating that miR-101 may enhance the sensitivity of cancer cells by downregulating Mcl-1 expression. | |||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [15] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| miR193b/MCL1 apoptosis signaling pathway | Regulation | N.A. | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | MCL-1 was significantly overexpressed in MCF-7/DOXR cells, suggesting that the MCL-1 might be essential for doxorubicin resistance in breast cancer. Further results showed that MCL-1 was directly regulated by miR-193b, which is in accordance with the prior finding in melanoma. There was a negative correlation between the expression levels of miR-193b and MCL-1 in MCF-7/DOXR cells. Doxorubicin-induced apoptosis was inhibited in MCF-7/DOXR cells cotransfected with MCL-1 expression vector and miR-193b mimic, indicating that MCL-1 plays a pivotal role in mediating miR-193b-modulated doxorubicin resistance in human breast cancer. | |||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [11] | |||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell migration | Inhibition | hsa04670 | ||
| In Vitro Model | HL60 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0002 |
| K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | The ectopic expression of miR-181b in k562/A02 and HL-60/ADM cells robustly suppressed endogenous HMGB1 and Mcl-1 expression both at mRNA and protein levels. Conversely, knockdown of miR-181b by miR-181b inhibitor markedly increased the expression of both HMGB1 and Mcl-1. Restoration of miR-181b increased the drug sensitivity of AML MDR cells by targeting HMGB1 and Mcl-1. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Negroid cervix epitheloid carcinoma [ICD-11: 2E66.Y] | [16] | |||
| Sensitive Disease | Negroid cervix epitheloid carcinoma [ICD-11: 2E66.Y] | |||
| Sensitive Drug | Etoposide | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
xCELLigence cell viability assay; Flow cytometry assay; Caspase-3 activity assay | |||
| Mechanism Description | microRNA hsa-miR29b potentiates etoposide toxicity in HeLa cells via down-regulation of Mcl-1. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Chronic lymphocytic leukemia [ICD-11: 2A82.0] | [17] | |||
| Sensitive Disease | Chronic lymphocytic leukemia [ICD-11: 2A82.0] | |||
| Sensitive Drug | Fludarabine | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | CLL B cells | Lymph | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | miR-181a and miR-181b directly inhibit the expression of BCL-2, MCL-1 and XIAP by binding to the target sequence, sensitizes CLL cells to fludarabine-induced apoptosis. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Chronic myeloid leukemia [ICD-11: 2A20.0] | [18] | |||
| Sensitive Disease | Chronic myeloid leukemia [ICD-11: 2A20.0] | |||
| Sensitive Drug | Imatinib | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| p53 signaling pathway | Regulation | N.A. | ||
| In Vitro Model | K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | BCL-2, MCL-1 and XIAP were the target genes of miR-130a. BCL-2, MCL-1, TCL-1 and XIAP protein levels were significantly higher in patients with drug-sensitive CML cells. Transfected miR-130a mimics significantly decreased the protein expression of BCL-1, MCL-1 and XIAP. Transfected miR-130a significantly increased the CML sensitivity to Gleevec. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Osteosarcoma [ICD-11: 2B51.0] | [19] | |||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Sensitive Drug | Methotrexate | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| U2OS cells | Bone | Homo sapiens (Human) | CVCL_0042 | |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | Overexpression of miR-29a suppressed MTX resistance and promoted cell apoptosis by downregulating MCL1 expression. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Papillary thyroid carcinoma [ICD-11: 2D10.1] | [21] | |||
| Resistant Disease | Papillary thyroid carcinoma [ICD-11: 2D10.1] | |||
| Resistant Drug | Vemurafenib | |||
| Molecule Alteration | Structural variation | Copy number gain |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | ||
| Experiment for Molecule Alteration |
Low throughput experiment assay | |||
| Mechanism Description | We found that MCL1 (myeloid cell leukemia 1, chromosome 1q) copy number gain is associated with resistance to vemurafenib treatment in metastatic BRAF V600E-PTC cells. MCL1, an anti-apoptotic member of the BCL2 family, is amplified in many cancers and plays a crucial role in tumor progression and metastasis, and likely in drug resistance. | |||
Preclinical Drug(s)
1 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Chronic lymphocytic leukemia [ICD-11: 2A82.0] | [22] | |||
| Sensitive Disease | Chronic lymphocytic leukemia [ICD-11: 2A82.0] | |||
| Sensitive Drug | JNK1 inhibitors | |||
| Molecule Alteration | Phosphorylation | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | B cell receptor signaling pathway | Inhibition | hsa04662 | |
| In Vitro Model | 3T3-msCD40L cells | Embryo | Homo sapiens (Human) | CVCL_1H10 |
| M2-10B4 cells | Bone marrow | Homo sapiens (Human) | CVCL_5794 | |
| In Vivo Model | NOG mice; Eu-TCL1-tg mice | Mus musculus | ||
| Experiment for Molecule Alteration |
Immunoblotting assay | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | JNK1 inhibition affects BCL2 and MCL1 expression in CLL;JNK1 inhibition reduces CLL cell viability preferentially in IGHV unmutated CLLs and overcomes stromal protective effects;JNK1 is a crucial downstream mediator of BCR signaling in CLL. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Whole blood | |
| The Specified Disease | Myelofibrosis | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.55E-01; Fold-change: -6.99E-02; Z-score: -2.48E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
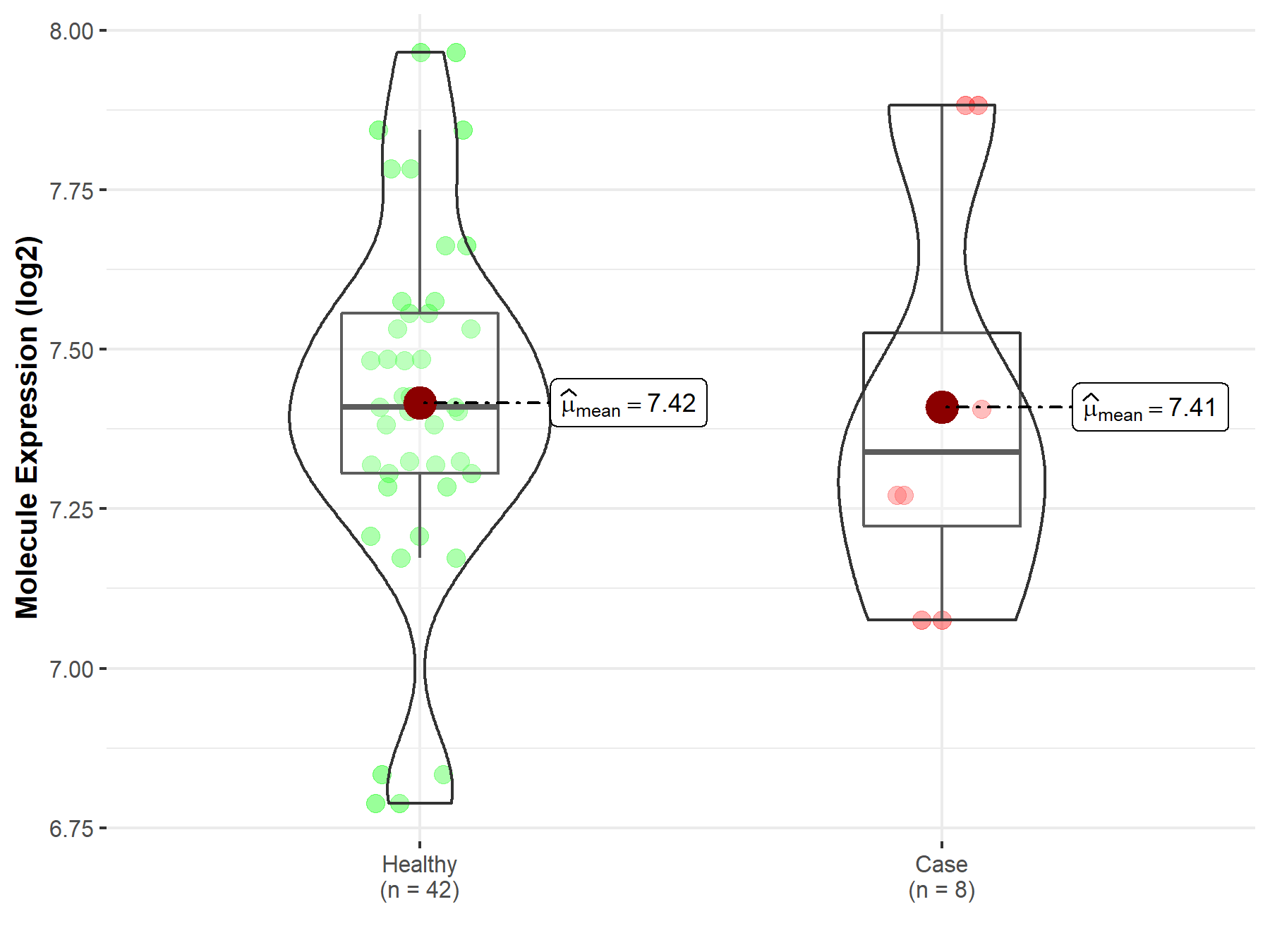
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Whole blood | |
| The Specified Disease | Polycythemia vera | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.63E-07; Fold-change: 3.40E-01; Z-score: 1.19E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
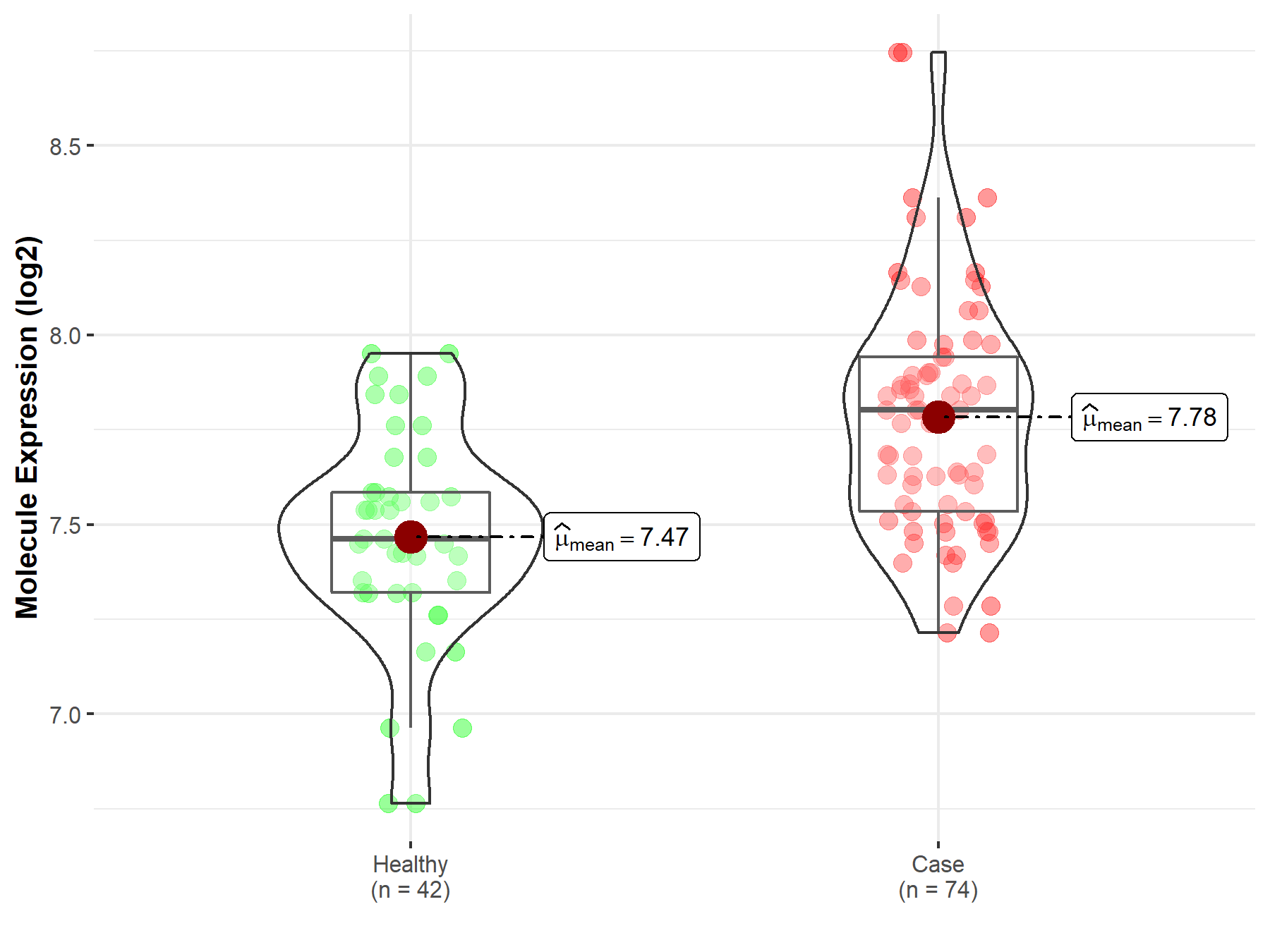
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Bone marrow | |
| The Specified Disease | Acute myeloid leukemia | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.25E-03; Fold-change: 2.47E-01; Z-score: 4.52E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
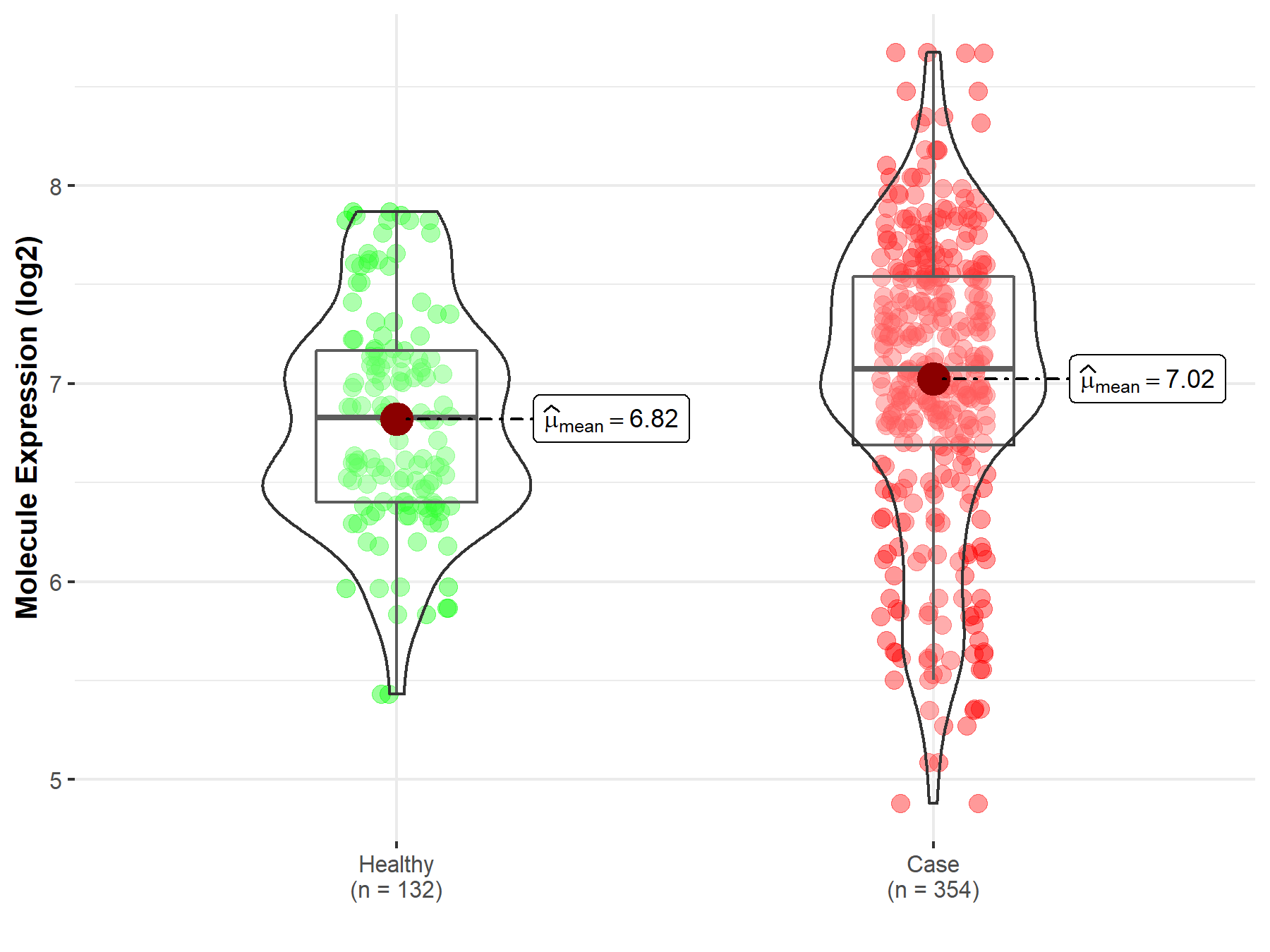
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Bone marrow | |
| The Specified Disease | Multiple myeloma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.65E-02; Fold-change: 3.74E-01; Z-score: 8.53E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
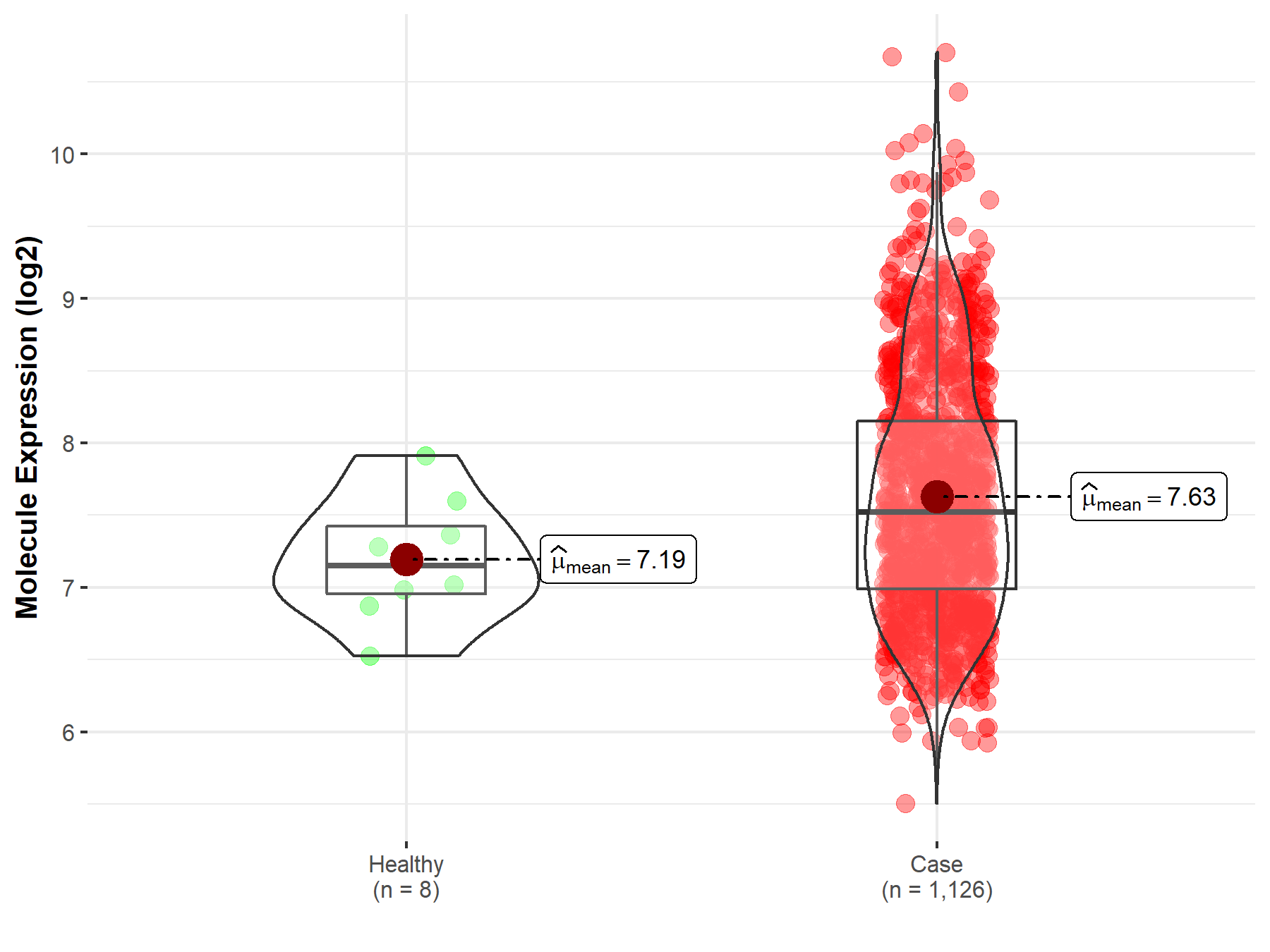
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Peripheral blood | |
| The Specified Disease | Multiple myeloma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.93E-01; Fold-change: 2.49E-01; Z-score: 4.11E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
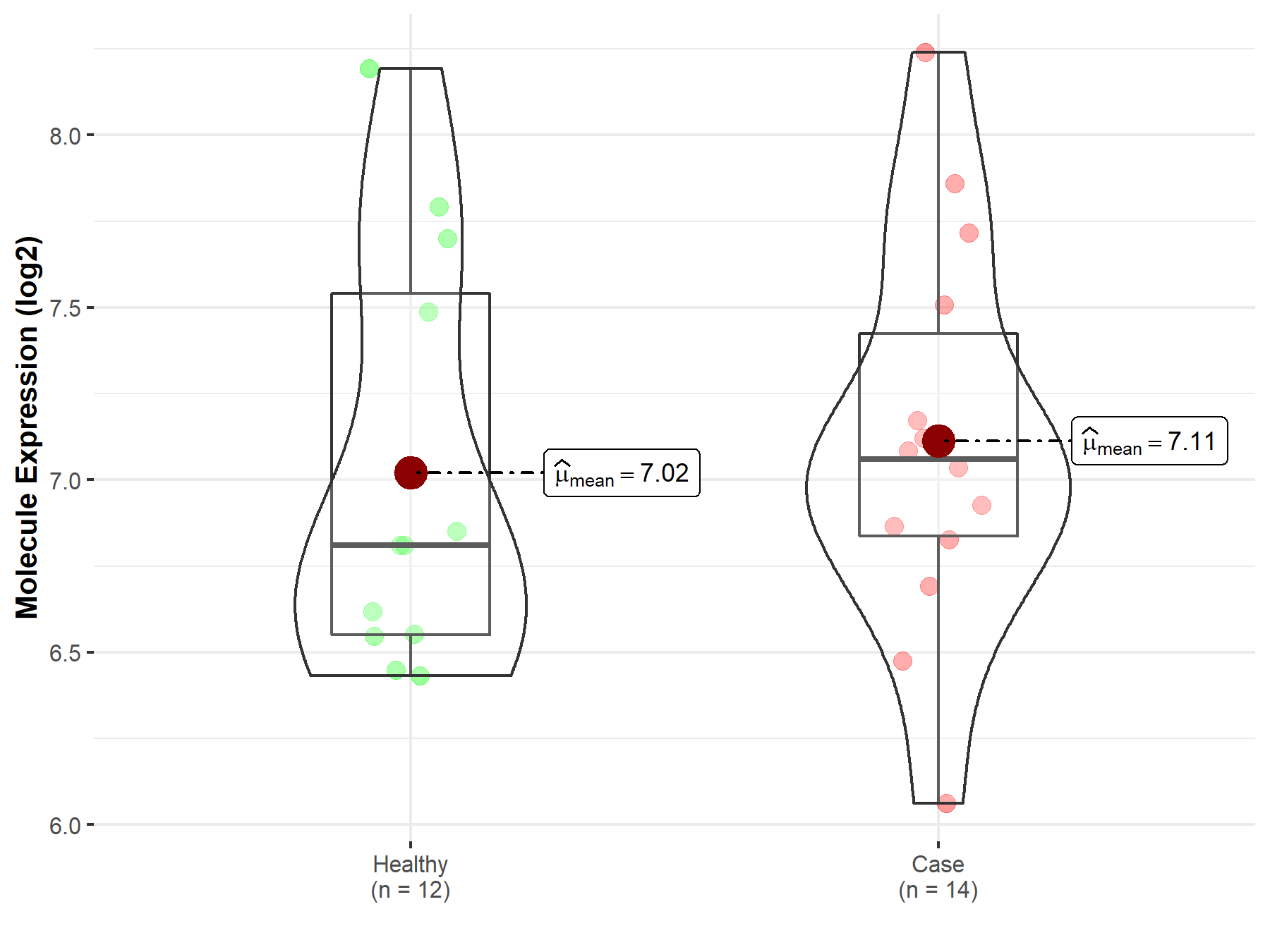
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Colon | |
| The Specified Disease | Colon cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.73E-07; Fold-change: -2.26E-01; Z-score: -2.85E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 4.76E-01; Fold-change: -7.21E-02; Z-score: -9.00E-02 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
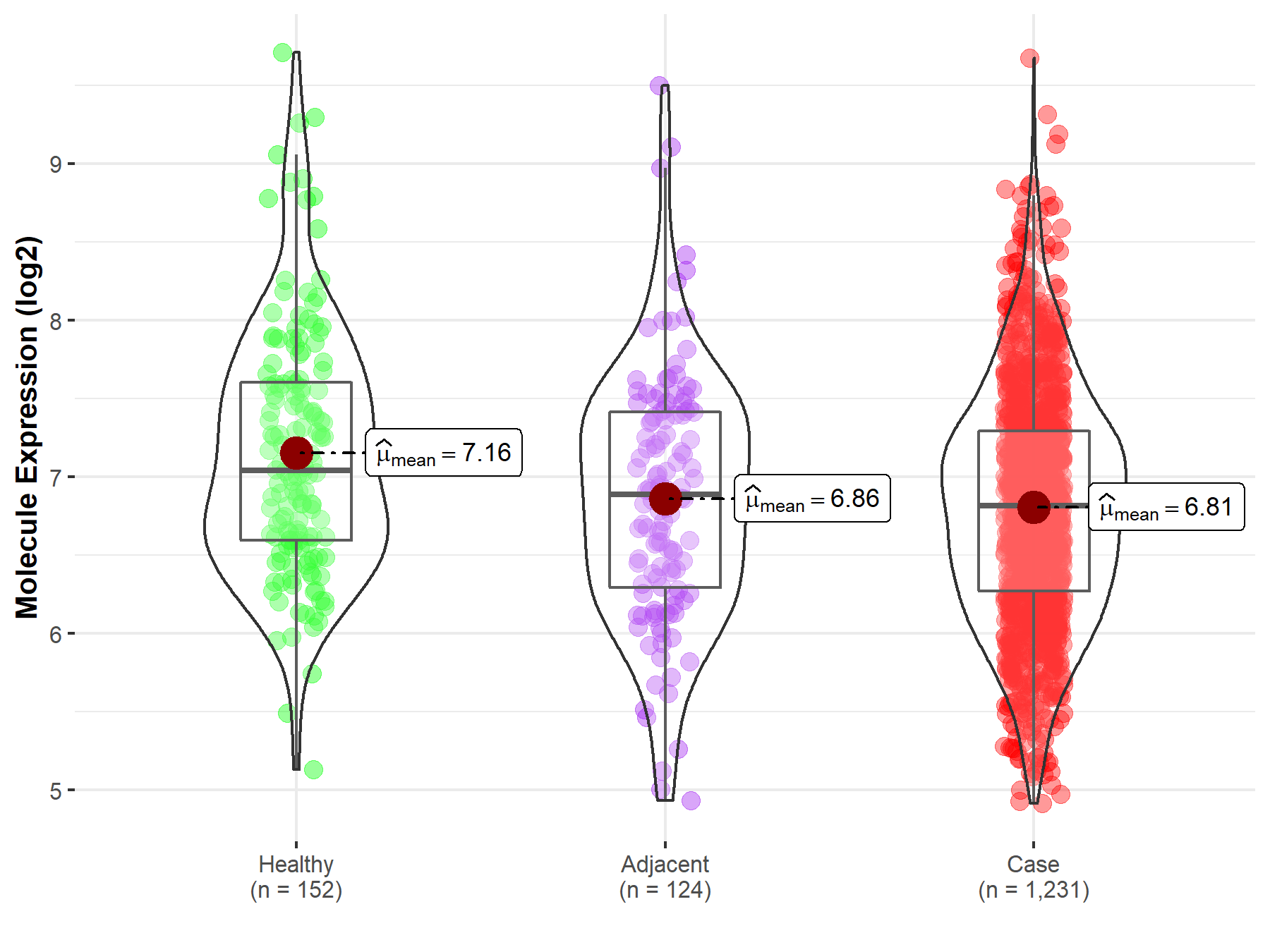
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Liver | |
| The Specified Disease | Liver cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.17E-05; Fold-change: -7.68E-01; Z-score: -7.62E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.87E-05; Fold-change: -3.64E-01; Z-score: -3.72E-01 | |
| The Expression Level of Disease Section Compare with the Other Disease Section | p-value: 2.85E-02; Fold-change: -9.91E-01; Z-score: -1.89E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
Molecule expression in tissue other than the diseased tissue of patients
|
||
| Disease-specific Molecule Abundances |
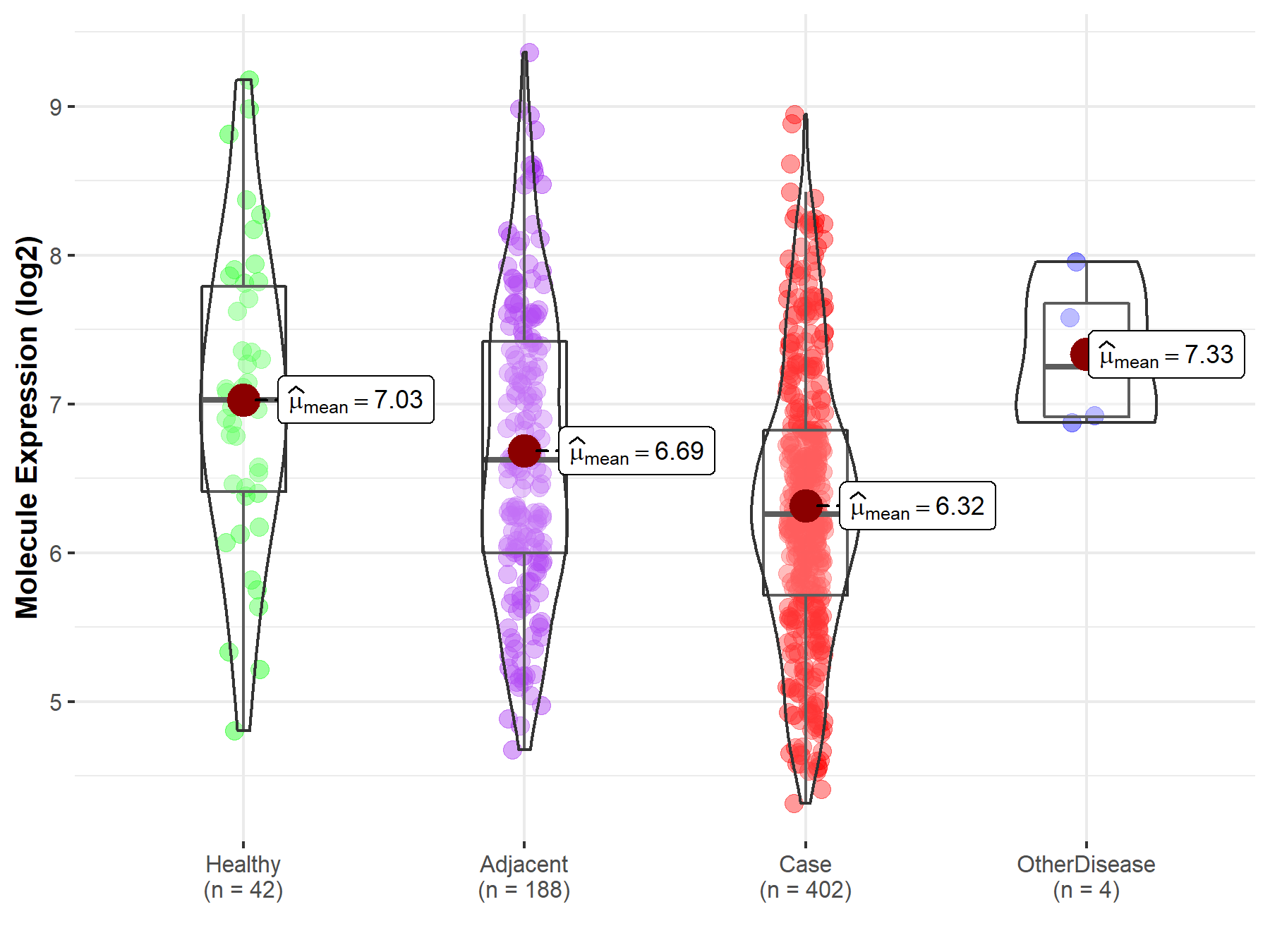
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Lung | |
| The Specified Disease | Lung cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.52E-41; Fold-change: -1.13E+00; Z-score: -1.48E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 5.47E-09; Fold-change: -5.76E-01; Z-score: -6.84E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
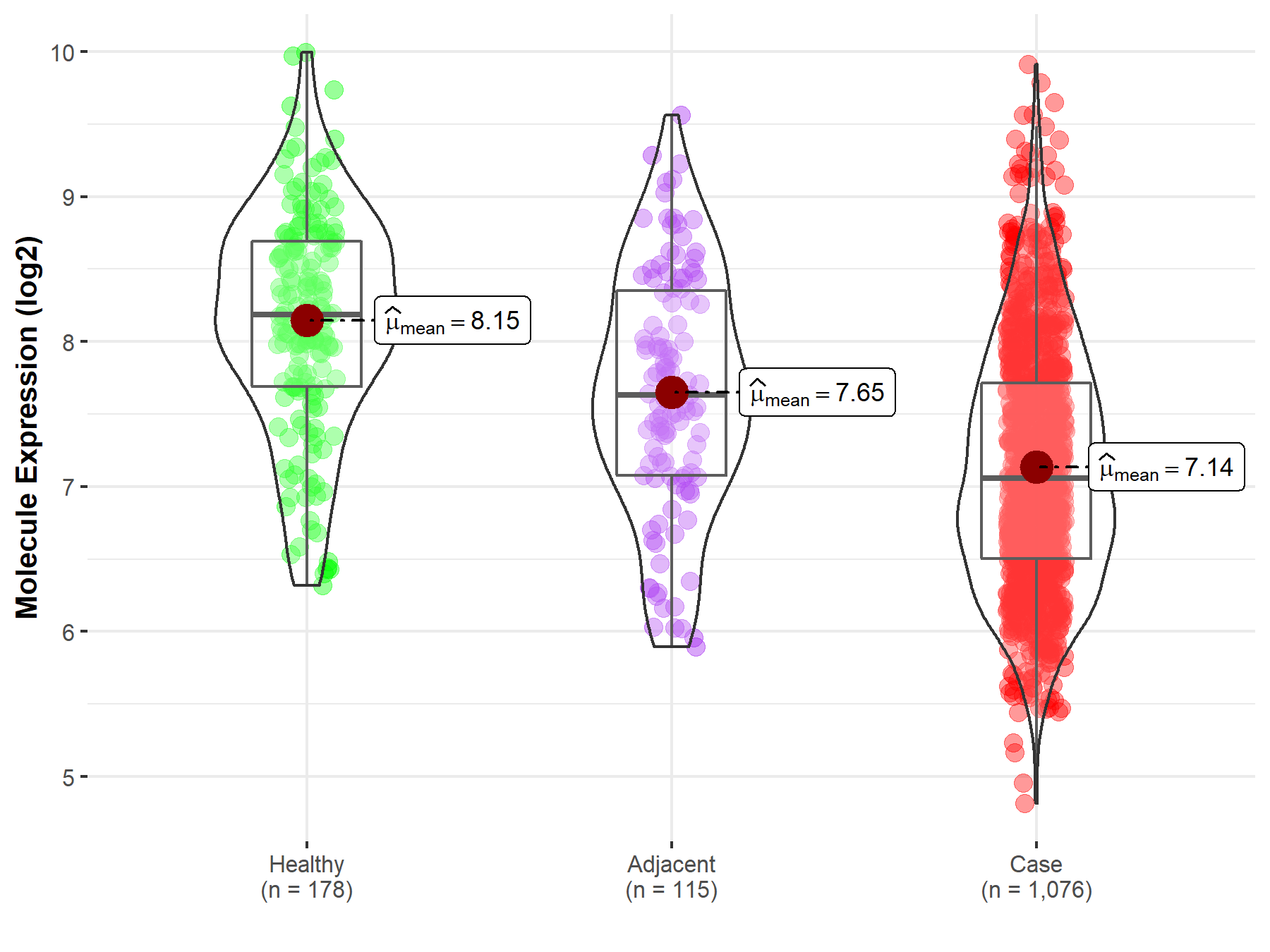
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.68E-05; Fold-change: 1.37E-01; Z-score: 1.27E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 8.23E-03; Fold-change: -3.14E-01; Z-score: -3.44E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
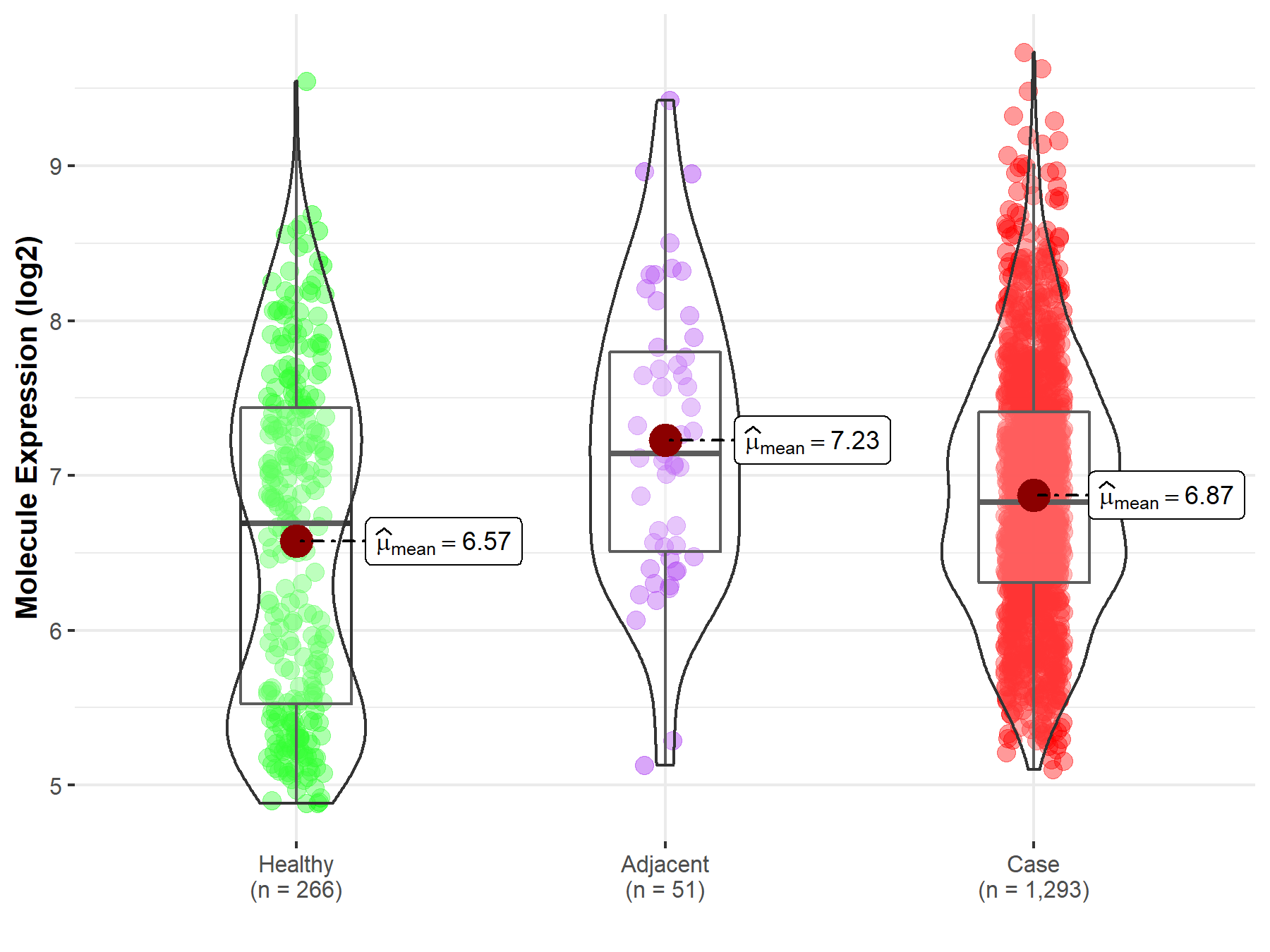
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Ovary | |
| The Specified Disease | Ovarian cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.87E-01; Fold-change: 1.09E-01; Z-score: 9.87E-02 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 3.93E-01; Fold-change: 7.52E-01; Z-score: 6.23E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Thyroid | |
| The Specified Disease | Thyroid cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.46E-01; Fold-change: -9.27E-02; Z-score: -1.28E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 9.85E-01; Fold-change: -2.77E-02; Z-score: -3.10E-02 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals


|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
