Drug Information
Drug (ID: DG00222) and It's Reported Resistant Information
| Name |
Voriconazole
|
||||
|---|---|---|---|---|---|
| Synonyms |
VCZ; Vfend; Pfizer brand of voriconazole; UK 109496; Voriconazole in combination with MGCD290; DRG-0301; UK 109,496; UK-109496; VFEND (TN); Vfend (TN); Vfend, Voriconazole; UK-109,496; Voriconazole [USAN:INN:BAN]; Voriconazole (JAN/USAN/INN); (2R,3S)-2-(2,4-difluorophenyl)-3-(5-fluoropyrimidin-4-yl)-1-(1,2,4-triazol-1-yl)butan-2-ol; (2R,3S)-2-(2,4-difluorophenyl)-3-(5-fluoropyrimidin-4-yl)-1-(1H-1,2,4-triazol-1-yl)butan-2-ol; (R-(R*,S*))-alpha-(2,4-difluorophenyl)-5-fluoro-beta-methyl-alpha-(1H-1,2,4-triazol-1-ylmethyl)-4-pyrimidineethanol; (alphaR,betaS)-alpha-(2,4-Difluorophenyl)-5-fluoro-beta-methyl-alpha-(1H-1,2,4-triazol-1-ylmethyl)-4-pyrimidineethanol; (alphaR,betaS)-alpha-(2,4-difluorophenyl)-5-fluoro-beta-methyl-alpha(1H-1,2,4-triazol-1-ylmethyl)-4-pyrimidineethanol; VRC
Click to Show/Hide
|
||||
| Indication |
In total 1 Indication(s)
|
||||
| Structure |
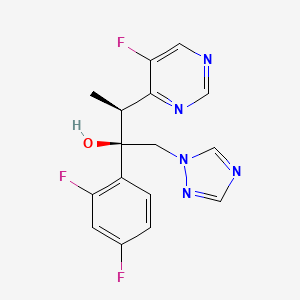
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Clinically Reported Resistance for This Drug
(5 diseases)
[2]
[3]
[5]
[6]
[7]
Disease(s) with Resistance Information Discovered by Cell Line Test for This Drug
(1 diseases)
[4]
|
||||
| Target | Candida Cytochrome P450 51 (Candi ERG11) | CP51_CANAL | [1] | ||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C16H14F3N5O
|
||||
| IsoSMILES |
C[C@@H](C1=NC=NC=C1F)[C@](CN2C=NC=N2)(C3=C(C=C(C=C3)F)F)O
|
||||
| InChI |
1S/C16H14F3N5O/c1-10(15-14(19)5-20-7-22-15)16(25,6-24-9-21-8-23-24)12-3-2-11(17)4-13(12)18/h2-5,7-10,25H,6H2,1H3/t10-,16+/m0/s1
|
||||
| InChIKey |
BCEHBSKCWLPMDN-MGPLVRAMSA-N
|
||||
| PubChem CID | |||||
| ChEBI ID | |||||
| TTD Drug ID | |||||
| VARIDT ID | |||||
| INTEDE ID | |||||
| DrugBank ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-01: Infectious/parasitic diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Sterol 14-alpha demethylase (CYP51C) | [1] | |||
| Resistant Disease | Aspergillus flavus infection [ICD-11: 1F20.1] | |||
| Molecule Alteration | Missense mutation | p.Y319H |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Aspergillus flavus strain | 5059 | ||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | A novel Y319H substitution in CYP51C associated with azole resistance in Aspergillus flavus. | |||
| Key Molecule: Sterol 14-alpha demethylase cyp51A (CYP51A) | [8] | |||
| Resistant Disease | Aspergillus fumigatus infection [ICD-11: 1F20.2] | |||
| Molecule Alteration | Missense mutation | p.G138C |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Aspergillus fumigatus strain | 746128 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Multivariate analysis of overall survival or disease-free survival assay | |||
| Mechanism Description | Each single mutationad a measurable effect on the affinity of the target enzyme for specific azole derivatives. | |||
| Key Molecule: Sterol 14-alpha demethylase cyp51A (CYP51A) | [9] | |||
| Resistant Disease | Aspergillus fumigatus infection [ICD-11: 1F20.2] | |||
| Molecule Alteration | Missense mutation | p.G448S |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Aspergillus fumigatus strain | 746128 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Resistance to voriconazole due to a G448S substitution in Aspergillus fumigatus in a patient with cerebral aspergillosis. | |||
| Key Molecule: Sterol 14-alpha demethylase cyp51A (CYP51A) | [10] | |||
| Resistant Disease | Aspergillus fumigatus infection [ICD-11: 1F20.2] | |||
| Molecule Alteration | Tandem repeat | TR53 (GAATCACGCGGTCCGATGTGTGCTGAGCCGAATGAAAGTTGTCTAATGTCTAGAATCACGCGGTCCGATGTGTGCTGAGCCGAATGAAAGTTGTCTAATGTCTA) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Aspergillus fumigatus strain | 746128 | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | The azole-resistant A. fumigatus strains were detected tandem repeats (TRs) in the promoter region. | |||
| Key Molecule: Sterol 14-alpha demethylase cyp51A (CYP51A) | [2] | |||
| Resistant Disease | Invasive aspergillosis [ICD-11: 1F20.6] | |||
| Molecule Alteration | Missense mutation | p.G54R |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Aspergillus fumigatus strain RIT | 746128 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
NCCLS M38-P microdilution methodology assay | |||
| Mechanism Description | Itraconazole resistance has been tightly linked to cyp51A mutations in the codon for Gly54, resulting in five different amino substitutions (G54k, G54V, G54R, G54E, and G54W). | |||
| Key Molecule: Sterol 14-alpha demethylase cyp51A (CYP51A) | [11] | |||
| Resistant Disease | Invasive aspergillosis [ICD-11: 1F20.6] | |||
| Molecule Alteration | Missense mutation | p.H147Y |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Aspergillus fumigatus strain | 746128 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
EUCAST method assay | |||
| Mechanism Description | Four novel mutations were found (H147Y, P216L, Y431C, and G434C). The isolate bearing the P216L mutation was resistant to itraconazole and posaconazole, whereas the isolates with Y431C and G434C showed pan-azole resistance phenotypes. | |||
| Key Molecule: Sterol 14-alpha demethylase cyp51A (CYP51A) | [11] | |||
| Resistant Disease | Invasive aspergillosis [ICD-11: 1F20.6] | |||
| Molecule Alteration | Missense mutation | p.G434C |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Aspergillus fumigatus strain | 746128 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
EUCAST method assay | |||
| Mechanism Description | Four novel mutations were found (H147Y, P216L, Y431C, and G434C). The isolate bearing the P216L mutation was resistant to itraconazole and posaconazole, whereas the isolates with Y431C and G434C showed pan-azole resistance phenotypes. | |||
| Key Molecule: Sterol 14-alpha demethylase cyp51A (CYP51A) | [11] | |||
| Resistant Disease | Invasive aspergillosis [ICD-11: 1F20.6] | |||
| Molecule Alteration | Missense mutation | p.Y431C |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Aspergillus fumigatus strain | 746128 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
EUCAST method assay | |||
| Mechanism Description | Four novel mutations were found (H147Y, P216L, Y431C, and G434C). The isolate bearing the P216L mutation was resistant to itraconazole and posaconazole, whereas the isolates with Y431C and G434C showed pan-azole resistance phenotypes. | |||
| Key Molecule: Sterol 14-alpha demethylase cyp51A (CYP51A) | [12] | |||
| Resistant Disease | Invasive aspergillosis [ICD-11: 1F20.6] | |||
| Molecule Alteration | Missense mutation | p.Y121F+p.T289A+p.G448S+p.M172I |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Aspergillus fumigatus strain TR463 | 746128 | ||
| Experiment for Molecule Alteration |
PCR analysis | |||
| Mechanism Description | In addition, to compare the susceptibility of TR463 with those of TR34 and TR46, the high resistance of TR463/Y121F/M172I/T289A/G448S was confirmed by MIC testing, displaying a pan-triazole-resistant phenotype to posaconazole, itraconazole, and voriconazole, indicating no in vitro activity of itraconazole and voriconazole (MIC, >16 mg/liter). | |||
| Key Molecule: Sterol 14-alpha demethylase (CYP51C) | [13] | |||
| Resistant Disease | Invasive aspergillosis [ICD-11: 1F20.6] | |||
| Molecule Alteration | Missense mutation | p.T788G |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Aspergillus flavus strain | 5059 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | The T788G mutation in the cyp51C gene confers voriconazole resistance in aspergillus flavus causing aspergillosis. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Lanosterol 14-alpha demethylase (ERG11) | [14] | |||
| Resistant Disease | Candida auris infection [ICD-11: 1F23.2] | |||
| Molecule Alteration | Missense mutation | p.Y132F |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Candida auris strain | 498019 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
CLSI broth microdilution method assay | |||
| Mechanism Description | Overall, among 45% (n = 20) of isolates that had Y132F and k143R substitutions, 16 showed cross-resistance to one or more azoles namely voriconazole, isavuconazole and posaconazole and four were pan-azole resistant. | |||
| Key Molecule: Lanosterol 14-alpha demethylase (ERG11) | [14] | |||
| Resistant Disease | Candida auris infection [ICD-11: 1F23.2] | |||
| Molecule Alteration | Missense mutation | p.K143R |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Candida auris strain | 498019 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
CLSI broth microdilution method assay | |||
| Mechanism Description | Overall, among 45% (n = 20) of isolates that had Y132F and k143R substitutions, 16 showed cross-resistance to one or more azoles namely voriconazole, isavuconazole and posaconazole and four were pan-azole resistant. | |||
| Key Molecule: Lanosterol 14-alpha demethylase (ERG11) | [3] | |||
| Resistant Disease | Recurrent oropharyngeal candidiasis [ICD-11: 1F23.6] | |||
| Molecule Alteration | Missense mutation | p.Y132H+p.G450E |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Candida albicans strain C572 | 5476 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Broth microdilution assay | |||
| Mechanism Description | Seventeen of the 38 isolates analyzed exhibited cross-resistance to fluconazole (MIC, >=64 ug/ml) and voriconazole (in the absence of established breakpoints, we labeled an isolate resistant to voriconazole if the MIC was >1 ug/ml). Sixteen of the 17 isolates (the exception was C587) exhibited the same pattern of mutations in ERG11; a substitution close to the N terminus of the protein (k128T, Y132H, or Y257H) together with a substitution towards the C terminus of the protein (G405F, G448V, G450E, or G464S). | |||
| Key Molecule: Lanosterol 14-alpha demethylase (ERG11) | [3] | |||
| Resistant Disease | Recurrent oropharyngeal candidiasis [ICD-11: 1F23.6] | |||
| Molecule Alteration | Missense mutation | p.G450E+p.G464S |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Candida albicans strain C530 | 5476 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Broth microdilution assay | |||
| Mechanism Description | Seventeen of the 38 isolates analyzed exhibited cross-resistance to fluconazole (MIC, >=64 ug/ml) and voriconazole (in the absence of established breakpoints, we labeled an isolate resistant to voriconazole if the MIC was >1 ug/ml). Sixteen of the 17 isolates (the exception was C587) exhibited the same pattern of mutations in ERG11; a substitution close to the N terminus of the protein (k128T, Y132H, or Y257H) together with a substitution towards the C terminus of the protein (G405F, G448V, G450E, or G464S). | |||
| Key Molecule: Lanosterol 14-alpha demethylase (ERG11) | [3] | |||
| Resistant Disease | Recurrent oropharyngeal candidiasis [ICD-11: 1F23.6] | |||
| Molecule Alteration | Missense mutation | p.Y132H+p.G448V |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Candida albicans strain C535 | 5476 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Broth microdilution assay | |||
| Mechanism Description | Seventeen of the 38 isolates analyzed exhibited cross-resistance to fluconazole (MIC, >=64 ug/ml) and voriconazole (in the absence of established breakpoints, we labeled an isolate resistant to voriconazole if the MIC was >1 ug/ml). Sixteen of the 17 isolates (the exception was C587) exhibited the same pattern of mutations in ERG11; a substitution close to the N terminus of the protein (k128T, Y132H, or Y257H) together with a substitution towards the C terminus of the protein (G405F, G448V, G450E, or G464S). | |||
| Key Molecule: Lanosterol 14-alpha demethylase (ERG11) | [3] | |||
| Resistant Disease | Recurrent oropharyngeal candidiasis [ICD-11: 1F23.6] | |||
| Molecule Alteration | Missense mutation | p.S405F |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Candida albicans strain C470 | 5476 | ||
| Candida albicans strain C478 | 5476 | |||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Broth microdilution assay | |||
| Mechanism Description | Seventeen of the 38 isolates analyzed exhibited cross-resistance to fluconazole (MIC, >=64 ug/ml) and voriconazole (in the absence of established breakpoints, we labeled an isolate resistant to voriconazole if the MIC was >1 ug/ml). Sixteen of the 17 isolates (the exception was C587) exhibited the same pattern of mutations in ERG11; a substitution close to the N terminus of the protein (k128T, Y132H, or Y257H) together with a substitution towards the C terminus of the protein (G405F, G448V, G450E, or G464S). | |||
| Key Molecule: Lanosterol 14-alpha demethylase (ERG11) | [3] | |||
| Resistant Disease | Recurrent oropharyngeal candidiasis [ICD-11: 1F23.6] | |||
| Molecule Alteration | Missense mutation | p.K128T+p.V452A |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Candida albicans strain C497 | 5476 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Broth microdilution assay | |||
| Mechanism Description | Seventeen of the 38 isolates analyzed exhibited cross-resistance to fluconazole (MIC, >=64 ug/ml) and voriconazole (in the absence of established breakpoints, we labeled an isolate resistant to voriconazole if the MIC was >1 ug/ml). Sixteen of the 17 isolates (the exception was C587) exhibited the same pattern of mutations in ERG11; a substitution close to the N terminus of the protein (k128T, Y132H, or Y257H) together with a substitution towards the C terminus of the protein (G405F, G448V, G450E, or G464S). | |||
| Key Molecule: Lanosterol 14-alpha demethylase (ERG11) | [3] | |||
| Resistant Disease | Recurrent oropharyngeal candidiasis [ICD-11: 1F23.6] | |||
| Molecule Alteration | Missense mutation | p.Y132H+p.S405F |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Candida albicans strain C600 | 5476 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Broth microdilution assay | |||
| Mechanism Description | Seventeen of the 38 isolates analyzed exhibited cross-resistance to fluconazole (MIC, >=64 ug/ml) and voriconazole (in the absence of established breakpoints, we labeled an isolate resistant to voriconazole if the MIC was >1 ug/ml). Sixteen of the 17 isolates (the exception was C587) exhibited the same pattern of mutations in ERG11; a substitution close to the N terminus of the protein (k128T, Y132H, or Y257H) together with a substitution towards the C terminus of the protein (G405F, G448V, G450E, or G464S). | |||
| Key Molecule: Lanosterol 14-alpha demethylase (ERG11) | [3] | |||
| Resistant Disease | Recurrent oropharyngeal candidiasis [ICD-11: 1F23.6] | |||
| Molecule Alteration | Missense mutation | p.G464S |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Candida albicans strain C587 | 5476 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Broth microdilution assay | |||
| Mechanism Description | Seventeen of the 38 isolates analyzed exhibited cross-resistance to fluconazole (MIC, >=64 ug/ml) and voriconazole (in the absence of established breakpoints, we labeled an isolate resistant to voriconazole if the MIC was >1 ug/ml). Sixteen of the 17 isolates (the exception was C587) exhibited the same pattern of mutations in ERG11; a substitution close to the N terminus of the protein (k128T, Y132H, or Y257H) together with a substitution towards the C terminus of the protein (G405F, G448V, G450E, or G464S). | |||
| Key Molecule: Lanosterol 14-alpha demethylase (ERG11) | [3] | |||
| Resistant Disease | Recurrent oropharyngeal candidiasis [ICD-11: 1F23.6] | |||
| Molecule Alteration | Missense mutation | p.G464S+p.K128T+p.R467I |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Candida albicans strain C477 | 5476 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Broth microdilution assay | |||
| Mechanism Description | Seventeen of the 38 isolates analyzed exhibited cross-resistance to fluconazole (MIC, >=64 ug/ml) and voriconazole (in the absence of established breakpoints, we labeled an isolate resistant to voriconazole if the MIC was >1 ug/ml). Sixteen of the 17 isolates (the exception was C587) exhibited the same pattern of mutations in ERG11; a substitution close to the N terminus of the protein (k128T, Y132H, or Y257H) together with a substitution towards the C terminus of the protein (G405F, G448V, G450E, or G464S). | |||
| Key Molecule: Lanosterol 14-alpha demethylase (ERG11) | [3] | |||
| Resistant Disease | Recurrent oropharyngeal candidiasis [ICD-11: 1F23.6] | |||
| Molecule Alteration | Missense mutation | p.Y257H+p.G464S |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Candida albicans strain C438 | 5476 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Broth microdilution assay | |||
| Mechanism Description | Seventeen of the 38 isolates analyzed exhibited cross-resistance to fluconazole (MIC, >=64 ug/ml) and voriconazole (in the absence of established breakpoints, we labeled an isolate resistant to voriconazole if the MIC was >1 ug/ml). Sixteen of the 17 isolates (the exception was C587) exhibited the same pattern of mutations in ERG11; a substitution close to the N terminus of the protein (k128T, Y132H, or Y257H) together with a substitution towards the C terminus of the protein (G405F, G448V, G450E, or G464S). | |||
| Key Molecule: Lanosterol 14-alpha demethylase (ERG11) | [3] | |||
| Resistant Disease | Recurrent oropharyngeal candidiasis [ICD-11: 1F23.6] | |||
| Molecule Alteration | Missense mutation | p.A61V+p.Y257H+p.G464S+p.G307S |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Candida albicans strain C440 | 5476 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Broth microdilution assay | |||
| Mechanism Description | Seventeen of the 38 isolates analyzed exhibited cross-resistance to fluconazole (MIC, >=64 ug/ml) and voriconazole (in the absence of established breakpoints, we labeled an isolate resistant to voriconazole if the MIC was >1 ug/ml). Sixteen of the 17 isolates (the exception was C587) exhibited the same pattern of mutations in ERG11; a substitution close to the N terminus of the protein (k128T, Y132H, or Y257H) together with a substitution towards the C terminus of the protein (G405F, G448V, G450E, or G464S). | |||
| Key Molecule: Lanosterol 14-alpha demethylase (ERG11) | [3] | |||
| Resistant Disease | Recurrent oropharyngeal candidiasis [ICD-11: 1F23.6] | |||
| Molecule Alteration | Missense mutation | p.Y257H+p.G464S+p.G307S |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Candida albicans strain C439 | 5476 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Broth microdilution assay | |||
| Mechanism Description | Seventeen of the 38 isolates analyzed exhibited cross-resistance to fluconazole (MIC, >=64 ug/ml) and voriconazole (in the absence of established breakpoints, we labeled an isolate resistant to voriconazole if the MIC was >1 ug/ml). Sixteen of the 17 isolates (the exception was C587) exhibited the same pattern of mutations in ERG11; a substitution close to the N terminus of the protein (k128T, Y132H, or Y257H) together with a substitution towards the C terminus of the protein (G405F, G448V, G450E, or G464S). | |||
| Key Molecule: Lanosterol 14-alpha demethylase (ERG11) | [3] | |||
| Resistant Disease | Recurrent oropharyngeal candidiasis [ICD-11: 1F23.6] | |||
| Molecule Alteration | Missense mutation | p.K143R |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Candida albicans strain C441 | 5476 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Broth microdilution assay | |||
| Mechanism Description | Seventeen of the 38 isolates analyzed exhibited cross-resistance to fluconazole (MIC, >=64 ug/ml) and voriconazole (in the absence of established breakpoints, we labeled an isolate resistant to voriconazole if the MIC was >1 ug/ml). Sixteen of the 17 isolates (the exception was C587) exhibited the same pattern of mutations in ERG11; a substitution close to the N terminus of the protein (k128T, Y132H, or Y257H) together with a substitution towards the C terminus of the protein (G405F, G448V, G450E, or G464S). | |||
| Key Molecule: Lanosterol 14-alpha demethylase (ERG11) | [3] | |||
| Resistant Disease | Recurrent oropharyngeal candidiasis [ICD-11: 1F23.6] | |||
| Molecule Alteration | Missense mutation | p.Y257H+p.Y132H+p.E266D |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Candida albicans strain C489 | 5476 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Broth microdilution assay | |||
| Mechanism Description | Seventeen of the 38 isolates analyzed exhibited cross-resistance to fluconazole (MIC, >=64 ug/ml) and voriconazole (in the absence of established breakpoints, we labeled an isolate resistant to voriconazole if the MIC was >1 ug/ml). Sixteen of the 17 isolates (the exception was C587) exhibited the same pattern of mutations in ERG11; a substitution close to the N terminus of the protein (k128T, Y132H, or Y257H) together with a substitution towards the C terminus of the protein (G405F, G448V, G450E, or G464S). | |||
| Key Molecule: Lanosterol 14-alpha demethylase (ERG11) | [3] | |||
| Resistant Disease | Recurrent oropharyngeal candidiasis [ICD-11: 1F23.6] | |||
| Molecule Alteration | Missense mutation | p.Y132H+p.G464S+p.H283R |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Candida albicans strain C507 | 5476 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Broth microdilution assay | |||
| Mechanism Description | Seventeen of the 38 isolates analyzed exhibited cross-resistance to fluconazole (MIC, >=64 ug/ml) and voriconazole (in the absence of established breakpoints, we labeled an isolate resistant to voriconazole if the MIC was >1 ug/ml). Sixteen of the 17 isolates (the exception was C587) exhibited the same pattern of mutations in ERG11; a substitution close to the N terminus of the protein (k128T, Y132H, or Y257H) together with a substitution towards the C terminus of the protein (G405F, G448V, G450E, or G464S). | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Cuticle protein (TERG_08771 ) | [5] | |||
| Resistant Disease | Tinea capitis [ICD-11: 1F28.0] | |||
| Molecule Alteration | SNP | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Trichophyton rubrum isolates | 5551 | ||
| Experiment for Drug Resistance |
Broth dilution method assay | |||
| Mechanism Description | The gene TERG_08771 harboring the highest SNPs density is found to be associated with resistance to voriconazole. | |||
| Key Molecule: Cuticle protein (TERG_08771 ) | [5] | |||
| Resistant Disease | Tinea corporis [ICD-11: 1F28.Y] | |||
| Molecule Alteration | SNP | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Trichophyton rubrum isolates | 5551 | ||
| Experiment for Drug Resistance |
Broth dilution method assay | |||
| Mechanism Description | The gene TERG_08771 harboring the highest SNPs density is found to be associated with resistance to voriconazole. | |||
| Key Molecule: Cuticle protein (TERG_08771 ) | [5] | |||
| Resistant Disease | Tinea inguinalis [ICD-11: 1F28.4] | |||
| Molecule Alteration | SNP | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Trichophyton rubrum isolates | 5551 | ||
| Experiment for Drug Resistance |
Broth dilution method assay | |||
| Mechanism Description | The gene TERG_08771 harboring the highest SNPs density is found to be associated with resistance to voriconazole. | |||
| Key Molecule: Cuticle protein (TERG_08771 ) | [5] | |||
| Resistant Disease | Tinea manus [ICD-11: 1F28.5] | |||
| Molecule Alteration | SNP | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Trichophyton rubrum isolates | 5551 | ||
| Experiment for Drug Resistance |
Broth dilution method assay | |||
| Mechanism Description | The gene TERG_08771 harboring the highest SNPs density is found to be associated with resistance to voriconazole. | |||
| Key Molecule: Cuticle protein (TERG_08771 ) | [5] | |||
| Resistant Disease | Tinea pedis [ICD-11: 1F28.2] | |||
| Molecule Alteration | SNP | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Trichophyton rubrum isolates | 5551 | ||
| Experiment for Drug Resistance |
Broth dilution method assay | |||
| Mechanism Description | The gene TERG_08771 harboring the highest SNPs density is found to be associated with resistance to voriconazole. | |||
| Key Molecule: Cuticle protein (TERG_08771 ) | [5] | |||
| Resistant Disease | Tinea unguium [ICD-11: 1F28.1] | |||
| Molecule Alteration | SNP | . |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Trichophyton rubrum isolates | 5551 | ||
| Experiment for Drug Resistance |
Broth dilution method assay | |||
| Mechanism Description | The gene TERG_08771 harboring the highest SNPs density is found to be associated with resistance to voriconazole. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Cytochrome P450 sterol 14 alpha-demethylase (CYP51) | [6] | |||
| Resistant Disease | Histoplasmosis [ICD-11: 1F2A.0] | |||
| Molecule Alteration | Missense mutation | p.Y136F |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Histoplasma capsulatum strain | 5037 | ||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
NCCLS method assay | |||
| Mechanism Description | In summary, fluconazole treatment of disseminated histoplasmosis in patients with AIDS was associated with induction of resistance to fluconazole and, to a lesser extent, to voriconazole. And the changes in susceptibility were due to tagert alterations which a single amino acid substitution in CYP51p at Y136 appeared to be responsible for the reduction in susceptibility seen in the relapse isolate. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Lanosterol 14-alpha demethylase (ERG11) | [7] | |||
| Resistant Disease | Mycotic vaginitis [ICD-11: 1F2Y.0] | |||
| Molecule Alteration | Missense mutation | p.Y33C |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Candida albicans strain | 5476 | ||
| Experiment for Molecule Alteration |
Northern blot analysis; DNA sequencing assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Resistance mechanisms that have been identified include overexpression of the MDR1 gene encoding a drug efflux pump, increased expression of the CDR1 and CDR2 genes, overexpression of the ERG11 gene coding for the FLU target enzyme, and alterations in the structure of Erg11p. | |||
| Key Molecule: Lanosterol 14-alpha demethylase (ERG11) | [7] | |||
| Resistant Disease | Mycotic vaginitis [ICD-11: 1F2Y.0] | |||
| Molecule Alteration | Missense mutation | p.Y39C |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Candida albicans strain | 5476 | ||
| Experiment for Molecule Alteration |
Northern blot analysis; DNA sequencing assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Resistance mechanisms that have been identified include overexpression of the MDR1 gene encoding a drug efflux pump, increased expression of the CDR1 and CDR2 genes, overexpression of the ERG11 gene coding for the FLU target enzyme, and alterations in the structure of Erg11p. | |||
| Key Molecule: Lanosterol 14-alpha demethylase (ERG11) | [7] | |||
| Resistant Disease | Mycotic vaginitis [ICD-11: 1F2Y.0] | |||
| Molecule Alteration | Missense mutation | p.K119L |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Candida albicans strain | 5476 | ||
| Experiment for Molecule Alteration |
Northern blot analysis; DNA sequencing assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Resistance mechanisms that have been identified include overexpression of the MDR1 gene encoding a drug efflux pump, increased expression of the CDR1 and CDR2 genes, overexpression of the ERG11 gene coding for the FLU target enzyme, and alterations in the structure of Erg11p. | |||
| Key Molecule: Lanosterol 14-alpha demethylase (ERG11) | [7] | |||
| Resistant Disease | Mycotic vaginitis [ICD-11: 1F2Y.0] | |||
| Molecule Alteration | Missense mutation | p.T494A |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Candida albicans strain | 5476 | ||
| Experiment for Molecule Alteration |
Northern blot analysis; DNA sequencing assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Resistance mechanisms that have been identified include overexpression of the MDR1 gene encoding a drug efflux pump, increased expression of the CDR1 and CDR2 genes, overexpression of the ERG11 gene coding for the FLU target enzyme, and alterations in the structure of Erg11p. | |||
| Key Molecule: Lanosterol 14-alpha demethylase (ERG11) | [7] | |||
| Resistant Disease | Mycotic vaginitis [ICD-11: 1F2Y.0] | |||
| Molecule Alteration | Missense mutation | p.L491V |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Candida albicans strain | 5476 | ||
| Experiment for Molecule Alteration |
Northern blot analysis; DNA sequencing assay | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Resistance mechanisms that have been identified include overexpression of the MDR1 gene encoding a drug efflux pump, increased expression of the CDR1 and CDR2 genes, overexpression of the ERG11 gene coding for the FLU target enzyme, and alterations in the structure of Erg11p. | |||
|
|
||||
| Key Molecule: Multidrug resistance protein 1 (ABCB1) | [7] | |||
| Resistant Disease | Mycotic vaginitis [ICD-11: 1F2Y.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Candida albicans strain | 5476 | ||
| Experiment for Molecule Alteration |
Northern blot analysis | |||
| Experiment for Drug Resistance |
Broth microdilution method assay | |||
| Mechanism Description | Resistance mechanisms that have been identified include overexpression of the MDR1 gene encoding a drug efflux pump, increased expression of the CDR1 and CDR2 genes, overexpression of the ERG11 gene coding for the FLU target enzyme, and alterations in the structure of Erg11p. | |||
ICD-12: Respiratory system diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Lanosterol 14-alpha demethylase (CYP51A1) | [4] | |||
| Resistant Disease | Cystic fibrosis [ICD-11: CA25.0] | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | S. minutisporum | 41687 | ||
| Experiment for Molecule Alteration |
Fluorescent reporter assay; GC-MS | |||
| Experiment for Drug Resistance |
Antifungal susceptibility assay; Membrane fluidity assay | |||
| Mechanism Description | SCFM increases Scedosporium/Lomentospora azole tolerance.Azole resistance is partially due to the efflux pump activity.SCFM leads to decrease in sterol membrane content and increase in membrane fluidity.Scedosporium/Lomentospora species undergo cellular adaptations in SCFM that favours their growth in face of the challenges imposed by azole antifungals. | |||
| Key Molecule: Ergosterol | [4] | |||
| Resistant Disease | Cystic fibrosis [ICD-11: CA25.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | S. minutisporum | 41687 | ||
| Experiment for Molecule Alteration |
Fluorescent reporter assay; GC-MS | |||
| Experiment for Drug Resistance |
Antifungal susceptibility assay; Membrane fluidity assay | |||
| Mechanism Description | SCFM increases Scedosporium/Lomentospora azole tolerance.Azole resistance is partially due to the efflux pump activity.SCFM leads to decrease in sterol membrane content and increase in membrane fluidity.Scedosporium/Lomentospora species undergo cellular adaptations in SCFM that favours their growth in face of the challenges imposed by azole antifungals. | |||
|
|
||||
| Key Molecule: Calcium-transporting ATPase type 2C member 1 | [4] | |||
| Resistant Disease | Cystic fibrosis [ICD-11: CA25.0] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | S. minutisporum | 41687 | ||
| Experiment for Molecule Alteration |
Fluorescent reporter assay; GC-MS | |||
| Experiment for Drug Resistance |
Antifungal susceptibility assay; Membrane fluidity assay | |||
| Mechanism Description | SCFM increases Scedosporium/Lomentospora azole tolerance.Azole resistance is partially due to the efflux pump activity.SCFM leads to decrease in sterol membrane content and increase in membrane fluidity.Scedosporium/Lomentospora species undergo cellular adaptations in SCFM that favours their growth in face of the challenges imposed by azole antifungals. | |||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
