Drug Information
Drug (ID: DG00062) and It's Reported Resistant Information
| Name |
Piperacillin
|
||||
|---|---|---|---|---|---|
| Synonyms |
PIPC; Peperacillin; Peracin; Pipercillin; Pipracil; Pipril; PIPERACILLIN SODIUM; Piperacillin Monosodium Salt; Piperacillin anhydrous; Cl-227193; Peracin (TN); Piperacillin (INN); Piperacillin (anhydrous); Pipracil, Piper; T-1220; Zobactin (TN); (2S,5R,6R)-6-[[(2R)-2-[(4-ethyl-2,3-dioxopiperazine-1-carbonyl)amino]-2-phenylacetyl]amino]-3,3-dimethyl-7-oxo-4-thia-1-azabicyclo[3.2.0]heptane-2-carboxylic acid; (2S,5R,6R)-6-{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}-3,3-dimethyl-7-oxo-4-thia-1-azabicyclo[3.2.0]heptane-2-carboxylic acid; (2S-(2alpha,5alpha,6beta(S*)))-6-(((((4-Ethyl-2,3-dioxopiperazin-1-yl)carbonyl)amino)phenylacetyl)amino)-3,3-dimethyl-7-oxo-4-thia-1-azabicyclo(3.2.0)heptane-2-carboxylic acid; 4-ethyl-2,3-dioxopiperazine carbonyl ampicillin; 6-(D-(-)-alpha-(4-Ethyl-2,3-dioxo-1-piperazinecarboxamido)phenylacetamido)penicillanicacid; 6beta-{(2R)-2-[(4-ethyl-2,3-dioxopiperazin-1-yl)carboxamido]-2-phenylacetamido}-2,2-dimethylpenam-3alpha-carboxylic acid
Click to Show/Hide
|
||||
| Indication |
In total 1 Indication(s)
|
||||
| Structure |
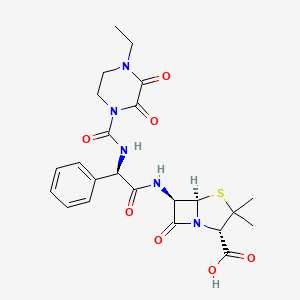
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Clinically Reported Resistance for This Drug
(4 diseases)
[3]
[3]
[5]
[6]
Disease(s) with Resistance Information Discovered by Cell Line Test for This Drug
(1 diseases)
[4]
|
||||
| Target | Bacterial Penicillin binding protein (Bact PBP) | NOUNIPROTAC | [1] | ||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C23H27N5O7S
|
||||
| IsoSMILES |
CCN1CCN(C(=O)C1=O)C(=O)N[C@H](C2=CC=CC=C2)C(=O)N[C@H]3[C@@H]4N(C3=O)[C@H](C(S4)(C)C)C(=O)O
|
||||
| InChI |
1S/C23H27N5O7S/c1-4-26-10-11-27(19(32)18(26)31)22(35)25-13(12-8-6-5-7-9-12)16(29)24-14-17(30)28-15(21(33)34)23(2,3)36-20(14)28/h5-9,13-15,20H,4,10-11H2,1-3H3,(H,24,29)(H,25,35)(H,33,34)/t13-,14-,15+,20-/m1/s1
|
||||
| InChIKey |
IVBHGBMCVLDMKU-GXNBUGAJSA-N
|
||||
| PubChem CID | |||||
| ChEBI ID | |||||
| TTD Drug ID | |||||
| VARIDT ID | |||||
| INTEDE ID | |||||
| DrugBank ID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-01: Infectious/parasitic diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Beta-lactamase (BLA) | [1], [2] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli HB101 | 634468 | ||
| Escherichia coli JM101 | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Mechanism Description | Beta-lactamases (Beta-lactamhydrolase, EC 3.5.2.6), responsible for most of the resistance to Beta-lactam antibiotics, are often plasmid mediated.The OXA-1 beta-lactamase gene is part of Tn2603, which is borne on the R plasmid RGN238. | |||
| Key Molecule: Beta-lactamase (BLA) | [2], [7] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.D240G |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli | 668369 | ||
| Escherichia coli Gre-1 | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | The first extended-spectrum Beta-lactamase (ESBL) of the CTX-M type (MEN-1/CTX-M-1) was reported at the beginning of the 1990s.CTX-M-27 differed from CTX-M-14 only by the substitution D240G and was the third CTX-M enzyme harbouring this mutation after CTX-M-15 and CTX-M-16. The Gly-240-harbouring enzyme CTX-M-27 conferred to Escherichia coli higher MICs of ceftazidime (MIC, 8 versus 1 mg/L) than did the Asp-240-harbouring CTX-M-14 enzyme. | |||
| Key Molecule: Beta-lactamase (BLA) | [2], [8], [9] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.D240G |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli DH10B | 316385 | ||
| Citrobacter freundii 2526/96 | 546 | |||
| Escherichia coli isolates | 562 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | We have reported recently the DNA sequence of another Beta-lactamase, CTX- M-15, from Indian enterobacterial isolates that were resistant to both cefotaxime and ceftazidime.CTX-M-15 has a single amino acid change [Asp-240-Gly (Ambler numbering)]7 compared with CTX-M-3. | |||
| Key Molecule: Metallo-beta-lactamase (VIM1) | [3] | |||
| Resistant Disease | Achromobacter xylosoxydans infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli | 668369 | ||
| Achromobacter xylosoxydans subsp. denitrificans AX-22 | 85698 | |||
| Escherichia coli MkD-135 | 562 | |||
| Pseudomonas aeruginosa 10145/3 | 287 | |||
| Experiment for Molecule Alteration |
DNA extraction and Sequencing assay | |||
| Experiment for Drug Resistance |
Macrodilution broth method assay | |||
| Mechanism Description | A. xylosoxydans AX22 exhibited broad-spectrum resistance to Beta-lactams and aminoglycosides. The Beta-lactam resistance pattern (including piperacillin, ceftazidime, and carbapenem resistance) was unusual for this species, and the high-level carbapenem resistance suggested the production of an acquired carbapenemase. In fact, carbapenemase activity was detected in a crude extract of AX22 (specific activity, 184 +/- 12 U/mg of protein), and this activity was reduced (>80%) after incubation of the crude extract with 2 mM EDTA, suggesting the presence of a metallo-Beta-lactamase determinant. | |||
| Key Molecule: Beta-lactamase (BLA) | [2], [10] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Missense mutation | p.V77A+p.D114N+p.S140A+p.N288D |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Citrobacter freundii strain 2524/96 | 546 | ||
| Citrobacter freundii strain 2525/96 | 546 | |||
| Citrobacter freundii strain 2526/96 | 546 | |||
| Escherichia coli strain 2527/96 | 562 | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | Sequencing has revealed that C. freundii isolates produced a new CTX-M-3 enzyme which is very closely related to the CTX-M-1/MEN-1 Beta-lactamase. | |||
| Key Molecule: KBL-1 protein (KBL-1) | [4] | |||
| Resistant Disease | Lactobacillus casei infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | . |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | S. maltophilia JUNP497 | N.A. | ||
| Mechanism Description | Recombinant KBL-1 protein had hydrolytic activities against all the beta-lactams tested, except for aztreonam (Table?3). Recombinant KBL-1 efficiently hydrolyzed the penicillins, including ampicillin, amoxicillin, penicillin G, and piperacillin with?kcat/km?values of 0.422 to 1.166. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Metallo-beta-lactamase (VIM1) | [3] | |||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | |||
| Molecule Alteration | Expression | Acquired |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli | 668369 | ||
| Achromobacter xylosoxydans subsp. denitrificans AX-22 | 85698 | |||
| Escherichia coli MkD-135 | 562 | |||
| Pseudomonas aeruginosa 10145/3 | 287 | |||
| Experiment for Molecule Alteration |
DNA extraction and Sequencing assay | |||
| Experiment for Drug Resistance |
Macrodilution broth method assay | |||
| Mechanism Description | Electroporation of Escherichia coli DH5alpha with the purified plasmid preparation yielded ampicillin-resistant transformants which contained a plasmid apparently identical to pAX22 (data not shown). DH5alpha(pAX22) produced carbapenemase activity (specific activity of crude extract, 202 +/- 14 U/mg of protein) and, compared to DH5alpha, exhibited a decreased susceptibility to several Beta-lactams. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: OXA-23 carbapenemase (BLA OXA-23) | [5] | |||
| Resistant Disease | Cutaneous bacterial infection [ICD-11: 1B21.4] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Acinetobacter baumannii isolates | 470 | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Broth microdilution method assay; Agar dilution method assay | |||
| Mechanism Description | The isolate was resistant to antibiotics other than ampicillin-sulbactam and colistin, suggesting drug resistance due to carbapenemase production by OXA-23.carbapenem resistance in the isolated carbapenem-resistant A. baumannii strain was at least partially conferred by bla OXA-23-like carbapenemase. | |||
ICD-12: Respiratory system diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Bcr/CflA family efflux transporter (BCML) | [6] | |||
| Resistant Disease | Klebsiella pneumoniae infection [ICD-11: CA40.1] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli DH10B | 316385 | ||
| Escherichia coli strain NCTC 50192 | 562 | |||
| Klebsiella pneumoniae strain ORI-1 | 573 | |||
| Experiment for Molecule Alteration |
PCR and hybridization experiments assay | |||
| Experiment for Drug Resistance |
Agar dilution technique assay | |||
| Mechanism Description | Klebsiella pneumoniae ORI-1 strain harbored a ca. 140-kb nontransferable plasmid, pTk1, that conferred an extended-spectrum cephalosporin resistance profile antagonized by the addition of clavulanic acid, tazobactam, or imipenem. The gene for GES-1 (Guiana extended-spectrum beta-lactamase) was cloned, and its protein was expressed in Escherichia coli DH10B, where this pI-5. 8 beta-lactamase of a ca. 31-kDa molecular mass conferred resistance to oxyimino cephalosporins (mostly to ceftazidime). GES-1 is weakly related to the other plasmid-located Ambler class A extended-spectrum beta-lactamases (ESBLs). | |||
| Key Molecule: Bcr/CflA family efflux transporter (BCML) | [6] | |||
| Resistant Disease | Klebsiella pneumoniae infection [ICD-11: CA40.1] | |||
| Molecule Alteration | Expression | Acquired |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli DH10B | 316385 | ||
| Escherichia coli strain NCTC 50192 | 562 | |||
| Klebsiella pneumoniae strain ORI-1 | 573 | |||
| Experiment for Molecule Alteration |
PCR and hybridization experiments assay | |||
| Experiment for Drug Resistance |
Agar dilution technique assay | |||
| Mechanism Description | Beta-Lactam MICs for k. pneumoniae ORI-1 and Escherichia coli DH10B harboring either the natural plasmid pTk1 or the recombinant plasmid pC1 were somewhat similar and might indicate the presence of an ESBL. In all cases, the ceftazidime MICs were higher than those of cefotaxime and aztreonam. Beta-Lactam MICs were always lowered by the addition of clavulanic acid or tazobactam, less so by sulbactam, and uncommonly by imipenem. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Bcr/CflA family efflux transporter (BCML) | [6] | |||
| Sensitive Disease | Klebsiella pneumoniae infection [ICD-11: CA40.1] | |||
| Molecule Alteration | Expression | Antagonism |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli DH10B | 316385 | ||
| Escherichia coli strain NCTC 50192 | 562 | |||
| Klebsiella pneumoniae strain ORI-1 | 573 | |||
| Experiment for Molecule Alteration |
PCR and hybridization experiments assay | |||
| Experiment for Drug Resistance |
Agar dilution technique assay | |||
| Mechanism Description | Inhibition studies, as measured by IC50 values with benzylpenicillin as the substrate, showed that GES-1 was inhibited by clavulanic acid (5 uM) and tazobactam (2.5 uM) and strongly inhibited by imipenem (0.1 uM). Beta-Lactam MICs were always lowered by the addition of clavulanic acid or tazobactam, less so by sulbactam, and uncommonly by imipenem. | |||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
