Molecule Information
General Information of the Molecule (ID: Mol01859)
| Name |
Cytochrome P450 family 1 subfamily A member 1 (CYP1A1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
CYP1A1
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
CYP1A1
|
||||
| Gene ID | |||||
| Location |
chr15:74,719,542-74,725,536[-]
|
||||
| Sequence |
MLFPISMSATEFLLASVIFCLVFWVIRASRPQVPKGLKNPPGPWGWPLIGHMLTLGKNPH
LALSRMSQQYGDVLQIRIGSTPVVVLSGLDTIRQALVRQGDDFKGRPDLYTFTLISNGQS MSFSPDSGPVWAARRRLAQNGLKSFSIASDPASSTSCYLEEHVSKEAEVLISTLQELMAG PGHFNPYRYVVVSVTNVICAICFGRRYDHNHQELLSLVNLNNNFGEVVGSGNPADFIPIL RYLPNPSLNAFKDLNEKFYSFMQKMVKEHYKTFEKGHIRDITDSLIEHCQEKQLDENANV QLSDEKIINIVLDLFGAGFDTVTTAISWSLMYLVMNPRVQRKIQEELDTVIGRSRRPRLS DRSHLPYMEAFILETFRHSSFVPFTIPHSTTRDTSLKGFYIPKGRCVFVNQWQINHDQKL WVNPSEFLPERFLTPDGAIDKVLSEKVIIFGMGKRKCIGETIARWEVFLFLAILLQRVEF SVPLGVKVDMTPIYGLTMKHACCEHFQMQLRS Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
A cytochrome P450 monooxygenase involved in the metabolism of various endogenous substrates, including fatty acids, steroid hormones and vitamins. Mechanistically, uses molecular oxygen inserting one oxygen atom into a substrate, and reducing the second into a water molecule, with two electrons provided by NADPH via cytochrome P450 reductase (NADPH--hemoprotein reductase). Catalyzes the hydroxylation of carbon-hydrogen bonds. Exhibits high catalytic activity for the formation of hydroxyestrogens from estrone (E1) and 17beta-estradiol (E2), namely 2-hydroxy E1 and E2, as well as D-ring hydroxylated E1 and E2 at the C15-alpha and C16-alpha positions. Displays different regioselectivities for polyunsaturated fatty acids (PUFA) hydroxylation. Catalyzes the epoxidation of double bonds of certain PUFA. Converts arachidonic acid toward epoxyeicosatrienoic acid (EET) regioisomers, 8,9-, 11,12-, and 14,15-EET, that function as lipid mediators in the vascular system. Displays an absolute stereoselectivity in the epoxidation of eicosapentaenoic acid (EPA) producing the 17(R),18(S) enantiomer. May play an important role in all-trans retinoic acid biosynthesis in extrahepatic tissues. Catalyzes two successive oxidative transformation of all-trans retinol to all-trans retinal and then to the active form all-trans retinoic acid. May also participate in eicosanoids metabolism by converting hydroperoxide species into oxo metabolites (lipoxygenase-like reaction, NADPH-independent).
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
3 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Acute Ischemic Stroke [ICD-11: 8B11.0] | [1] | |||
| Resistant Disease | Acute Ischemic Stroke [ICD-11: 8B11.0] | |||
| Resistant Drug | Clopidogrel | |||
| Molecule Alteration | Missense mutation | CYP3A5 (rs776746) GG + AG and CYP2C19*2 (rs4244285) AA + AG genotypes |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Experiment for Molecule Alteration |
Matrix-assisted laser desorption/ionization time of flight mass spectrometry assay | |||
| Experiment for Drug Resistance |
Platelet aggregation test assay | |||
| Mechanism Description | The estimated risk of clopidogrel resistance was significantly higher in patients with CYP3A5 (rs776746) GG and CYP2C19*2 (rs4244285) AA, as compared to patients harboring CYP3A5 (rs776746) AA and CYP2C19*2 (rs4244285) GG. These data suggest that these two CYP genetic variants together significantly contributed to clopidogrel resistance. The relative risk conferred by the combinations of CYP3A5 GG and CYP2C19*2 AA was considered as a high-risk variable, with assigned as one, and other combinations of CYP3A5 and CYP2C19*2 as a low-risk variable, with assigned as zero. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [2] | |||
| Metabolic Type | Mitochondrial metabolism | |||
| Resistant Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Resistant Drug | Cytarabine | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cushing syndrome | Activation | hsa04934 | |
| In Vitro Model | SHI-1 cells | Bone marrow | Homo sapiens (Human) | CVCL_2191 |
| Skm1 cells | Blood | Homo sapiens (Human) | CVCL_0098 | |
| U937 cells | Blood | Homo sapiens (Human) | CVCL_0007 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Cell viability assay | |||
| Mechanism Description | The data analysis reveals that the AHR signaling pathway is activated in AML patients. Furthermore, there is a correlation between the expressions of AHR and mitochondrial oxidative phosphorylation genes.In vitroexperiments show that enhancing AHR expression in AML cells increases mitochondrial oxidative phosphorylation and induces resistance to cytarabine. Conversely, reducing AHR expression in AML cells decreases cytarabine resistance. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Pediculosis [ICD-11: 1G00.0] | [3] | |||
| Resistant Disease | Pediculosis [ICD-11: 1G00.0] | |||
| Resistant Drug | Ivermectin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Pediculus humanus | 121225 | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Disk diffusion assay | |||
| Mechanism Description | Phylogenetic relatedness of P450 and ABC transporter genes over-transcribed following ivermectin exposure.Knockdown of CYP9AG2 P450 and ABCC4 transporter gene expression by RNA interference and subsequent increase in the sensitivity of lice to ivermectin. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 08

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Whole blood | |
| The Specified Disease | Cardioembolic Stroke | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.24E-02; Fold-change: 2.60E-02; Z-score: 1.93E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Peripheral blood | |
| The Specified Disease | Ischemic stroke | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.00E-01; Fold-change: 1.96E-02; Z-score: 1.29E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
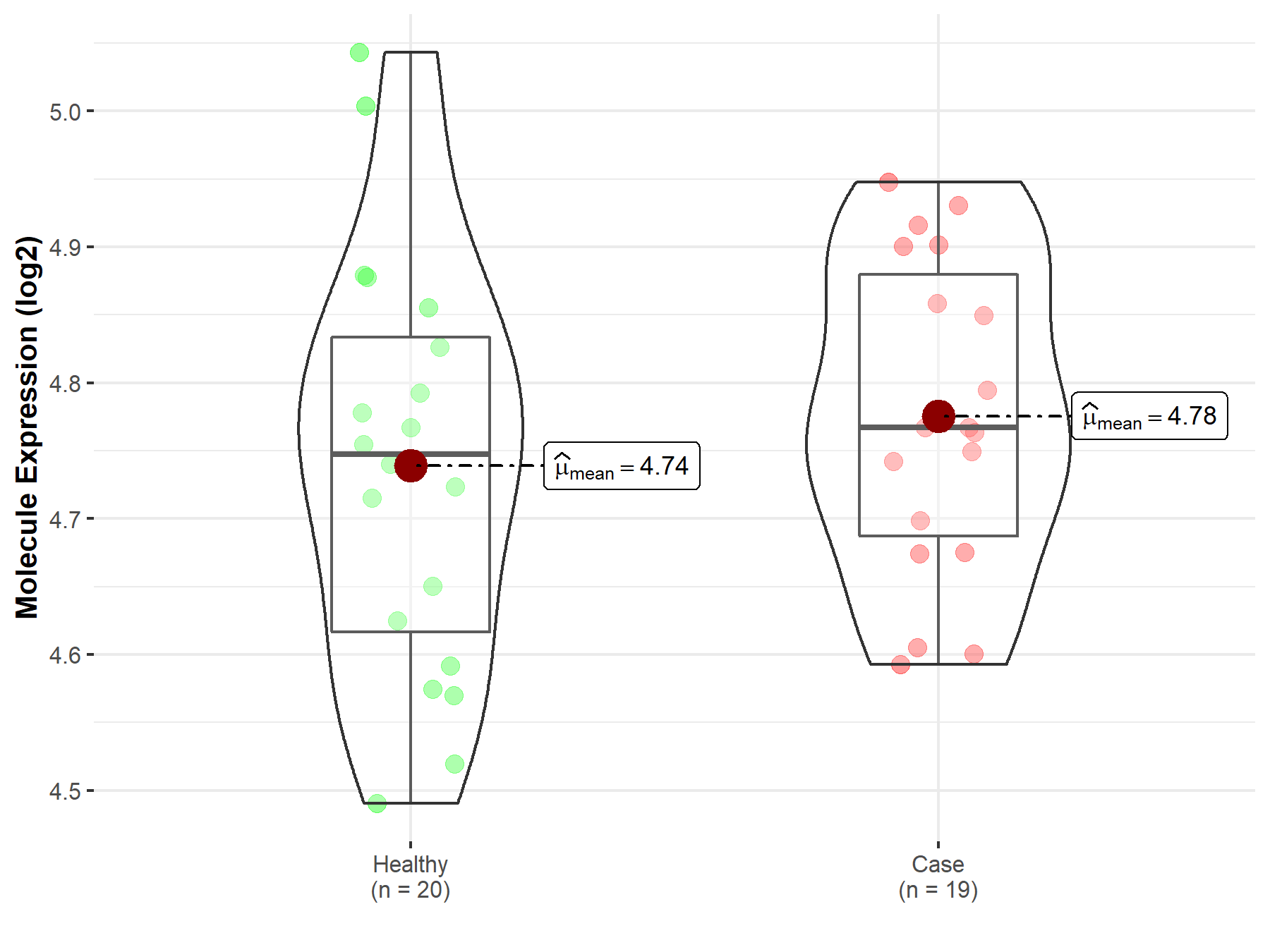
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals


|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
