Molecule Information
General Information of the Molecule (ID: Mol01296)
| Name |
Metastasis associated lung adenocarcinoma transcript 1 (MALAT1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
MALAT1
Click to Show/Hide
|
||||
| Molecule Type |
LncRNA
|
||||
| Gene Name |
MALAT1
|
||||
| Gene ID | |||||
| Location |
chr11:65497688-65506516[+]
|
||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
15 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [1] | |||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Gastric cancer [ICD-11: 2B72] | |||
| The Specified Disease | Stomach adenocarcinoma | |||
| The Studied Tissue | Stomach | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.77E-52 Fold-change: 2.18E+00 Z-score: 1.68E+01 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell autophagy | Activation | hsa04140 | |
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| BGC823 cells | Gastric | Homo sapiens (Human) | CVCL_3360 | |
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | MALAT1 acts as a competing endogenous RNA for miR23b-3p and attenuates the inhibitory effect of miR23b-3p on ATG12, leading to chemo-induced autophagy and chemoresistance in GC cells. MALAT1 regulates autophagy via ATG12. | |||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [6] | |||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | |
| SPC-A1 cells | Lung | Homo sapiens (Human) | CVCL_6955 | |
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; CCK8 assay; Colony formation assay; Flow cytometry assay | |||
| Mechanism Description | MALAT1 could alter chemo-resistance (Cisplatin, Adriamycin, Gefitinib and Paclitaxel) of NSCLC cells by targeting miR-197-3p and regulating p120-ctn expression, which might assist in improvement of chemo-therapies for NSCLC. | |||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [7] | |||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell autophagy | Activation | hsa04140 | |
| In Vitro Model | AGS cells | Gastric | Homo sapiens (Human) | CVCL_0139 |
| HGC27 cells | Gastric | Homo sapiens (Human) | CVCL_1279 | |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | LncRNA MALAT1 potentiates autophagy associated cisplatin resistance by suppressing the microRNA 30b/autophagy related gene 5 axis in gastric cancer. | |||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [1] | |||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| BGC823 cells | Gastric | Homo sapiens (Human) | CVCL_3360 | |
| Experiment for Molecule Alteration |
RT-PCR; Luciferase reporter assay; Pull down assay | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | MALAT1 acts as a competing endogenous RNA for miR23b-3p and attenuates the inhibitory effect of miR23b-3p on ATG12, leading to chemo-induced autophagy and chemoresistance in GC cells. | |||
| Disease Class: Lung cancer [ICD-11: 2C25.5] | [8] | |||
| Resistant Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell colony | Activation | hsa05200 | ||
| Cell viability | Activation | hsa05200 | ||
| STAT3 signaling pathway | Activation | hsa04550 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay; TUNEL assay | |||
| Mechanism Description | LncRNA-MALAT1 contributes to the cisplatin-resistance of lung cancer by upregulating MRP1 and MDR1 via STAT3 activation. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Liver cancer [ICD-11: 2C12.6] | [2] | |||
| Sensitive Disease | Liver cancer [ICD-11: 2C12.6] | |||
| Sensitive Drug | Fluorouracil | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Liver cancer [ICD-11: 2C12] | |||
| The Specified Disease | Cholangiocarcinoma | |||
| The Studied Tissue | Bile duct | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.43E-07 Fold-change: 2.16E+00 Z-score: 6.36E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell colony | Activation | hsa05200 | ||
| Cell viability | Activation | hsa05200 | ||
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| BEL-7402 cells | Liver | Homo sapiens (Human) | CVCL_5492 | |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| HCCLM3 cells | Liver | Homo sapiens (Human) | CVCL_6832 | |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| SMMC7721 cells | Uterus | Homo sapiens (Human) | CVCL_0534 | |
| MHCC97-L cells | Liver | Homo sapiens (Human) | CVCL_4973 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
CCK8 analysis; EdU analysis; Boyden chamber assay; Transwell assay; Flow cytometry assay | |||
| Mechanism Description | MALAT1 deficiency related increase in sensitivity of liver cancer cells was associated with regulation of NF-kB. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [1] | |||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Resistant Drug | Fluorouracil | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| BGC823 cells | Gastric | Homo sapiens (Human) | CVCL_3360 | |
| Experiment for Molecule Alteration |
RT-PCR; Luciferase reporter assay; Pull down assay | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | MALAT1 acts as a competing endogenous RNA for miR23b-3p and attenuates the inhibitory effect of miR23b-3p on ATG12, leading to chemo-induced autophagy and chemoresistance in GC cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [3] | |||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Sensitive Drug | Crizotinib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | |||
| The Specified Disease | Lung adenocarcinoma | |||
| The Studied Tissue | Lung | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.47E-03 Fold-change: -9.20E-01 Z-score: -2.68E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell colony | Inhibition | hsa05200 | ||
| Cell invasion | Inhibition | hsa05200 | ||
| Cell viability | Inhibition | hsa05200 | ||
| ULk1 signaling pathway | Inhibition | hsa04211 | ||
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| In Vivo Model | Nude mouse model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; TUNEL assay; Flow cytometry assay | |||
| Mechanism Description | Silencing of LncRNA-HOTAIR decreases drug resistance of Non-Small Cell Lung Cancer cells by inactivating autophagy via suppressing the phosphorylation of ULk1. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Glioblastoma [ICD-11: 2A00.02] | [4] | |||
| Sensitive Disease | Glioblastoma [ICD-11: 2A00.02] | |||
| Sensitive Drug | Temozolomide | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Brain cancer [ICD-11: 2A00] | |||
| The Specified Disease | Glioblastoma multiforme | |||
| The Studied Tissue | Brain | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.44E-13 Fold-change: -2.24E+00 Z-score: -7.32E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell colony | Inhibition | hsa05200 | ||
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; TUNEL assay; Flow cytometry assay | |||
| Mechanism Description | The endogenous protein level of GSk3beta and MGMT was significantly suppressed by combination of MALAT1 knockdown and miR-101 overexpression and the promoter methylation of MGMT was largely promoted by the combination of MALAT1 knockdown and miR-101 overexpression. | |||
| Disease Class: Glioblastoma [ICD-11: 2A00.02] | [18] | |||
| Sensitive Disease | Glioblastoma [ICD-11: 2A00.02] | |||
| Sensitive Drug | Temozolomide | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| U87 cells | Brain | Homo sapiens (Human) | CVCL_0022 | |
| LN-18 cells | Brain | Homo sapiens (Human) | CVCL_0392 | |
| T98G cells | Brain | Homo sapiens (Human) | CVCL_0556 | |
| U87-luc2 | Brain | Homo sapiens (Human) | CVCL_5J12 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
XTT assay; CellTiter-Glo Luminescent Cell Viability Assay | |||
| Mechanism Description | Targeted nanocomplex carrying siRNA against MALAT1 sensitizes glioblastoma to temozolomide. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Glioblastoma [ICD-11: 2A00.02] | [16], [17] | |||
| Resistant Disease | Glioblastoma [ICD-11: 2A00.02] | |||
| Resistant Drug | Temozolomide | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | miR203-TS signaling pathway | Regulation | N.A. | |
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| U87 cells | Brain | Homo sapiens (Human) | CVCL_0022 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometric analysis | |||
| Mechanism Description | LncRNA MALAT1 inhibition re-sensitized TMZ resistant cells through up-regulating miR203 and down-regulating TS expression. MALAT1 decreased the sensitivity of resistant glioma cell lines to TMZ by upregulating ZEB1. | |||
| Disease Class: Glioblastoma [ICD-11: 2A00.02] | [4] | |||
| Resistant Disease | Glioblastoma [ICD-11: 2A00.02] | |||
| Resistant Drug | Temozolomide | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell colony | Activation | hsa05200 | ||
| Cell viability | Activation | hsa05200 | ||
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
MTT assay; TUNEL assay; Flow cytometry assay | |||
| Mechanism Description | The endogenous protein level of GSk3beta and MGMT was significantly suppressed by combination of MALAT1 knockdown and miR-101 overexpression and the promoter methylation of MGMT was largely promoted by the combination of MALAT1 knockdown and miR-101 overexpression. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Cervical cancer [ICD-11: 2C77.0] | [5] | |||
| Sensitive Disease | Cervical cancer [ICD-11: 2C77.0] | |||
| Sensitive Drug | Metformin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Cervical cancer [ICD-11: 2C77] | |||
| The Specified Disease | Cervical & endocervical cancer | |||
| The Studied Tissue | Cervix Uteri | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.06E-02 Fold-change: -2.59E+00 Z-score: -3.07E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | |
| Cell migration | Inhibition | hsa04670 | ||
| In Vitro Model | Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 |
| Siha cells | Cervix uteri | Homo sapiens (Human) | CVCL_0032 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Wound-healing assay; Transwell assay | |||
| Mechanism Description | Metformin inhibits the expression of MALAT1 and upregulates miR-142-3p in cervical cancer cells. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Prostate cancer [ICD-11: 2C82.0] | [9] | |||
| Resistant Disease | Prostate cancer [ICD-11: 2C82.0] | |||
| Resistant Drug | Docetaxel | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell invasion | Activation | hsa05200 | ||
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | DU-145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 |
| PC3 cells | Prostate | Homo sapiens (Human) | CVCL_0035 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Scratch Wound healing assay; Transwell Invasion assay; Flow cytometry assay | |||
| Mechanism Description | Knockdown of MALAT1 in DTX-resistant PCa cells up-regulated miR-145-5p as well as suppressed AkAP12 expression, further inhibited cell viability and induced apoptosis. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [6] | |||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Resistant Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | |
| SPC-A1 cells | Lung | Homo sapiens (Human) | CVCL_6955 | |
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; CCK8 assay; Colony formation assay; Flow cytometry assay | |||
| Mechanism Description | MALAT1 could alter chemo-resistance (Cisplatin, Adriamycin, Gefitinib and Paclitaxel) of NSCLC cells by targeting miR-197-3p and regulating p120-ctn expression, which might assist in improvement of chemo-therapies for NSCLC. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lung cancer [ICD-11: 2C25.5] | [10] | |||
| Resistant Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Resistant Drug | Gefitinib | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Activation | hsa05200 | |
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Colony formation assay | |||
| Mechanism Description | LncRNA MALAT1 promoted the proliferation and gefitinib resistance of lung cancer cells by sponging miR-200a, which regulates expression of ZEB1 in the A549 cells. | |||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [6] | |||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Resistant Drug | Gefitinib | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | |
| SPC-A1 cells | Lung | Homo sapiens (Human) | CVCL_6955 | |
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; CCK8 assay; Colony formation assay; Flow cytometry assay | |||
| Mechanism Description | MALAT1 could alter chemo-resistance (Cisplatin, Adriamycin, Gefitinib and Paclitaxel) of NSCLC cells by targeting miR-197-3p and regulating p120-ctn expression, which might assist in improvement of chemo-therapies for NSCLC. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [11] | |||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Sensitive Drug | Gefitinib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell colony | Inhibition | hsa05200 | ||
| Cell viability | Inhibition | hsa05200 | ||
| STAT3 signaling pathway | Inhibition | hsa04550 | ||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 |
| In Vivo Model | BALB/c nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | PPI might down-regulate MALAT1 expression and inactivate STAT3 signaling pathway and could serve a promising therapeutic agent for gefitinib-resistant NSCLC. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Pancreatic cancer [ICD-11: 2C10.3] | [12] | |||
| Resistant Disease | Pancreatic cancer [ICD-11: 2C10.3] | |||
| Resistant Drug | Gemcitabine | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell migration | Activation | hsa04670 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | AsPC-1 cells | Pancreas | Homo sapiens (Human) | CVCL_0152 |
| CFPAC1 cells | Pancreas | Homo sapiens (Human) | CVCL_1119 | |
| Experiment for Molecule Alteration |
qPCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | MALAT-1 could increase the proportion of pancreatic CSCs, maintain self-renewing capacity, decrease the chemosensitivity to anticancer drugs, and accelerate tumor angiogenesis in vitro, and promote tumorigenicity of pancreatic cancer cells in vivo. The underlying mechanisms may involve in increased expression of self-renewal related factors Sox2. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Chronic myeloid leukemia [ICD-11: 2A20.0] | [13] | |||
| Resistant Disease | Chronic myeloid leukemia [ICD-11: 2A20.0] | |||
| Resistant Drug | Imatinib | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | KG-1 cells | Bone marrow | Homo sapiens (Human) | CVCL_0374 |
| K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | LncRNA MALAT1 promotes cell proliferation and imatinib resistance by suppressing miR-328 in chronic myelogenous leukemia. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [14] | |||
| Resistant Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Resistant Drug | Insulin recombinant | |||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 |
| Experiment for Molecule Alteration |
qRT-PCR; RNA pull down assay; RIP experiments assay; Co-IP; Western bloting analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | MALAT1 binding competes with the interaction between sirtuin1 (SIRT1) and DBC1, which then releases SIRT1 and enhances its deacetylation activity. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [15] | |||
| Resistant Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Resistant Drug | Oxaliplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | HT29 Cells | Colon | Homo sapiens (Human) | CVCL_A8EZ |
| SW480 cells | Colon | Homo sapiens (Human) | CVCL_0546 | |
| FHC cells | Colon | Homo sapiens (Human) | CVCL_3688 | |
| SW620 cells | Colon | Homo sapiens (Human) | CVCL_0547 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Boyden chambers cell migration and invasion assays | |||
| Mechanism Description | MALAT1 tethers EZH2 to CDH1 promoter and suppresses miR218 during oxaliplatin treatment, which finally promotes colorectal cancer cell EMT, metastasis, and chemoresistance. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [6] | |||
| Resistant Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Resistant Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | |
| SPC-A1 cells | Lung | Homo sapiens (Human) | CVCL_6955 | |
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; CCK8 assay; Colony formation assay; Flow cytometry assay | |||
| Mechanism Description | MALAT1 could alter chemo-resistance (Cisplatin, Adriamycin, Gefitinib and Paclitaxel) of NSCLC cells by targeting miR-197-3p and regulating p120-ctn expression, which might assist in improvement of chemo-therapies for NSCLC. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Prostate cancer [ICD-11: 2C82.0] | [14] | |||
| Resistant Disease | Prostate cancer [ICD-11: 2C82.0] | |||
| Resistant Drug | Quercetin | |||
| Molecule Alteration | Down-regulation | Expression |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | PC-3 cells | Prostate | Homo sapiens (Human) | CVCL_0035 |
| In Vivo Model | Male BALB/c nude mice xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Overexpression assay; qRT-PCR; Western bloting analysis; Immunohistochemistry analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | MALAT1 Overexpression in PC cells resulted in the resistance against quercetin treatment. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [1] | |||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Sensitive Drug | Vincristine | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| BGC823 cells | Gastric | Homo sapiens (Human) | CVCL_3360 | |
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | MALAT1 acts as a competing endogenous RNA for miR23b-3p and attenuates the inhibitory effect of miR23b-3p on ATG12, leading to chemo-induced autophagy and chemoresistance in GC cells. MALAT1 promotes autophagy-associated chemoresistance of GC cells via sequestration of miR23b-3p. | |||
Investigative Drug(s)
4 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Type 2 diabetes mellitus [ICD-11: 5A11.0] | [14] | |||
| Resistant Disease | Type 2 diabetes mellitus [ICD-11: 5A11.0] | |||
| Resistant Drug | D-Glucose | |||
| Molecule Alteration | Up-regulation | Interaction |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | HK-2 cells | Kidney | Homo sapiens (Human) | CVCL_0302 |
| In Vivo Model | Male C57BL/6 mice | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR; Western bloting analysis; ELISA assay; RIP experiments assay; RNA pull down assay; Dual luciferase assay | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | LncRNA MALAT1 interacts with transcription factor Foxo1 to represses SIRT1 transcription in high glucose incubated HK-2 cells, which promotes high glucose-induced HK-2 cells injury. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Ischemic cardiovascular disease [ICD-11: BA51.0] | [14] | |||
| Resistant Disease | Ischemic cardiovascular disease [ICD-11: BA51.0] | |||
| Resistant Drug | Heliox | |||
| Molecule Alteration | Up-regulation | Interaction |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | HCAECs | N.A. | N.A. | N.A. |
| In Vivo Model | Hind limb ischemia rat model | Rattus norvegicus | ||
| Experiment for Molecule Alteration |
Overexpression assay; Knockdown assay; Luciferase assay | |||
| Experiment for Drug Resistance |
CellTiter-Glo assay; dual-luciferase reporter assay | |||
| Mechanism Description | Hyperbaric oxygen boosts long noncoding RNA MALAT1 exosome secretion to suppress microRNA-92a expression in therapeutic angiogenesis. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Periodontitis [ICD-11: DA0C.0] | [14] | |||
| Resistant Disease | Periodontitis [ICD-11: DA0C.0] | |||
| Resistant Drug | Lipopolysaccharide | |||
| Molecule Alteration | Up-regulation | Interaction |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Primary human gingival fibroblast cells | N.A. | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
qRT-PCR; Luciferase assay; RIP experiments assay; Mimic; Overexpression assay; Enzyme-linked immunosorbent assay; Western bloting analysis | |||
| Mechanism Description | LncRNA MALAT1 regulates inflammatory cytokine production in lipopolysaccharide-stimulated human gingival fibroblasts through sponging miR-20a and activating TLR4 pathway. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Colorectal cancer [ICD-11: 2B91.1] | [19] | |||
| Resistant Disease | Colorectal cancer [ICD-11: 2B91.1] | |||
| Resistant Drug | Oxymatrine | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell invasion | Activation | hsa05200 | ||
| Cell migration | Activation | hsa04670 | ||
| In Vitro Model | HT29 Cells | Colon | Homo sapiens (Human) | CVCL_A8EZ |
| SW480 cells | Colon | Homo sapiens (Human) | CVCL_0546 | |
| Experiment for Molecule Alteration |
RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | Chronic oxymatrine treatment induces resistance and epithelial-mesenchymal transition through targeting the long non-coding RNA MALAT1 in colorectal cancer cells. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Brain | |
| The Specified Disease | Brain lower grade glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.50E-164; Fold-change: 1.11E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
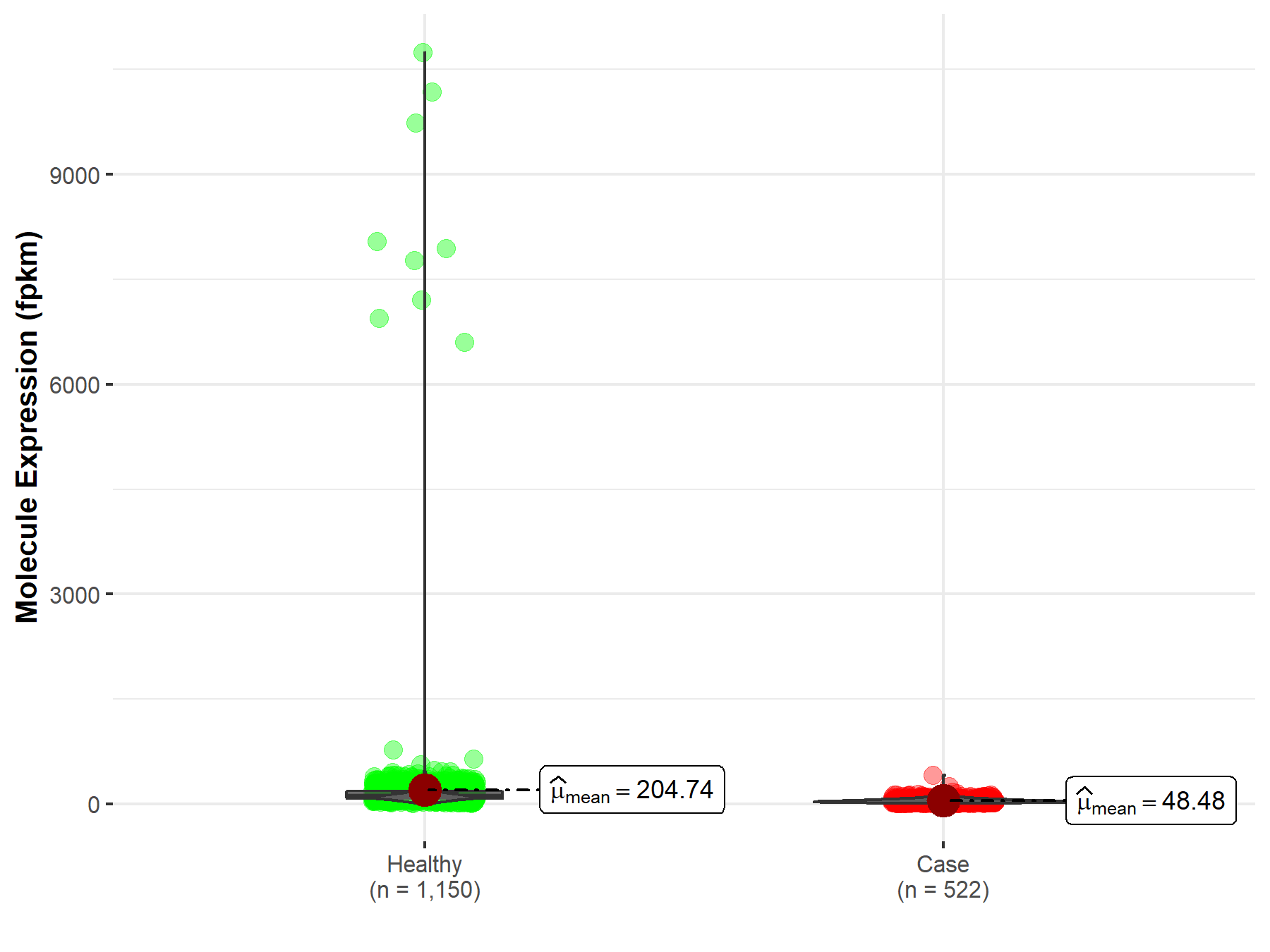
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brain | |
| The Specified Disease | Glioblastoma multiforme | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.89E-93; Fold-change: 1.38E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
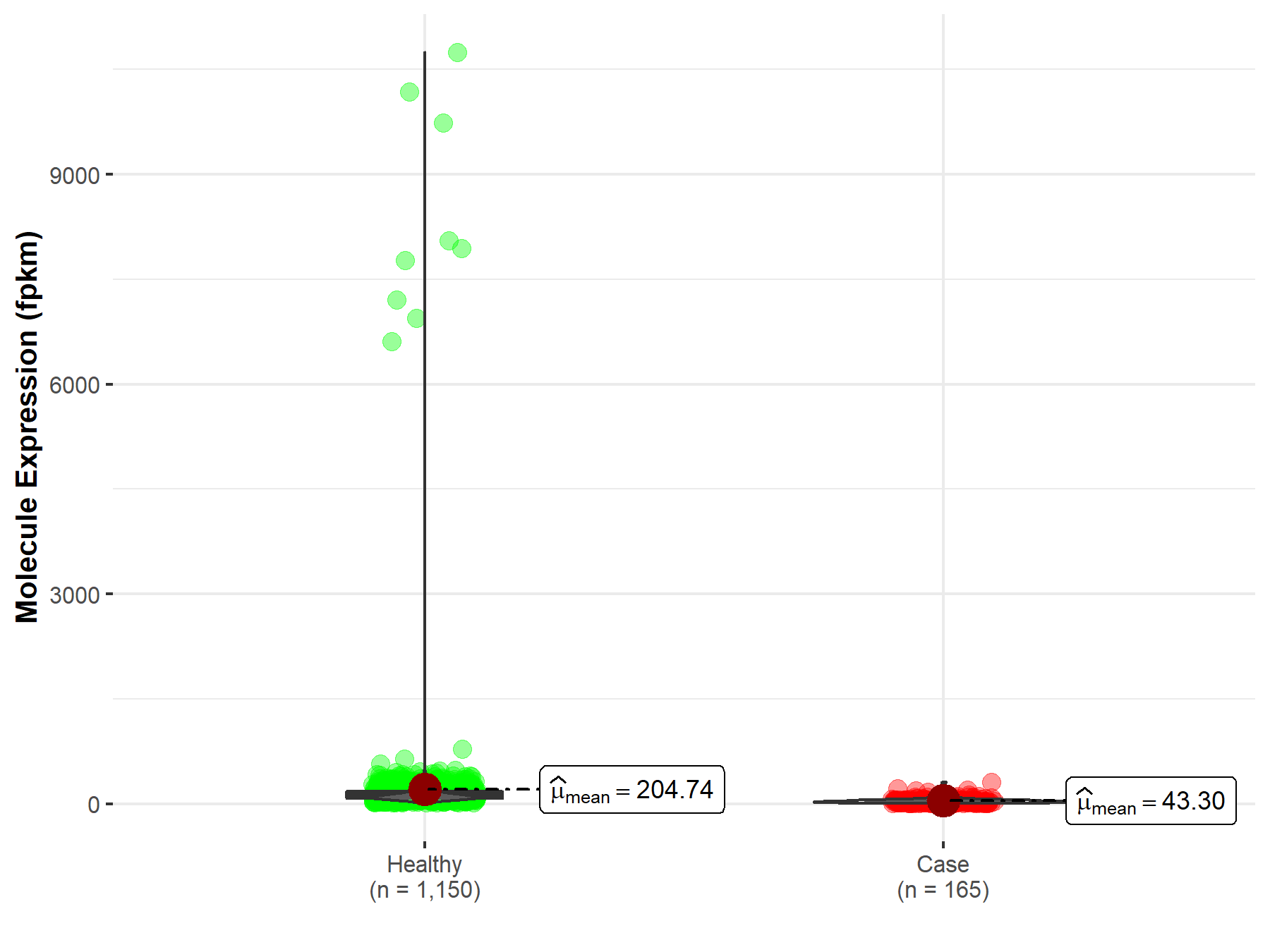
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Stomach | |
| The Specified Disease | Stomach adenocarcinoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.85E-80; Fold-change: -1.26E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
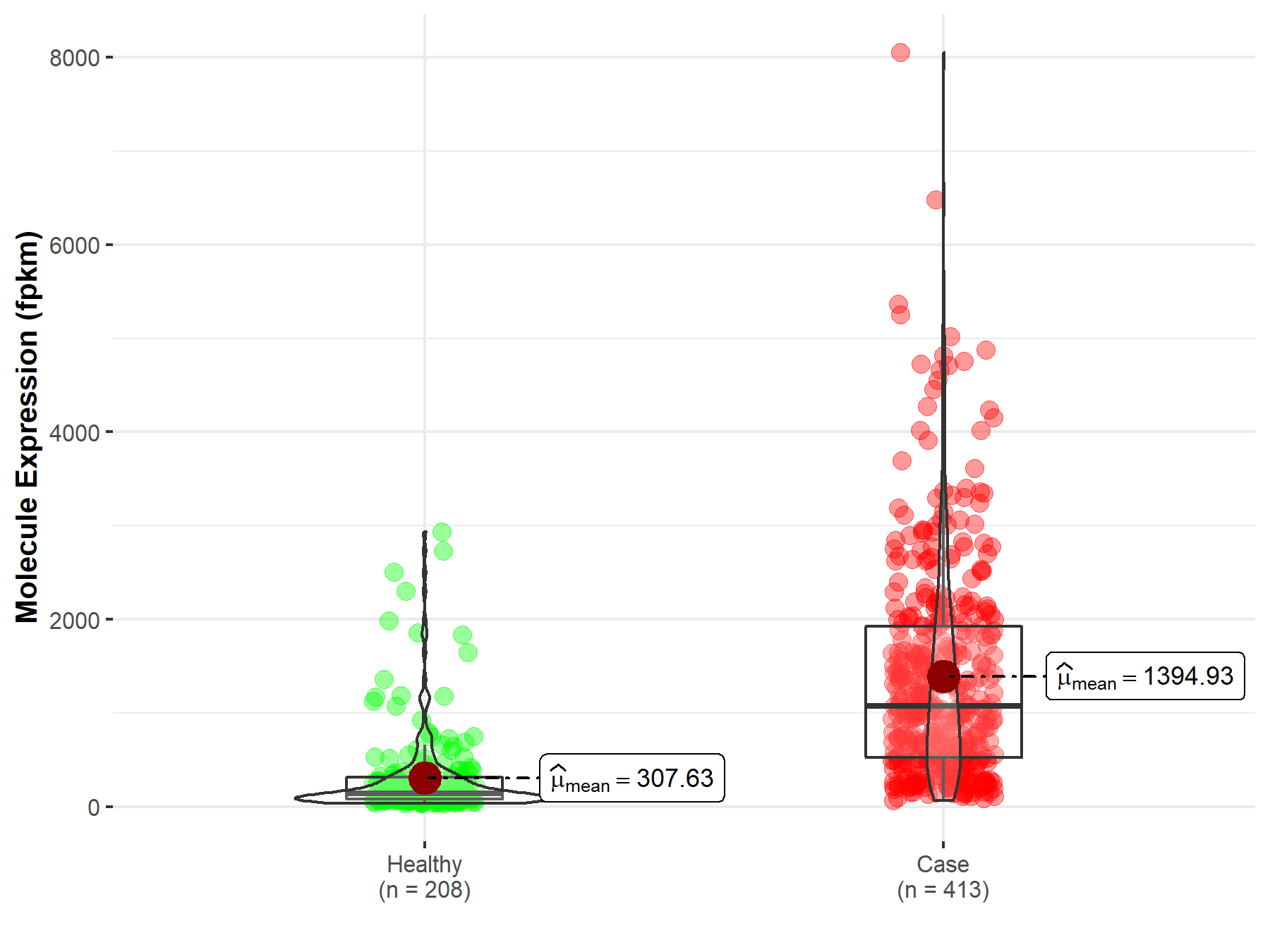
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Rectum | |
| The Specified Disease | Rectum adenocarcinoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.83E-02; Fold-change: -4.90E-02 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Pancreas | |
| The Specified Disease | Pancreatic adenocarcinoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.07E-38; Fold-change: 9.09E-02 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
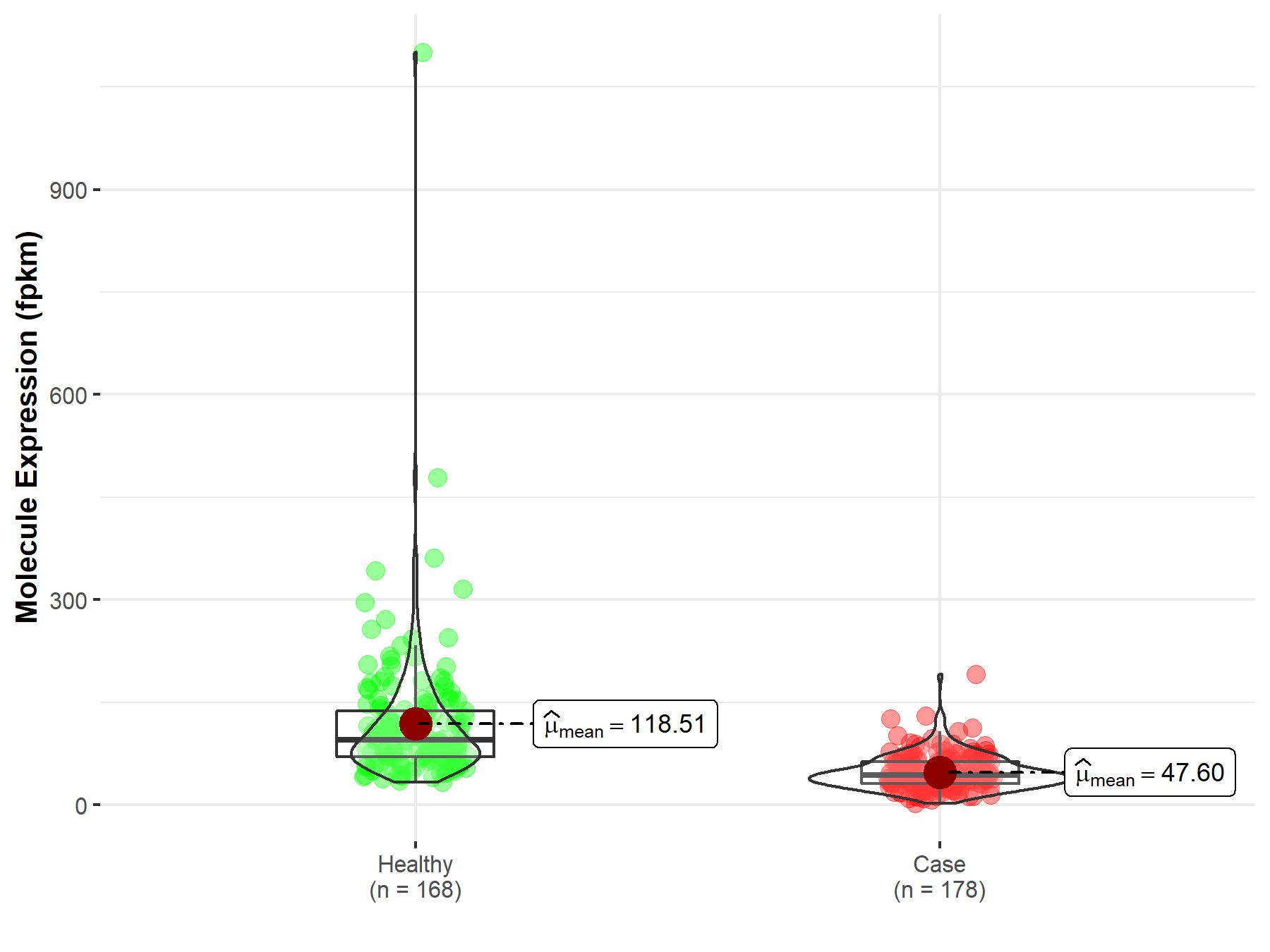
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Bile duct | |
| The Specified Disease | Cholangiocarcinoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.79E-08; Fold-change: -2.05E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Liver | |
| The Specified Disease | Liver hepatocellular carcinoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 7.49E-13; Fold-change: 8.56E-02 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Lung | |
| The Specified Disease | Lung adenocarcinoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.37E-48; Fold-change: 1.07E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
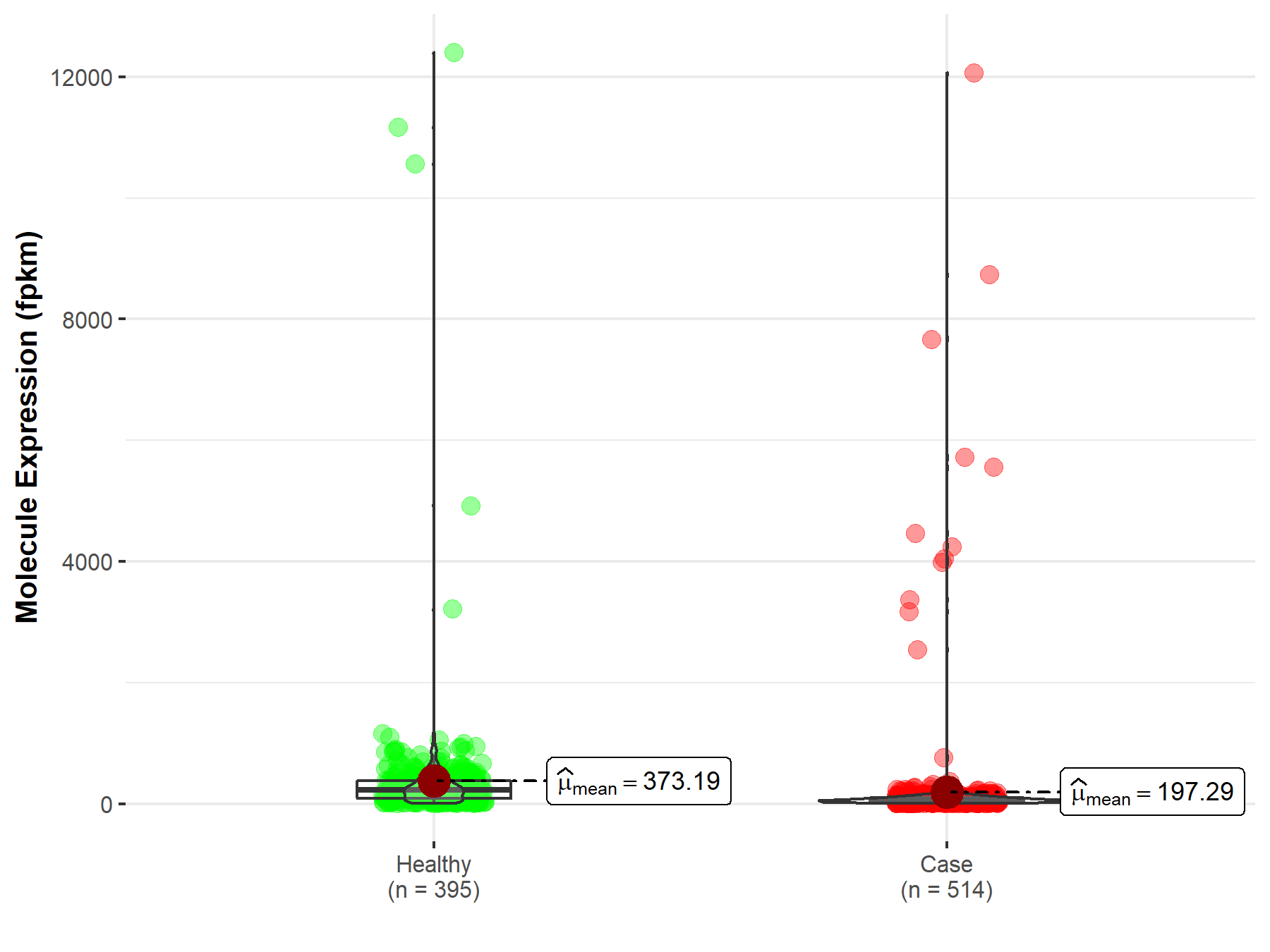
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Lung | |
| The Specified Disease | Lung squamous cell carcinoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.11E-90; Fold-change: 1.42E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
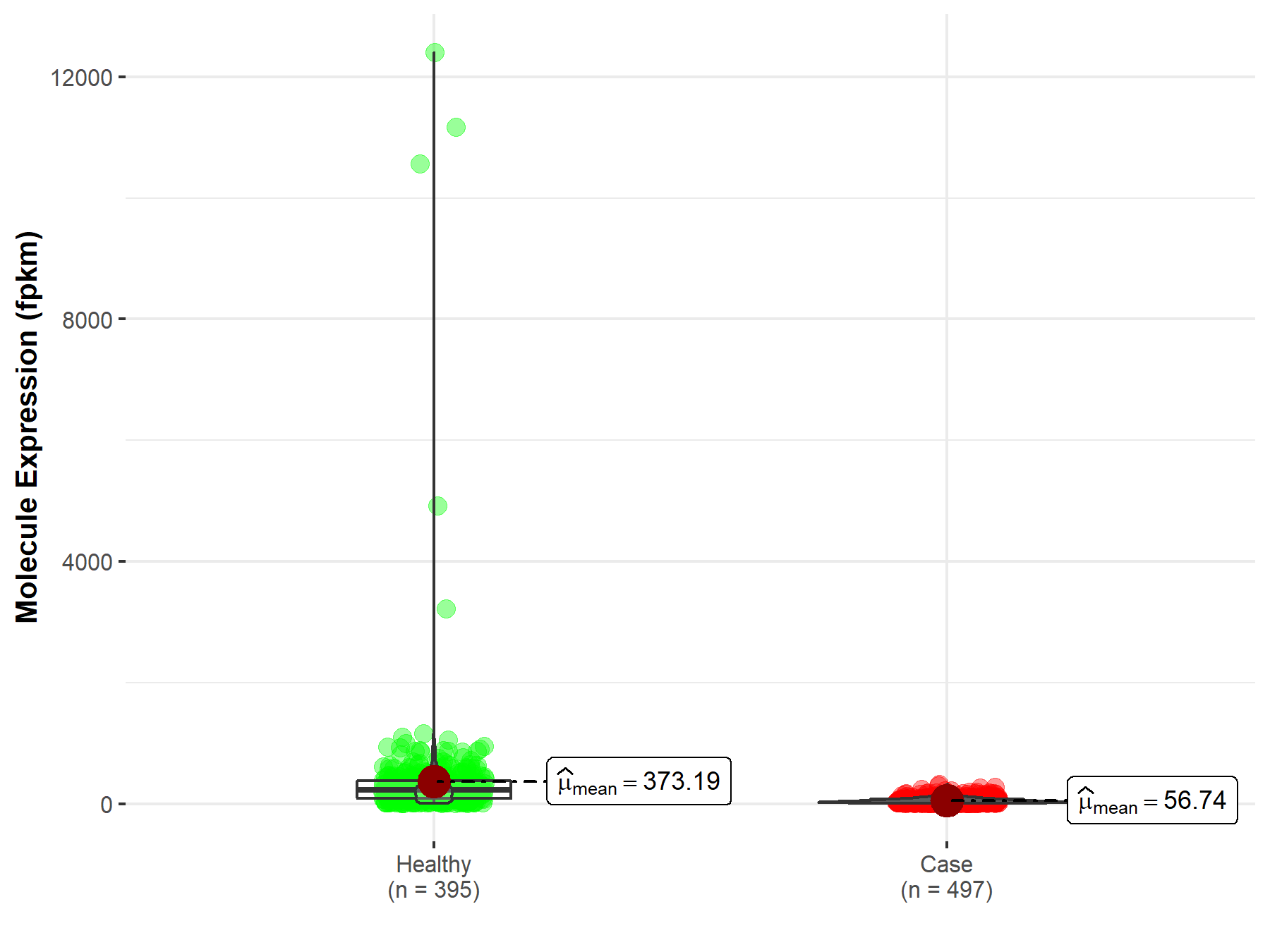
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Cervix uteri | |
| The Specified Disease | Cervical and endocervical cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.23E-16; Fold-change: 1.41E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Prostate | |
| The Specified Disease | Prostate adenocarcinoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.12E-33; Fold-change: 1.05E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
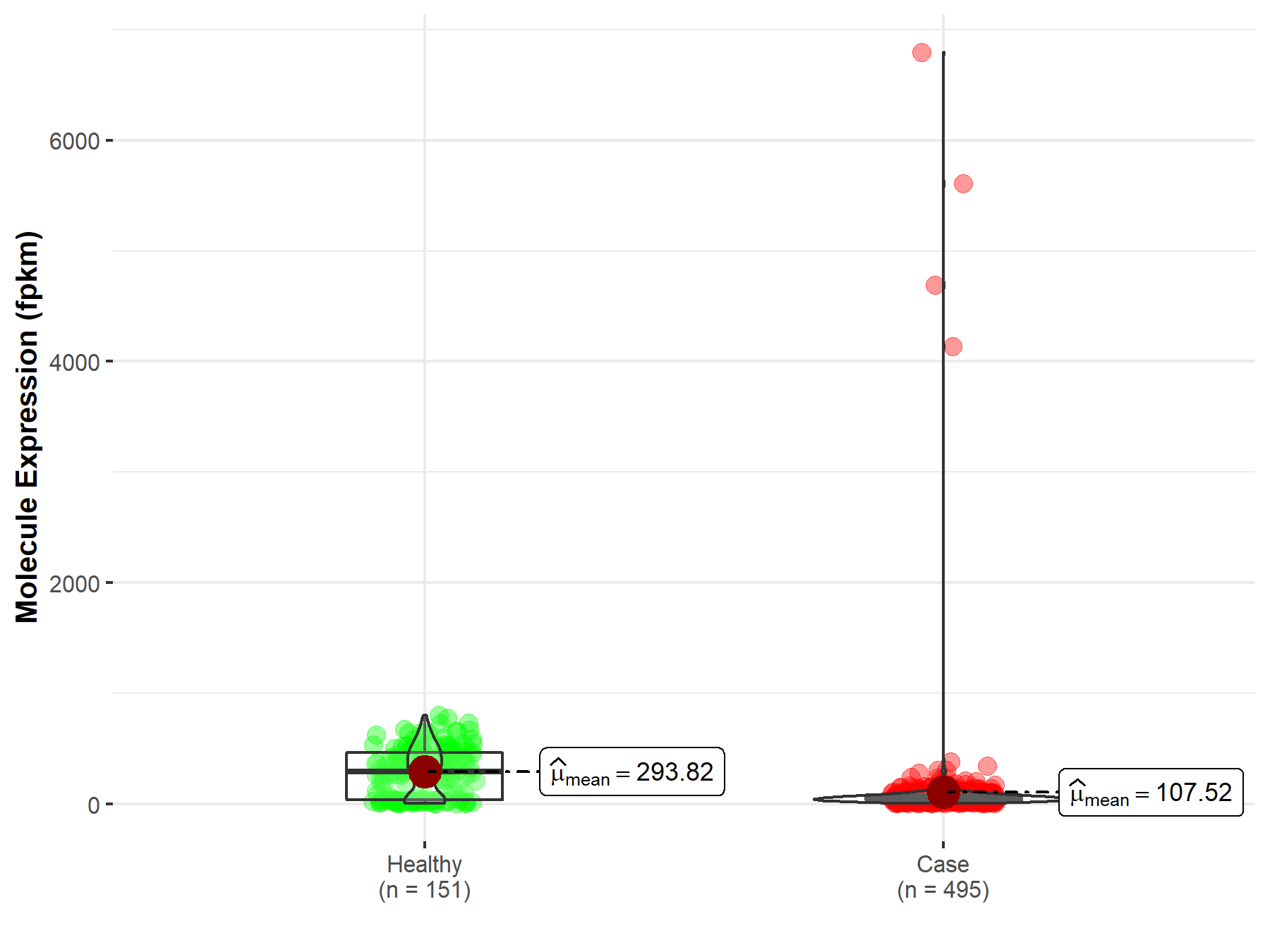
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

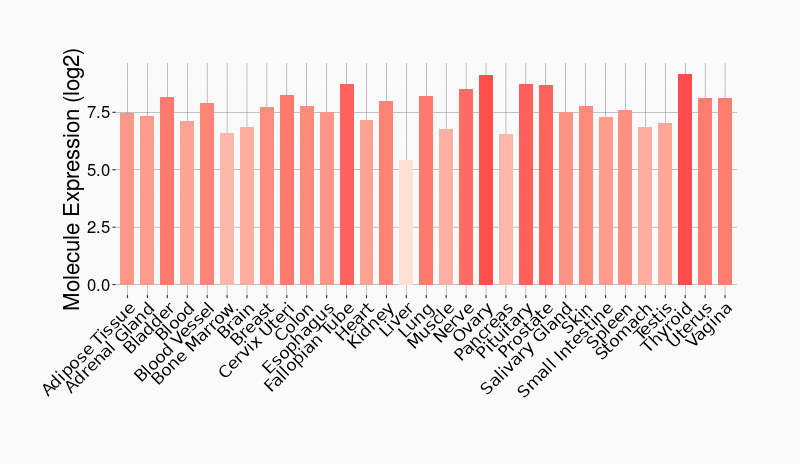
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
