Molecule Information
General Information of the Molecule (ID: Mol00717)
| Name |
Zinc finger E-box-binding homeobox 1 (ZEB1)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
NIL-2-A zinc finger protein; Negative regulator of IL2; Transcription factor 8; TCF-8; AREB6; TCF8
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
ZEB1
|
||||
| Gene ID | |||||
| Location |
chr10:31318495-31529814[+]
|
||||
| Sequence |
MADGPRCKRRKQANPRRNNVTNYNTVVETNSDSDDEDKLHIVEEESVTDAADCEGVPEDD
LPTDQTVLPGRSSEREGNAKNCWEDDRKEGQEILGPEAQADEAGCTVKDDECESDAENEQ NHDPNVEEFLQQQDTAVIFPEAPEEDQRQGTPEASGHDENGTPDAFSQLLTCPYCDRGYK RFTSLKEHIKYRHEKNEDNFSCSLCSYTFAYRTQLERHMTSHKSGRDQRHVTQSGCNRKF KCTECGKAFKYKHHLKEHLRIHSGEKPYECPNCKKRFSHSGSYSSHISSKKCISLIPVNG RPRTGLKTSQCSSPSLSASPGSPTRPQIRQKIENKPLQEQLSVNQIKTEPVDYEFKPIVV ASGINCSTPLQNGVFTGGGPLQATSSPQGMVQAVVLPTVGLVSPISINLSDIQNVLKVAV DGNVIRQVLENNQANLASKEQETINASPIQQGGHSVISAISLPLVDQDGTTKIIINYSLE QPSQLQVVPQNLKKENPVATNSCKSEKLPEDLTVKSEKDKSFEGGVNDSTCLLCDDCPGD INALPELKHYDLKQPTQPPPLPAAEAEKPESSVSSATGDGNLSPSQPPLKNLLSLLKAYY ALNAQPSAEELSKIADSVNLPLDVVKKWFEKMQAGQISVQSSEPSSPEPGKVNIPAKNND QPQSANANEPQDSTVNLQSPLKMTNSPVLPVGSTTNGSRSSTPSPSPLNLSSSRNTQGYL YTAEGAQEEPQVEPLDLSLPKQQGELLERSTITSVYQNSVYSVQEEPLNLSCAKKEPQKD SCVTDSEPVVNVIPPSANPINIAIPTVTAQLPTIVAIADQNSVPCLRALAANKQTILIPQ VAYTYSTTVSPAVQEPPLKVIQPNGNQDERQDTSSEGVSNVEDQNDSDSTPPKKKMRKTE NGMYACDLCDKIFQKSSSLLRHKYEHTGKRPHECGICKKAFKHKHHLIEHMRLHSGEKPY QCDKCGKRFSHSGSYSQHMNHRYSYCKREAEERDSTEQEEAGPEILSNEHVGARASPSQG DSDERESLTREEDEDSEKEEEEEDKEMEELQEEKECEKPQGDEEEEEEEEEVEEEEVEEA ENEGEEAKTEGLMKDDRAESQASSLGQKVGESSEQVSEEKTNEA Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Acts as a transcriptional repressor. Inhibits interleukin-2 (IL-2) gene expression. Enhances or represses the promoter activity of the ATP1A1 gene depending on the quantity of cDNA and on the cell type. Represses E-cadherin promoter and induces an epithelial-mesenchymal transition (EMT) by recruiting SMARCA4/BRG1. Represses BCL6 transcription in the presence of the corepressor CTBP1. Positively regulates neuronal differentiation. Represses RCOR1 transcription activation during neurogenesis. Represses transcription by binding to the E box (5'-CANNTG-3'). Promotes tumorigenicity by repressing stemness-inhibiting microRNAs.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
15 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Endometrial cancer [ICD-11: 2C76.1] | [1] | |||
| Sensitive Disease | Endometrial cancer [ICD-11: 2C76.1] | |||
| Sensitive Drug | Epothilone B | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Endometrial cancer [ICD-11: 2C76] | |||
| The Specified Disease | Endometrial cancer | |||
| The Studied Tissue | Uterus | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.45E-43 Fold-change: -1.36E+00 Z-score: -1.98E+01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell migration | Inhibition | hsa04670 | |
| In Vitro Model | Hec50 cells | Endometrium | Homo sapiens (Human) | CVCL_2929 |
| Experiment for Molecule Alteration |
Immunoblotting analysis | |||
| Experiment for Drug Resistance |
ELISA assay | |||
| Mechanism Description | Low or absent miR-200c results in aberrant expression of ZEB1 and consequent repression of E-cadherin. Reinstatement of miR-200c to such cells restores E-cadherin and dramatically reduces migration and invasion. One such gene, class IIIbeta-tubulin (TUBB3), which encodes a tubulin isotype normally found only in neuronal cells, is a direct target of miR-200c. Restoration of miR-200c increases sensitivity to microtubule-targeting agents by up to 85%. Since expression of TUBB3 is a common mechanism of resistance to microtubule-binding chemotherapeutic agents in many types of solid tumors, the ability of miR-200c to restore chemosensitivity to such agents may be explained by its ability to reduce TUBB3. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Endometrial cancer [ICD-11: 2C76.1] | [1] | |||
| Sensitive Disease | Endometrial cancer [ICD-11: 2C76.1] | |||
| Sensitive Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Endometrial cancer [ICD-11: 2C76] | |||
| The Specified Disease | Endometrial cancer | |||
| The Studied Tissue | Uterus | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.45E-43 Fold-change: -1.36E+00 Z-score: -1.98E+01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell migration | Inhibition | hsa04670 | |
| In Vitro Model | Hec50 cells | Endometrium | Homo sapiens (Human) | CVCL_2929 |
| Experiment for Molecule Alteration |
Immunoblotting analysis | |||
| Experiment for Drug Resistance |
ELISA assay | |||
| Mechanism Description | Low or absent miR-200c results in aberrant expression of ZEB1 and consequent repression of E-cadherin. Reinstatement of miR-200c to such cells restores E-cadherin and dramatically reduces migration and invasion. One such gene, class IIIbeta-tubulin (TUBB3), which encodes a tubulin isotype normally found only in neuronal cells, is a direct target of miR-200c. Restoration of miR-200c increases sensitivity to microtubule-targeting agents by up to 85%. Since expression of TUBB3 is a common mechanism of resistance to microtubule-binding chemotherapeutic agents in many types of solid tumors, the ability of miR-200c to restore chemosensitivity to such agents may be explained by its ability to reduce TUBB3. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Ovarian cancer [ICD-11: 2C73.0] | [22] | |||
| Resistant Disease | Ovarian cancer [ICD-11: 2C73.0] | |||
| Resistant Drug | Paclitaxel | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | SkOV3 cells | Ovary | Homo sapiens (Human) | CVCL_0532 |
| Experiment for Molecule Alteration |
Immunoblotting analysis | |||
| Experiment for Drug Resistance |
TUNEL assay | |||
| Mechanism Description | miR-106a and miR-591 have important roles in conferring PTX resistance to ovarian cancer cells. Modulation of these microRNAs resensitizes PTX-resistant cancer cells by targeting BCL10, caspase-7, and ZEB1. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Endometrial cancer [ICD-11: 2C76.1] | [1] | |||
| Sensitive Disease | Endometrial cancer [ICD-11: 2C76.1] | |||
| Sensitive Drug | Vincristine | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Endometrial cancer [ICD-11: 2C76] | |||
| The Specified Disease | Endometrial cancer | |||
| The Studied Tissue | Uterus | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.45E-43 Fold-change: -1.36E+00 Z-score: -1.98E+01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell migration | Inhibition | hsa04670 | |
| In Vitro Model | Hec50 cells | Endometrium | Homo sapiens (Human) | CVCL_2929 |
| Experiment for Molecule Alteration |
Immunoblotting analysis | |||
| Experiment for Drug Resistance |
ELISA assay | |||
| Mechanism Description | Low or absent miR-200c results in aberrant expression of ZEB1 and consequent repression of E-cadherin. Reinstatement of miR-200c to such cells restores E-cadherin and dramatically reduces migration and invasion. One such gene, class IIIbeta-tubulin (TUBB3), which encodes a tubulin isotype normally found only in neuronal cells, is a direct target of miR-200c. Restoration of miR-200c increases sensitivity to microtubule-targeting agents by up to 85%. Since expression of TUBB3 is a common mechanism of resistance to microtubule-binding chemotherapeutic agents in many types of solid tumors, the ability of miR-200c to restore chemosensitivity to such agents may be explained by its ability to reduce TUBB3. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [2] | |||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Sensitive Drug | Intedanib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | |||
| The Specified Disease | Non-small cell lung cancer | |||
| The Studied Tissue | Lung tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.74E-02 Fold-change: -1.61E-01 Z-score: -2.40E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | PC3 cells | Prostate | Homo sapiens (Human) | CVCL_0035 |
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |
| NCI-H1650 cells | Lung | Homo sapiens (Human) | CVCL_1483 | |
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |
| NCI-H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 | |
| PC-14 cells | Lung | Homo sapiens (Human) | CVCL_1640 | |
| EBC-1 cells | Lung | Homo sapiens (Human) | CVCL_2891 | |
| LC-1/sq cells | Lung | Homo sapiens (Human) | CVCL_3008 | |
| LC-2/ad cells | Lung | Homo sapiens (Human) | CVCL_1373 | |
| Lk-2 cells | Lung | Homo sapiens (Human) | CVCL_1377 | |
| NCI-HCC827 cells | Lung | Homo sapiens (Human) | CVCL_2063 | |
| PC-1 cells | Pancreas | Homo sapiens (Human) | CVCL_S978 | |
| PC-10 cells | Lung | Homo sapiens (Human) | CVCL_7088 | |
| QG56 cells | Lung | Homo sapiens (Human) | CVCL_6943 | |
| RERF-LCkJ cells | Lung | Homo sapiens (Human) | CVCL_1654 | |
| SQ5 cells | Lung | Homo sapiens (Human) | CVCL_8273 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | miR-200b and miR-141 associated with epithelial-mesenchymal transition (EMT) are predictive biomarkers and therapeutic targets of nintedanib in NSCLC cells. nintedanib inhibited EMT and reversed the resistance to EGFR-TkI with TGF-beta-induced EMT through miR-200 family induction in NSCLC cells. low expression of miR-200b and miR-141, resulting in high level of ZEB1 and low level of E-cadherin, was associated with the resistance to nintedanib in NSCLC cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lung cancer [ICD-11: 2C25.5] | [3] | |||
| Sensitive Disease | Lung cancer [ICD-11: 2C25.5] | |||
| Sensitive Drug | Crizotinib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | |||
| The Specified Disease | Lung cancer | |||
| The Studied Tissue | Lung tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.74E-02 Fold-change: -1.61E-01 Z-score: -2.40E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | |
| NCI-2228 cells | Lung | Homo sapiens (Human) | CVCL_1543 | |
| NCI-2228/CRI cells | Lung | Homo sapiens (Human) | CVCL_1543 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR200c regulates crizotinib-resistant ALk-positive lung cancer cells by reversing epithelial-mesenchymal transition via targeting ZEB1. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [4] | |||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Sensitive Drug | Gefitinib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | |||
| The Specified Disease | Non-small cell lung cancer | |||
| The Studied Tissue | Lung tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.74E-02 Fold-change: -1.61E-01 Z-score: -2.40E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell migration | Inhibition | hsa04670 | |
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 |
| PC9-ZD cells | Lung | Homo sapiens (Human) | CVCL_V337 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Annexin-V/PI assay; Wound healing assay | |||
| Mechanism Description | miR200c enhances sensitivity of drug-resistant non-small cell lung cancer to gefitinib by suppression of PI3k/Akt signaling pathway and inhibites cell migration via targeting ZEB1. | |||
|
|
||||
| Disease Class: Non-small cell lung cancer [ICD-11: 2C25.Y] | [10] | |||
| Sensitive Disease | Non-small cell lung cancer [ICD-11: 2C25.Y] | |||
| Sensitive Drug | Gefitinib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Lung cancer [ICD-11: 2C25] | |||
| The Specified Disease | Lung cancer | |||
| The Studied Tissue | Lung tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.29E-79 Fold-change: -2.78E-01 Z-score: -2.61E+01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| MEK/ERK signaling pathway | Regulation | N.A. | ||
| PI3K/AKT signaling pathway | Regulation | N.A. | ||
| In Vitro Model | H1975 cells | Lung | Homo sapiens (Human) | CVCL_1511 |
| A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 | |
| H460 cells | Lung | Homo sapiens (Human) | CVCL_0459 | |
| H1299 cells | Lung | Homo sapiens (Human) | CVCL_0060 | |
| PC9 cells | Lung | Homo sapiens (Human) | CVCL_B260 | |
| H23 cells | Lung | Homo sapiens (Human) | CVCL_1547 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Ectopic expression of miR-200c resulted in partial restoration of gefitinib sensitivity in NSCLC cells with ZEB1 downrerulating. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Glioblastoma [ICD-11: 2A00.02] | [5] | |||
| Resistant Disease | Glioblastoma [ICD-11: 2A00.02] | |||
| Resistant Drug | Temozolomide | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Brain cancer [ICD-11: 2A00] | |||
| The Specified Disease | Glioblastoma | |||
| The Studied Tissue | Nervous tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.13E-14 Fold-change: 1.21E+00 Z-score: 1.64E+01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| U87 cells | Brain | Homo sapiens (Human) | CVCL_0022 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | MALAT1 decreased the sensitivity of resistant glioma cell lines to TMZ by upregulating ZEB1. | |||
|
|
||||
| Disease Class: Malignant glioma [ICD-11: 2A00.2] | [6] | |||
| Resistant Disease | Malignant glioma [ICD-11: 2A00.2] | |||
| Resistant Drug | Temozolomide | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Brain cancer [ICD-11: 2A00] | |||
| The Specified Disease | Brain cancer | |||
| The Studied Tissue | Nervous tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.67E-109 Fold-change: 1.73E-01 Z-score: 2.47E+01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell viability | Activation | hsa05200 | ||
| Wnt/beta-catenin signaling pathway | Activation | hsa04310 | ||
| In Vitro Model | U251 cells | Brain | Homo sapiens (Human) | CVCL_0021 |
| M059J cells | Brain | Homo sapiens (Human) | CVCL_0400 | |
| Experiment for Molecule Alteration |
Western blot analysis; RNAi assay | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Silencing of H19 decreases chemoresistance through suppressing EMT via the Wnt/beta-Catenin pathway. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastric carcinoma [ICD-11: 2B72.Z] | [7] | |||
| Sensitive Disease | Gastric carcinoma [ICD-11: 2B72.Z] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Gastric cancer [ICD-11: 2B72] | |||
| The Specified Disease | Gastric cancer | |||
| The Studied Tissue | Gastric tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.04E-01 Fold-change: -4.33E-02 Z-score: -1.36E-01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | |
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | SGC7901 cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | microRNA-574-3p regulates epithelial mesenchymal transition and cisplatin resistance via targeting ZEB1 in human gastric carcinoma cells. | |||
| Disease Class: Prostate cancer [ICD-11: 2C82.0] | [8] | |||
| Sensitive Disease | Prostate cancer [ICD-11: 2C82.0] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Prostate cancer [ICD-11: 2C82] | |||
| The Specified Disease | Prostate cancer | |||
| The Studied Tissue | Prostate | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.99E-03 Fold-change: -1.37E-01 Z-score: -3.47E+00 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | |
| In Vitro Model | DU-145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 |
| LNCaP cells | Prostate | Homo sapiens (Human) | CVCL_0395 | |
| HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 | |
| Experiment for Molecule Alteration |
RT-PCR; Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-128 binded to the 3'UTR of ZEB1 and inhibited its expression. And ZEB1 (+) PCa chemoresistance and invasion, while miR-128 could reverse that by down-regulated ZEB1. These indicated that miR-128-mediated sensitizing chemoresistance and inhibiting invasion of PCa cells by directly targeting ZEB1. | |||
| Disease Class: Epithelial ovarian cancer [ICD-11: 2B5D.0] | [14] | |||
| Sensitive Disease | Epithelial ovarian cancer [ICD-11: 2B5D.0] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | SkOV3 cells | Ovary | Homo sapiens (Human) | CVCL_0532 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Colony formation assay; Flow cytometric apoptosis assay | |||
| Mechanism Description | Down-regulation of miR429 contributes to the development of drug resistance in epithelial ovarian cancer by targeting ZEB1. | |||
|
|
||||
| Disease Class: Nasopharyngeal carcinoma [ICD-11: 2B6B.0] | [13] | |||
| Sensitive Disease | Nasopharyngeal carcinoma [ICD-11: 2B6B.0] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | HNE1 cells | Nasopharynx | Homo sapiens (Human) | CVCL_0308 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | The upregulation of miR-139-5p significantly increases DDP-induced apoptosis in NPC cells and modulates ZEB1 expression. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Cervical cancer [ICD-11: 2C77.0] | [11] | |||
| Resistant Disease | Cervical cancer [ICD-11: 2C77.0] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell proliferation | Activation | hsa05200 | ||
| Cell viability | Activation | hsa05200 | ||
| HOXD/AS1/miR130a-3p/ZEB1 signaling pathway | Regulation | N.A. | ||
| In Vitro Model | Hela cells | Cervix uteri | Homo sapiens (Human) | CVCL_0030 |
| Caski cells | Uterus | Homo sapiens (Human) | CVCL_1100 | |
| Experiment for Molecule Alteration |
Western blot analysis; RIP assay; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay; Transwell assay | |||
| Mechanism Description | HOXD-AS1 enhanced chemoresistance of cisplatin-resistant CC cells by modulating miR-130a-3p/ZEB1 axis expression. | |||
|
|
||||
| Disease Class: Osteosarcoma [ICD-11: 2B51.0] | [12] | |||
| Resistant Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell viability | Activation | hsa05200 | ||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometry assay | |||
| Mechanism Description | Overexpression of ZEB1 reversed the miR-340-induced alleviation of chemoresistance in drug-resistant OS cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [9] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Breast cancer [ICD-11: 2C60] | |||
| The Specified Disease | Breast cancer | |||
| The Studied Tissue | Breast tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.98E-29 Fold-change: -1.63E-01 Z-score: -1.24E+01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | |
| p53 signaling pathway | Activation | hsa04115 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| BT549 cells | Breast | Homo sapiens (Human) | CVCL_1092 | |
| HCC70 cells | Breast | Homo sapiens (Human) | CVCL_1270 | |
| Hs-578T cells | Breast | Homo sapiens (Human) | CVCL_0332 | |
| MDA-MB-361 cells | Breast | Homo sapiens (Human) | CVCL_0620 | |
| CAMA-1 cells | Breast | Homo sapiens (Human) | CVCL_1115 | |
| MCF-10-2A cells | Breast | Homo sapiens (Human) | CVCL_3743 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Celltiter-blue cell viability assay | |||
| Mechanism Description | The up-regulation of the miR-200b and miR-200c diminishes EMT by directly targeting the transcriptional repressor ZEB1 leading to up-regulation of E-cadherin. Restoration of E-cadherin expression increases the sensitivity of cancer cells to chemotherapeutic agents. Disruption of ZEB1-histone deacetylase repressor complexes and down-regulation of histone deacetylase, in particular SIRT1, positively affect the p53 apoptotic pathway leading to the increased sensitivity of breast cancer cells to chemotherapy and radiotherapy. | |||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [18] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell viability | Inhibition | hsa05200 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| MDA-MB-468 cells | Breast | Homo sapiens (Human) | CVCL_0419 | |
| MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Overexpression of miR-708-3p dramatically inhibits breast cancer cell lung metastasis and the expression of ZEB1, CDH2 and vimentin was significantly decreased in miR-708-3p-overexpressing cells at both the mRNA and protein levels compared to that in vector control cells. | |||
|
|
||||
| Disease Class: Adrenocortical carcinoma [ICD-11: 2D11.0] | [19] | |||
| Sensitive Disease | Adrenocortical carcinoma [ICD-11: 2D11.0] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | H295R cells | Kidney | Homo sapiens (Human) | CVCL_0458 |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
Cell Growth Assay; Flow cytometry assay | |||
| Mechanism Description | Restoration of miR-431 in vitro decreased the half maximal inhibitory concentrations of doxorubicin and mitotane, with markedly increased apoptosis via downregulating ZEB1. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [17] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | PTEN/AKT signaling pathway | Activation | hsa05235 | |
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MCF-7/ADR cells | Breast | Homo sapiens (Human) | CVCL_1452 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | The loss of miRNA-200c correlates with the acquired resistance of breast cancer cells to ADR,the loss of miRNA-200c correlated with decreased levels of E-cadherin and PTEN, and increased levels of ZEB1 and phospho-Akt (p-Akt) in ADR-resistant breast cancer cells (MCF-7/ADR cells). miRNA-200c inhibited Akt signaling through its effects on E-cadherin and PTEN, resulting in the inhibition of ADR resistance in breast cancer cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Prostate cancer [ICD-11: 2C82.0] | [15], [16] | |||
| Sensitive Disease | Prostate cancer [ICD-11: 2C82.0] | |||
| Sensitive Drug | Docetaxel | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell viability | Inhibition | hsa05200 | ||
| ZEB1 signaling pathway | Inhibition | hsa05215 | ||
| In Vitro Model | DU-145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 |
| PC3 cells | Prostate | Homo sapiens (Human) | CVCL_0035 | |
| PrEC cells | Prostate | Homo sapiens (Human) | CVCL_0061 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometric analysis | |||
| Mechanism Description | miR27b and miR34a enhance docetaxel sensitivity of prostate cancer cells through inhibiting epithelial-to-mesenchymal transition by targeting ZEB1. And microRNA-204 modulates chemosensitivity and apoptosis of prostate cancer cells by targeting ZEB1. Suppression of ZEB1 could effectively improve miR204 deficiency-triggered chemoresistance in PC cells. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Lung adenocarcinoma [ICD-11: 2C25.0] | [20] | |||
| Resistant Disease | Lung adenocarcinoma [ICD-11: 2C25.0] | |||
| Resistant Drug | Etoposide | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | A549 cells | Lung | Homo sapiens (Human) | CVCL_0023 |
| Experiment for Molecule Alteration |
Western blot assay | |||
| Experiment for Drug Resistance |
Cell viability assay | |||
| Mechanism Description | M. arginini?and?M. salivarium, promoted the initiation of EMT and simultaneous suppression of the p53 tumor suppressor in A549 lung cancer cells. This led to an increase of cancer cell motility, resistance to the antitumor drug etoposide concomitantly with decreased autophagy. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Pancreatic cancer [ICD-11: 2C10.3] | [21] | |||
| Sensitive Disease | Pancreatic cancer [ICD-11: 2C10.3] | |||
| Sensitive Drug | Gemcitabine | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MIA PaCa-2 cells | Pancreas | Homo sapiens (Human) | CVCL_0428 |
| PANC-1 cells | Pancreas | Homo sapiens (Human) | CVCL_0480 | |
| AsPC-1 cells | Pancreas | Homo sapiens (Human) | CVCL_0152 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
WST-1 assay | |||
| Mechanism Description | Re-expression of miR-200 in gemcitabine-resistant cells showed partial reversal of EMT characteristics as documented by increased expression of E-cadherin and decreased expression of vimentin, ZEB1, and slug. These results suggest that miR-200 family regulates the expression of ZEB1, slug, E-cadherin, and vimentin and that the re-expression of miR-200 could be useful for the reversal of EMT phenotype to mesenchymal-epithelial transition (MET). re-expression of miR-200 by transfection studies or treatment of gemcitabine-resistant cells with either DIM or isoflavone resulted in the down-regulation of ZEB1, slug, and vimentin, which was consistent with morphological reversal of EMT phenotype leading to epithelial morphology. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast adenocarcinoma [ICD-11: 2C60.1] | [19] | |||
| Sensitive Disease | Breast adenocarcinoma [ICD-11: 2C60.1] | |||
| Sensitive Drug | Mitotane | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | H295R cells | Kidney | Homo sapiens (Human) | CVCL_0458 |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
Cell Growth Assay; Flow cytometry assay | |||
| Mechanism Description | Restoration of miR-431 in vitro decreased the half maximal inhibitory concentrations of doxorubicin and mitotane, with markedly increased apoptosis via downregulating ZEB1. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [23] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Tamoxifen | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| HEK293T cells | Kidney | Homo sapiens (Human) | CVCL_0063 | |
| MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | In MDA-MB-231 cells, down-regulated LncRNA-ROR could inhibit the EMT of breast cancer cells and enhance the sensibility of breast cancer cells to tamoxifen by increasing miR205 expression and suppressing the expressions of ZEB1 and ZEB2. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: HER2 positive breast cancer [ICD-11: 2C60.8] | [24] | |||
| Resistant Disease | HER2 positive breast cancer [ICD-11: 2C60.8] | |||
| Resistant Drug | Trastuzumab | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Inhibition | hsa04210 | |
| Cell invasion | Activation | hsa05200 | ||
| Cell migration | Activation | hsa04670 | ||
| Cell proliferation | Activation | hsa05200 | ||
| TGF-beta signaling pathway | Regulation | N.A. | ||
| In Vitro Model | SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | Lnc-ATB is up-regulated in TR breast cancer tissues and TR SkBR-3 cells. Up-regulation of lnc-ATB account for the trastuzumab resistance and high invasiveness of TR SkBR-3 cells. miR-200c is down-regulated and inverse correlated with lnc-ATB in TR breast cancer tissues and TR SkBR-3 cells. Lnc-ATB functions as a ceRNA by competitively biding miR-200c in TR SkBR-3 cells. Lnc-ATB up-regulates and positive correlates with ZEB1 and ZNF217 levels. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [25] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Trastuzumab | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell invasion | Inhibition | hsa05200 | |
| TGF-Beta/ZEB1 signaling pathway | Inhibition | hsa04350 | ||
| In Vitro Model | SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-200c, which was the most significantly downregulated miRNA in trastuzumab-resistant cells, restored trastuzumab sensitivity and suppressed invasion of breast cancer cells by concurrently targeting ZNF217, a transcriptional activator of TGF-beta, and ZEB1, a known mediator of TGF-beta signaling. Restoration of miR-200c, silencing of ZEB1 or ZNF217 or blockade of TGF-beta signaling increased trastuzumab sensitivity and suppressed invasiveness of breast cancer cells. | |||
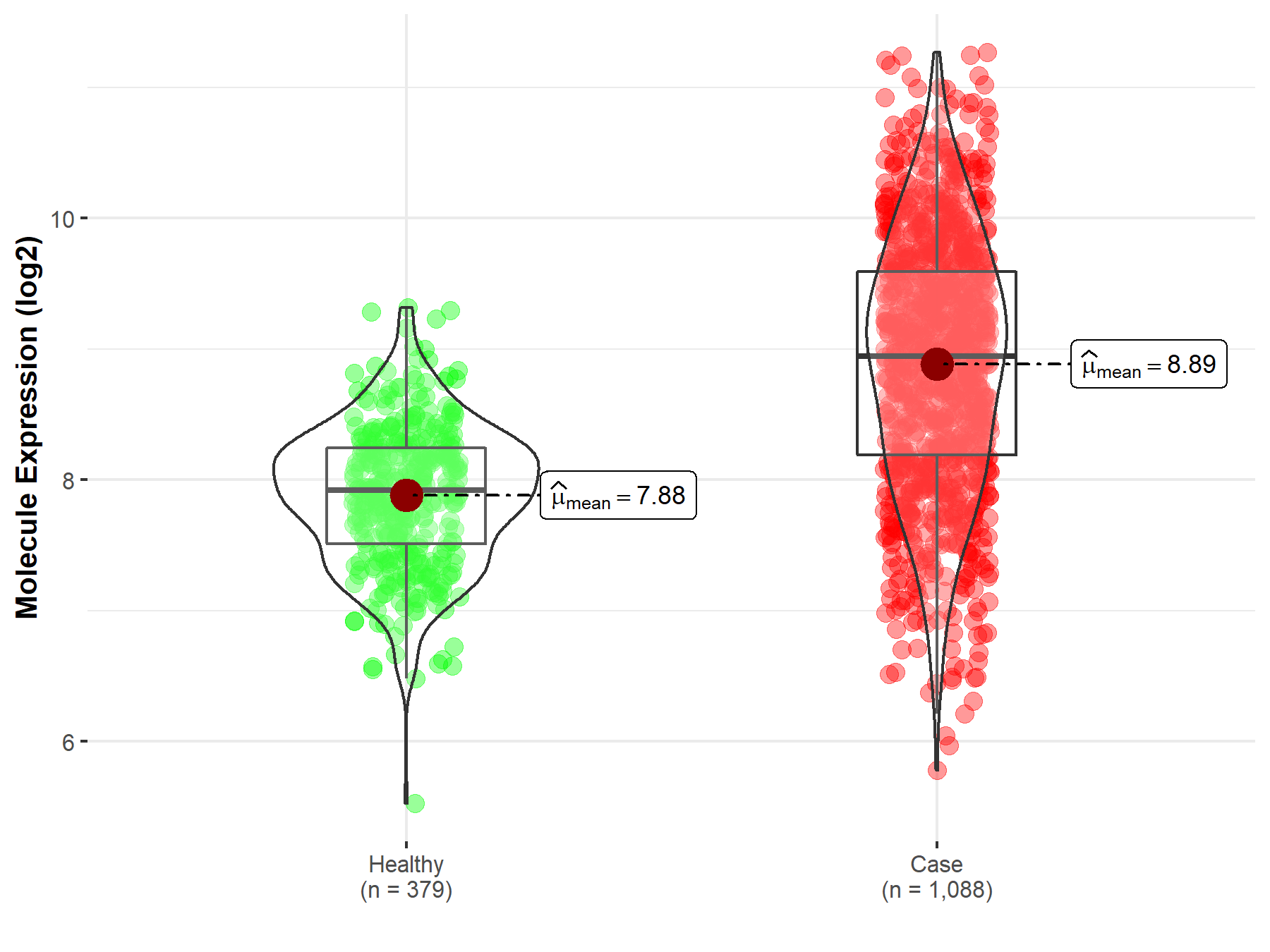
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Nervous tissue | |
| The Specified Disease | Brain cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.67E-109; Fold-change: 1.02E+00; Z-score: 1.89E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.43E-01; Fold-change: 1.71E+00; Z-score: 2.86E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| The Studied Tissue | White matter | |
| The Specified Disease | Glioma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.34E-03; Fold-change: 1.10E+00; Z-score: 1.25E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Brainstem tissue | |
| The Specified Disease | Neuroectodermal tumor | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.81E-09; Fold-change: 1.54E+00; Z-score: 4.10E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Gastric tissue | |
| The Specified Disease | Gastric cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.04E-01; Fold-change: 8.80E-01; Z-score: 3.54E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.80E-07; Fold-change: 3.85E-01; Z-score: 1.05E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
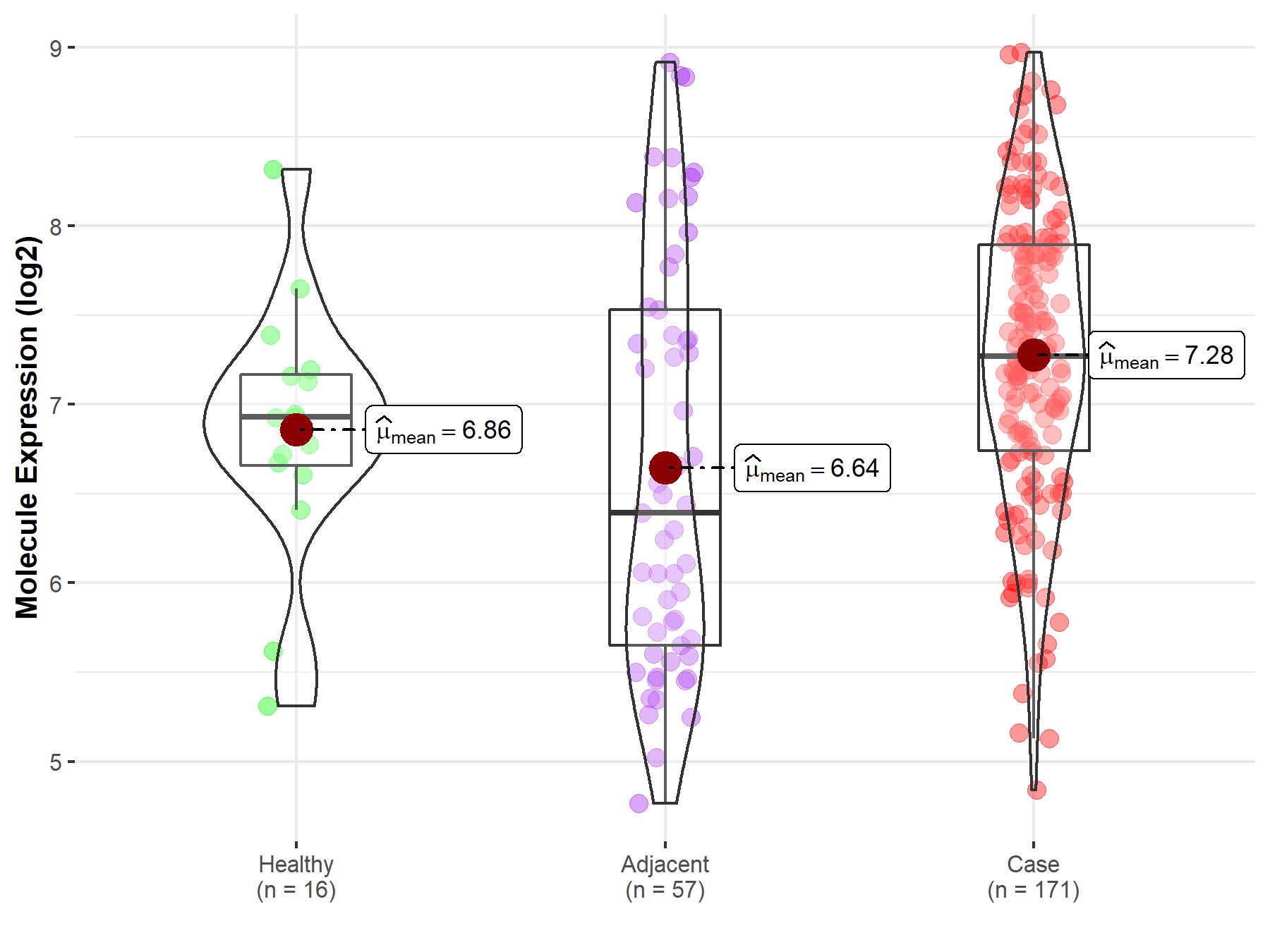
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Pancreas | |
| The Specified Disease | Pancreatic cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.99E-02; Fold-change: 3.41E-01; Z-score: 4.78E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.53E-04; Fold-change: 8.77E-01; Z-score: 7.64E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
|
Click to View the Clearer Original Diagram |
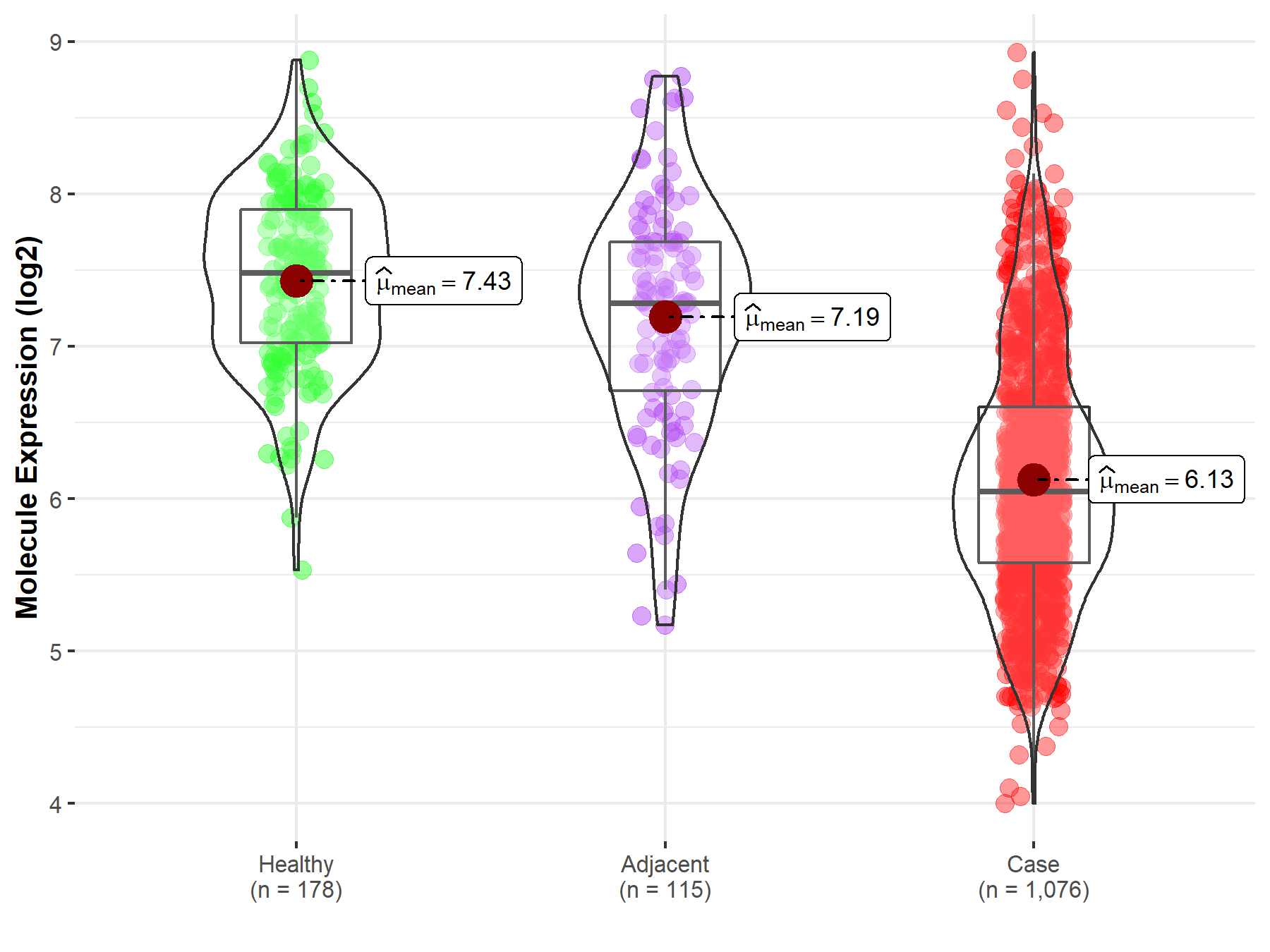
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Lung | |
| The Specified Disease | Lung cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.29E-79; Fold-change: -1.43E+00; Z-score: -2.47E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 6.19E-29; Fold-change: -1.24E+00; Z-score: -1.62E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.98E-29; Fold-change: -9.32E-01; Z-score: -1.01E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.35E-04; Fold-change: -8.67E-01; Z-score: -8.24E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
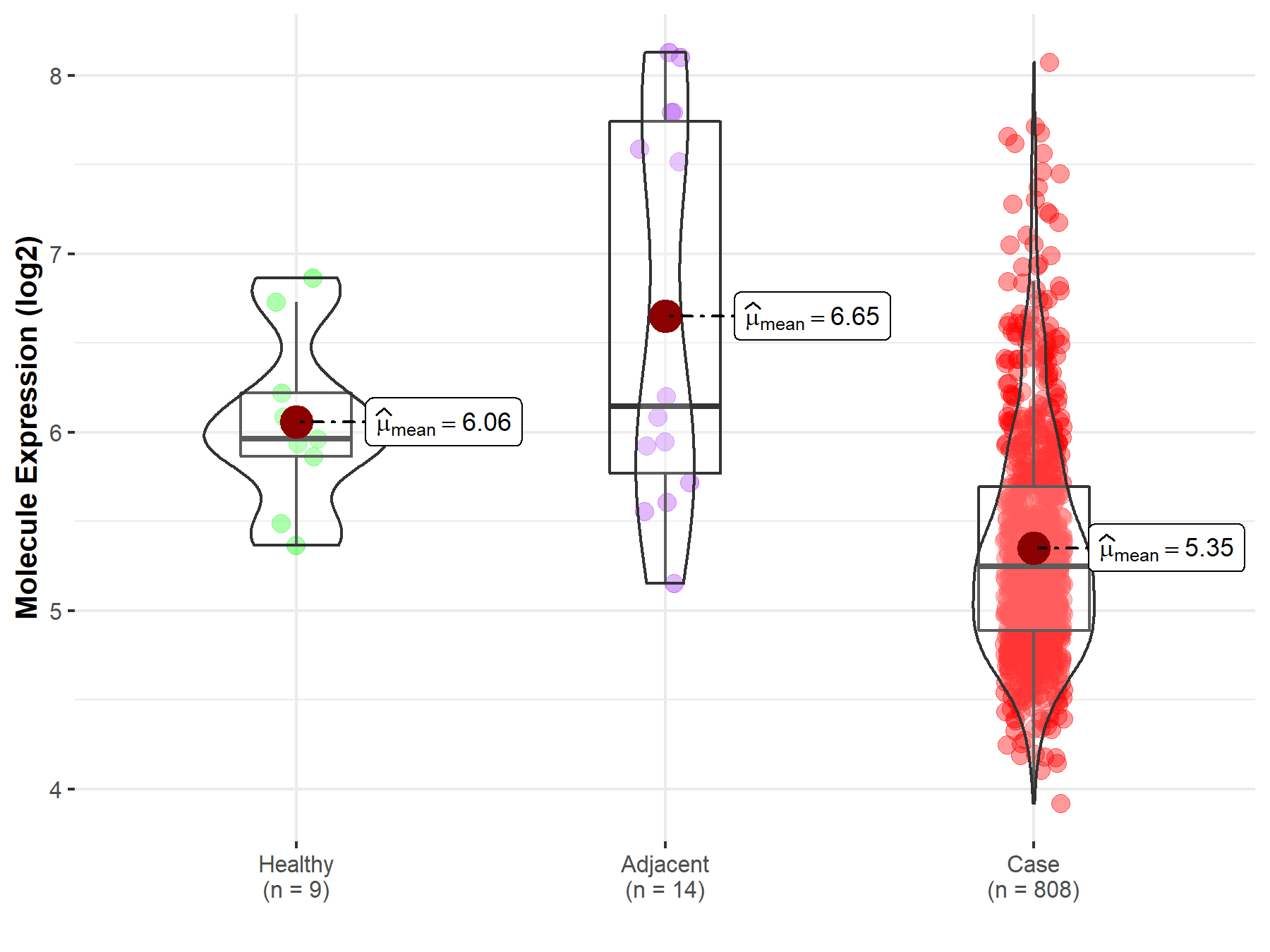
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Ovary | |
| The Specified Disease | Ovarian cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.75E-03; Fold-change: -7.16E-01; Z-score: -1.44E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 6.45E-04; Fold-change: -8.97E-01; Z-score: -8.23E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Cervix uteri | |
| The Specified Disease | Cervical cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.70E-02; Fold-change: 1.01E-01; Z-score: 3.69E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Prostate | |
| The Specified Disease | Prostate cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.99E-03; Fold-change: -6.94E-01; Z-score: -8.54E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Adrenal cortex | |
| The Specified Disease | Adrenocortical carcinoma | |
| The Expression Level of Disease Section Compare with the Other Disease Section | p-value: 1.09E-02; Fold-change: 1.45E-01; Z-score: 5.54E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in tissue other than the diseased tissue of patients
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
