Molecule Information
General Information of the Molecule (ID: Mol00242)
| Name |
ATM interactor (ATMIN)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
ATM/ATR-substrate CHK2-interacting zinc finger protein; ASCIZ; Zinc finger protein 822; KIAA0431; ZNF822
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
ATMIN
|
||||
| Gene ID | |||||
| Location |
chr16:81035842-81047350[+]
|
||||
| Sequence |
MAASEAAAAAGSAALAAGARAVPAATTGAAAAASGPWVPPGPRLRGSRPRPAGATQQPAV
PAPPAGELIQPSVSELSRAVRTNILCTVRGCGKILPNSPALNMHLVKSHRLQDGIVNPTI RKDLKTGPKFYCCPIEGCPRGPERPFSQFSLVKQHFMKMHAEKKHKCSKCSNSYGTEWDL KRHAEDCGKTFRCTCGCPYASRTALQSHIYRTGHEIPAEHRDPPSKKRKMENCAQNQKLS NKTIESLNNQPIPRPDTQELEASEIKLEPSFEDSCGSNTDKQTLTTPPRYPQKLLLPKPK VALVKLPVMQFSVMPVFVPTADSSAQPVVLGVDQGSATGAVHLMPLSVGTLILGLDSEAC SLKESLPLFKIANPIAGEPISTGVQVNFGKSPSNPLQELGNTCQKNSISSINVQTDLSYA SQNFIPSAQWATADSSVSSCSQTDLSFDSQVSLPISVHTQTFLPSSKVTSSIAAQTDAFM DTCFQSGGVSRETQTSGIESPTDDHVQMDQAGMCGDIFESVHSSYNVATGNIISNSLVAE TVTHSLLPQNEPKTLNQDIEKSAPIINFSAQNSMLPSQNMTDNQTQTIDLLSDLENILSS NLPAQTLDHRSLLSDTNPGPDTQLPSGPAQNPGIDFDIEEFFSASNIQTQTEESELSTMT TEPVLESLDIETQTDFLLADTSAQSYGCRGNSNFLGLEMFDTQTQTDLNFFLDSSPHLPL GSILKHSSFSVSTDSSDTETQTEGVSTAKNIPALESKVQLNSTETQTMSSGFETLGSLFF TSNETQTAMDDFLLADLAWNTMESQFSSVETQTSAEPHTVSNF Click to Show/Hide
|
||||
| Function |
Transcription factor. Plays a crucial role in cell survival and RAD51 foci formation in response to methylating DNA damage. Involved in regulating the activity of ATM in the absence of DNA damage. May play a role in stabilizing ATM. Binds to the DYNLL1 promoter and activates its transcription.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Investigative Drug(s)
1 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [1] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Iridium | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Breast cancer [ICD-11: 2C60] | |||
| The Specified Disease | Breast cancer | |||
| The Studied Tissue | Breast tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.00E-15 Fold-change: -3.70E-02 Z-score: -8.01E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| HCC1937 cells | Breast | Homo sapiens (Human) | CVCL_0290 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-124 may be involved in DNA repair by directly targeting ATMIN and PARP1, suggesting that multiple DNA repair pathways are affected by miR-124 and therefore manipulation of miR-124 level/activity may improve the efficacy of chemotherapies that induce DNA damage. repression of ATMIN (+) the HR repair defect induced by miR-124, and restoration of ATMIN reversed the effect of miR-124 overexpression in breast cancer cells. Therefore, it is intriguing to further speculate which of the multiple roles of ATMIN is specifically affected in breast carcinogenesis. On the other hand, PARP1-mediated processes play a role in oncogenesis, cancer progression, and therapeutic resistance. | |||
| Disease Class: Osteosarcoma [ICD-11: 2B51.0] | [1] | |||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Sensitive Drug | Iridium | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| HCC1937 cells | Breast | Homo sapiens (Human) | CVCL_0290 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-124 may be involved in DNA repair by directly targeting ATMIN and PARP1, suggesting that multiple DNA repair pathways are affected by miR-124 and therefore manipulation of miR-124 level/activity may improve the efficacy of chemotherapies that induce DNA damage. repression of ATMIN (+) the HR repair defect induced by miR-124, and restoration of ATMIN reversed the effect of miR-124 overexpression in breast cancer cells. Therefore, it is intriguing to further speculate which of the multiple roles of ATMIN is specifically affected in breast carcinogenesis. On the other hand, PARP1-mediated processes play a role in oncogenesis, cancer progression, and therapeutic resistance. | |||
Approved Drug(s)
2 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Osteosarcoma [ICD-11: 2B51.0] | [1] | |||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| HCC1937 cells | Breast | Homo sapiens (Human) | CVCL_0290 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-124 may be involved in DNA repair by directly targeting ATMIN and PARP1, suggesting that multiple DNA repair pathways are affected by miR-124 and therefore manipulation of miR-124 level/activity may improve the efficacy of chemotherapies that induce DNA damage. repression of ATMIN (+) the HR repair defect induced by miR-124, and restoration of ATMIN reversed the effect of miR-124 overexpression in breast cancer cells. Therefore, it is intriguing to further speculate which of the multiple roles of ATMIN is specifically affected in breast carcinogenesis. On the other hand, PARP1-mediated processes play a role in oncogenesis, cancer progression, and therapeutic resistance. | |||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [1] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| HCC1937 cells | Breast | Homo sapiens (Human) | CVCL_0290 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-124 may be involved in DNA repair by directly targeting ATMIN and PARP1, suggesting that multiple DNA repair pathways are affected by miR-124 and therefore manipulation of miR-124 level/activity may improve the efficacy of chemotherapies that induce DNA damage. repression of ATMIN (+) the HR repair defect induced by miR-124, and restoration of ATMIN reversed the effect of miR-124 overexpression in breast cancer cells. Therefore, it is intriguing to further speculate which of the multiple roles of ATMIN is specifically affected in breast carcinogenesis. On the other hand, PARP1-mediated processes play a role in oncogenesis, cancer progression, and therapeutic resistance. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Osteosarcoma [ICD-11: 2B51.0] | [1] | |||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Sensitive Drug | Etoposide | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| HCC1937 cells | Breast | Homo sapiens (Human) | CVCL_0290 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-124 may be involved in DNA repair by directly targeting ATMIN and PARP1, suggesting that multiple DNA repair pathways are affected by miR-124 and therefore manipulation of miR-124 level/activity may improve the efficacy of chemotherapies that induce DNA damage. repression of ATMIN (+) the HR repair defect induced by miR-124, and restoration of ATMIN reversed the effect of miR-124 overexpression in breast cancer cells. Therefore, it is intriguing to further speculate which of the multiple roles of ATMIN is specifically affected in breast carcinogenesis. On the other hand, PARP1-mediated processes play a role in oncogenesis, cancer progression, and therapeutic resistance. | |||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [1] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Etoposide | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| HCC1937 cells | Breast | Homo sapiens (Human) | CVCL_0290 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-124 may be involved in DNA repair by directly targeting ATMIN and PARP1, suggesting that multiple DNA repair pathways are affected by miR-124 and therefore manipulation of miR-124 level/activity may improve the efficacy of chemotherapies that induce DNA damage. repression of ATMIN (+) the HR repair defect induced by miR-124, and restoration of ATMIN reversed the effect of miR-124 overexpression in breast cancer cells. Therefore, it is intriguing to further speculate which of the multiple roles of ATMIN is specifically affected in breast carcinogenesis. On the other hand, PARP1-mediated processes play a role in oncogenesis, cancer progression, and therapeutic resistance. | |||
Clinical Trial Drug(s)
1 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Osteosarcoma [ICD-11: 2B51.0] | [1] | |||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Sensitive Drug | Camptothecin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| HCC1937 cells | Breast | Homo sapiens (Human) | CVCL_0290 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-124 may be involved in DNA repair by directly targeting ATMIN and PARP1, suggesting that multiple DNA repair pathways are affected by miR-124 and therefore manipulation of miR-124 level/activity may improve the efficacy of chemotherapies that induce DNA damage. repression of ATMIN (+) the HR repair defect induced by miR-124, and restoration of ATMIN reversed the effect of miR-124 overexpression in breast cancer cells. Therefore, it is intriguing to further speculate which of the multiple roles of ATMIN is specifically affected in breast carcinogenesis. On the other hand, PARP1-mediated processes play a role in oncogenesis, cancer progression, and therapeutic resistance. | |||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [1] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Camptothecin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell proliferation | Inhibition | hsa05200 | ||
| PI3K/AKT signaling pathway | Inhibition | hsa04151 | ||
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| ZR75-1 cells | Breast | Homo sapiens (Human) | CVCL_0588 | |
| HCC1937 cells | Breast | Homo sapiens (Human) | CVCL_0290 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | miR-124 may be involved in DNA repair by directly targeting ATMIN and PARP1, suggesting that multiple DNA repair pathways are affected by miR-124 and therefore manipulation of miR-124 level/activity may improve the efficacy of chemotherapies that induce DNA damage. repression of ATMIN (+) the HR repair defect induced by miR-124, and restoration of ATMIN reversed the effect of miR-124 overexpression in breast cancer cells. Therefore, it is intriguing to further speculate which of the multiple roles of ATMIN is specifically affected in breast carcinogenesis. On the other hand, PARP1-mediated processes play a role in oncogenesis, cancer progression, and therapeutic resistance. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.00E-15; Fold-change: -2.60E-01; Z-score: -7.04E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.28E-01; Fold-change: -5.24E-02; Z-score: -1.05E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
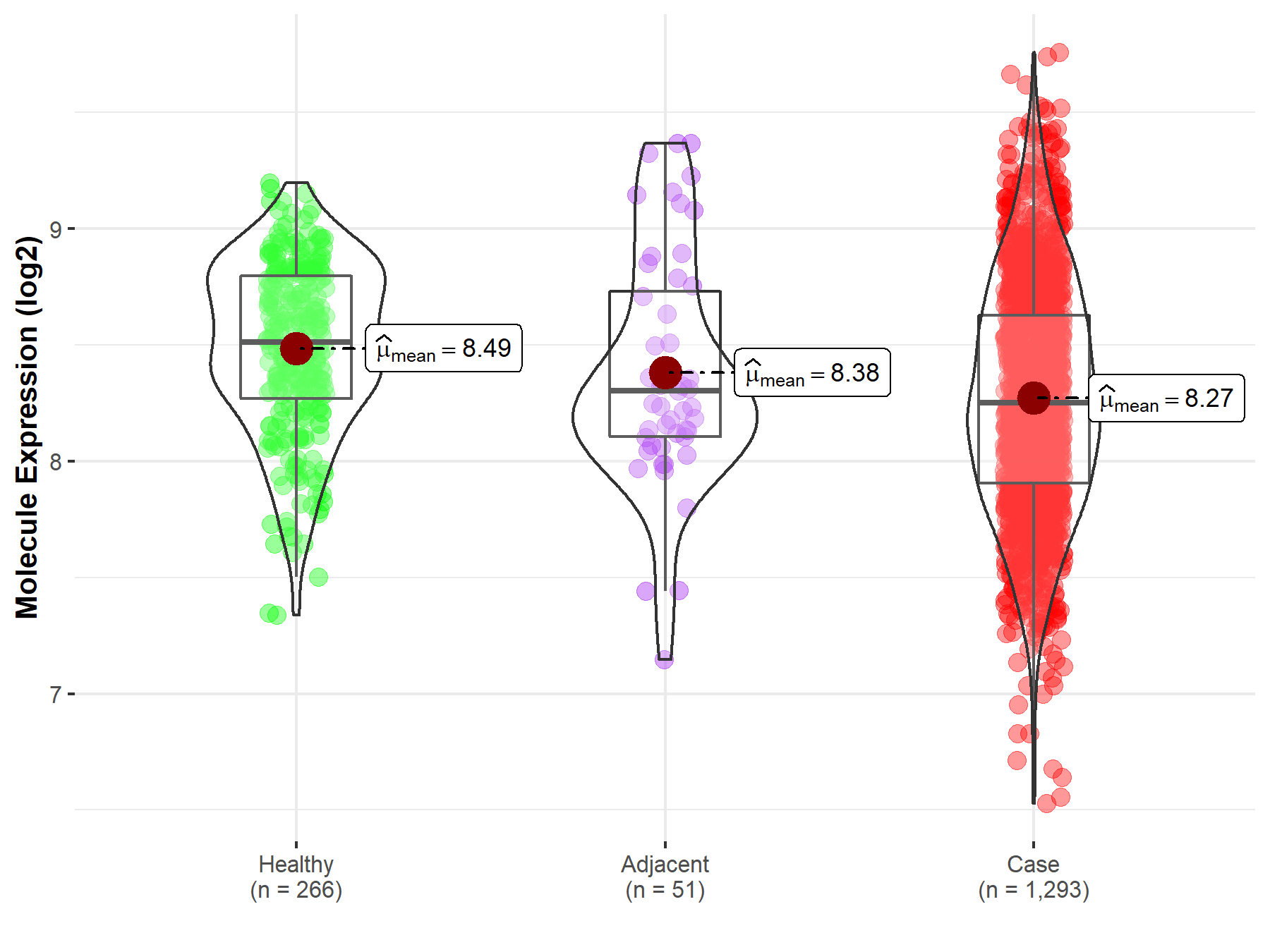
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

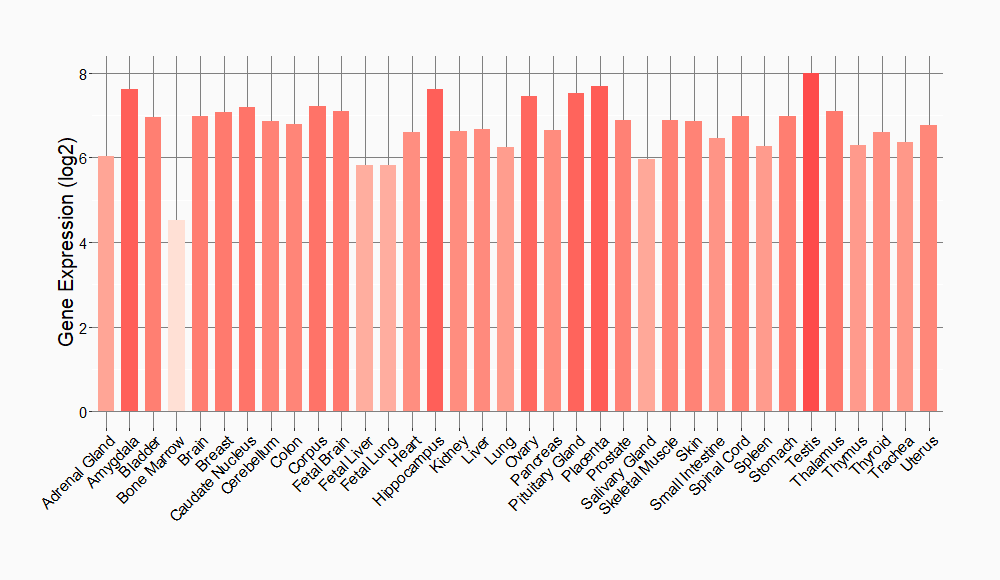
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
