Molecule Information
General Information of the Molecule (ID: Mol00200)
| Name |
Cytosolic purine 5'-nucleotidase (NT5C2)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Cytosolic 5'-nucleotidase II; cN-II; Cytosolic IMP/GMP-specific 5'-nucleotidase; Cytosolic nucleoside phosphotransferase 5'N; High Km 5'-nucleotidase; NT5B; NT5CP; PNT5
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
NT5C2
|
||||
| Gene ID | |||||
| Location |
chr10:103087185-103277605[-]
|
||||
| Sequence |
MSTSWSDRLQNAADMPANMDKHALKKYRREAYHRVFVNRSLAMEKIKCFGFDMDYTLAVY
KSPEYESLGFELTVERLVSIGYPQELLSFAYDSTFPTRGLVFDTLYGNLLKVDAYGNLLV CAHGFNFIRGPETREQYPNKFIQRDDTERFYILNTLFNLPETYLLACLVDFFTNCPRYTS CETGFKDGDLFMSYRSMFQDVRDAVDWVHYKGSLKEKTVENLEKYVVKDGKLPLLLSRMK EVGKVFLATNSDYKYTDKIMTYLFDFPHGPKPGSSHRPWQSYFDLILVDARKPLFFGEGT VLRQVDTKTGKLKIGTYTGPLQHGIVYSGGSSDTICDLLGAKGKDILYIGDHIFGDILKS KKRQGWRTFLVIPELAQELHVWTDKSSLFEELQSLDIFLAELYKHLDSSSNERPDISSIQ RRIKKVTHDMDMCYGMMGSLFRSGSRQTLFASQVMRYADLYAASFINLLYYPFSYLFRAA HVLMPHESTVEHTHVDINEMESPLATRNRTSVDFKDTDYKRHQLTRSISEIKPPNLFPLA PQEITHCHDEDDDEEEEEEEE Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Broad specificity cytosolic 5'-nucleotidase that catalyzes the dephosphorylation of 6-hydroxypurine nucleoside 5'-monophosphates. In addition, possesses a phosphotransferase activity by which it can transfer a phosphate from a donor nucleoside monophosphate to an acceptor nucleoside, preferably inosine, deoxyinosine and guanosine. Has the highest activities for IMP and GMP followed by dIMP, dGMP and XMP. Could also catalyze the transfer of phosphates from pyrimidine monophosphates but with lower efficiency. Through these activities regulates the purine nucleoside/nucleotide pools within the cell.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
3 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Leukemia [ICD-11: 2B33.6] | [1] | |||
| Resistant Disease | Leukemia [ICD-11: 2B33.6] | |||
| Resistant Drug | Cytarabine | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Mechanism Description | Since monophosphorilated intermediate of cytarabine activation is reduced by cytosolic 5'-nucleotidases NT5C2 and NT5C3, the activity level of this enzyme may represent one of the factors affecting the clinical outcome of cytarabine therapy. Increased expression of NT5C2 has been correlated with resistance to cytarabine chemotherapy and to a lower survival rate in a hundred patients undergoing cytarabine chemotherapy. An increase in the NT5C2 has emerged as a mechanism of resistance to cytarabine. Patients with AML and low expression level of NT5C2 have a better overall survival after treatment with cytarabine than patients with high expression. NT5C2 is implicated in pharmacokinetic of cytarabine has been associated with poor clinical outcome. | |||
| Disease Class: Lymphoma [ICD-11: 2A90- 2A85] | [1] | |||
| Resistant Disease | Lymphoma [ICD-11: 2A90- 2A85] | |||
| Resistant Drug | Cytarabine | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Mechanism Description | Since monophosphorilated intermediate of cytarabine activation is reduced by cytosolic 5'-nucleotidases NT5C2 and NT5C3, the activity level of this enzyme may represent one of the factors affecting the clinical outcome of cytarabine therapy. Increased expression of NT5C2 has been correlated with resistance to cytarabine chemotherapy and to a lower survival rate in a hundred patients undergoing cytarabine chemotherapy. An increase in the NT5C2 has emerged as a mechanism of resistance to cytarabine. Patients with AML and low expression level of NT5C2 have a better overall survival after treatment with cytarabine than patients with high expression. NT5C2 is implicated in pharmacokinetic of cytarabine has been associated with poor clinical outcome. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [2], [3] | ||||||||||||
| Resistant Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Resistant Drug | Mercaptopurine | ||||||||||||
| Molecule Alteration | Missense mutation | p.R238W (c.c712t) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.70 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.84 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
M
-
G
-
S
-
S
-
H
-
H
-
H
-
H
-10
|
-
H
-
H
-
S
-
S
-
G
-
L
-
V
-
P
-
R
-
G
0
|
-
S
-
M
-
S
T
T
S
S
W
W
S
S
D
D
R
R
L
L
10
|
Q
Q
N
N
A
A
A
A
D
D
M
M
P
P
A
A
N
N
M
M
20
|
D
D
K
K
H
H
A
A
L
L
K
K
K
K
Y
Y
R
R
R
R
30
|
E
E
A
A
Y
Y
H
H
R
R
V
V
F
F
V
V
N
N
R
R
40
|
S
S
L
L
A
A
M
M
E
E
K
K
I
I
K
K
C
C
F
F
50
|
G
G
F
F
D
D
M
M
D
D
Y
Y
T
T
L
L
A
A
V
V
60
|
Y
Y
K
K
S
S
P
P
E
E
Y
Y
E
E
S
S
L
L
G
G
70
|
F
F
E
E
L
L
T
T
V
V
E
E
R
R
L
L
V
V
S
S
80
|
I
I
G
G
Y
Y
P
P
Q
Q
E
E
L
L
L
L
S
S
F
F
90
|
A
A
Y
Y
D
D
S
S
T
T
F
F
P
P
T
T
R
R
G
G
100
|
L
L
V
V
F
F
D
D
T
T
L
L
Y
Y
G
G
N
N
L
L
110
|
L
L
K
K
V
V
D
D
A
A
Y
Y
G
G
N
N
L
L
L
L
120
|
V
V
C
C
A
A
H
H
G
G
F
F
N
N
F
F
I
I
R
R
130
|
G
G
P
P
E
E
T
T
R
R
E
E
Q
Q
Y
Y
P
P
N
N
140
|
K
K
F
F
I
I
Q
Q
R
R
D
D
D
D
T
T
E
E
R
R
150
|
F
F
Y
Y
I
I
L
L
N
N
T
T
L
L
F
F
N
N
L
L
160
|
P
P
E
E
T
T
Y
Y
L
L
L
L
A
A
C
C
L
L
V
V
170
|
D
D
F
F
F
F
T
T
N
N
C
C
P
P
R
R
Y
Y
T
T
180
|
S
S
C
C
E
E
T
T
G
G
F
F
K
K
D
D
G
G
D
D
190
|
L
L
F
F
M
M
S
S
Y
Y
R
R
S
S
M
M
F
F
Q
Q
200
|
D
D
V
V
R
R
D
D
A
A
V
V
D
D
W
W
V
V
H
H
210
|
Y
Y
K
K
G
G
S
S
L
L
K
K
E
E
K
K
T
T
V
V
220
|
E
E
N
N
L
L
E
E
K
K
Y
Y
V
V
V
V
K
K
D
D
230
|
G
G
K
K
L
L
P
P
L
L
L
L
L
L
S
S
R
W
M
M
240
|
K
K
E
E
V
V
G
G
K
K
V
V
F
F
L
L
A
A
T
T
250
|
N
N
S
S
D
D
Y
Y
K
K
Y
Y
T
T
D
D
K
K
I
I
260
|
M
M
T
T
Y
Y
L
L
F
F
D
D
F
F
P
P
H
H
G
G
270
|
P
P
K
K
P
P
G
G
S
S
S
S
H
H
R
R
P
P
W
W
280
|
Q
Q
S
S
Y
Y
F
F
D
D
L
L
I
I
L
L
V
V
D
D
290
|
A
A
R
R
K
K
P
P
L
L
F
F
F
F
G
G
E
E
G
G
300
|
T
T
V
V
L
L
R
R
Q
Q
V
V
D
D
T
T
K
K
T
T
310
|
G
G
K
K
L
L
K
K
I
I
G
G
T
T
Y
Y
T
T
G
G
320
|
P
P
L
L
Q
Q
H
H
G
G
I
I
V
V
Y
Y
S
S
G
G
330
|
G
G
S
S
S
S
D
D
T
T
I
I
C
C
D
D
L
L
L
L
340
|
G
G
A
A
K
K
G
G
K
K
D
D
I
I
L
L
Y
Y
I
I
350
|
G
G
D
D
H
H
I
I
F
F
G
G
D
D
I
I
L
L
K
K
360
|
S
S
K
K
K
K
R
R
Q
Q
G
G
W
W
R
R
T
T
F
F
370
|
L
L
V
V
I
I
P
P
E
E
L
L
A
A
Q
Q
E
E
L
L
380
|
H
H
V
V
W
W
T
T
D
D
K
K
S
S
S
S
L
L
F
F
390
|
E
E
E
E
L
L
Q
Q
S
S
L
L
D
D
I
I
F
F
L
L
400
|
A
A
E
E
L
L
Y
Y
K
K
H
H
L
L
D
D
S
S
S
S
410
|
S
S
N
N
E
E
R
R
P
P
D
D
I
I
S
S
S
S
I
I
420
|
Q
Q
R
R
R
R
I
I
K
K
K
K
V
V
T
T
H
H
D
D
430
|
M
M
D
D
M
M
C
C
Y
Y
G
G
M
M
M
M
G
G
S
S
440
|
L
L
F
F
R
R
S
S
G
G
S
S
R
R
Q
Q
T
T
L
L
450
|
F
F
A
A
S
S
Q
Q
V
V
M
M
R
R
Y
Y
A
A
D
D
460
|
L
L
Y
Y
A
A
A
A
S
S
F
F
I
I
N
N
L
L
L
L
470
|
Y
Y
Y
Y
P
P
F
F
S
S
Y
Y
L
L
F
F
R
R
A
A
480
|
A
A
H
H
V
V
L
L
M
M
P
P
H
H
E
E
S
S
-
T
490
|
-
V
-
E
-
H
-
T
-
H
-
V
-
D
-
I
-
N
-
E
500
|
-
M
-
E
-
S
-
P
-
L
-
A
-
T
-
R
-
N
-
R
510
|
-
T
-
S
-
V
-
D
-
F
-
K
-
D
-
T
-
D
-
Y
520
|
-
K
-
R
-
H
-
Q
-
L
-
T
-
R
-
S
-
I
-
S
530
|
-
E
-
I
-
K
-
P
-
P
-
N
-
L
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Exome sequencing assay; Transcriptome sequencing assay; Whole genome sequencing assay; Sanger Sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Flow cytometry assay | ||||||||||||
| Mechanism Description | Several of these alterations are known to induce a more stem cell-like state (eg, IkZF1) or confer resistance directly to specific chemotherapy agents such as CREBBP and glucocorticoids and mutations in the 5-nucleotidase gene NT5C2 and nucleoside a.logs. Many relapse-acquired lesions are enriched in specific pathways, including B-cell development (IkZF1), tumor suppression (TP53),34 Ras signaling, chromatin modification (CREBBP, SETD2),17 and drug metabolism (NT5C2). | ||||||||||||
| Disease Class: Acute lymphocytic leukemia [ICD-11: 2B33.0] | [4] | ||||||||||||
| Resistant Disease | Acute lymphocytic leukemia [ICD-11: 2B33.0] | ||||||||||||
| Resistant Drug | Mercaptopurine | ||||||||||||
| Molecule Alteration | Missense mutation | p.R367Q |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 2.30 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.50 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
G
-
S
-
S
-
H
-
H
-
H
-
H
-10
|
-
H
-
H
-
S
-
S
-
G
-
L
-
V
-
P
-
R
-
G
0
|
-
S
M
M
S
S
T
T
S
S
W
W
S
S
D
D
R
R
L
L
10
|
Q
Q
N
N
A
A
A
A
D
D
M
M
P
P
A
A
N
N
M
M
20
|
D
D
K
K
H
H
A
A
L
L
K
K
K
K
Y
Y
R
R
R
R
30
|
E
E
A
A
Y
Y
H
H
R
R
V
V
F
F
V
V
N
N
R
R
40
|
S
S
L
L
A
A
M
M
E
E
K
K
I
I
K
K
C
C
F
F
50
|
G
G
F
F
D
N
M
M
D
D
Y
Y
T
T
L
L
A
A
V
V
60
|
Y
Y
K
K
S
S
P
P
E
E
Y
Y
E
E
S
S
L
L
G
G
70
|
F
F
E
E
L
L
T
T
V
V
E
E
R
R
L
L
V
V
S
S
80
|
I
I
G
G
Y
Y
P
P
Q
Q
E
E
L
L
L
L
S
S
F
F
90
|
A
A
Y
Y
D
D
S
S
T
T
F
F
P
P
T
T
R
R
G
G
100
|
L
L
V
V
F
F
D
D
T
T
L
L
Y
Y
G
G
N
N
L
L
110
|
L
L
K
K
V
V
D
D
A
A
Y
Y
G
G
N
N
L
L
L
L
120
|
V
V
C
C
A
A
H
H
G
G
F
F
N
N
F
F
I
I
R
R
130
|
G
G
P
P
E
E
T
T
R
R
E
E
Q
Q
Y
Y
P
P
N
N
140
|
K
K
F
F
I
I
Q
Q
R
R
D
D
D
D
T
T
E
E
R
R
150
|
F
F
Y
Y
I
I
L
L
N
N
T
T
L
L
F
F
N
N
L
L
160
|
P
P
E
E
T
T
Y
Y
L
L
L
L
A
A
C
C
L
L
V
V
170
|
D
D
F
F
F
F
T
T
N
N
C
C
P
P
R
R
Y
Y
T
T
180
|
S
S
C
C
E
E
T
T
G
G
F
F
K
K
D
D
G
G
D
D
190
|
L
L
F
F
M
M
S
S
Y
Y
R
R
S
S
M
M
F
F
Q
Q
200
|
D
D
V
V
R
R
D
D
A
A
V
V
D
D
W
W
V
V
H
H
210
|
Y
Y
K
K
G
G
S
S
L
L
K
K
E
E
K
K
T
T
V
V
220
|
E
E
N
N
L
L
E
E
K
K
Y
Y
V
V
V
V
K
K
D
D
230
|
G
G
K
K
L
L
P
P
L
L
L
L
L
L
S
S
R
R
M
M
240
|
K
K
E
E
V
V
G
G
K
K
V
V
F
F
L
L
A
A
T
T
250
|
N
N
S
S
D
D
Y
Y
K
K
Y
Y
T
T
D
D
K
K
I
I
260
|
M
M
T
T
Y
Y
L
L
F
F
D
D
F
F
P
P
H
H
G
G
270
|
P
P
K
K
P
P
G
G
S
S
S
S
H
H
R
R
P
P
W
W
280
|
Q
Q
S
S
Y
Y
F
F
D
D
L
L
I
I
L
L
V
V
D
D
290
|
A
A
R
R
K
K
P
P
L
L
F
F
F
F
G
G
E
E
G
G
300
|
T
T
V
V
L
L
R
R
Q
Q
V
V
D
D
T
T
K
K
T
T
310
|
G
G
K
K
L
L
K
K
I
I
G
G
T
T
Y
Y
T
T
G
G
320
|
P
P
L
L
Q
Q
H
H
G
G
I
I
V
V
Y
Y
S
S
G
G
330
|
G
G
S
S
S
S
D
D
T
T
I
I
C
C
D
D
L
L
L
L
340
|
G
G
A
A
K
K
G
G
K
K
D
D
I
I
L
L
Y
Y
I
I
350
|
G
G
D
D
H
H
I
I
F
F
G
G
D
D
I
I
L
L
K
K
360
|
S
S
K
K
K
K
R
R
Q
Q
G
G
W
W
R
Q
T
T
F
F
370
|
L
L
V
V
I
I
P
P
E
E
L
L
A
A
Q
Q
E
E
L
L
380
|
H
H
V
V
W
W
T
T
D
D
K
K
S
S
S
S
L
L
F
F
390
|
E
E
E
E
L
L
Q
Q
S
S
L
L
D
D
I
I
F
F
L
L
400
|
A
A
E
E
L
L
Y
Y
K
K
H
H
L
L
D
D
S
S
S
S
410
|
S
S
N
N
E
E
R
R
P
P
D
D
I
I
S
S
S
S
I
I
420
|
Q
Q
R
R
R
R
I
I
K
K
K
K
V
V
T
T
H
H
D
D
430
|
M
M
D
D
M
M
C
C
Y
Y
G
G
M
M
M
M
G
G
S
S
440
|
L
L
F
F
R
R
S
S
G
G
S
S
R
R
Q
Q
T
T
L
L
450
|
F
F
A
A
S
S
Q
Q
V
V
M
M
R
R
Y
Y
A
A
D
D
460
|
L
L
Y
Y
A
A
A
A
S
S
F
F
I
I
N
N
L
L
L
L
470
|
Y
Y
Y
Y
P
P
F
F
S
S
Y
Y
L
L
F
F
R
R
A
A
480
|
A
A
H
H
V
V
L
L
M
M
P
P
H
H
E
E
S
S
T
T
490
|
V
V
E
E
H
H
T
T
H
H
V
V
D
D
I
I
N
N
E
E
500
|
M
M
E
E
S
S
P
P
L
L
A
A
T
T
R
R
N
N
R
R
510
|
T
T
S
S
V
V
D
D
F
F
K
K
D
D
T
T
D
D
Y
Y
520
|
K
K
R
R
H
H
Q
Q
L
L
T
T
R
R
S
S
I
I
S
S
530
|
E
E
I
I
K
K
P
P
P
P
N
N
L
L
F
F
P
P
L
L
540
|
A
A
P
P
Q
Q
E
E
I
I
T
T
H
H
C
C
H
H
D
D
550
|
E
E
D
D
D
D
D
D
E
E
E
E
E
E
E
E
E
E
E
E
560
|
E
E
E
E
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
Exome sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Conality analyses assay | ||||||||||||
| Mechanism Description | These two NT5C2 mutations (R367Q, D407V) occur as recurrent mutational hotspots in relapse-ALL and they have been functionally validated. These mutations increase the NT5C2 inosine-5-monophosphate-nucleotidase activity; and therefore lead to resistance to one of the chemotherapeutic drugs, 6-mercaptopurine. | ||||||||||||
| Disease Class: Acute lymphocytic leukemia [ICD-11: 2B33.0] | [5] | ||||||||||||
| Resistant Disease | Acute lymphocytic leukemia [ICD-11: 2B33.0] | ||||||||||||
| Resistant Drug | Mercaptopurine | ||||||||||||
| Molecule Alteration | Mutation | . |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
Genome sequencing assay; Whole-exome sequencing assay | ||||||||||||
| Mechanism Description | Recent sequencing studies of T-ALL have confirmed the presence of these mutations as well as novel recurrent mutations in the tumor suppressor CNOT3, ribosomal proteins(RPL5 and RPL10) and in the setting of relapse, the NT5C2 gene, which inactivates nucleoside-analogue chemotherapy drugs. | ||||||||||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [2], [3] | ||||||||||||
| Resistant Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Resistant Drug | Mercaptopurine | ||||||||||||
| Molecule Alteration | Missense mutation | p.S445F (c.c1334t) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Exome sequencing assay; Transcriptome sequencing assay; Whole genome sequencing assay; Sanger Sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Flow cytometry assay | ||||||||||||
| Mechanism Description | Several of these alterations are known to induce a more stem cell-like state (eg, IkZF1) or confer resistance directly to specific chemotherapy agents such as CREBBP and glucocorticoids and mutations in the 5-nucleotidase gene NT5C2 and nucleoside a.logs. Many relapse-acquired lesions are enriched in specific pathways, including B-cell development (IkZF1), tumor suppression (TP53),34 Ras signaling, chromatin modification (CREBBP, SETD2),17 and drug metabolism (NT5C2). | ||||||||||||
| Disease Class: Acute lymphocytic leukemia [ICD-11: 2B33.0] | [4] | ||||||||||||
| Resistant Disease | Acute lymphocytic leukemia [ICD-11: 2B33.0] | ||||||||||||
| Resistant Drug | Mercaptopurine | ||||||||||||
| Molecule Alteration | Missense mutation | p.D407V |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
Exome sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Conality analyses assay | ||||||||||||
| Mechanism Description | These two NT5C2 mutations (R367Q, D407V) occur as recurrent mutational hotspots in relapse-ALL and they have been functionally validated. These mutations increase the NT5C2 inosine-5-monophosphate-nucleotidase activity; and therefore lead to resistance to one of the chemotherapeutic drugs, 6-mercaptopurine. | ||||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [2], [3] | ||||||||||||
| Resistant Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Resistant Drug | Thioguanine | ||||||||||||
| Molecule Alteration | Missense mutation | p.R238W (c.c712t) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.70 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.84 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
M
-
G
-
S
-
S
-
H
-
H
-
H
-
H
-10
|
-
H
-
H
-
S
-
S
-
G
-
L
-
V
-
P
-
R
-
G
0
|
-
S
-
M
-
S
T
T
S
S
W
W
S
S
D
D
R
R
L
L
10
|
Q
Q
N
N
A
A
A
A
D
D
M
M
P
P
A
A
N
N
M
M
20
|
D
D
K
K
H
H
A
A
L
L
K
K
K
K
Y
Y
R
R
R
R
30
|
E
E
A
A
Y
Y
H
H
R
R
V
V
F
F
V
V
N
N
R
R
40
|
S
S
L
L
A
A
M
M
E
E
K
K
I
I
K
K
C
C
F
F
50
|
G
G
F
F
D
D
M
M
D
D
Y
Y
T
T
L
L
A
A
V
V
60
|
Y
Y
K
K
S
S
P
P
E
E
Y
Y
E
E
S
S
L
L
G
G
70
|
F
F
E
E
L
L
T
T
V
V
E
E
R
R
L
L
V
V
S
S
80
|
I
I
G
G
Y
Y
P
P
Q
Q
E
E
L
L
L
L
S
S
F
F
90
|
A
A
Y
Y
D
D
S
S
T
T
F
F
P
P
T
T
R
R
G
G
100
|
L
L
V
V
F
F
D
D
T
T
L
L
Y
Y
G
G
N
N
L
L
110
|
L
L
K
K
V
V
D
D
A
A
Y
Y
G
G
N
N
L
L
L
L
120
|
V
V
C
C
A
A
H
H
G
G
F
F
N
N
F
F
I
I
R
R
130
|
G
G
P
P
E
E
T
T
R
R
E
E
Q
Q
Y
Y
P
P
N
N
140
|
K
K
F
F
I
I
Q
Q
R
R
D
D
D
D
T
T
E
E
R
R
150
|
F
F
Y
Y
I
I
L
L
N
N
T
T
L
L
F
F
N
N
L
L
160
|
P
P
E
E
T
T
Y
Y
L
L
L
L
A
A
C
C
L
L
V
V
170
|
D
D
F
F
F
F
T
T
N
N
C
C
P
P
R
R
Y
Y
T
T
180
|
S
S
C
C
E
E
T
T
G
G
F
F
K
K
D
D
G
G
D
D
190
|
L
L
F
F
M
M
S
S
Y
Y
R
R
S
S
M
M
F
F
Q
Q
200
|
D
D
V
V
R
R
D
D
A
A
V
V
D
D
W
W
V
V
H
H
210
|
Y
Y
K
K
G
G
S
S
L
L
K
K
E
E
K
K
T
T
V
V
220
|
E
E
N
N
L
L
E
E
K
K
Y
Y
V
V
V
V
K
K
D
D
230
|
G
G
K
K
L
L
P
P
L
L
L
L
L
L
S
S
R
W
M
M
240
|
K
K
E
E
V
V
G
G
K
K
V
V
F
F
L
L
A
A
T
T
250
|
N
N
S
S
D
D
Y
Y
K
K
Y
Y
T
T
D
D
K
K
I
I
260
|
M
M
T
T
Y
Y
L
L
F
F
D
D
F
F
P
P
H
H
G
G
270
|
P
P
K
K
P
P
G
G
S
S
S
S
H
H
R
R
P
P
W
W
280
|
Q
Q
S
S
Y
Y
F
F
D
D
L
L
I
I
L
L
V
V
D
D
290
|
A
A
R
R
K
K
P
P
L
L
F
F
F
F
G
G
E
E
G
G
300
|
T
T
V
V
L
L
R
R
Q
Q
V
V
D
D
T
T
K
K
T
T
310
|
G
G
K
K
L
L
K
K
I
I
G
G
T
T
Y
Y
T
T
G
G
320
|
P
P
L
L
Q
Q
H
H
G
G
I
I
V
V
Y
Y
S
S
G
G
330
|
G
G
S
S
S
S
D
D
T
T
I
I
C
C
D
D
L
L
L
L
340
|
G
G
A
A
K
K
G
G
K
K
D
D
I
I
L
L
Y
Y
I
I
350
|
G
G
D
D
H
H
I
I
F
F
G
G
D
D
I
I
L
L
K
K
360
|
S
S
K
K
K
K
R
R
Q
Q
G
G
W
W
R
R
T
T
F
F
370
|
L
L
V
V
I
I
P
P
E
E
L
L
A
A
Q
Q
E
E
L
L
380
|
H
H
V
V
W
W
T
T
D
D
K
K
S
S
S
S
L
L
F
F
390
|
E
E
E
E
L
L
Q
Q
S
S
L
L
D
D
I
I
F
F
L
L
400
|
A
A
E
E
L
L
Y
Y
K
K
H
H
L
L
D
D
S
S
S
S
410
|
S
S
N
N
E
E
R
R
P
P
D
D
I
I
S
S
S
S
I
I
420
|
Q
Q
R
R
R
R
I
I
K
K
K
K
V
V
T
T
H
H
D
D
430
|
M
M
D
D
M
M
C
C
Y
Y
G
G
M
M
M
M
G
G
S
S
440
|
L
L
F
F
R
R
S
S
G
G
S
S
R
R
Q
Q
T
T
L
L
450
|
F
F
A
A
S
S
Q
Q
V
V
M
M
R
R
Y
Y
A
A
D
D
460
|
L
L
Y
Y
A
A
A
A
S
S
F
F
I
I
N
N
L
L
L
L
470
|
Y
Y
Y
Y
P
P
F
F
S
S
Y
Y
L
L
F
F
R
R
A
A
480
|
A
A
H
H
V
V
L
L
M
M
P
P
H
H
E
E
S
S
-
T
490
|
-
V
-
E
-
H
-
T
-
H
-
V
-
D
-
I
-
N
-
E
500
|
-
M
-
E
-
S
-
P
-
L
-
A
-
T
-
R
-
N
-
R
510
|
-
T
-
S
-
V
-
D
-
F
-
K
-
D
-
T
-
D
-
Y
520
|
-
K
-
R
-
H
-
Q
-
L
-
T
-
R
-
S
-
I
-
S
530
|
-
E
-
I
-
K
-
P
-
P
-
N
-
L
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Exome sequencing assay; Transcriptome sequencing assay; Whole genome sequencing assay; Sanger Sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Flow cytometry assay | ||||||||||||
| Mechanism Description | Several of these alterations are known to induce a more stem cell-like state (eg, IkZF1) or confer resistance directly to specific chemotherapy agents such as CREBBP and glucocorticoids and mutations in the 5-nucleotidase gene NT5C2 and nucleoside a.logs. Many relapse-acquired lesions are enriched in specific pathways, including B-cell development (IkZF1), tumor suppression (TP53),34 Ras signaling, chromatin modification (CREBBP, SETD2),17 and drug metabolism (NT5C2). | ||||||||||||
| Disease Class: Acute lymphocytic leukemia [ICD-11: 2B33.0] | [5] | ||||||||||||
| Resistant Disease | Acute lymphocytic leukemia [ICD-11: 2B33.0] | ||||||||||||
| Resistant Drug | Thioguanine | ||||||||||||
| Molecule Alteration | Mutation | . |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
Genome sequencing assay; Whole-exome sequencing assay | ||||||||||||
| Mechanism Description | Recent sequencing studies of T-ALL have confirmed the presence of these mutations as well as novel recurrent mutations in the tumor suppressor CNOT3, ribosomal proteins(RPL5 and RPL10) and in the setting of relapse, the NT5C2 gene, which inactivates nucleoside-analogue chemotherapy drugs. | ||||||||||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [2], [3] | ||||||||||||
| Resistant Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Resistant Drug | Thioguanine | ||||||||||||
| Molecule Alteration | Missense mutation | p.S445F (c.c1334t) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Exome sequencing assay; Transcriptome sequencing assay; Whole genome sequencing assay; Sanger Sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Flow cytometry assay | ||||||||||||
| Mechanism Description | Several of these alterations are known to induce a more stem cell-like state (eg, IkZF1) or confer resistance directly to specific chemotherapy agents such as CREBBP and glucocorticoids and mutations in the 5-nucleotidase gene NT5C2 and nucleoside a.logs. Many relapse-acquired lesions are enriched in specific pathways, including B-cell development (IkZF1), tumor suppression (TP53),34 Ras signaling, chromatin modification (CREBBP, SETD2),17 and drug metabolism (NT5C2). | ||||||||||||
Investigative Drug(s)
1 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Acute lymphocytic leukemia [ICD-11: 2B33.0] | [6] | ||||||||||||
| Resistant Disease | Acute lymphocytic leukemia [ICD-11: 2B33.0] | ||||||||||||
| Resistant Drug | Thiopurine | ||||||||||||
| Molecule Alteration | Missense mutation | p.R238W |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.70 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 1.84 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
-
M
-
G
-
S
-
S
-
H
-
H
-
H
-
H
-10
|
-
H
-
H
-
S
-
S
-
G
-
L
-
V
-
P
-
R
-
G
0
|
-
S
-
M
-
S
T
T
S
S
W
W
S
S
D
D
R
R
L
L
10
|
Q
Q
N
N
A
A
A
A
D
D
M
M
P
P
A
A
N
N
M
M
20
|
D
D
K
K
H
H
A
A
L
L
K
K
K
K
Y
Y
R
R
R
R
30
|
E
E
A
A
Y
Y
H
H
R
R
V
V
F
F
V
V
N
N
R
R
40
|
S
S
L
L
A
A
M
M
E
E
K
K
I
I
K
K
C
C
F
F
50
|
G
G
F
F
D
D
M
M
D
D
Y
Y
T
T
L
L
A
A
V
V
60
|
Y
Y
K
K
S
S
P
P
E
E
Y
Y
E
E
S
S
L
L
G
G
70
|
F
F
E
E
L
L
T
T
V
V
E
E
R
R
L
L
V
V
S
S
80
|
I
I
G
G
Y
Y
P
P
Q
Q
E
E
L
L
L
L
S
S
F
F
90
|
A
A
Y
Y
D
D
S
S
T
T
F
F
P
P
T
T
R
R
G
G
100
|
L
L
V
V
F
F
D
D
T
T
L
L
Y
Y
G
G
N
N
L
L
110
|
L
L
K
K
V
V
D
D
A
A
Y
Y
G
G
N
N
L
L
L
L
120
|
V
V
C
C
A
A
H
H
G
G
F
F
N
N
F
F
I
I
R
R
130
|
G
G
P
P
E
E
T
T
R
R
E
E
Q
Q
Y
Y
P
P
N
N
140
|
K
K
F
F
I
I
Q
Q
R
R
D
D
D
D
T
T
E
E
R
R
150
|
F
F
Y
Y
I
I
L
L
N
N
T
T
L
L
F
F
N
N
L
L
160
|
P
P
E
E
T
T
Y
Y
L
L
L
L
A
A
C
C
L
L
V
V
170
|
D
D
F
F
F
F
T
T
N
N
C
C
P
P
R
R
Y
Y
T
T
180
|
S
S
C
C
E
E
T
T
G
G
F
F
K
K
D
D
G
G
D
D
190
|
L
L
F
F
M
M
S
S
Y
Y
R
R
S
S
M
M
F
F
Q
Q
200
|
D
D
V
V
R
R
D
D
A
A
V
V
D
D
W
W
V
V
H
H
210
|
Y
Y
K
K
G
G
S
S
L
L
K
K
E
E
K
K
T
T
V
V
220
|
E
E
N
N
L
L
E
E
K
K
Y
Y
V
V
V
V
K
K
D
D
230
|
G
G
K
K
L
L
P
P
L
L
L
L
L
L
S
S
R
W
M
M
240
|
K
K
E
E
V
V
G
G
K
K
V
V
F
F
L
L
A
A
T
T
250
|
N
N
S
S
D
D
Y
Y
K
K
Y
Y
T
T
D
D
K
K
I
I
260
|
M
M
T
T
Y
Y
L
L
F
F
D
D
F
F
P
P
H
H
G
G
270
|
P
P
K
K
P
P
G
G
S
S
S
S
H
H
R
R
P
P
W
W
280
|
Q
Q
S
S
Y
Y
F
F
D
D
L
L
I
I
L
L
V
V
D
D
290
|
A
A
R
R
K
K
P
P
L
L
F
F
F
F
G
G
E
E
G
G
300
|
T
T
V
V
L
L
R
R
Q
Q
V
V
D
D
T
T
K
K
T
T
310
|
G
G
K
K
L
L
K
K
I
I
G
G
T
T
Y
Y
T
T
G
G
320
|
P
P
L
L
Q
Q
H
H
G
G
I
I
V
V
Y
Y
S
S
G
G
330
|
G
G
S
S
S
S
D
D
T
T
I
I
C
C
D
D
L
L
L
L
340
|
G
G
A
A
K
K
G
G
K
K
D
D
I
I
L
L
Y
Y
I
I
350
|
G
G
D
D
H
H
I
I
F
F
G
G
D
D
I
I
L
L
K
K
360
|
S
S
K
K
K
K
R
R
Q
Q
G
G
W
W
R
R
T
T
F
F
370
|
L
L
V
V
I
I
P
P
E
E
L
L
A
A
Q
Q
E
E
L
L
380
|
H
H
V
V
W
W
T
T
D
D
K
K
S
S
S
S
L
L
F
F
390
|
E
E
E
E
L
L
Q
Q
S
S
L
L
D
D
I
I
F
F
L
L
400
|
A
A
E
E
L
L
Y
Y
K
K
H
H
L
L
D
D
S
S
S
S
410
|
S
S
N
N
E
E
R
R
P
P
D
D
I
I
S
S
S
S
I
I
420
|
Q
Q
R
R
R
R
I
I
K
K
K
K
V
V
T
T
H
H
D
D
430
|
M
M
D
D
M
M
C
C
Y
Y
G
G
M
M
M
M
G
G
S
S
440
|
L
L
F
F
R
R
S
S
G
G
S
S
R
R
Q
Q
T
T
L
L
450
|
F
F
A
A
S
S
Q
Q
V
V
M
M
R
R
Y
Y
A
A
D
D
460
|
L
L
Y
Y
A
A
A
A
S
S
F
F
I
I
N
N
L
L
L
L
470
|
Y
Y
Y
Y
P
P
F
F
S
S
Y
Y
L
L
F
F
R
R
A
A
480
|
A
A
H
H
V
V
L
L
M
M
P
P
H
H
E
E
S
S
-
T
490
|
-
V
-
E
-
H
-
T
-
H
-
V
-
D
-
I
-
N
-
E
500
|
-
M
-
E
-
S
-
P
-
L
-
A
-
T
-
R
-
N
-
R
510
|
-
T
-
S
-
V
-
D
-
F
-
K
-
D
-
T
-
D
-
Y
520
|
-
K
-
R
-
H
-
Q
-
L
-
T
-
R
-
S
-
I
-
S
530
|
-
E
-
I
-
K
-
P
-
P
-
N
-
L
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| Experiment for Molecule Alteration |
Whole-exome sequencing assay; Whole-genome sequencing assay | ||||||||||||
| Experiment for Drug Resistance |
Flow cytometric analysis assay; MTT assay | ||||||||||||
| Mechanism Description | Finally, genomic profiling of diagnostic and relapsed leukemias has identified relapse-associated mutations in the 5'-nucleotidase, cytosolic II(NT5C2) gene as drivers of resistance to thiopurine chemotherapy in about 20% of T-ALL and 5% of B-precursor ALL cases at relapse. | ||||||||||||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Bone marrow | |
| The Specified Disease | Acute myeloid leukemia | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.82E-01; Fold-change: 2.07E-02; Z-score: 9.93E-02 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
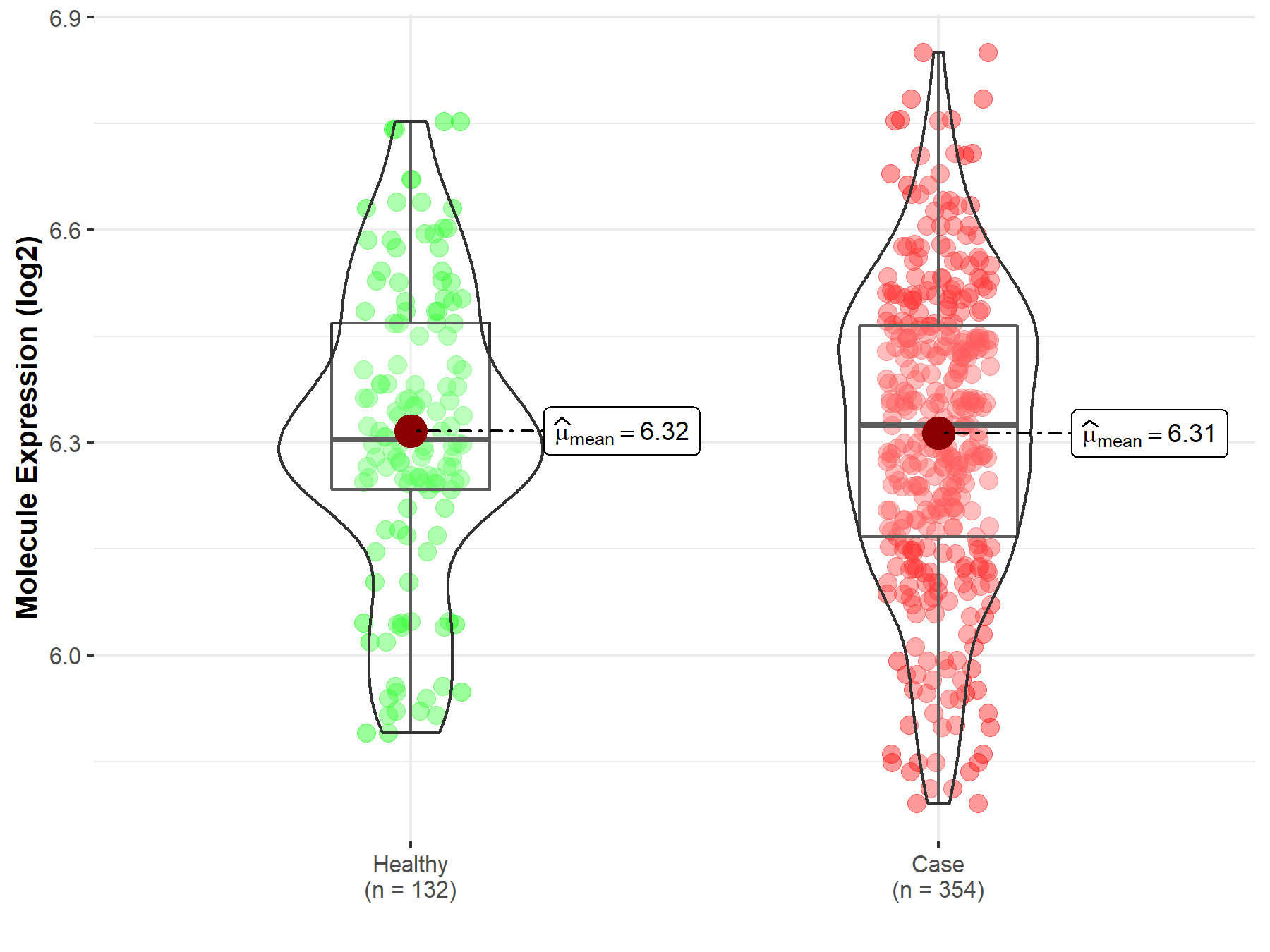
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Tonsil tissue | |
| The Specified Disease | Lymphoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.62E-01; Fold-change: -5.89E-02; Z-score: -5.52E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
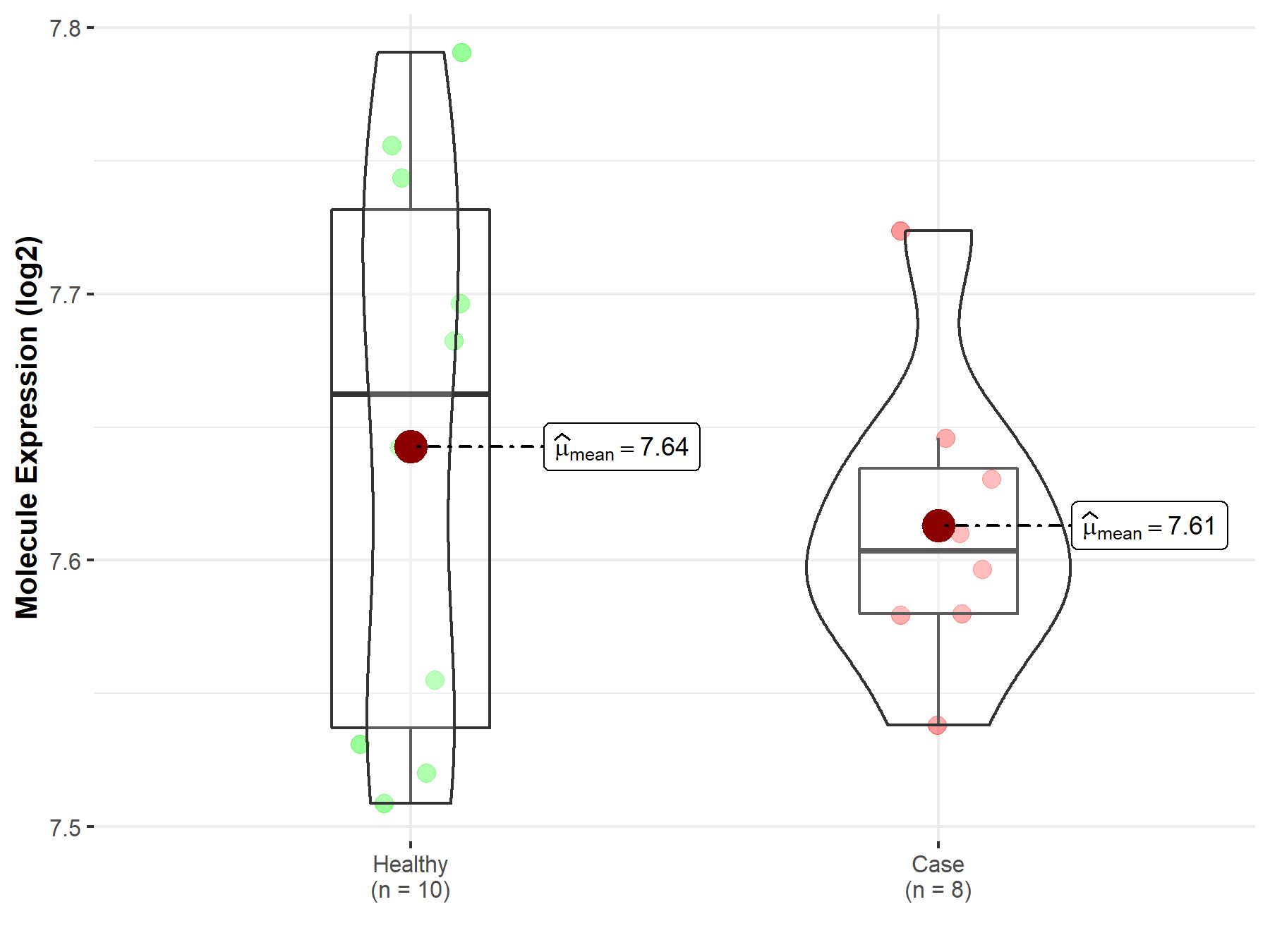
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

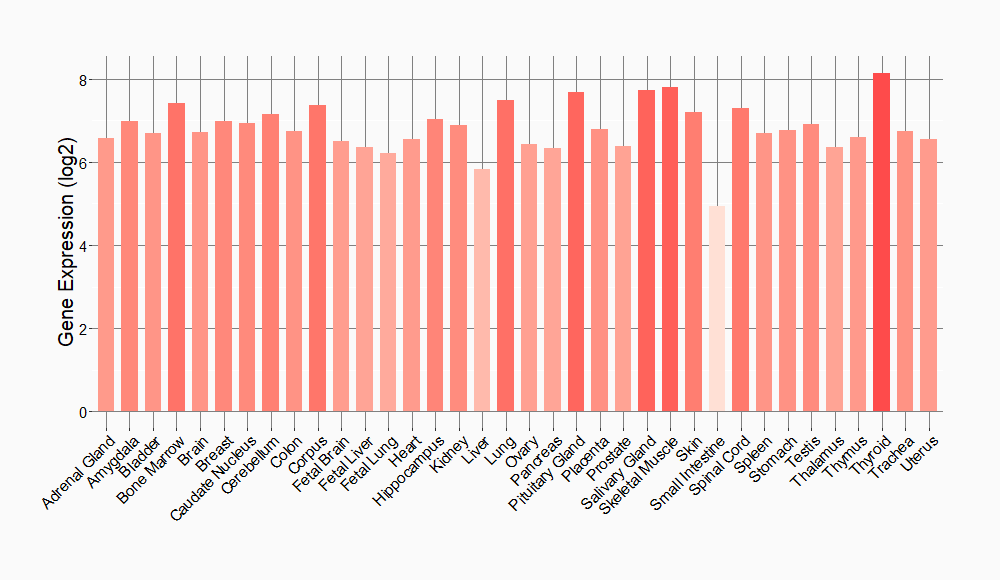
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
