Molecule Information
General Information of the Molecule (ID: Mol00147)
| Name |
Tyrosine-protein phosphatase non-receptor type 11 (PTPN11)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Protein-tyrosine phosphatase 1D; PTP-1D; Protein-tyrosine phosphatase 2C; PTP-2C; SH-PTP2; SHP-2; Shp2; SH-PTP3; PTP2C; SHPTP2
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
PTPN11
|
||||
| Gene ID | |||||
| Location |
chr12:112418351-112509918[+]
|
||||
| Sequence |
MTSRRWFHPNITGVEAENLLLTRGVDGSFLARPSKSNPGDFTLSVRRNGAVTHIKIQNTG
DYYDLYGGEKFATLAELVQYYMEHHGQLKEKNGDVIELKYPLNCADPTSERWFHGHLSGK EAEKLLTEKGKHGSFLVRESQSHPGDFVLSVRTGDDKGESNDGKSKVTHVMIRCQELKYD VGGGERFDSLTDLVEHYKKNPMVETLGTVLQLKQPLNTTRINAAEIESRVRELSKLAETT DKVKQGFWEEFETLQQQECKLLYSRKEGQRQENKNKNRYKNILPFDHTRVVLHDGDPNEP VSDYINANIIMPEFETKCNNSKPKKSYIATQGCLQNTVNDFWRMVFQENSRVIVMTTKEV ERGKSKCVKYWPDEYALKEYGVMRVRNVKESAAHDYTLRELKLSKVGQGNTERTVWQYHF RTWPDHGVPSDPGGVLDFLEEVHHKQESIMDAGPVVVHCSAGIGRTGTFIVIDILIDIIR EKGVDCDIDVPKTIQMVRSQRSGMVQTEAQYRFIYMAVQHYIETLQRRIEEEQKSKRKGH EYTNIKYSLADQTSGDQSPLPPCTPTPPCAEMREDSARVYENVGLMQQQKSFR Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Acts downstream of various receptor and cytoplasmic protein tyrosine kinases to participate in the signal transduction from the cell surface to the nucleus. Positively regulates MAPK signal transduction pathway. Dephosphorylates GAB1, ARHGAP35 and EGFR. Dephosphorylates ROCK2 at 'Tyr-722' resulting in stimulation of its RhoA binding activity. Dephosphorylates CDC73. Dephosphorylates SOX9 on tyrosine residues, leading to inactivate SOX9 and promote ossification.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
1 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Hepatocellular carcinoma [ICD-11: 2C12.2] | [1] | |||
| Resistant Disease | Hepatocellular carcinoma [ICD-11: 2C12.2] | |||
| Resistant Drug | Doxorubicin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Liver cancer [ICD-11: 2C12] | |||
| The Specified Disease | Hepatocellular carcinoma | |||
| The Studied Tissue | Liver tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.92E-03 Fold-change: -6.39E-02 Z-score: -2.73E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | STAT3 signaling pathway | Activation | hsa04550 | |
| In Vitro Model | Huh-7 cells | Liver | Homo sapiens (Human) | CVCL_0336 |
| HepG2 cells | Liver | Homo sapiens (Human) | CVCL_0027 | |
| Hep3B cells | Liver | Homo sapiens (Human) | CVCL_0326 | |
| QGY-7703 cells | Liver | Homo sapiens (Human) | CVCL_6715 | |
| SMMC7721 cells | Uterus | Homo sapiens (Human) | CVCL_0534 | |
| 97H cells | Liver | Homo sapiens (Human) | N.A. | |
| PLC cells | Liver | Homo sapiens (Human) | CVCL_0485 | |
| Experiment for Molecule Alteration |
Western blot analysis; Luciferase reporter assay | |||
| Experiment for Drug Resistance |
Flow cytometric analysis; Spheroid formation assay | |||
| Mechanism Description | miR589-5p promotes the cancer stem cell characteristics and chemoresistance via targeting multiple negative regulators of STAT3 signaling pathway, including SOCS2, SOCS5, PTPN1 and PTPN11, leading to constitutive activation of STAT3 signaling. | |||
Preclinical Drug(s)
3 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Juvenile myelomonocytic leukaemia [ICD-11: 2A42.0] | [2] | ||||||||||||
| Sensitive Disease | Juvenile myelomonocytic leukaemia [ICD-11: 2A42.0] | ||||||||||||
| Sensitive Drug | Cryptotanshinone | ||||||||||||
| Molecule Alteration | Missense mutation | p.E76K (c.226G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.70 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.62 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
0
|
S
G
M
M
T
T
S
S
R
R
R
R
W
W
F
F
H
H
P
P
10
|
N
N
I
I
T
T
G
G
V
V
E
E
A
A
E
E
N
N
L
L
20
|
L
L
L
L
T
T
R
R
G
G
V
V
D
D
G
G
S
S
F
F
30
|
L
L
A
A
R
R
P
P
S
S
K
K
S
S
N
N
P
P
G
G
40
|
D
D
F
F
T
T
L
L
S
S
V
V
R
R
R
R
N
N
G
G
50
|
A
A
V
V
T
T
H
H
I
I
K
K
I
I
Q
Q
N
N
T
T
60
|
G
G
D
D
Y
Y
Y
Y
D
D
L
L
Y
Y
G
G
G
G
E
E
70
|
K
K
F
F
A
A
T
T
L
L
A
A
E
K
L
L
V
V
Q
Q
80
|
Y
Y
Y
Y
M
M
E
E
H
H
H
H
G
G
Q
Q
L
L
K
K
90
|
E
E
K
K
N
N
G
G
D
D
V
V
I
I
E
E
L
L
K
K
100
|
Y
Y
P
P
L
L
N
N
C
C
A
A
D
D
P
P
T
T
S
S
110
|
E
E
R
R
W
W
F
F
H
H
G
G
H
H
L
L
S
S
G
G
120
|
K
K
E
E
A
A
E
E
K
K
L
L
L
L
T
T
E
E
K
K
130
|
G
G
K
K
H
H
G
G
S
S
F
F
L
L
V
V
R
R
E
E
140
|
S
S
Q
Q
S
S
H
H
P
P
G
G
D
D
F
F
V
V
L
L
150
|
S
S
V
V
R
R
T
T
G
G
D
D
D
D
K
K
G
G
E
E
160
|
S
S
N
N
D
D
G
G
K
K
S
S
K
K
V
V
T
T
H
H
170
|
V
V
M
M
I
I
R
R
C
C
Q
Q
E
E
L
L
K
K
Y
Y
180
|
D
D
V
V
G
G
G
G
G
G
E
E
R
R
F
F
D
D
S
S
190
|
L
L
T
T
D
D
L
L
V
V
E
E
H
H
Y
Y
K
K
K
K
200
|
N
N
P
P
M
M
V
V
E
E
T
T
L
L
G
G
T
T
V
V
210
|
L
L
Q
Q
L
L
K
K
Q
Q
P
P
L
L
N
N
T
T
T
T
220
|
R
R
I
I
N
N
A
A
A
A
E
E
I
I
E
E
S
S
R
R
230
|
V
V
R
R
E
E
L
L
S
S
K
K
L
L
A
A
E
E
T
T
240
|
T
T
D
D
K
K
V
V
K
K
Q
Q
G
G
F
F
W
W
E
E
250
|
E
E
F
F
E
E
T
T
L
L
Q
Q
Q
Q
Q
Q
E
E
C
C
260
|
K
K
L
L
L
L
Y
Y
S
S
R
R
K
K
E
E
G
G
Q
Q
270
|
R
R
Q
Q
E
E
N
N
K
K
N
N
K
K
N
N
R
R
Y
Y
280
|
K
K
N
N
I
I
L
L
P
P
F
F
D
D
H
H
T
T
R
R
290
|
V
V
V
V
L
L
H
H
D
D
G
G
D
D
P
P
N
N
E
E
300
|
P
P
V
V
S
S
D
D
Y
Y
I
I
N
N
A
A
N
N
I
I
310
|
I
I
M
M
P
P
E
E
F
F
E
E
T
T
K
K
C
C
N
N
320
|
N
N
S
S
K
K
P
P
K
K
K
K
S
S
Y
Y
I
I
A
A
330
|
T
T
Q
Q
G
G
C
C
L
L
Q
Q
N
N
T
T
V
V
N
N
340
|
D
D
F
F
W
W
R
R
M
M
V
V
F
F
Q
Q
E
E
N
N
350
|
S
S
R
R
V
V
I
I
V
V
M
M
T
T
T
T
K
K
E
E
360
|
V
V
E
E
R
R
G
G
K
K
S
S
K
K
C
C
V
V
K
K
370
|
Y
Y
W
W
P
P
D
D
E
E
Y
Y
A
A
L
L
K
K
E
E
380
|
Y
Y
G
G
V
V
M
M
R
R
V
V
R
R
N
N
V
V
K
K
390
|
E
E
S
S
A
A
A
A
H
H
D
D
Y
Y
T
T
L
L
R
R
400
|
E
E
L
L
K
K
L
L
S
S
K
K
V
V
G
G
Q
Q
G
G
410
|
N
N
T
T
E
E
R
R
T
T
V
V
W
W
Q
Q
Y
Y
H
H
420
|
F
F
R
R
T
T
W
W
P
P
D
D
H
H
G
G
V
V
P
P
430
|
S
S
D
D
P
P
G
G
G
G
V
V
L
L
D
D
F
F
L
L
440
|
E
E
E
E
V
V
H
H
H
H
K
K
Q
Q
E
E
S
S
I
I
450
|
M
M
D
D
A
A
G
G
P
P
V
V
V
V
V
V
H
H
C
C
460
|
S
S
A
A
G
G
I
I
G
G
R
R
T
T
G
G
T
T
F
F
470
|
I
I
V
V
I
I
D
D
I
I
L
L
I
I
D
D
I
I
I
I
480
|
R
R
E
E
K
K
G
G
V
V
D
D
C
C
D
D
I
I
D
D
490
|
V
V
P
P
K
K
T
T
I
I
Q
Q
M
M
V
V
R
R
S
S
500
|
Q
Q
R
R
S
S
G
G
M
M
V
V
Q
Q
T
T
E
E
A
A
510
|
Q
Q
Y
Y
R
R
F
F
I
I
Y
Y
M
M
A
A
V
V
Q
Q
520
|
H
H
Y
Y
I
I
E
E
T
T
L
L
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
CellTiter 96 aqueous one solution cell proliferation assay | ||||||||||||
| Mechanism Description | The missense mutation p.E76K (c.226G>A) in gene PTPN11 cause the sensitivity of Cryptotanshinone by unusual activation of pro-survival pathway | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Hematologic Cancer [ICD-11: MG24.Y] | [3] | ||||||||||||
| Sensitive Disease | Hematologic Cancer [ICD-11: MG24.Y] | ||||||||||||
| Sensitive Drug | II-B08 | ||||||||||||
| Molecule Alteration | Missense mutation | p.E76K (c.226G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.70 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.62 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
0
|
S
G
M
M
T
T
S
S
R
R
R
R
W
W
F
F
H
H
P
P
10
|
N
N
I
I
T
T
G
G
V
V
E
E
A
A
E
E
N
N
L
L
20
|
L
L
L
L
T
T
R
R
G
G
V
V
D
D
G
G
S
S
F
F
30
|
L
L
A
A
R
R
P
P
S
S
K
K
S
S
N
N
P
P
G
G
40
|
D
D
F
F
T
T
L
L
S
S
V
V
R
R
R
R
N
N
G
G
50
|
A
A
V
V
T
T
H
H
I
I
K
K
I
I
Q
Q
N
N
T
T
60
|
G
G
D
D
Y
Y
Y
Y
D
D
L
L
Y
Y
G
G
G
G
E
E
70
|
K
K
F
F
A
A
T
T
L
L
A
A
E
K
L
L
V
V
Q
Q
80
|
Y
Y
Y
Y
M
M
E
E
H
H
H
H
G
G
Q
Q
L
L
K
K
90
|
E
E
K
K
N
N
G
G
D
D
V
V
I
I
E
E
L
L
K
K
100
|
Y
Y
P
P
L
L
N
N
C
C
A
A
D
D
P
P
T
T
S
S
110
|
E
E
R
R
W
W
F
F
H
H
G
G
H
H
L
L
S
S
G
G
120
|
K
K
E
E
A
A
E
E
K
K
L
L
L
L
T
T
E
E
K
K
130
|
G
G
K
K
H
H
G
G
S
S
F
F
L
L
V
V
R
R
E
E
140
|
S
S
Q
Q
S
S
H
H
P
P
G
G
D
D
F
F
V
V
L
L
150
|
S
S
V
V
R
R
T
T
G
G
D
D
D
D
K
K
G
G
E
E
160
|
S
S
N
N
D
D
G
G
K
K
S
S
K
K
V
V
T
T
H
H
170
|
V
V
M
M
I
I
R
R
C
C
Q
Q
E
E
L
L
K
K
Y
Y
180
|
D
D
V
V
G
G
G
G
G
G
E
E
R
R
F
F
D
D
S
S
190
|
L
L
T
T
D
D
L
L
V
V
E
E
H
H
Y
Y
K
K
K
K
200
|
N
N
P
P
M
M
V
V
E
E
T
T
L
L
G
G
T
T
V
V
210
|
L
L
Q
Q
L
L
K
K
Q
Q
P
P
L
L
N
N
T
T
T
T
220
|
R
R
I
I
N
N
A
A
A
A
E
E
I
I
E
E
S
S
R
R
230
|
V
V
R
R
E
E
L
L
S
S
K
K
L
L
A
A
E
E
T
T
240
|
T
T
D
D
K
K
V
V
K
K
Q
Q
G
G
F
F
W
W
E
E
250
|
E
E
F
F
E
E
T
T
L
L
Q
Q
Q
Q
Q
Q
E
E
C
C
260
|
K
K
L
L
L
L
Y
Y
S
S
R
R
K
K
E
E
G
G
Q
Q
270
|
R
R
Q
Q
E
E
N
N
K
K
N
N
K
K
N
N
R
R
Y
Y
280
|
K
K
N
N
I
I
L
L
P
P
F
F
D
D
H
H
T
T
R
R
290
|
V
V
V
V
L
L
H
H
D
D
G
G
D
D
P
P
N
N
E
E
300
|
P
P
V
V
S
S
D
D
Y
Y
I
I
N
N
A
A
N
N
I
I
310
|
I
I
M
M
P
P
E
E
F
F
E
E
T
T
K
K
C
C
N
N
320
|
N
N
S
S
K
K
P
P
K
K
K
K
S
S
Y
Y
I
I
A
A
330
|
T
T
Q
Q
G
G
C
C
L
L
Q
Q
N
N
T
T
V
V
N
N
340
|
D
D
F
F
W
W
R
R
M
M
V
V
F
F
Q
Q
E
E
N
N
350
|
S
S
R
R
V
V
I
I
V
V
M
M
T
T
T
T
K
K
E
E
360
|
V
V
E
E
R
R
G
G
K
K
S
S
K
K
C
C
V
V
K
K
370
|
Y
Y
W
W
P
P
D
D
E
E
Y
Y
A
A
L
L
K
K
E
E
380
|
Y
Y
G
G
V
V
M
M
R
R
V
V
R
R
N
N
V
V
K
K
390
|
E
E
S
S
A
A
A
A
H
H
D
D
Y
Y
T
T
L
L
R
R
400
|
E
E
L
L
K
K
L
L
S
S
K
K
V
V
G
G
Q
Q
G
G
410
|
N
N
T
T
E
E
R
R
T
T
V
V
W
W
Q
Q
Y
Y
H
H
420
|
F
F
R
R
T
T
W
W
P
P
D
D
H
H
G
G
V
V
P
P
430
|
S
S
D
D
P
P
G
G
G
G
V
V
L
L
D
D
F
F
L
L
440
|
E
E
E
E
V
V
H
H
H
H
K
K
Q
Q
E
E
S
S
I
I
450
|
M
M
D
D
A
A
G
G
P
P
V
V
V
V
V
V
H
H
C
C
460
|
S
S
A
A
G
G
I
I
G
G
R
R
T
T
G
G
T
T
F
F
470
|
I
I
V
V
I
I
D
D
I
I
L
L
I
I
D
D
I
I
I
I
480
|
R
R
E
E
K
K
G
G
V
V
D
D
C
C
D
D
I
I
D
D
490
|
V
V
P
P
K
K
T
T
I
I
Q
Q
M
M
V
V
R
R
S
S
500
|
Q
Q
R
R
S
S
G
G
M
M
V
V
Q
Q
T
T
E
E
A
A
510
|
Q
Q
Y
Y
R
R
F
F
I
I
Y
Y
M
M
A
A
V
V
Q
Q
520
|
H
H
Y
Y
I
I
E
E
T
T
L
L
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| In Vitro Model | HEK293 cells | Kidney | Homo sapiens (Human) | CVCL_0045 | |||||||||
| Experiment for Molecule Alteration |
Western blot analysis | ||||||||||||
| Experiment for Drug Resistance |
3H-thymidine incorporation assay | ||||||||||||
| Mechanism Description | The missense mutation p.E76K (c.226G>A) in gene PTPN11 cause the sensitivity of II-B08 by unusual activation of pro-survival pathway | ||||||||||||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [4] | ||||||||||||
| Resistant Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Resistant Drug | SHP099 | ||||||||||||
| Molecule Alteration | Missense mutation | p.E76K (c.226G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.70 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.62 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
0
|
S
G
M
M
T
T
S
S
R
R
R
R
W
W
F
F
H
H
P
P
10
|
N
N
I
I
T
T
G
G
V
V
E
E
A
A
E
E
N
N
L
L
20
|
L
L
L
L
T
T
R
R
G
G
V
V
D
D
G
G
S
S
F
F
30
|
L
L
A
A
R
R
P
P
S
S
K
K
S
S
N
N
P
P
G
G
40
|
D
D
F
F
T
T
L
L
S
S
V
V
R
R
R
R
N
N
G
G
50
|
A
A
V
V
T
T
H
H
I
I
K
K
I
I
Q
Q
N
N
T
T
60
|
G
G
D
D
Y
Y
Y
Y
D
D
L
L
Y
Y
G
G
G
G
E
E
70
|
K
K
F
F
A
A
T
T
L
L
A
A
E
K
L
L
V
V
Q
Q
80
|
Y
Y
Y
Y
M
M
E
E
H
H
H
H
G
G
Q
Q
L
L
K
K
90
|
E
E
K
K
N
N
G
G
D
D
V
V
I
I
E
E
L
L
K
K
100
|
Y
Y
P
P
L
L
N
N
C
C
A
A
D
D
P
P
T
T
S
S
110
|
E
E
R
R
W
W
F
F
H
H
G
G
H
H
L
L
S
S
G
G
120
|
K
K
E
E
A
A
E
E
K
K
L
L
L
L
T
T
E
E
K
K
130
|
G
G
K
K
H
H
G
G
S
S
F
F
L
L
V
V
R
R
E
E
140
|
S
S
Q
Q
S
S
H
H
P
P
G
G
D
D
F
F
V
V
L
L
150
|
S
S
V
V
R
R
T
T
G
G
D
D
D
D
K
K
G
G
E
E
160
|
S
S
N
N
D
D
G
G
K
K
S
S
K
K
V
V
T
T
H
H
170
|
V
V
M
M
I
I
R
R
C
C
Q
Q
E
E
L
L
K
K
Y
Y
180
|
D
D
V
V
G
G
G
G
G
G
E
E
R
R
F
F
D
D
S
S
190
|
L
L
T
T
D
D
L
L
V
V
E
E
H
H
Y
Y
K
K
K
K
200
|
N
N
P
P
M
M
V
V
E
E
T
T
L
L
G
G
T
T
V
V
210
|
L
L
Q
Q
L
L
K
K
Q
Q
P
P
L
L
N
N
T
T
T
T
220
|
R
R
I
I
N
N
A
A
A
A
E
E
I
I
E
E
S
S
R
R
230
|
V
V
R
R
E
E
L
L
S
S
K
K
L
L
A
A
E
E
T
T
240
|
T
T
D
D
K
K
V
V
K
K
Q
Q
G
G
F
F
W
W
E
E
250
|
E
E
F
F
E
E
T
T
L
L
Q
Q
Q
Q
Q
Q
E
E
C
C
260
|
K
K
L
L
L
L
Y
Y
S
S
R
R
K
K
E
E
G
G
Q
Q
270
|
R
R
Q
Q
E
E
N
N
K
K
N
N
K
K
N
N
R
R
Y
Y
280
|
K
K
N
N
I
I
L
L
P
P
F
F
D
D
H
H
T
T
R
R
290
|
V
V
V
V
L
L
H
H
D
D
G
G
D
D
P
P
N
N
E
E
300
|
P
P
V
V
S
S
D
D
Y
Y
I
I
N
N
A
A
N
N
I
I
310
|
I
I
M
M
P
P
E
E
F
F
E
E
T
T
K
K
C
C
N
N
320
|
N
N
S
S
K
K
P
P
K
K
K
K
S
S
Y
Y
I
I
A
A
330
|
T
T
Q
Q
G
G
C
C
L
L
Q
Q
N
N
T
T
V
V
N
N
340
|
D
D
F
F
W
W
R
R
M
M
V
V
F
F
Q
Q
E
E
N
N
350
|
S
S
R
R
V
V
I
I
V
V
M
M
T
T
T
T
K
K
E
E
360
|
V
V
E
E
R
R
G
G
K
K
S
S
K
K
C
C
V
V
K
K
370
|
Y
Y
W
W
P
P
D
D
E
E
Y
Y
A
A
L
L
K
K
E
E
380
|
Y
Y
G
G
V
V
M
M
R
R
V
V
R
R
N
N
V
V
K
K
390
|
E
E
S
S
A
A
A
A
H
H
D
D
Y
Y
T
T
L
L
R
R
400
|
E
E
L
L
K
K
L
L
S
S
K
K
V
V
G
G
Q
Q
G
G
410
|
N
N
T
T
E
E
R
R
T
T
V
V
W
W
Q
Q
Y
Y
H
H
420
|
F
F
R
R
T
T
W
W
P
P
D
D
H
H
G
G
V
V
P
P
430
|
S
S
D
D
P
P
G
G
G
G
V
V
L
L
D
D
F
F
L
L
440
|
E
E
E
E
V
V
H
H
H
H
K
K
Q
Q
E
E
S
S
I
I
450
|
M
M
D
D
A
A
G
G
P
P
V
V
V
V
V
V
H
H
C
C
460
|
S
S
A
A
G
G
I
I
G
G
R
R
T
T
G
G
T
T
F
F
470
|
I
I
V
V
I
I
D
D
I
I
L
L
I
I
D
D
I
I
I
I
480
|
R
R
E
E
K
K
G
G
V
V
D
D
C
C
D
D
I
I
D
D
490
|
V
V
P
P
K
K
T
T
I
I
Q
Q
M
M
V
V
R
R
S
S
500
|
Q
Q
R
R
S
S
G
G
M
M
V
V
Q
Q
T
T
E
E
A
A
510
|
Q
Q
Y
Y
R
R
F
F
I
I
Y
Y
M
M
A
A
V
V
Q
Q
520
|
H
H
Y
Y
I
I
E
E
T
T
L
L
|
|||||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | TF-1 cells | Bone marrow | Homo sapiens (Human) | CVCL_0559 | |||||||||
| Mechanism Description | The SHP2 variant carrying the potent activating mutation E76K exhibits an open-state structure, which induces drastic domain reorganization to expose the active site and eliminate the binding pocket for the allosteric inhibitor SHP099. | ||||||||||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [4] | ||||||||||||
| Resistant Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Resistant Drug | SHP099 | ||||||||||||
| Molecule Alteration | Missense mutation | p.D61Y (c.181G>T) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | TF-1 cells | Bone marrow | Homo sapiens (Human) | CVCL_0559 | |||||||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [4] | ||||||||||||
| Resistant Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | ||||||||||||
| Resistant Drug | SHP099 | ||||||||||||
| Molecule Alteration | Missense mutation | p.A72V (c.215C>T) |
|||||||||||
| Experimental Note | Identified from the Human Clinical Data | ||||||||||||
| In Vitro Model | TF-1 cells | Bone marrow | Homo sapiens (Human) | CVCL_0559 | |||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Acute myeloid leukemia [ICD-11: 2A60.0] | [4] | |||
| Sensitive Disease | Acute myeloid leukemia [ICD-11: 2A60.0] | |||
| Sensitive Drug | SHP099 | |||
| Molecule Alteration | Missense mutation | p.E69K (c.205G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | TF-1 cells | Bone marrow | Homo sapiens (Human) | CVCL_0559 |
| Disease Class: Thyroid gland cancer [ICD-11: 2D10.0] | [5] | |||
| Sensitive Disease | Thyroid gland cancer [ICD-11: 2D10.0] | |||
| Sensitive Drug | SHP099 | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | MAPK/ERK signaling pathway | Inhibition | hsa04011 | |
| In Vitro Model | K1 cells | Thyroid | Homo sapiens (Human) | CVCL_2537 |
| BCPAP cells | Thyroid | Homo sapiens (Human) | CVCL_0153 | |
| KTC-1 cells | Thyroid | Homo sapiens (Human) | CVCL_6300 | |
| 8305C cells | Thyroid | Homo sapiens (Human) | CVCL_1053 | |
| BHT-101 cells | Thyroid gland | Homo sapiens (Human) | CVCL_1085 | |
| In Vivo Model | Mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot assay; RT-qPCR | |||
| Experiment for Drug Resistance |
Cell proliferation capacity assay | |||
| Mechanism Description | In treating thyroid cancer cells with vemurafenib, we have detected reactivation of the MAPK/ERK signaling pathway as a result of the release of multiple receptor tyrosine kinases (RTKs) from the negative feedback of ERK phosphorylation. SHP2 is an important target protein downstream of the RTK signaling pathway. Decreasing it through SHP2 knockdown or the use of an inhibitor of SHP2 (SHP099) was found to significantly increase the early sensitivity and reverse the late resistance to vemurafenib in BRAFV600E mutant thyroid cancer cells.Blocking SHP2 reverses the reactivation of the MAPK/ERK signaling pathway caused by the activation of RTKs and improves the sensitivity of thyroid cancer to vemurafenib. | |||
Investigative Drug(s)
2 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Acute lymphocytic leukemia [ICD-11: 2B33.0] | [6] | ||||||||||||
| Sensitive Disease | Acute lymphocytic leukemia [ICD-11: 2B33.0] | ||||||||||||
| Sensitive Drug | PD98059 | ||||||||||||
| Molecule Alteration | Missense mutation | p.E76K (c.226G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.70 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.62 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
0
|
S
G
M
M
T
T
S
S
R
R
R
R
W
W
F
F
H
H
P
P
10
|
N
N
I
I
T
T
G
G
V
V
E
E
A
A
E
E
N
N
L
L
20
|
L
L
L
L
T
T
R
R
G
G
V
V
D
D
G
G
S
S
F
F
30
|
L
L
A
A
R
R
P
P
S
S
K
K
S
S
N
N
P
P
G
G
40
|
D
D
F
F
T
T
L
L
S
S
V
V
R
R
R
R
N
N
G
G
50
|
A
A
V
V
T
T
H
H
I
I
K
K
I
I
Q
Q
N
N
T
T
60
|
G
G
D
D
Y
Y
Y
Y
D
D
L
L
Y
Y
G
G
G
G
E
E
70
|
K
K
F
F
A
A
T
T
L
L
A
A
E
K
L
L
V
V
Q
Q
80
|
Y
Y
Y
Y
M
M
E
E
H
H
H
H
G
G
Q
Q
L
L
K
K
90
|
E
E
K
K
N
N
G
G
D
D
V
V
I
I
E
E
L
L
K
K
100
|
Y
Y
P
P
L
L
N
N
C
C
A
A
D
D
P
P
T
T
S
S
110
|
E
E
R
R
W
W
F
F
H
H
G
G
H
H
L
L
S
S
G
G
120
|
K
K
E
E
A
A
E
E
K
K
L
L
L
L
T
T
E
E
K
K
130
|
G
G
K
K
H
H
G
G
S
S
F
F
L
L
V
V
R
R
E
E
140
|
S
S
Q
Q
S
S
H
H
P
P
G
G
D
D
F
F
V
V
L
L
150
|
S
S
V
V
R
R
T
T
G
G
D
D
D
D
K
K
G
G
E
E
160
|
S
S
N
N
D
D
G
G
K
K
S
S
K
K
V
V
T
T
H
H
170
|
V
V
M
M
I
I
R
R
C
C
Q
Q
E
E
L
L
K
K
Y
Y
180
|
D
D
V
V
G
G
G
G
G
G
E
E
R
R
F
F
D
D
S
S
190
|
L
L
T
T
D
D
L
L
V
V
E
E
H
H
Y
Y
K
K
K
K
200
|
N
N
P
P
M
M
V
V
E
E
T
T
L
L
G
G
T
T
V
V
210
|
L
L
Q
Q
L
L
K
K
Q
Q
P
P
L
L
N
N
T
T
T
T
220
|
R
R
I
I
N
N
A
A
A
A
E
E
I
I
E
E
S
S
R
R
230
|
V
V
R
R
E
E
L
L
S
S
K
K
L
L
A
A
E
E
T
T
240
|
T
T
D
D
K
K
V
V
K
K
Q
Q
G
G
F
F
W
W
E
E
250
|
E
E
F
F
E
E
T
T
L
L
Q
Q
Q
Q
Q
Q
E
E
C
C
260
|
K
K
L
L
L
L
Y
Y
S
S
R
R
K
K
E
E
G
G
Q
Q
270
|
R
R
Q
Q
E
E
N
N
K
K
N
N
K
K
N
N
R
R
Y
Y
280
|
K
K
N
N
I
I
L
L
P
P
F
F
D
D
H
H
T
T
R
R
290
|
V
V
V
V
L
L
H
H
D
D
G
G
D
D
P
P
N
N
E
E
300
|
P
P
V
V
S
S
D
D
Y
Y
I
I
N
N
A
A
N
N
I
I
310
|
I
I
M
M
P
P
E
E
F
F
E
E
T
T
K
K
C
C
N
N
320
|
N
N
S
S
K
K
P
P
K
K
K
K
S
S
Y
Y
I
I
A
A
330
|
T
T
Q
Q
G
G
C
C
L
L
Q
Q
N
N
T
T
V
V
N
N
340
|
D
D
F
F
W
W
R
R
M
M
V
V
F
F
Q
Q
E
E
N
N
350
|
S
S
R
R
V
V
I
I
V
V
M
M
T
T
T
T
K
K
E
E
360
|
V
V
E
E
R
R
G
G
K
K
S
S
K
K
C
C
V
V
K
K
370
|
Y
Y
W
W
P
P
D
D
E
E
Y
Y
A
A
L
L
K
K
E
E
380
|
Y
Y
G
G
V
V
M
M
R
R
V
V
R
R
N
N
V
V
K
K
390
|
E
E
S
S
A
A
A
A
H
H
D
D
Y
Y
T
T
L
L
R
R
400
|
E
E
L
L
K
K
L
L
S
S
K
K
V
V
G
G
Q
Q
G
G
410
|
N
N
T
T
E
E
R
R
T
T
V
V
W
W
Q
Q
Y
Y
H
H
420
|
F
F
R
R
T
T
W
W
P
P
D
D
H
H
G
G
V
V
P
P
430
|
S
S
D
D
P
P
G
G
G
G
V
V
L
L
D
D
F
F
L
L
440
|
E
E
E
E
V
V
H
H
H
H
K
K
Q
Q
E
E
S
S
I
I
450
|
M
M
D
D
A
A
G
G
P
P
V
V
V
V
V
V
H
H
C
C
460
|
S
S
A
A
G
G
I
I
G
G
R
R
T
T
G
G
T
T
F
F
470
|
I
I
V
V
I
I
D
D
I
I
L
L
I
I
D
D
I
I
I
I
480
|
R
R
E
E
K
K
G
G
V
V
D
D
C
C
D
D
I
I
D
D
490
|
V
V
P
P
K
K
T
T
I
I
Q
Q
M
M
V
V
R
R
S
S
500
|
Q
Q
R
R
S
S
G
G
M
M
V
V
Q
Q
T
T
E
E
A
A
510
|
Q
Q
Y
Y
R
R
F
F
I
I
Y
Y
M
M
A
A
V
V
Q
Q
520
|
H
H
Y
Y
I
I
E
E
T
T
L
L
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Experiment for Drug Resistance |
Flow cytometry assay | ||||||||||||
| Mechanism Description | The missense mutation p.E76K (c.226G>A) in gene PTPN11 cause the sensitivity of PD98059 by unusual activation of pro-survival pathway | ||||||||||||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | |||||||||||||
|
|
|||||||||||||
| Disease Class: Acute lymphocytic leukemia [ICD-11: 2B33.0] | [6] | ||||||||||||
| Sensitive Disease | Acute lymphocytic leukemia [ICD-11: 2B33.0] | ||||||||||||
| Sensitive Drug | U0126 | ||||||||||||
| Molecule Alteration | Missense mutation | p.E76K (c.226G>A) |
|||||||||||
| Wild Type Structure | Method: X-ray diffraction | Resolution: 1.70 Å | |||||||||||
| Mutant Type Structure | Method: X-ray diffraction | Resolution: 2.62 Å | |||||||||||
| Download The Information of Sequence | Download The Structure File | ||||||||||||
-
0
|
S
G
M
M
T
T
S
S
R
R
R
R
W
W
F
F
H
H
P
P
10
|
N
N
I
I
T
T
G
G
V
V
E
E
A
A
E
E
N
N
L
L
20
|
L
L
L
L
T
T
R
R
G
G
V
V
D
D
G
G
S
S
F
F
30
|
L
L
A
A
R
R
P
P
S
S
K
K
S
S
N
N
P
P
G
G
40
|
D
D
F
F
T
T
L
L
S
S
V
V
R
R
R
R
N
N
G
G
50
|
A
A
V
V
T
T
H
H
I
I
K
K
I
I
Q
Q
N
N
T
T
60
|
G
G
D
D
Y
Y
Y
Y
D
D
L
L
Y
Y
G
G
G
G
E
E
70
|
K
K
F
F
A
A
T
T
L
L
A
A
E
K
L
L
V
V
Q
Q
80
|
Y
Y
Y
Y
M
M
E
E
H
H
H
H
G
G
Q
Q
L
L
K
K
90
|
E
E
K
K
N
N
G
G
D
D
V
V
I
I
E
E
L
L
K
K
100
|
Y
Y
P
P
L
L
N
N
C
C
A
A
D
D
P
P
T
T
S
S
110
|
E
E
R
R
W
W
F
F
H
H
G
G
H
H
L
L
S
S
G
G
120
|
K
K
E
E
A
A
E
E
K
K
L
L
L
L
T
T
E
E
K
K
130
|
G
G
K
K
H
H
G
G
S
S
F
F
L
L
V
V
R
R
E
E
140
|
S
S
Q
Q
S
S
H
H
P
P
G
G
D
D
F
F
V
V
L
L
150
|
S
S
V
V
R
R
T
T
G
G
D
D
D
D
K
K
G
G
E
E
160
|
S
S
N
N
D
D
G
G
K
K
S
S
K
K
V
V
T
T
H
H
170
|
V
V
M
M
I
I
R
R
C
C
Q
Q
E
E
L
L
K
K
Y
Y
180
|
D
D
V
V
G
G
G
G
G
G
E
E
R
R
F
F
D
D
S
S
190
|
L
L
T
T
D
D
L
L
V
V
E
E
H
H
Y
Y
K
K
K
K
200
|
N
N
P
P
M
M
V
V
E
E
T
T
L
L
G
G
T
T
V
V
210
|
L
L
Q
Q
L
L
K
K
Q
Q
P
P
L
L
N
N
T
T
T
T
220
|
R
R
I
I
N
N
A
A
A
A
E
E
I
I
E
E
S
S
R
R
230
|
V
V
R
R
E
E
L
L
S
S
K
K
L
L
A
A
E
E
T
T
240
|
T
T
D
D
K
K
V
V
K
K
Q
Q
G
G
F
F
W
W
E
E
250
|
E
E
F
F
E
E
T
T
L
L
Q
Q
Q
Q
Q
Q
E
E
C
C
260
|
K
K
L
L
L
L
Y
Y
S
S
R
R
K
K
E
E
G
G
Q
Q
270
|
R
R
Q
Q
E
E
N
N
K
K
N
N
K
K
N
N
R
R
Y
Y
280
|
K
K
N
N
I
I
L
L
P
P
F
F
D
D
H
H
T
T
R
R
290
|
V
V
V
V
L
L
H
H
D
D
G
G
D
D
P
P
N
N
E
E
300
|
P
P
V
V
S
S
D
D
Y
Y
I
I
N
N
A
A
N
N
I
I
310
|
I
I
M
M
P
P
E
E
F
F
E
E
T
T
K
K
C
C
N
N
320
|
N
N
S
S
K
K
P
P
K
K
K
K
S
S
Y
Y
I
I
A
A
330
|
T
T
Q
Q
G
G
C
C
L
L
Q
Q
N
N
T
T
V
V
N
N
340
|
D
D
F
F
W
W
R
R
M
M
V
V
F
F
Q
Q
E
E
N
N
350
|
S
S
R
R
V
V
I
I
V
V
M
M
T
T
T
T
K
K
E
E
360
|
V
V
E
E
R
R
G
G
K
K
S
S
K
K
C
C
V
V
K
K
370
|
Y
Y
W
W
P
P
D
D
E
E
Y
Y
A
A
L
L
K
K
E
E
380
|
Y
Y
G
G
V
V
M
M
R
R
V
V
R
R
N
N
V
V
K
K
390
|
E
E
S
S
A
A
A
A
H
H
D
D
Y
Y
T
T
L
L
R
R
400
|
E
E
L
L
K
K
L
L
S
S
K
K
V
V
G
G
Q
Q
G
G
410
|
N
N
T
T
E
E
R
R
T
T
V
V
W
W
Q
Q
Y
Y
H
H
420
|
F
F
R
R
T
T
W
W
P
P
D
D
H
H
G
G
V
V
P
P
430
|
S
S
D
D
P
P
G
G
G
G
V
V
L
L
D
D
F
F
L
L
440
|
E
E
E
E
V
V
H
H
H
H
K
K
Q
Q
E
E
S
S
I
I
450
|
M
M
D
D
A
A
G
G
P
P
V
V
V
V
V
V
H
H
C
C
460
|
S
S
A
A
G
G
I
I
G
G
R
R
T
T
G
G
T
T
F
F
470
|
I
I
V
V
I
I
D
D
I
I
L
L
I
I
D
D
I
I
I
I
480
|
R
R
E
E
K
K
G
G
V
V
D
D
C
C
D
D
I
I
D
D
490
|
V
V
P
P
K
K
T
T
I
I
Q
Q
M
M
V
V
R
R
S
S
500
|
Q
Q
R
R
S
S
G
G
M
M
V
V
Q
Q
T
T
E
E
A
A
510
|
Q
Q
Y
Y
R
R
F
F
I
I
Y
Y
M
M
A
A
V
V
Q
Q
520
|
H
H
Y
Y
I
I
E
E
T
T
L
L
|
|||||||||||||
| Experimental Note | Revealed Based on the Cell Line Data | ||||||||||||
| Experiment for Molecule Alteration |
Immunoblotting analysis | ||||||||||||
| Experiment for Drug Resistance |
Flow cytometry assay | ||||||||||||
| Mechanism Description | The missense mutation p.E76K (c.226G>A) in gene PTPN11 cause the sensitivity of U0126 by unusual activation of pro-survival pathway | ||||||||||||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Bone marrow | |
| The Specified Disease | Acute myeloid leukemia | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 8.25E-23; Fold-change: 2.78E-01; Z-score: 1.24E+00 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
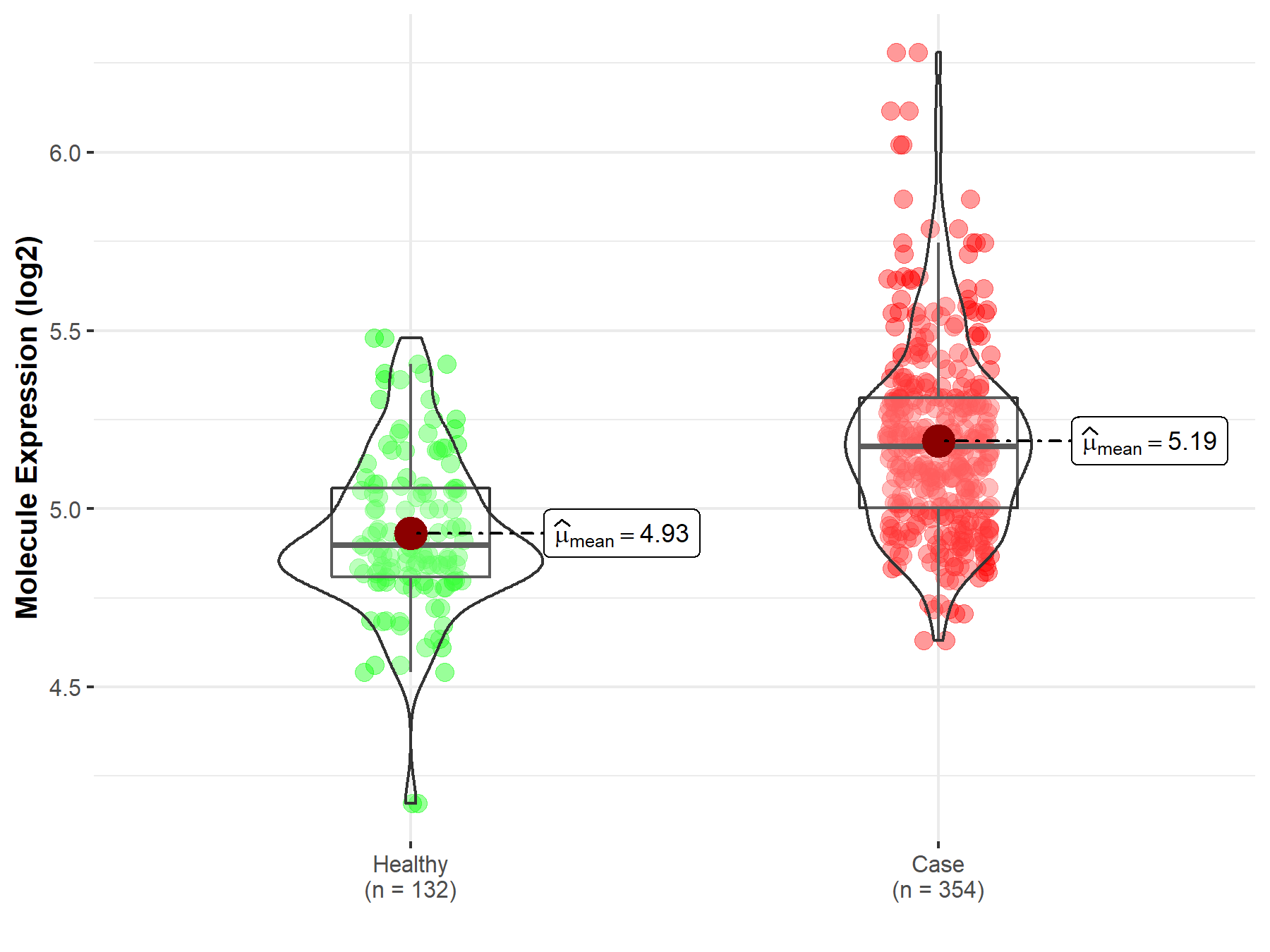
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Liver | |
| The Specified Disease | Liver cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.46E-01; Fold-change: 9.97E-02; Z-score: 1.41E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 7.56E-18; Fold-change: 4.40E-01; Z-score: 8.85E-01 | |
| The Expression Level of Disease Section Compare with the Other Disease Section | p-value: 5.62E-01; Fold-change: 2.16E-01; Z-score: 2.50E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
Molecule expression in tissue other than the diseased tissue of patients
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

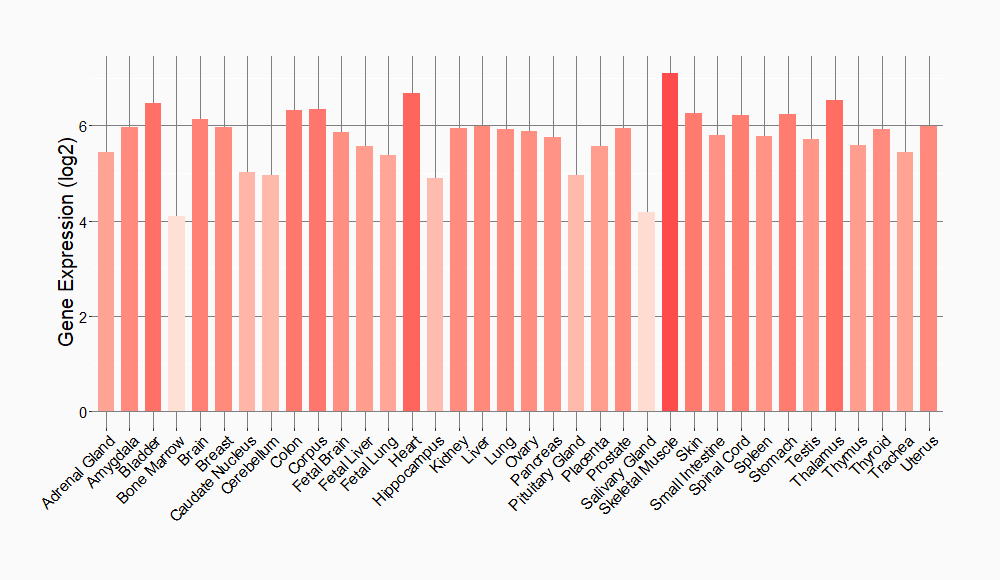
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
