Molecule Information
General Information of the Molecule (ID: Mol00137)
| Name |
Platelet-derived growth factor receptor alpha (PDGFRA)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Molecule Type |
Protein
|
||||
| Gene Name |
PDGFRA
|
||||
| Gene ID | |||||
| Location |
chr4:54229280-54298245[+]
|
||||
| Sequence |
MGTSHPAFLVLGCLLTGLSLILCQLSLPSILPNENEKVVQLNSSFSLRCFGESEVSWQYP
MSEEESSDVEIRNEENNSGLFVTVLEVSSASAAHTGLYTCYYNHTQTEENELEGRHIYIY VPDPDVAFVPLGMTDYLVIVEDDDSAIIPCRTTDPETPVTLHNSEGVVPASYDSRQGFNG TFTVGPYICEATVKGKKFQTIPFNVYALKATSELDLEMEALKTVYKSGETIVVTCAVFNN EVVDLQWTYPGEVKGKGITMLEEIKVPSIKLVYTLTVPEATVKDSGDYECAARQATREVK EMKKVTISVHEKGFIEIKPTFSQLEAVNLHEVKHFVVEVRAYPPPRISWLKNNLTLIENL TEITTDVEKIQEIRYRSKLKLIRAKEEDSGHYTIVAQNEDAVKSYTFELLTQVPSSILDL VDDHHGSTGGQTVRCTAEGTPLPDIEWMICKDIKKCNNETSWTILANNVSNIITEIHSRD RSTVEGRVTFAKVEETIAVRCLAKNLLGAENRELKLVAPTLRSELTVAAAVLVLLVIVII SLIVLVVIWKQKPRYEIRWRVIESISPDGHEYIYVDPMQLPYDSRWEFPRDGLVLGRVLG SGAFGKVVEGTAYGLSRSQPVMKVAVKMLKPTARSSEKQALMSELKIMTHLGPHLNIVNL LGACTKSGPIYIITEYCFYGDLVNYLHKNRDSFLSHHPEKPKKELDIFGLNPADESTRSY VILSFENNGDYMDMKQADTTQYVPMLERKEVSKYSDIQRSLYDRPASYKKKSMLDSEVKN LLSDDNSEGLTLLDLLSFTYQVARGMEFLASKNCVHRDLAARNVLLAQGKIVKICDFGLA RDIMHDSNYVSKGSTFLPVKWMAPESIFDNLYTTLSDVWSYGILLWEIFSLGGTPYPGMM VDSTFYNKIKSGYRMAKPDHATSEVYEIMVKCWNSEPEKRPSFYHLSEIVENLLPGQYKK SYEKIHLDFLKSDHPAVARMRVDSDNAYIGVTYKNEEDKLKDWEGGLDEQRLSADSGYII PLPDIDPVPEEEDLGKRNRHSSQTSEESAIETGSSSSTFIKREDETIEDIDMMDDIGIDS SDLVEDSFL Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Tyrosine-protein kinase that acts as a cell-surface receptor for PDGFA, PDGFB and PDGFC and plays an essential role in the regulation of embryonic development, cell proliferation, survival and chemotaxis. Depending on the context, promotes or inhibits cell proliferation and cell migration. Plays an important role in the differentiation of bone marrow-derived mesenchymal stem cells. Required for normal skeleton development and cephalic closure during embryonic development. Required for normal development of the mucosa lining the gastrointestinal tract, and for recruitment of mesenchymal cells and normal development of intestinal villi. Plays a role in cell migration and chemotaxis in wound healing. Plays a role in platelet activation, secretion of agonists from platelet granules, and in thrombin-induced platelet aggregation. Binding of its cognate ligands - homodimeric PDGFA, homodimeric PDGFB, heterodimers formed by PDGFA and PDGFB or homodimeric PDGFC -leads to the activation of several signaling cascades; the response depends on the nature of the bound ligand and is modulated by the formation of heterodimers between PDGFRA and PDGFRB. Phosphorylates PIK3R1, PLCG1, and PTPN11. Activation of PLCG1 leads to the production of the cellular signaling molecules diacylglycerol and inositol 1,4,5-trisphosphate, mobilization of cytosolic Ca(2+) and the activation of protein kinase C. Phosphorylates PIK3R1, the regulatory subunit of phosphatidylinositol 3-kinase, and thereby mediates activation of the AKT1 signaling pathway. Mediates activation of HRAS and of the MAP kinases MAPK1/ERK2 and/or MAPK3/ERK1. Promotes activation of STAT family members STAT1, STAT3 and STAT5A and/or STAT5B. Receptor signaling is down-regulated by protein phosphatases that dephosphorylate the receptor and its down-stream effectors, and by rapid internalization of the activated receptor.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
9 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Renal cell carcinoma [ICD-11: 2C90.0] | [1] | |||
| Resistant Disease | Renal cell carcinoma [ICD-11: 2C90.0] | |||
| Resistant Drug | Sunitinib | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Kidney cancer [ICD-11: 2C90] | |||
| The Specified Disease | Renal cell carcinoma | |||
| The Studied Tissue | Blood | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.44E-02 Fold-change: 4.10E-01 Z-score: 2.85E+00 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Caki-2 cells | Kidney | Homo sapiens (Human) | CVCL_0235 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay | |||
| Mechanism Description | High miR-942 levels in MRCC cells up-regulates MMP-9 and VEGF secretion to enhance endothelial migration and sunitinib resistance. | |||
| Disease Class: Gastrointestinal stromal cancer [ICD-11: 2B5B.1] | [11] | |||
| Resistant Disease | Gastrointestinal stromal cancer [ICD-11: 2B5B.1] | |||
| Resistant Drug | Sunitinib | |||
| Molecule Alteration | Missense mutation | p.D842V |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Experiment for Molecule Alteration |
Next-generation sequencing assay; Circulating-free DNA assay | |||
| Experiment for Drug Resistance |
Computerized tomography assay | |||
| Mechanism Description | We were able to identify primary kIT mutations in all plasma samples. Additional mutations, including kIT exon 17 S821F and PDGFRA exon 18 D842V, were detected in the patient-matched plasma samples during follow-up and appeared to result in decreased sensitivity to TkIs. Our results demonstrate an approach by which primary and secondary mutations are readily detected in blood-derived circulating tumor DNA from patients with GIST. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | [2] | |||
| Resistant Disease | Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | |||
| Resistant Drug | Avapritinib | |||
| Molecule Alteration | Missense mutation | p.V658A |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | U373 cells | Brain | Homo sapiens (Human) | CVCL_2219 |
| NOMO1 cells | Bone marrow | Homo sapiens (Human) | CVCL_1609 | |
| Trsh1 cells | Stomach | Homo sapiens (Human) | N.A. | |
| Experiment for Molecule Alteration |
Whole genome sequencing assay | |||
| Experiment for Drug Resistance |
SRB assay | |||
| Mechanism Description | Tumor and plasma biopsies in 6 of 7 patients with PDGFRA primary mutations who progressed on avapritinib or imatinib had secondary resistance mutations within PDGFRA exons 13, 14, and 15 that interfere with avapritinib binding. Secondary PDGFRA mutations causing V658A, N659K, Y676C, and G680R substitutions were found in 2 or more patients each, representing recurrent mechanisms of PDGFRA GIST drug resistance. | |||
| Disease Class: Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | [2] | |||
| Resistant Disease | Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | |||
| Resistant Drug | Avapritinib | |||
| Molecule Alteration | Missense mutation | p.N659K |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | U373 cells | Brain | Homo sapiens (Human) | CVCL_2219 |
| NOMO1 cells | Bone marrow | Homo sapiens (Human) | CVCL_1609 | |
| Trsh1 cells | Stomach | Homo sapiens (Human) | N.A. | |
| Experiment for Molecule Alteration |
Whole genome sequencing assay | |||
| Experiment for Drug Resistance |
SRB assay | |||
| Mechanism Description | Tumor and plasma biopsies in 6 of 7 patients with PDGFRA primary mutations who progressed on avapritinib or imatinib had secondary resistance mutations within PDGFRA exons 13, 14, and 15 that interfere with avapritinib binding. Secondary PDGFRA mutations causing V658A, N659K, Y676C, and G680R substitutions were found in 2 or more patients each, representing recurrent mechanisms of PDGFRA GIST drug resistance. | |||
| Disease Class: Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | [2] | |||
| Resistant Disease | Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | |||
| Resistant Drug | Avapritinib | |||
| Molecule Alteration | Missense mutation | p.Y676C |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | U373 cells | Brain | Homo sapiens (Human) | CVCL_2219 |
| NOMO1 cells | Bone marrow | Homo sapiens (Human) | CVCL_1609 | |
| Trsh1 cells | Stomach | Homo sapiens (Human) | N.A. | |
| Experiment for Molecule Alteration |
Whole genome sequencing assay | |||
| Experiment for Drug Resistance |
SRB assay | |||
| Mechanism Description | Tumor and plasma biopsies in 6 of 7 patients with PDGFRA primary mutations who progressed on avapritinib or imatinib had secondary resistance mutations within PDGFRA exons 13, 14, and 15 that interfere with avapritinib binding. Secondary PDGFRA mutations causing V658A, N659K, Y676C, and G680R substitutions were found in 2 or more patients each, representing recurrent mechanisms of PDGFRA GIST drug resistance. | |||
| Disease Class: Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | [2] | |||
| Resistant Disease | Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | |||
| Resistant Drug | Avapritinib | |||
| Molecule Alteration | Missense mutation | p.G680R |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | U373 cells | Brain | Homo sapiens (Human) | CVCL_2219 |
| NOMO1 cells | Bone marrow | Homo sapiens (Human) | CVCL_1609 | |
| Trsh1 cells | Stomach | Homo sapiens (Human) | N.A. | |
| Experiment for Molecule Alteration |
Whole genome sequencing assay | |||
| Experiment for Drug Resistance |
SRB assay | |||
| Mechanism Description | Tumor and plasma biopsies in 6 of 7 patients with PDGFRA primary mutations who progressed on avapritinib or imatinib had secondary resistance mutations within PDGFRA exons 13, 14, and 15 that interfere with avapritinib binding. Secondary PDGFRA mutations causing V658A, N659K, Y676C, and G680R substitutions were found in 2 or more patients each, representing recurrent mechanisms of PDGFRA GIST drug resistance. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [3] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Avapritinib | |||
| Molecule Alteration | Missense mutation | p.D842V (c.2525A>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Kasumi-1 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0589 |
| HMC-1.2 cells | Blood | Homo sapiens (Human) | CVCL_H205 | |
| P815 cells | N.A. | Mus musculus (Mouse) | CVCL_2154 | |
| M-07e cells | Peripheral blood | Homo sapiens (Human) | CVCL_2106 | |
| HMC-1.1 cells | Peripheral blood | Homo sapiens (Human) | CVCL_H206 | |
| Chinese hamster ovary (CHO)-K1 cells | Ovary | Cricetulus griseus (Chinese hamster) (Cricetulus barabensis griseus) | CVCL_0214 | |
| In Vivo Model | BALB/c nude mouse PDX model | Mus musculus | ||
| Experiment for Molecule Alteration |
Immunoblotting analysis | |||
| Experiment for Drug Resistance |
Enzyme-linked immunosorbent assay; Cellular proliferation test assay | |||
| Disease Class: Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | [3] | |||
| Sensitive Disease | Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | |||
| Sensitive Drug | Avapritinib | |||
| Molecule Alteration | Missense mutation | p.D842V (c.2525A>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Kasumi-1 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0589 |
| HMC-1.2 cells | Blood | Homo sapiens (Human) | CVCL_H205 | |
| P815 cells | N.A. | Mus musculus (Mouse) | CVCL_2154 | |
| M-07e cells | Peripheral blood | Homo sapiens (Human) | CVCL_2106 | |
| HMC-1.1 cells | Peripheral blood | Homo sapiens (Human) | CVCL_H206 | |
| Chinese hamster ovary (CHO)-K1 cells | Ovary | Cricetulus griseus (Chinese hamster) (Cricetulus barabensis griseus) | CVCL_0214 | |
| In Vivo Model | BALB/c nude mouse PDX model | Mus musculus | ||
| Experiment for Molecule Alteration |
Immunoblotting analysis | |||
| Experiment for Drug Resistance |
Enzyme-linked immunosorbent assay; Cellular proliferation test assay | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastrointestinal stromal cancer [ICD-11: 2B5B.1] | [4] | |||
| Resistant Disease | Gastrointestinal stromal cancer [ICD-11: 2B5B.1] | |||
| Resistant Drug | Imatinib | |||
| Molecule Alteration | Missense mutation | p.D842_D846>G |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | MAPK/STAT3 signaling pathway | Activation | hsa01521 | |
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | ||
| Experiment for Molecule Alteration |
Sanger sequencing assay | |||
| Experiment for Drug Resistance |
Radiological response evaluation assay; Pathological response evaluation assay | |||
| Mechanism Description | The most common PDGFRA mutation, a D842_D846delinsG shows primary resistance to imatinib in the patients. | |||
| Disease Class: Gastrointestinal stromal cancer [ICD-11: 2B5B.1] | [4] | |||
| Resistant Disease | Gastrointestinal stromal cancer [ICD-11: 2B5B.1] | |||
| Resistant Drug | Imatinib | |||
| Molecule Alteration | Missense mutation | p.I843_S847>T |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | MAPK/STAT3 signaling pathway | Activation | hsa01521 | |
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | ||
| Experiment for Molecule Alteration |
Sanger sequencing assay | |||
| Experiment for Drug Resistance |
Radiological response evaluation assay; Pathological response evaluation assay | |||
| Mechanism Description | The most common PDGFRA mutation, a D842V substitution in exon 18, shows primary resistance to imatinib in in vitro and in vivo studies. | |||
| Disease Class: Gastrointestinal stromal cancer [ICD-11: 2B5B.1] | [5], [6], [7] | |||
| Resistant Disease | Gastrointestinal stromal cancer [ICD-11: 2B5B.1] | |||
| Resistant Drug | Imatinib | |||
| Molecule Alteration | Missense mutation | p.D842V |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | MAPK/STAT3 signaling pathway | Activation | hsa01521 | |
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| In Vivo Model | A retrospective survey in conducting clinical studies | Homo sapiens | ||
| Experiment for Molecule Alteration |
Sanger sequencing assay | |||
| Experiment for Drug Resistance |
Radiological response evaluation assay; Pathological response evaluation assay | |||
| Mechanism Description | The most common PDGFRA mutation, a I843_S847delinsT shows primary resistance to imatinib in the patients. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [8] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Midostaurin | |||
| Molecule Alteration | Missense mutation | p.D842V (c.2525A>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 |
| THP-1 cells | Blood | Homo sapiens (Human) | CVCL_0006 | |
| Kasumi-1 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0589 | |
| H1703 cells | Lung | Homo sapiens (Human) | CVCL_1490 | |
| HCT-116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| HMC-1.2 cells | Blood | Homo sapiens (Human) | CVCL_H205 | |
| P815 cells | N.A. | Mus musculus (Mouse) | CVCL_2154 | |
| MV-4-11 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0064 | |
| HMC-1.1 cells | Peripheral blood | Homo sapiens (Human) | CVCL_H206 | |
| EOL1 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0258 | |
| CHO-K1 cells | Ovary | Cricetulus griseus (Chinese hamster) (Cricetulus barabensis griseus) | CVCL_0214 | |
| In Vivo Model | Female Hsd:Athymic Nude-Foxn1nu nude mouse xenograft model | Mus musculus | ||
| Experiment for Drug Resistance |
IC50 assay | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [8] | |||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Resistant Drug | Regorafenib | |||
| Molecule Alteration | Missense mutation | p.D842V (c.2525A>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 |
| THP-1 cells | Blood | Homo sapiens (Human) | CVCL_0006 | |
| Kasumi-1 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0589 | |
| H1703 cells | Lung | Homo sapiens (Human) | CVCL_1490 | |
| HCT-116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| HMC-1.2 cells | Blood | Homo sapiens (Human) | CVCL_H205 | |
| P815 cells | N.A. | Mus musculus (Mouse) | CVCL_2154 | |
| MV-4-11 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0064 | |
| HMC-1.1 cells | Peripheral blood | Homo sapiens (Human) | CVCL_H206 | |
| EOL1 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0258 | |
| CHO-K1 cells | Ovary | Cricetulus griseus (Chinese hamster) (Cricetulus barabensis griseus) | CVCL_0214 | |
| In Vivo Model | Female Hsd:Athymic Nude-Foxn1nu nude mouse xenograft model | Mus musculus | ||
| Experiment for Drug Resistance |
IC50 assay | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | [9] | |||
| Sensitive Disease | Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | |||
| Sensitive Drug | Regorafenib | |||
| Molecule Alteration | IF-deletion | p.C814_S854 (c.2440_2562) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Disease Class: Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | [10] | |||
| Sensitive Disease | Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | |||
| Sensitive Drug | Regorafenib | |||
| Molecule Alteration | Missense mutation | p.D842V (c.2525A>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Gastrointestinal tract | N.A. | ||
| Experiment for Drug Resistance |
CT scan assay; MRI assay | |||
| Disease Class: Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | [9] | |||
| Sensitive Disease | Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | |||
| Sensitive Drug | Regorafenib | |||
| Molecule Alteration | Missense mutation | p.Y894C |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Disease Class: Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | [9] | |||
| Sensitive Disease | Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | |||
| Sensitive Drug | Regorafenib | |||
| Molecule Alteration | IF-deletion | p.K552_G596 (c.1654_1788) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Disease Class: Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | [9] | |||
| Sensitive Disease | Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | |||
| Sensitive Drug | Regorafenib | |||
| Molecule Alteration | IF-deletion | p.P631_G668 (c.1891_2004) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Disease Class: Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | [9] | |||
| Sensitive Disease | Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | |||
| Sensitive Drug | Regorafenib | |||
| Molecule Alteration | Missense mutation | p.R748G (c.2242A>G) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [8] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Ripretinib | |||
| Molecule Alteration | Missense mutation | p.D842V (c.2525A>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 |
| THP-1 cells | Blood | Homo sapiens (Human) | CVCL_0006 | |
| Kasumi-1 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0589 | |
| H1703 cells | Lung | Homo sapiens (Human) | CVCL_1490 | |
| HCT-116 cells | Colon | Homo sapiens (Human) | CVCL_0291 | |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| HMC-1.2 cells | Blood | Homo sapiens (Human) | CVCL_H205 | |
| P815 cells | N.A. | Mus musculus (Mouse) | CVCL_2154 | |
| MV-4-11 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0064 | |
| HMC-1.1 cells | Peripheral blood | Homo sapiens (Human) | CVCL_H206 | |
| EOL1 cells | Peripheral blood | Homo sapiens (Human) | CVCL_0258 | |
| CHO-K1 cells | Ovary | Cricetulus griseus (Chinese hamster) (Cricetulus barabensis griseus) | CVCL_0214 | |
| In Vivo Model | Female Hsd:Athymic Nude-Foxn1nu nude mouse xenograft model | Mus musculus | ||
| Experiment for Drug Resistance |
IC50 assay | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [12] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Tamoxifen | |||
| Molecule Alteration | Missense mutation | p.D714E |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Angiogenic potential | Inhibition | hsa04370 | |
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Circulating-free DNA assay; Whole exome sequencing assay | |||
| Mechanism Description | Quantification of allele fractions in plasma identified increased representation of mutant alleles in association with emergence of therapy resistance. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [13] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Trametinib | |||
| Molecule Alteration | Missense mutation | p.Y288C (c.863A>G) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Presto blue assay | |||
| Mechanism Description | PDGFRA Y288C induces constitutive phosphorylation of Akt, ERK1/2, and STAT3. PDGFRA Y288C is resistant to PDGFR inhibitors, such as crenolanib, but sensitive to PI3K/mTOR and MEK inhibitors, such as omipalisib, consistent with pathway activation results. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [12] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Trastuzumab | |||
| Molecule Alteration | Missense mutation | p.D714E |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | AKT signaling pathway | Activation | hsa04151 | |
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Circulating-free DNA assay; Whole exome sequencing assay | |||
| Mechanism Description | Quantification of allele fractions in plasma identified increased representation of mutant alleles in association with emergence of therapy resistance. | |||
Clinical Trial Drug(s)
2 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [13] | |||
| Resistant Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Resistant Drug | Crenolanib | |||
| Molecule Alteration | Missense mutation | p.Y288C (c.863A>G) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Presto blue assay | |||
| Mechanism Description | PDGFRA Y288C induces constitutive phosphorylation of Akt, ERK1/2, and STAT3. PDGFRA Y288C is resistant to PDGFR inhibitors, such as crenolanib, but sensitive to PI3K/mTOR and MEK inhibitors, such as omipalisib, consistent with pathway activation results. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [13] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Crenolanib | |||
| Molecule Alteration | Missense mutation | p.D842V (c.2525A>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | BaF3 cells | Bone | Mus musculus (Mouse) | CVCL_0161 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
XTT assay | |||
| Disease Class: Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | [14] | |||
| Sensitive Disease | Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | |||
| Sensitive Drug | Crenolanib | |||
| Molecule Alteration | Missense mutation | p.D842V (c.2525A>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [13] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Crenolanib | |||
| Molecule Alteration | Missense mutation | p.V561D (c.1682T>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Presto blue assay | |||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [15] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Crenolanib | |||
| Molecule Alteration | Missense mutation | p.P577S (c.1729C>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Mechanism Description | The missense mutation p.P577S (c.1729C>T) in gene PDGFRA cause the sensitivity of Crenolanib by aberration of the drug's therapeutic target | |||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [15] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Crenolanib | |||
| Molecule Alteration | Missense mutation | p.V658A (c.1973T>C) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Mechanism Description | The missense mutation p.V658A (c.1973T>C) in gene PDGFRA cause the sensitivity of Crenolanib by aberration of the drug's therapeutic target | |||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [15] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Crenolanib | |||
| Molecule Alteration | Missense mutation | p.R841K (c.2522G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Mechanism Description | The missense mutation p.R841K (c.2522G>A) in gene PDGFRA cause the sensitivity of Crenolanib by aberration of the drug's therapeutic target | |||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [15] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Crenolanib | |||
| Molecule Alteration | Missense mutation | p.D842Y (c.2524G>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Mechanism Description | The missense mutation p.D842Y (c.2524G>T) in gene PDGFRA cause the sensitivity of Crenolanib by aberration of the drug's therapeutic target | |||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [15] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Crenolanib | |||
| Molecule Alteration | Missense mutation | p.H845Y (c.2533C>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Mechanism Description | The missense mutation p.H845Y (c.2533C>T) in gene PDGFRA cause the sensitivity of Crenolanib by aberration of the drug's therapeutic target | |||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [15] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Crenolanib | |||
| Molecule Alteration | Missense mutation | p.G853D (c.2558G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Mechanism Description | The missense mutation p.G853D (c.2558G>A) in gene PDGFRA cause the sensitivity of Crenolanib by aberration of the drug's therapeutic target | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [15] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Crenolanib | |||
| Molecule Alteration | Missense mutation | p.P577S (c.1729C>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Mechanism Description | The missense mutation p.P577S (c.1729C>T) in gene PDGFRA cause the sensitivity of Crenolanib by aberration of the drug's therapeutic target | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [15] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Crenolanib | |||
| Molecule Alteration | Missense mutation | p.V658A (c.1973T>C) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Mechanism Description | The missense mutation p.V658A (c.1973T>C) in gene PDGFRA cause the sensitivity of Crenolanib by aberration of the drug's therapeutic target | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [15] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Crenolanib | |||
| Molecule Alteration | Missense mutation | p.R841K (c.2522G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Mechanism Description | The missense mutation p.R841K (c.2522G>A) in gene PDGFRA cause the sensitivity of Crenolanib by aberration of the drug's therapeutic target | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [15] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Crenolanib | |||
| Molecule Alteration | Missense mutation | p.H845Y (c.2533C>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Mechanism Description | The missense mutation p.H845Y (c.2533C>T) in gene PDGFRA cause the sensitivity of Crenolanib by aberration of the drug's therapeutic target | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [15] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Crenolanib | |||
| Molecule Alteration | Missense mutation | p.G853D (c.2558G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Mechanism Description | The missense mutation p.G853D (c.2558G>A) in gene PDGFRA cause the sensitivity of Crenolanib by aberration of the drug's therapeutic target | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [15] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Crenolanib | |||
| Molecule Alteration | Missense mutation | p.P577S (c.1729C>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Mechanism Description | The missense mutation p.P577S (c.1729C>T) in gene PDGFRA cause the sensitivity of Crenolanib by aberration of the drug's therapeutic target | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [15] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Crenolanib | |||
| Molecule Alteration | Missense mutation | p.V658A (c.1973T>C) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Mechanism Description | The missense mutation p.V658A (c.1973T>C) in gene PDGFRA cause the sensitivity of Crenolanib by aberration of the drug's therapeutic target | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [15] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Crenolanib | |||
| Molecule Alteration | Missense mutation | p.R841K (c.2522G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Mechanism Description | The missense mutation p.R841K (c.2522G>A) in gene PDGFRA cause the sensitivity of Crenolanib by aberration of the drug's therapeutic target | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [15] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Crenolanib | |||
| Molecule Alteration | Missense mutation | p.H845Y (c.2533C>T) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Mechanism Description | The missense mutation p.H845Y (c.2533C>T) in gene PDGFRA cause the sensitivity of Crenolanib by aberration of the drug's therapeutic target | |||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [15] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Crenolanib | |||
| Molecule Alteration | Missense mutation | p.G853D (c.2558G>A) |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | 293T cells | Breast | Homo sapiens (Human) | CVCL_0063 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK-8 assay | |||
| Mechanism Description | The missense mutation p.G853D (c.2558G>A) in gene PDGFRA cause the sensitivity of Crenolanib by aberration of the drug's therapeutic target | |||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [16] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Crenolanib | |||
| Molecule Alteration | Missense mutation | p.N659K (c.1977C>G) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
Biochemical assessment of PDGFRA/KIT kinase activity assay | |||
| Experiment for Drug Resistance |
XTT assay | |||
| Mechanism Description | The missense mutation p.N659K (c.1977C>G) in gene PDGFRA cause the sensitivity of Crenolanib by aberration of the drug's therapeutic target | |||
| Disease Class: Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | [16] | |||
| Sensitive Disease | Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | |||
| Sensitive Drug | Crenolanib | |||
| Molecule Alteration | Missense mutation | p.D842Y (c.2524G>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
Biochemical assessment of PDGFRA/KIT kinase activity assay | |||
| Experiment for Drug Resistance |
XTT assay | |||
| Mechanism Description | The missense mutation p.D842Y (c.2524G>T) in gene PDGFRA cause the sensitivity of Crenolanib by aberration of the drug's therapeutic target | |||
| Disease Class: Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | [16] | |||
| Sensitive Disease | Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | |||
| Sensitive Drug | Crenolanib | |||
| Molecule Alteration | Missense mutation | p.D842I (c.2524_2525delGAinsAT) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
Biochemical assessment of PDGFRA/KIT kinase activity assay | |||
| Experiment for Drug Resistance |
XTT assay | |||
| Mechanism Description | The missense mutation p.D842I (c.2524_2525delGAinsAT) in gene PDGFRA cause the sensitivity of Crenolanib by aberration of the drug's therapeutic target | |||
| Disease Class: Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | [16] | |||
| Sensitive Disease | Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | |||
| Sensitive Drug | Crenolanib | |||
| Molecule Alteration | Complex-indel | p.D842_I843delinsVM (c.2524_2529delinsGTAATG) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
Biochemical assessment of PDGFRA/KIT kinase activity assay | |||
| Experiment for Drug Resistance |
XTT assay | |||
| Mechanism Description | The complex-indel p.D842_I843delinsVM (c.2524_2529delinsGTAATG) in gene PDGFRA cause the sensitivity of Crenolanib by aberration of the drug's therapeutic target. | |||
| Disease Class: Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | [16] | |||
| Sensitive Disease | Gastrointestinal stromal tumor [ICD-11: 2B5B.0] | |||
| Sensitive Drug | Crenolanib | |||
| Molecule Alteration | IF-deletion | p.I843delI (c.2529_2531delCAT) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
Biochemical assessment of PDGFRA/KIT kinase activity assay | |||
| Experiment for Drug Resistance |
XTT assay | |||
| Mechanism Description | The if-deletion p.I843delI (c.2529_2531delCAT) in gene PDGFRA cause the sensitivity of Crenolanib by aberration of the drug's therapeutic target. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [13] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Omipalisib | |||
| Molecule Alteration | Missense mutation | p.Y288C (c.863A>G) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Presto blue assay | |||
| Mechanism Description | PDGFRA Y288C induces constitutive phosphorylation of Akt, ERK1/2, and STAT3. PDGFRA Y288C is resistant to PDGFR inhibitors, such as crenolanib, but sensitive to PI3K/mTOR and MEK inhibitors, such as omipalisib and trametinib, consistent with pathway activation results. | |||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [13] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Omipalisib | |||
| Molecule Alteration | Missense mutation | p.V561D (c.1682T>A) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Presto blue assay | |||
| Disease Class: Solid tumour/cancer [ICD-11: 2A00-2F9Z] | [13] | |||
| Sensitive Disease | Solid tumour/cancer [ICD-11: 2A00-2F9Z] | |||
| Sensitive Drug | Omipalisib | |||
| Molecule Alteration | Missense mutation | p.D842V (c.2525A>T) |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF10A cells | Breast | Homo sapiens (Human) | CVCL_0598 |
| Ba/F3 cells | Colon | Homo sapiens (Human) | CVCL_0161 | |
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Presto blue assay | |||
| Mechanism Description | PDGFRA Y288C induces constitutive phosphorylation of Akt, ERK1/2, and STAT3. PDGFRA Y288C is resistant to PDGFR inhibitors, such as crenolanib, but sensitive to PI3K/mTOR and MEK inhibitors, such as omipalisib, consistent with pathway activation results. | |||
Investigative Drug(s)
1 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [12] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Tamoxifen/Trastuzumab | |||
| Molecule Alteration | Missense mutation | p.D714E |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | ALK signaling pathway | Activation | hsa05200 | |
| In Vitro Model | Plasma | Blood | Homo sapiens (Human) | N.A. |
| Experiment for Molecule Alteration |
Circulating-free DNA assay; Whole exome sequencing assay | |||
| Mechanism Description | Quantification of allele fractions in plasma identified increased representation of mutant alleles in association with emergence of therapy resistance. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Skin | |
| The Specified Disease | Melanoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.96E-01; Fold-change: -2.00E-01; Z-score: -2.93E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
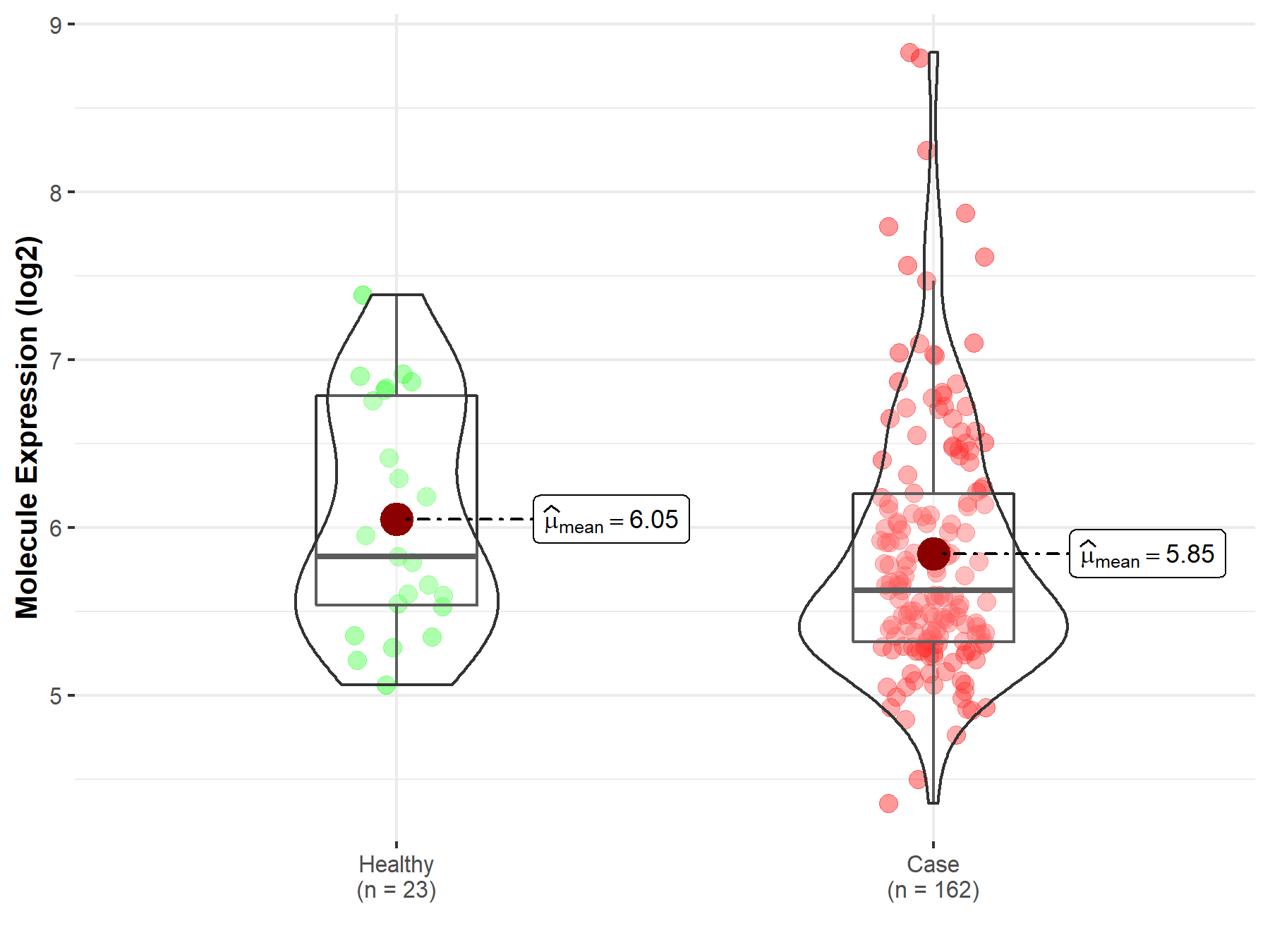
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 3.48E-63; Fold-change: -4.08E-01; Z-score: -1.52E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 1.14E-09; Fold-change: -3.72E-01; Z-score: -1.14E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
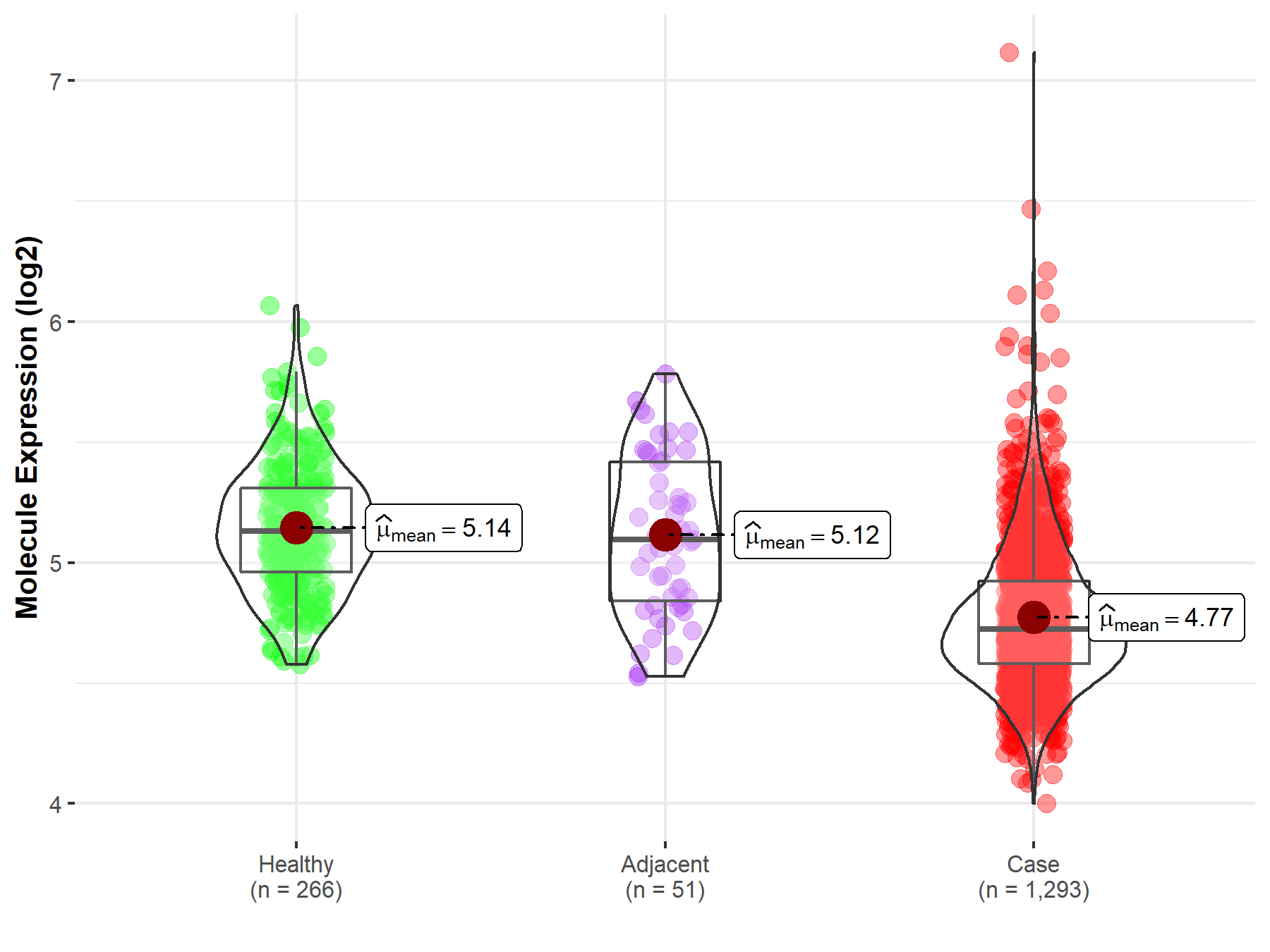
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Kidney | |
| The Specified Disease | Kidney cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.48E-03; Fold-change: -3.79E-01; Z-score: -1.26E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 9.43E-03; Fold-change: -1.19E-01; Z-score: -6.22E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
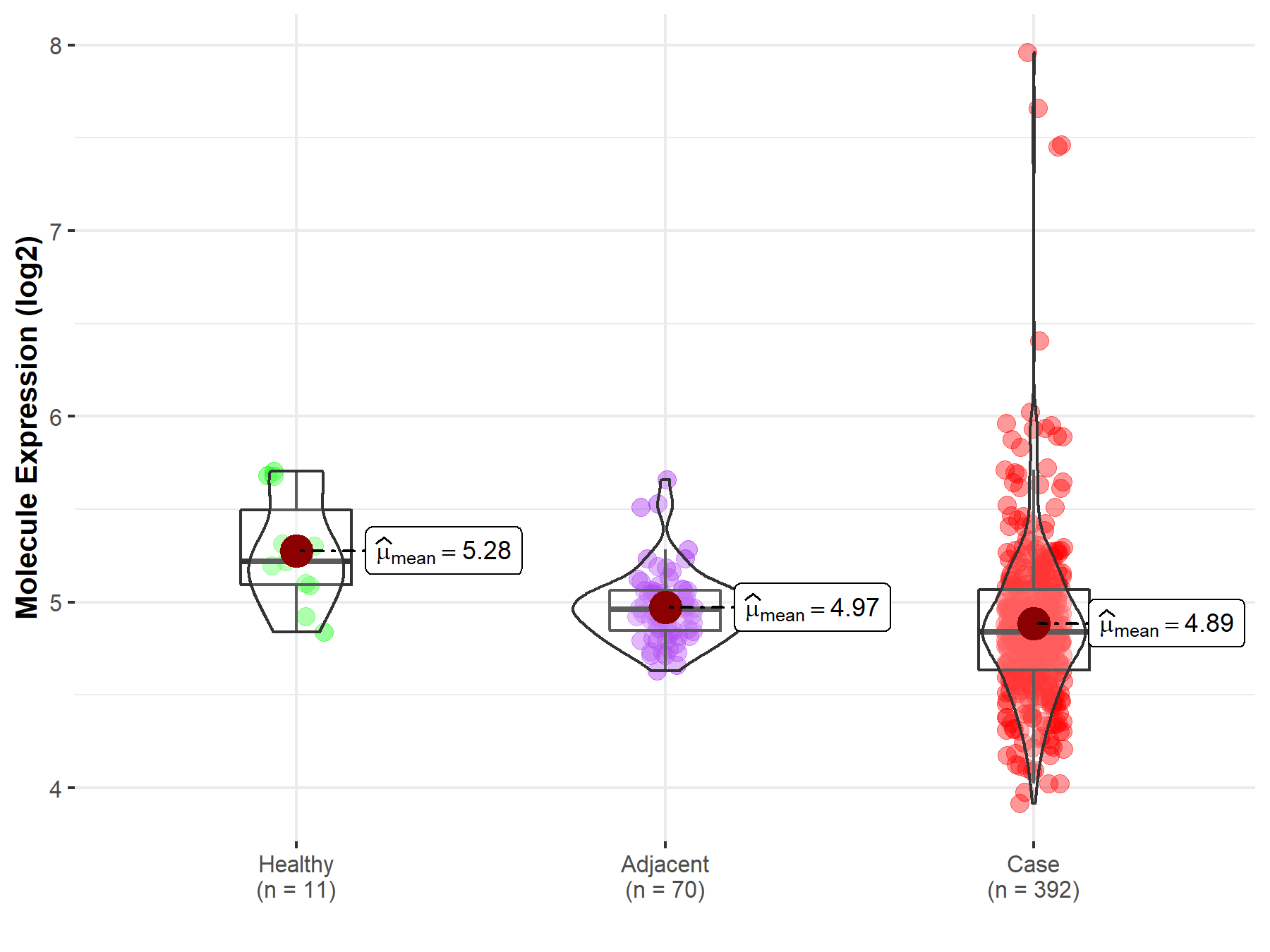
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

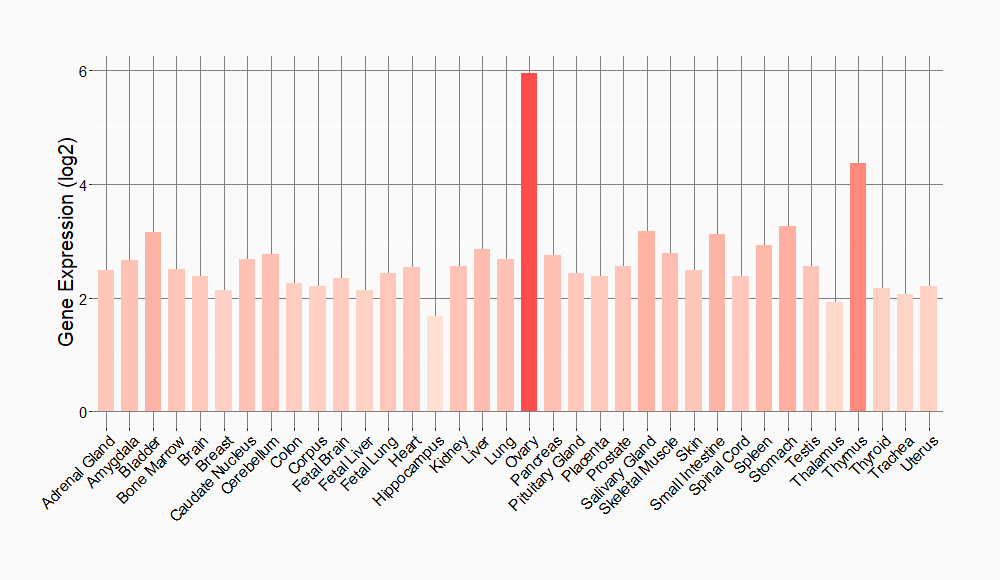
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
