Drug Information
Drug (ID: DG00369) and It's Reported Resistant Information
| Name |
Gentamicin C
|
||||
|---|---|---|---|---|---|
| Synonyms |
Gentamicin C; 11097-82-8; (2R,3S,4R,5R)-2-[(1S,2R,3R,4S,6R)-4,6-diamino-3-[(2R,3R,6S)-3-amino-6-(aminomethyl)oxan-2-yl]oxy-2-hydroxycyclohexyl]oxy-5-methyl-4-(methylamino)oxane-3,5-diol; Gentamicin C complex
Click to Show/Hide
|
||||
| Structure |
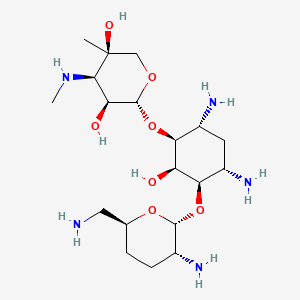
|
||||
| Drug Resistance Disease(s) |
Disease(s) with Clinically Reported Resistance for This Drug
(2 diseases)
[1]
[2]
Disease(s) with Resistance Information Validated by in-vivo Model for This Drug
(1 diseases)
[3]
|
||||
| Click to Show/Hide the Molecular Information and External Link(s) of This Drug | |||||
| Formula |
C19H39N5O7
|
||||
| IsoSMILES |
C[C@@]1(CO[C@@H]([C@H]([C@H]1NC)O)O[C@H]2[C@@H](C[C@@H]([C@H]([C@H]2O)O[C@@H]3[C@@H](CC[C@H](O3)CN)N)N)N)O
|
||||
| InChI |
1S/C19H39N5O7/c1-19(27)7-28-18(13(26)16(19)24-2)31-15-11(23)5-10(22)14(12(15)25)30-17-9(21)4-3-8(6-20)29-17/h8-18,24-27H,3-7,20-23H2,1-2H3/t8-,9+,10-,11+,12+,13-,14+,15-,16+,17+,18+,19-/m0/s1
|
||||
| InChIKey |
VEGXETMJINRLTH-ALRICIOSSA-N
|
||||
| PubChem CID | |||||
Type(s) of Resistant Mechanism of This Drug
Drug Resistance Data Categorized by Their Corresponding Diseases
ICD-01: Infectious/parasitic diseases
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: 16S rRNA (guanine(1405)-N(7))-methyltransferase (RMTA) | [4] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Methylation | p.M7G1405 |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli BL21(DE3) | 469008 | ||
| Experiment for Molecule Alteration |
Protein-RNA footprinting assay | |||
| Experiment for Drug Resistance |
Isothermal titration calorimetry assay | |||
| Mechanism Description | Sgm methylates G1405 in 16S rRNA to m7G, thereby rendering the ribosome resistant to 4, 6-disubstituted deoxystreptamine aminoglycosides. | |||
|
|
||||
| Key Molecule: Aminoglycoside N(3)-acetyltransferase (AACC2) | [5], [6], [7] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli HB101 | 634468 | ||
| Pseudomonas aeruginosa PAe1100 | 287 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay | |||
| Mechanism Description | The AAC(3)-II AGRP is characterized by resistance to gentamicin, tobramycin, dibekacin, netilmicin, 2'-N-ethylnetilmicin, 6'-N-ethylnetilmicin, and sisomicin. | |||
| Key Molecule: Aminoglycoside adenyltransferase 2''-Ia (ANT2I) | [8], [9] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Acinetobacter baumannii AB5075 | 1116234 | ||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Etest assay | |||
| Mechanism Description | ANT(2")-Ia confers resistance by magnesium-dependent transfer of a nucleoside monophosphate (AMP) to the 2"-hydroxyl of aminoglycoside substrates containing a 2-deoxystreptamine core. | |||
| Key Molecule: AAC(6')-Ib family aminoglycoside 6'-N-acetyltransferase (AAC6IB) | [1] | |||
| Resistant Disease | Pseudomonas aeruginosa infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli DH10B | 316385 | ||
| Escherichia coli HB101 | 634468 | |||
| Pseudomonas aeruginosa ATCC 27853 | 287 | |||
| Escherichia coli JM109 | 562 | |||
| Escherichia coli k-12 | 83333 | |||
| Pseudomonas aeruginosa Pa695 | 287 | |||
| Experiment for Molecule Alteration |
PCR experiments assay | |||
| Experiment for Drug Resistance |
Disk diffusion method assay | |||
| Mechanism Description | The fusion product was functional, as was the product of each gene cloned separately: AAC(3)-I, despite the deletion of the four last amino acids, and AAC(6"), which carried three amino acid changes compared with the most homologous sequence. The AAC(3)-I protein conferred an expected gentamicin and fortimicin resistance, and the AAC(6"), despite the Leu-119-Ser substitution, yielded resistance to kanamycin, tobramycin, and dibekacin, but slightly affected netilmicin and amikacin, and had no apparent effect on gentamicin. The fusion product conveyed a large profile of resistance, combining the AAC(6") activity with a higher level of gentamicin resistance without accompanying fortimicin resistance. | |||
| Key Molecule: Gentamicin 3'-acetyltransferase (AACC1) | [10], [11] | |||
| Resistant Disease | Bacterial infection [ICD-11: 1A00-1C4Z] | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli strain BN | 562 | ||
| Escherichia coli strain J62 | 562 | |||
| Escherichia coli strain k12 W3110 | 83333 | |||
| Experiment for Molecule Alteration |
Whole genome sequence assay | |||
| Experiment for Drug Resistance |
Agar dilution method assay; disk diffusion test assay | |||
| Mechanism Description | The most common mechanisms of resistance to aminoglycoside-aminocyclitol (AG) antibiotics in bacteria are exerted by enzymatic modification which results in failure of their binding to ribosomal targets and in prevention of uptake by the cell. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Aminoglycoside 2'-N-acetyltransferase (A2NA) | [3] | |||
| Resistant Disease | Escherichia coli infection [ICD-11: 1A03.0] | |||
| Molecule Alteration | Expression | Acquired |
||
| Experimental Note | Discovered Using In-vivo Testing Model | |||
| In Vitro Model | Escherichia coli strain DH5a | 668369 | ||
| Escherichia coli strain XLI-Blue | 562 | |||
| Providencia stuartii strain PR50 | 588 | |||
| Providencia stuartii strain SCH75082831A | 588 | |||
| Experiment for Molecule Alteration |
DNA sequencing assay | |||
| Experiment for Drug Resistance |
Microdilution plates assay | |||
| Mechanism Description | E.coli DH5alpha/pR 1000 demonstrated an AAC(2')-Ia resistance profile,with gentamicin, tobramycin, netilmicin, and 6'-Nethylnetilmicin MICs increased over those seen with E.coli DH5alpha. In addition, E.coli DH5alpha/pR 1000 did not show an elevated 2'-N-ethylnetilmicin MIC (MIC was 0.25ug/ml). Therefore, pR1000 encoded an enzyme capable of acetylating 6'-N-ethylnetilmicin but not 2'-N-ethylnetilmicin, suggesting 2'-N-acetyltransferase activity. DH5alpha/pSCH4500, which contains a subcloned 1.3-kb fragment, also demonstrated an AAC(2')-Ia resistance profile. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Key Molecule: Aminoglycoside 2'-N-acetyltransferase (A2NA) | [2] | |||
| Resistant Disease | Mycobacterium fortuitum infection [ICD-11: 1B2Z.2] | |||
| Molecule Alteration | Expression | Inherence |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli XL1-Blue | 562 | ||
| Streptomyces lividans strain 1326 | 1200984 | |||
| Mycolicibacterium fortuitum strain FC1k | 1766 | |||
| Mycolicibacterium smegmatis strain mc2 155 | 246196 | |||
| Experiment for Molecule Alteration |
Southern blot hybridizations assay | |||
| Experiment for Drug Resistance |
Twofold dilution of antibiotics assay | |||
| Mechanism Description | Thirty-four environmental and clinical isolates belonging to theM. fortuitumcomplex were chosen for the present study. The MICs of gentamicin varied, ranging from 2 to 16mg/ml. Crude extracts of all 34 strains were shown to have AAC activity. Acetylation of gentamicin, tobramycin, and kanamycins A and B was found for all the strains, showing a substrate profile consistent with the presence of an AAC(3) activity. Environmental isolateM. fortuitumFC1k was chosen for further studies because of its high level of AAC activity and the level of resistance to gentamicin (MIC, 16mg/ml). | |||
| Key Molecule: Aminoglycoside 2'-N-acetyltransferase (A2NA) | [2] | |||
| Resistant Disease | Mycobacterium smegmatis infection [ICD-11: 1B2Z.3] | |||
| Molecule Alteration | Expression | Acquired |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | Escherichia coli XL1-Blue | 562 | ||
| Streptomyces lividans strain 1326 | 1200984 | |||
| Mycolicibacterium fortuitum strain FC1k | 1766 | |||
| Mycolicibacterium smegmatis strain mc2 155 | 246196 | |||
| Experiment for Molecule Alteration |
Southern blot hybridizations assay | |||
| Experiment for Drug Resistance |
Twofold dilution of antibiotics assay | |||
| Mechanism Description | The aac(2')-Ib gene cloned in a mycobacterial plasmid and introduced in Mycobacterium smegmatis conferred resistance to gentamicin, tobramycin, dibekacin, netilmicin, and 6'-N-ethylnetilmicin. | |||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
