In the field of “Search for Protein Structure Alteration”, user can find molecule entries by searching drug name, disease name, molecule name, resistance mechanism name and so on among the entire textual component of DRESIS. Query can be submitted by entering keywords into the main searching frame. The resulting webpage displays profiles of all the molecules directly related to the search term, including Molecule Name, Gene Name, Molecule Type and Molecule information link.
In order to facilitate a more customized input query, the wild characters of “*” and “?” are also supported.
(1) If search “ALK”, find a single API entry which is named “ALK tyrosine kinase receptor (ALK), Homo sapiens”;
(2) If search: “Lung cancer”, finds 12 entries with molecules which related to lung cancer;
(3) If search: “FGF*”, finds 4 entries with molecule names;
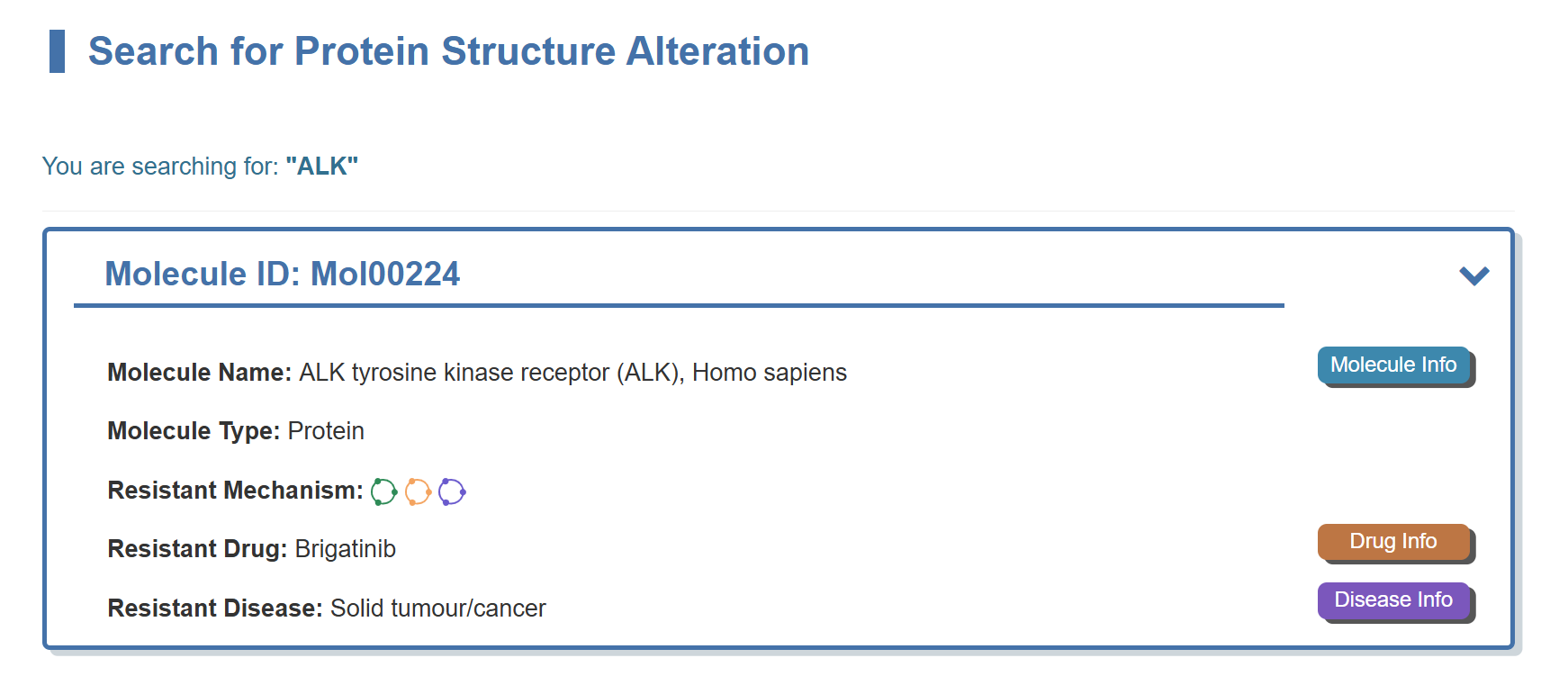
For example, if you want to know the detail information of MOLERE for ALK, you can search “ALK” in the “Search for Protein Structure Alteration” field.
Search result of the “ALK” shows the information of Molecule ID, Molecule Name, Gene Name, Gene ID, Molecule Type, Resistant Mechanism, Representative Molecule Response Drug, Representative Molecule Response Disease and External buttons. The “Molecule Info” button links to the detailed molecule information page of ALK. The “Drug Info” button links to the detailed drug information page of Brigatinib. The “Disease Info” button links to the detailed disease information page of Solid tumour/cancer.
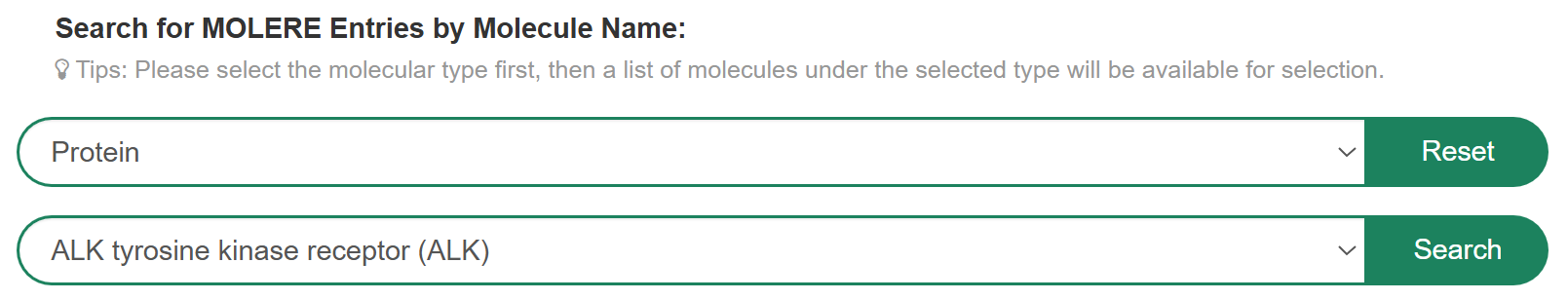
If you want to know the detail information of MOLERE belonging to a specific molecule type, you can select “Molecule Type” in the “Search for MOLERE Entries by Molecular Name” field, then select a molecule to search for detail information. For example, if you want to know drug resistance information for ALK tyrosine kinase receptor belonging to protein, you can select “Protein” at “Step 1” and “ALK tyrosine kinase receptor (ALK)” at “Step 2”.
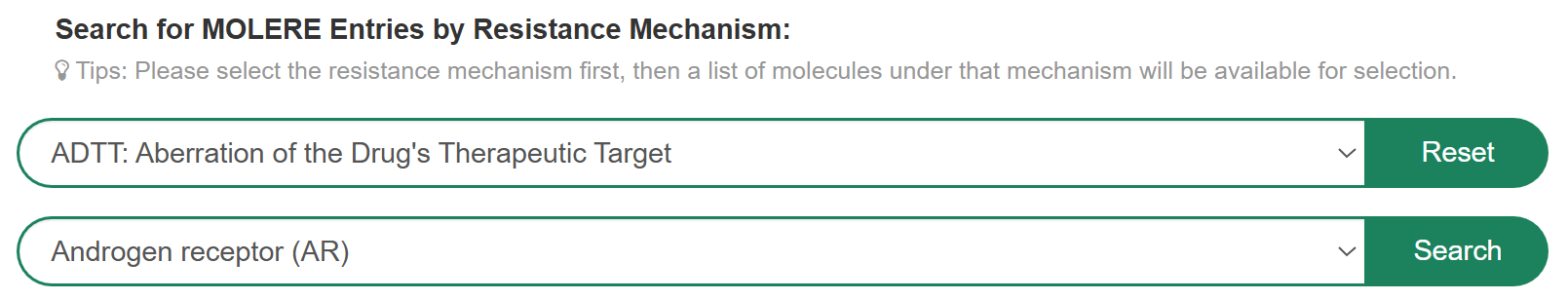
If you want to know the detail information of MOLERE based on the resistance mechanism, you can select “Resistance Mechanism” in the “Search for MOLERE Entries by Resistance Mechanism” field, then select a molecule to search for detail information. For example, if you want to know drug resistance information for androgen receptor based on the resistance mechanism of ADTT (Aberration of the Drug's Therapeutic Target), you can select “ADTT: Aberration of the Drug's Therapeutic Target” at “Step 1” and “Androgen receptor(AR)” in the “Step 2”.
If you want to know the detail information of MOLERE based on the disease, you can select “Disease Class” in the “Search for MOLERE Entries by Disease Name” field, then select a molecule to search for detail information. For example, if you want to know drug resistance information for ALK which is related to the resistance of Brain cancer, you can select “2A00: Brain cancer” at “Step 1” and “ALK tyrosine kinase receptor (ALK)” in the “Step 2”.
1.1. By clicking the “Molecule Info” button, the detailed molecule information page for particular molecule will be displayed.
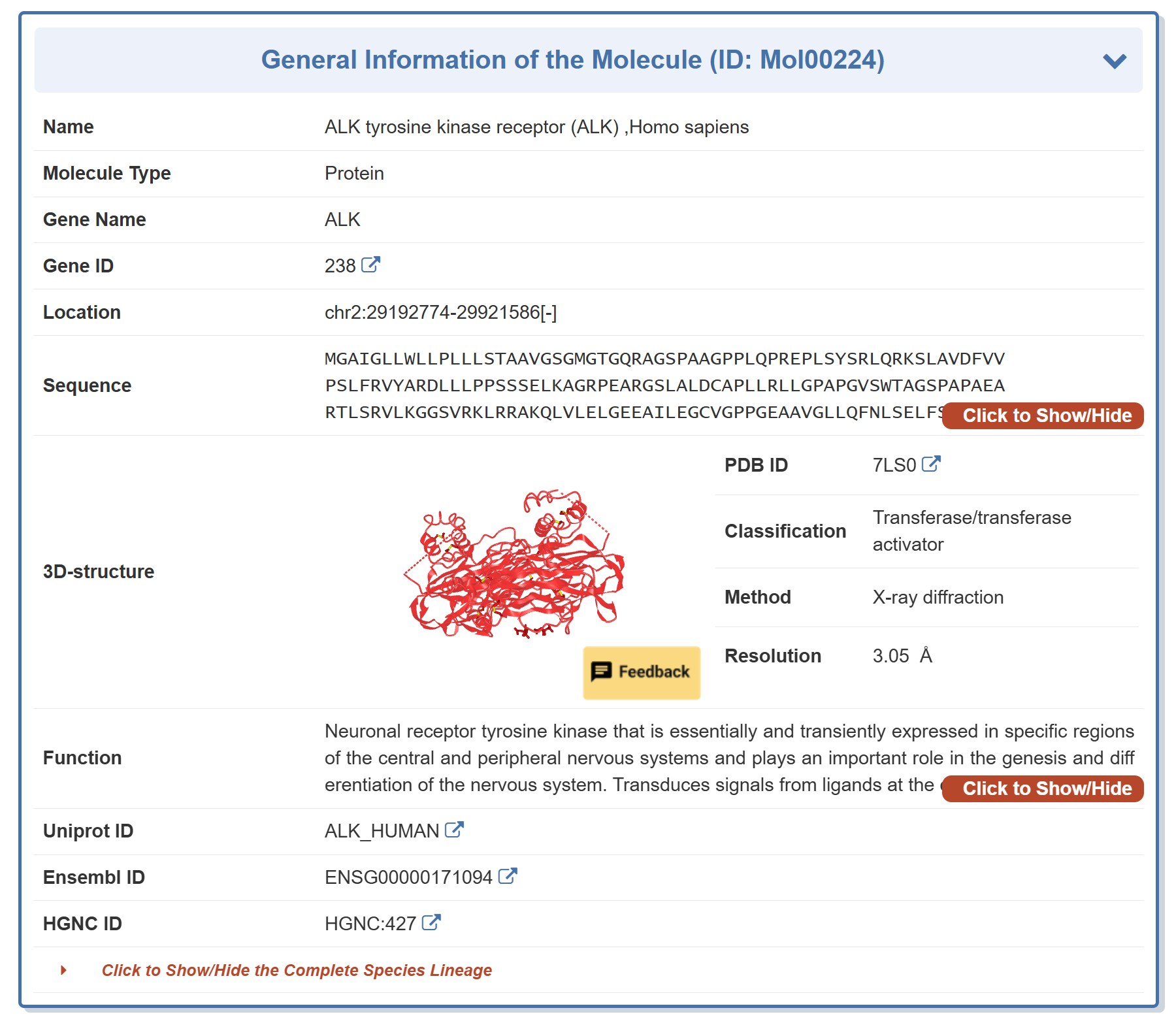
Take ALK as an example, “General Information of the Molecule” section displays the general information of ALK tyrosine kinase receptor including its Name, Synonyms, Molecule Type, Gene Name, Gene ID, Location, Sequence, 3D-structure, Function, Uniprot ID, Ensemble ID, HGNC ID, PDB ID,Classification, Method and Resolution.
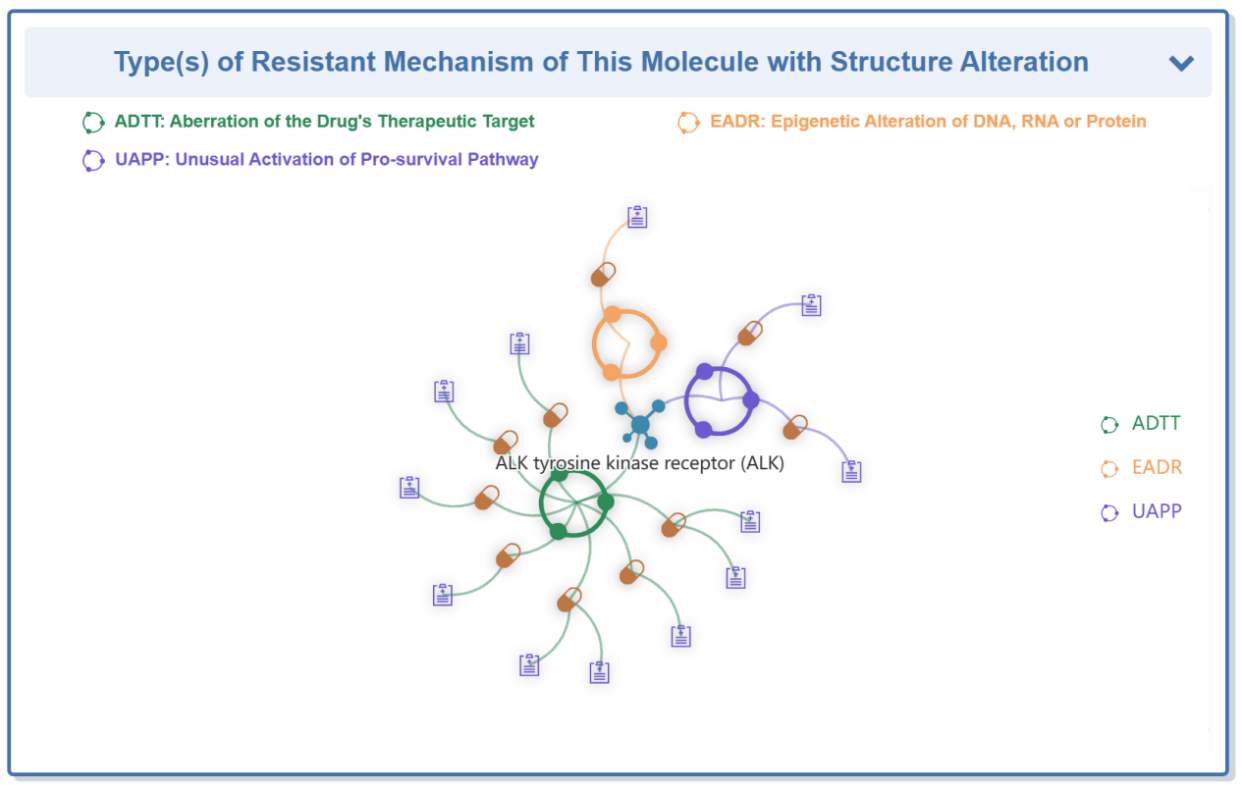
“Type(s) of Resistant Mechanism of this Molecule” section displays three types of interactions between drug, disease and mechanism.
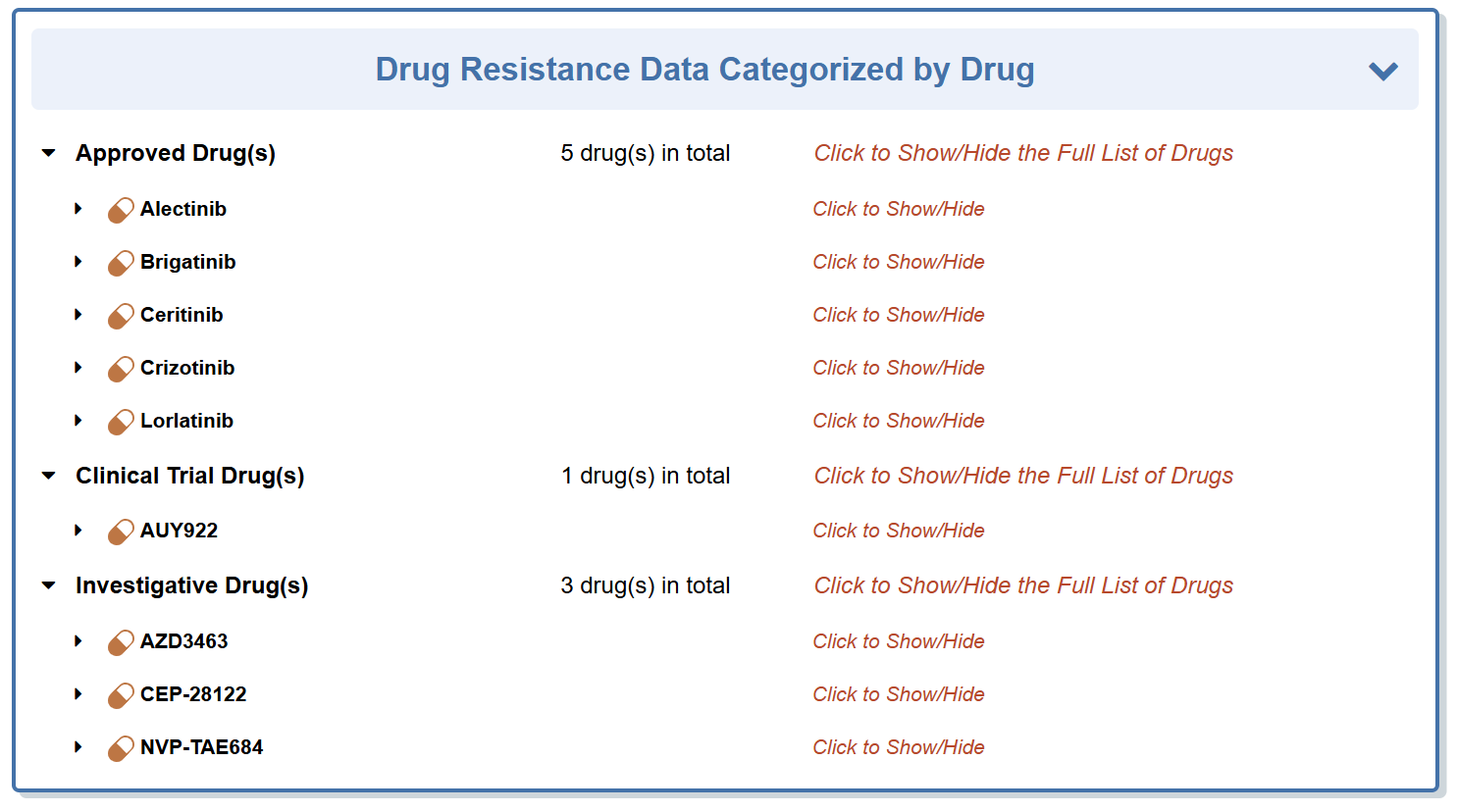
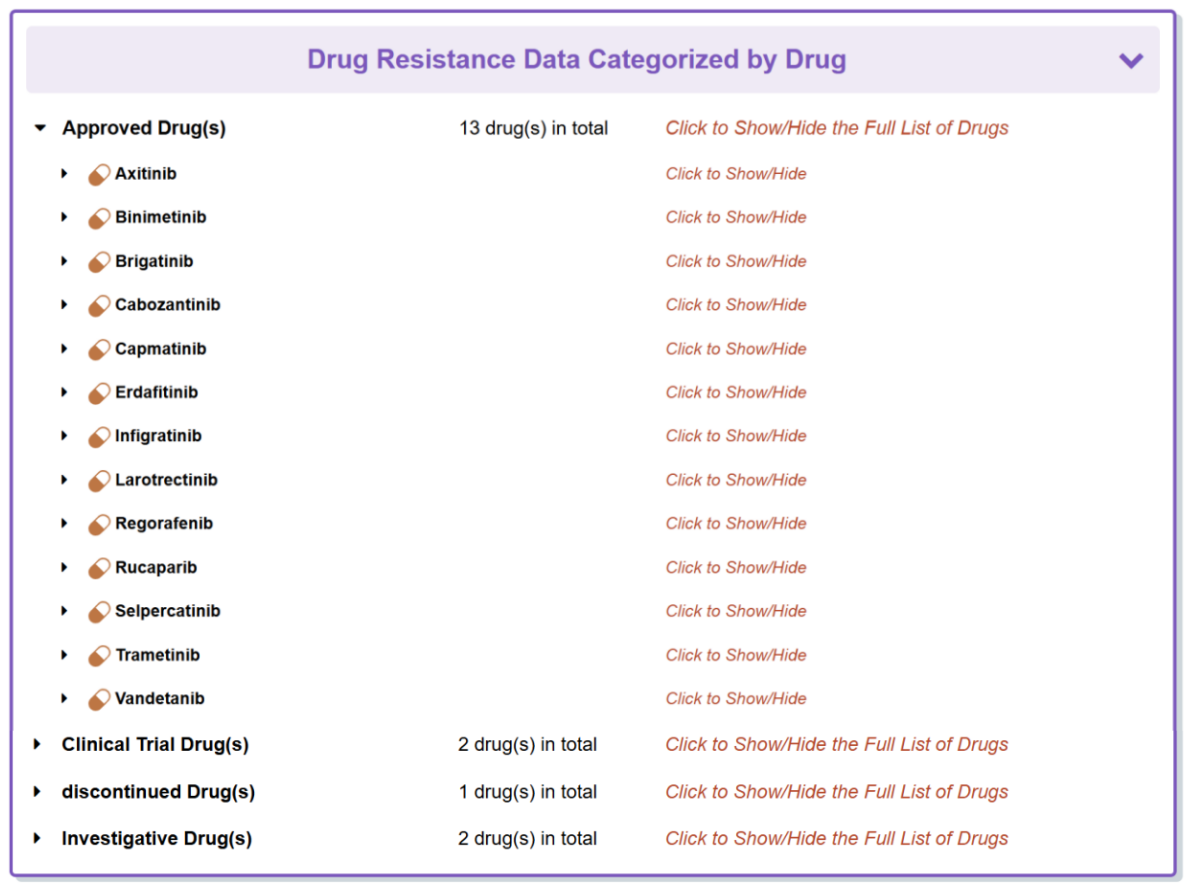
“Comprehensive Drug Resistance Data Categorized by Drug” section classifies all the drugs that are responded to this molecule.
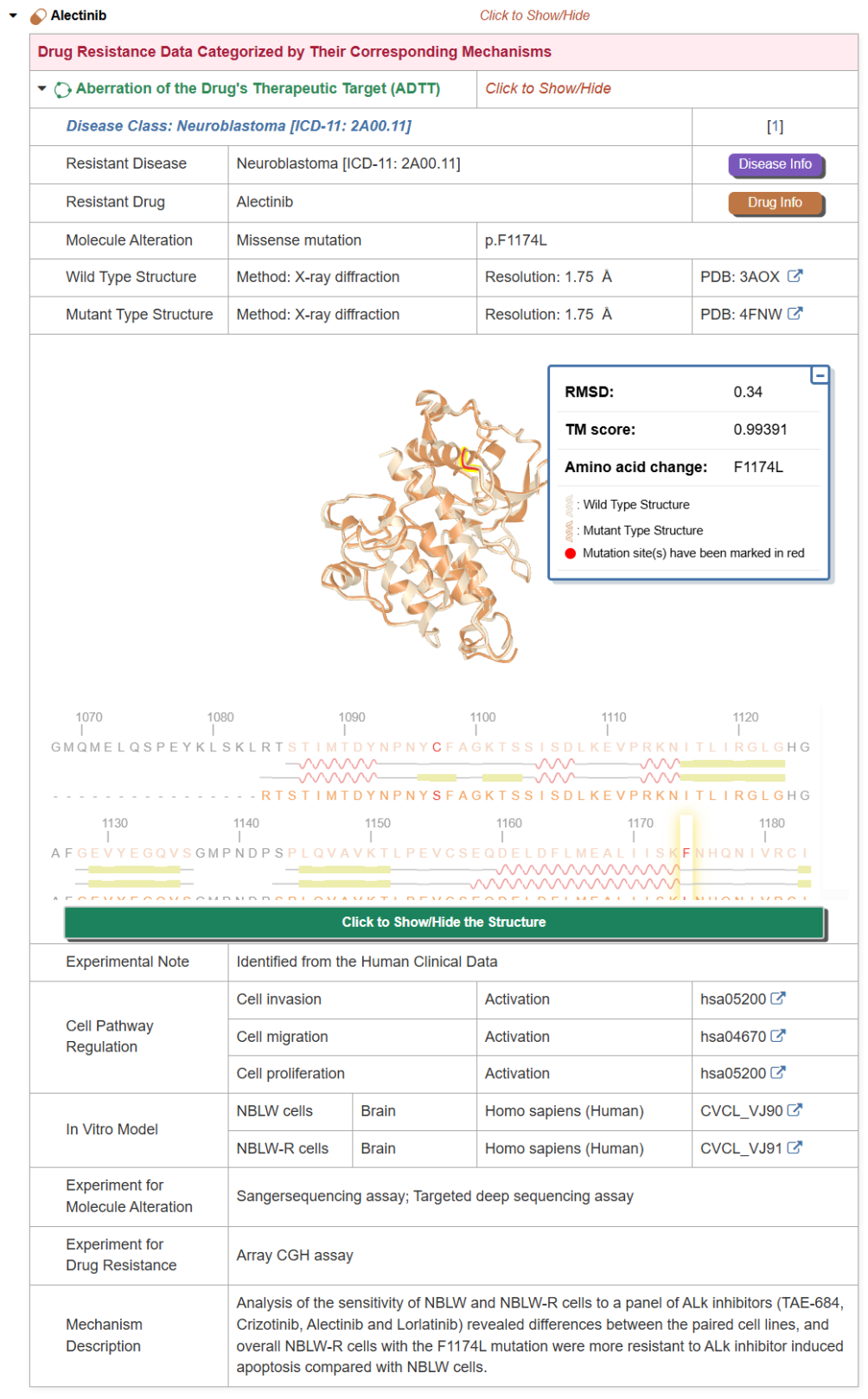
The "Drug Resistance Data Categorized by Their Corresponding Mechanisms" section displays drug resistance data, including Resistance Mechanism, Disease Class, Resistant Disease, Resistant Drug, Molecule Alteration, Wild Type Structure, Mutant Type Structure, Experimental Note, Cell Pathway Regulation, In Vitro Model, In Vivo Model, Experiment for Molecule Alteration, Experiment for Drug Resistance and Mechanism Description.
1.2. By clicking the “Drug Info” button, the detailed drug information page for particular “Representative Molecule Response Drug” will be displayed.
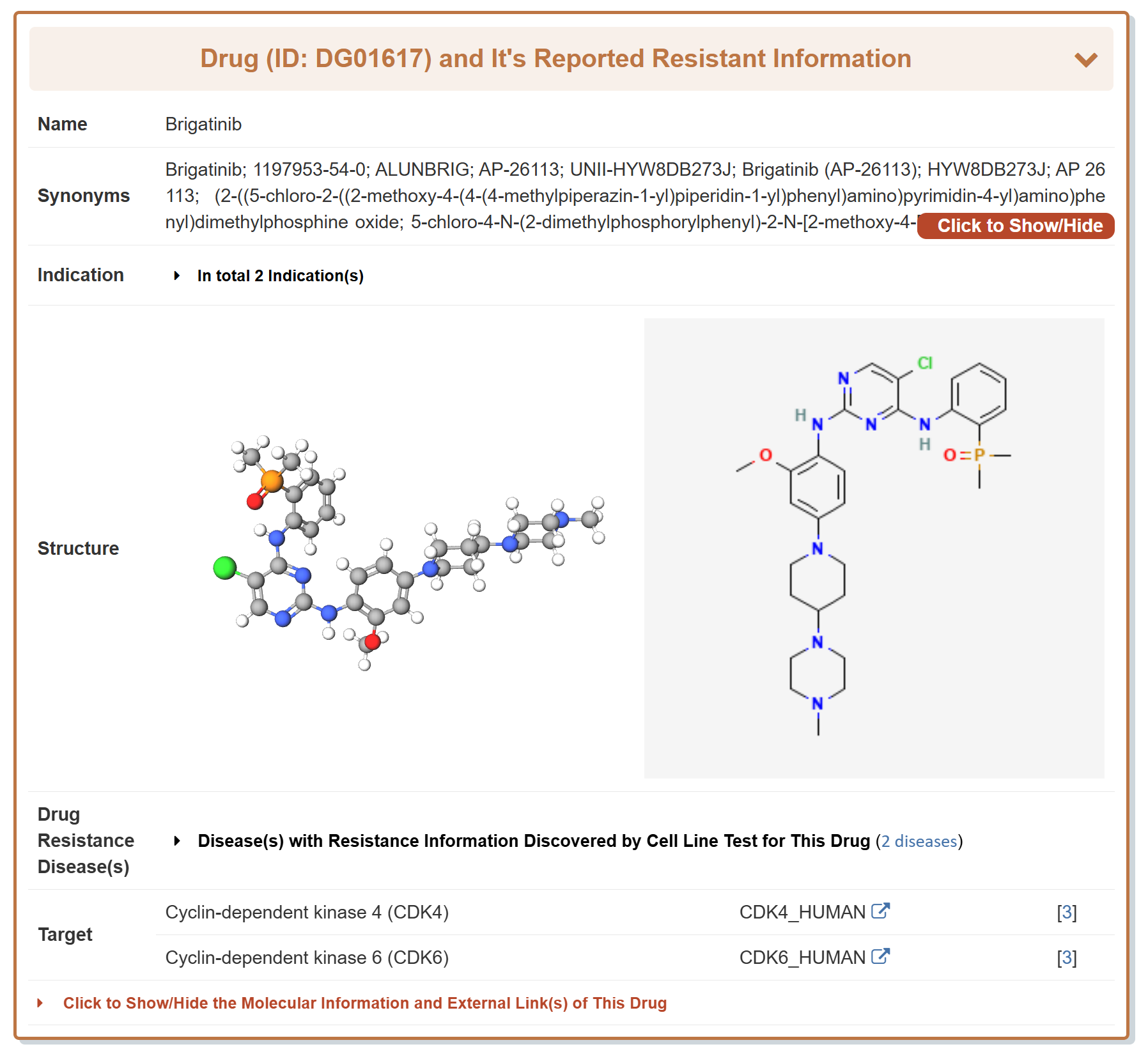
Take ALK as an example, “General Information of the Drug” section displays the general information of Brigatinib which responds to ALK, including its Name, Synonyms, Indication, Structure, Resistant Disease, Target, Molecular Information and External Link(s).
“Type(s) of Resistant Mechanism of This Drug due to Structure Alteration” section displays two types of interactions between drug, disease and mechanism.
“Drug Resistance Data Categorized by Their Corresponding Diseases” section displays all disease that confer resistance to Brigatinib.

1.3. By clicking the “Disease Info” button, the detailed disease information page for particular “Representative Molecule Response Disease” will be displayed.
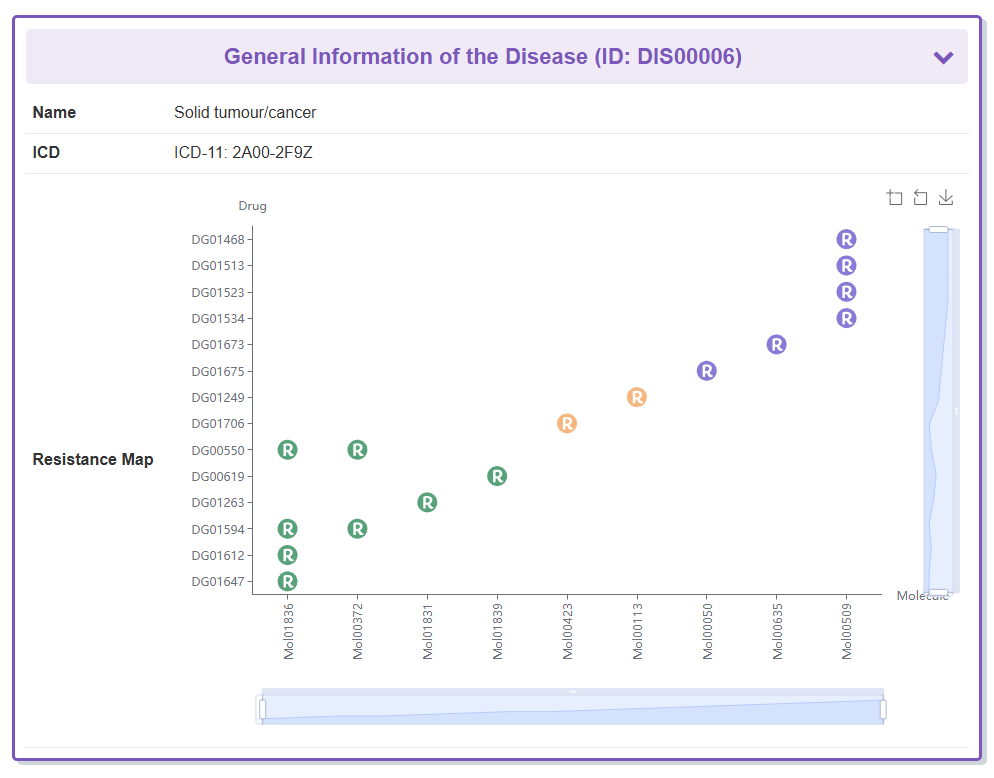
Take ALK as an example, “General Information of the Disease” section displays the general information of Solid tumour/cancer which ALK related to, including its Name, ICD and Resistance Map.
“Type(s) of Resistant Mechanism of This Disease with Structure Alteration” section displays four types of interactions between drug, disease and mechanism.
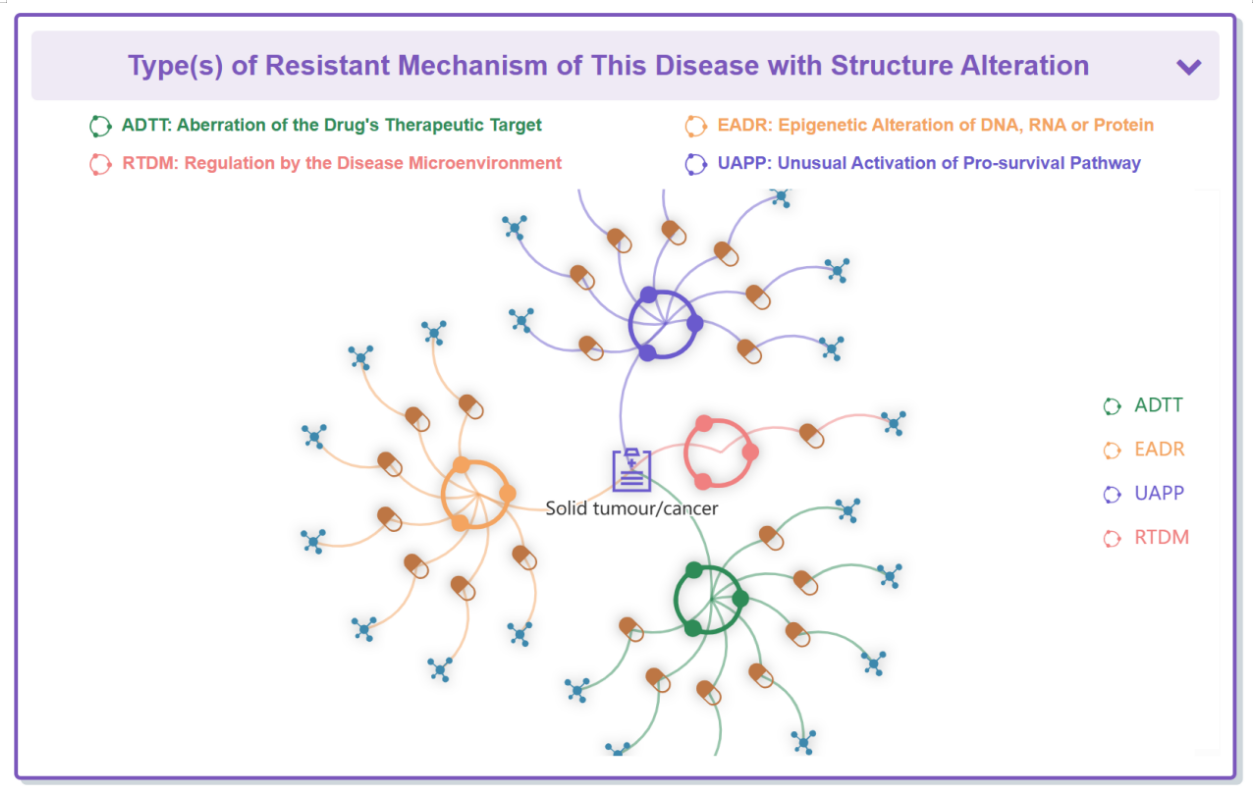
“Drug Resistance Data Categorized by Drug” section classifies all the drugs that respond to this disease.
In the field of “Search for Disease Metabolic Reprogramming”, user can find drug entries by searching drug name, disease name, molecule name, resistance mechanism name, and so on among the entire textual component of DRESIS. Query can be submitted by entering keywords into the main searching frame. The resulting webpage displays profiles of all the drugs directly related to the search term, including Drug name, Highest Status, Metabolic Type, Resistant Disease, Resistant Molecule and Drug information link.
In order to facilitate a more customized input query, the wild characters of “*” and “?” are also supported.
(1) If search “Oxaliplatin”, find a single API entry which is named “Oxaliplatin”;
(2) If search: “Colorectal cancer”, finds 7 entries with drug names;
(3) If search: “Oxalipla*”, find a single entry which is named “Oxaliplatin”;
For example, if you want to know the detail information of DRUGRE for Oxaliplatin, you can search “Oxaliplatin” in the “Search for Disease Metabolic Reprogramming” field.
Search result of the “Oxaliplatin” shows the information of Drug ID, Drug Name, Drug Clinical Status, Drug Indication, Drug Target, Metabolic Type, Representative Drug Response Molecule, Representative Drug Response Disease and External buttons. The “Drug Info” button links to the detailed drug information page of Oxaliplatin. The “Molecule Info” button links to the detailed molecule information page of Histone H3. The “Disease Info” button links to the detailed disease information page of Oral squamous cell carcinoma.

If you want to know the detail information of DRUGRE based on the resistance mechanism, you can select “Resistance Mechanism” in the “Search for DRUGRE Entries by Resistance Mechanism” field, then select a drug to search for detail information. For example, if you want to know drug resistance information for Anastrozole based on the resistance mechanism of MRAP (Metabolic Reprogramming via Altered Pathways), you can select “MRAP: Metabolic Reprogramming via Altered Pathways” at “Step 1” and “Anastrozole” in the “Step 2”.

If you want to know the detail information of DISERE based on the resistance mechanism, you can select “Resistance Mechanism” in the “Search for DISERE Entries by Resistance Mechanism” field, then select a disease to search for detail information. For example, if you want to know drug resistance information for Brain cancer based on the resistance mechanism of MRAP (Metabolic Reprogramming via Altered Pathways), you can select “MRAP: Metabolic Reprogramming via Altered Pathways” at “Step 1” and “2A00: Brain cancer” in the “Step 2”.

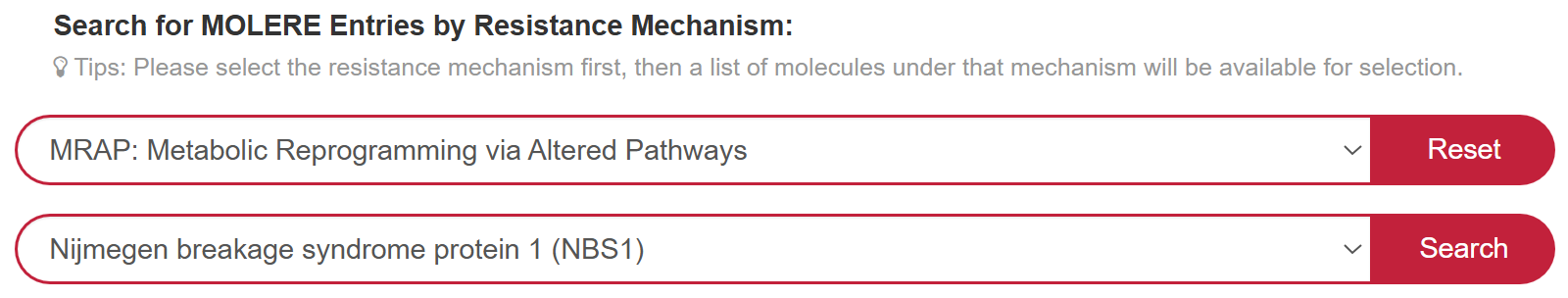
If you want to know the detail information of MOLERE based on the resistance mechanism, you can select “Resistance Mechanism” in the “Search for MOLERE Entries by Resistance Mechanism” field, then select a molecule to search for detail information. For example, if you want to know drug resistance information for NBS1 based on the resistance mechanism of MRAP (Metabolic Reprogramming via Altered Pathways), you can select “MRAP: Metabolic Reprogramming via Altered Pathways” at “Step 1” and “Nijmegen breakage syndrome protein 1(NBS1)” in the “Step 2”.
2.1. By clicking the “Drug Info” button, the detailed drug information page for particular drug will be displayed.
Take Doxorubicin as an example, “General Information of the Drug” section displays the general information of Doxorubicin including its Name, Synonyms, Indication, Structure, Resistant Disease, Target, Molecular Information and External Link(s).
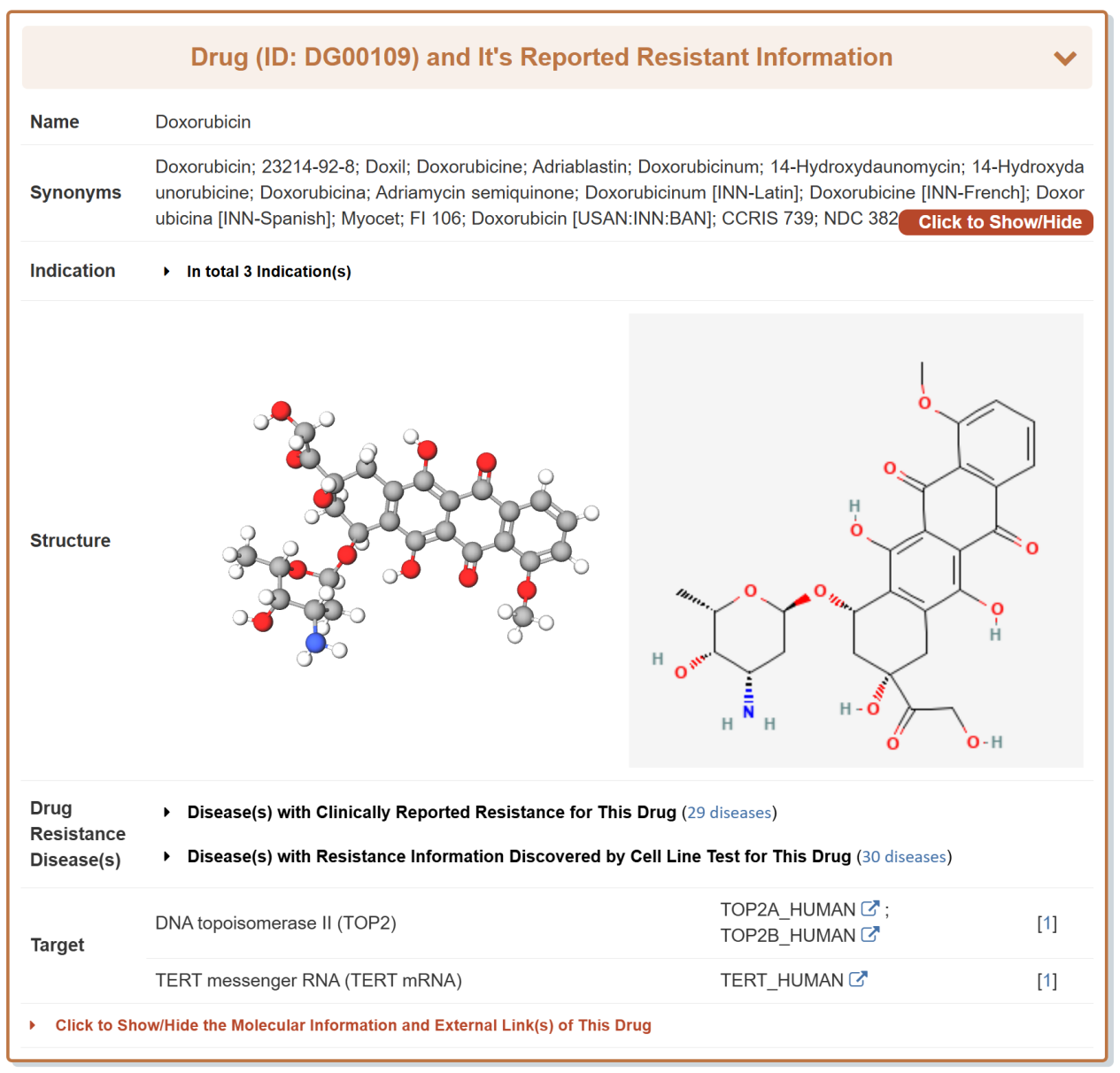
“Disease Metabolic Reprogramming of This Drug” section displays a single type of interactions between drug, disease and mechanism.
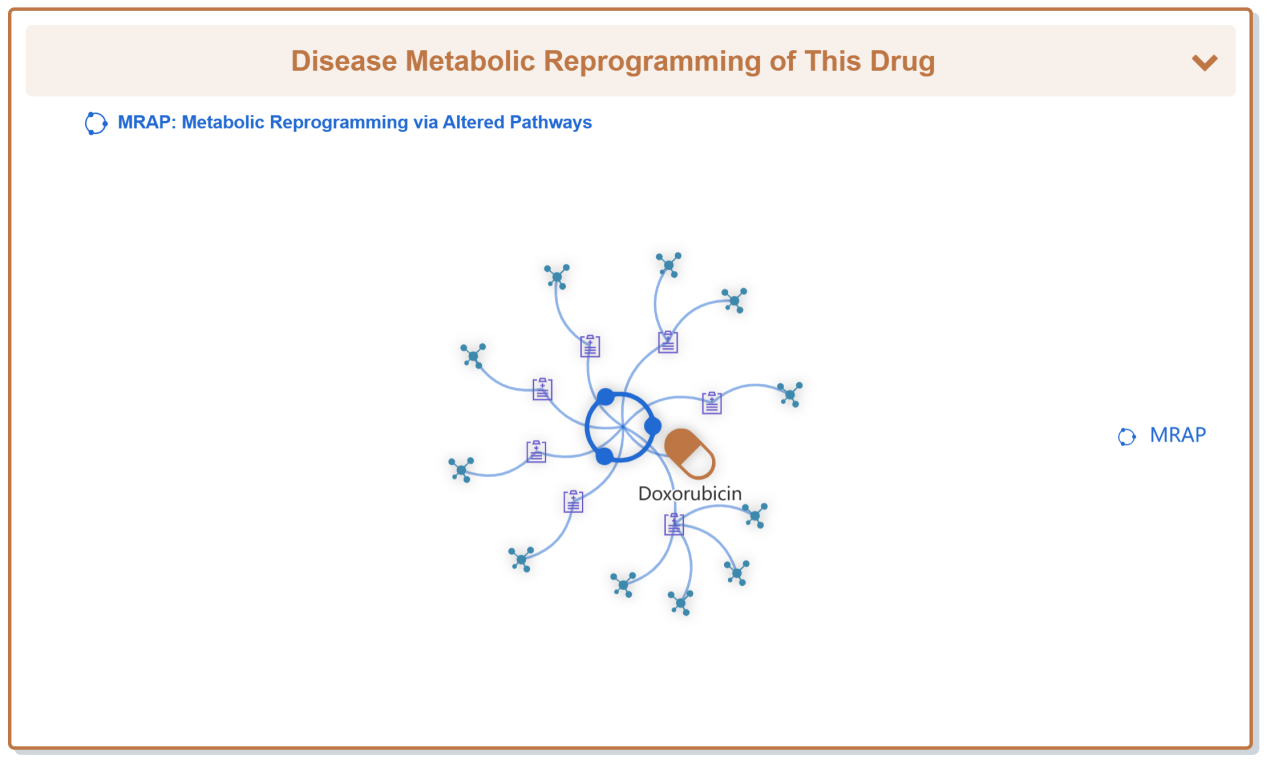
“Drug Resistance Data Categorized by Their Corresponding Diseases” section displays all kinds of disease which confer resistance to Doxorubicin.
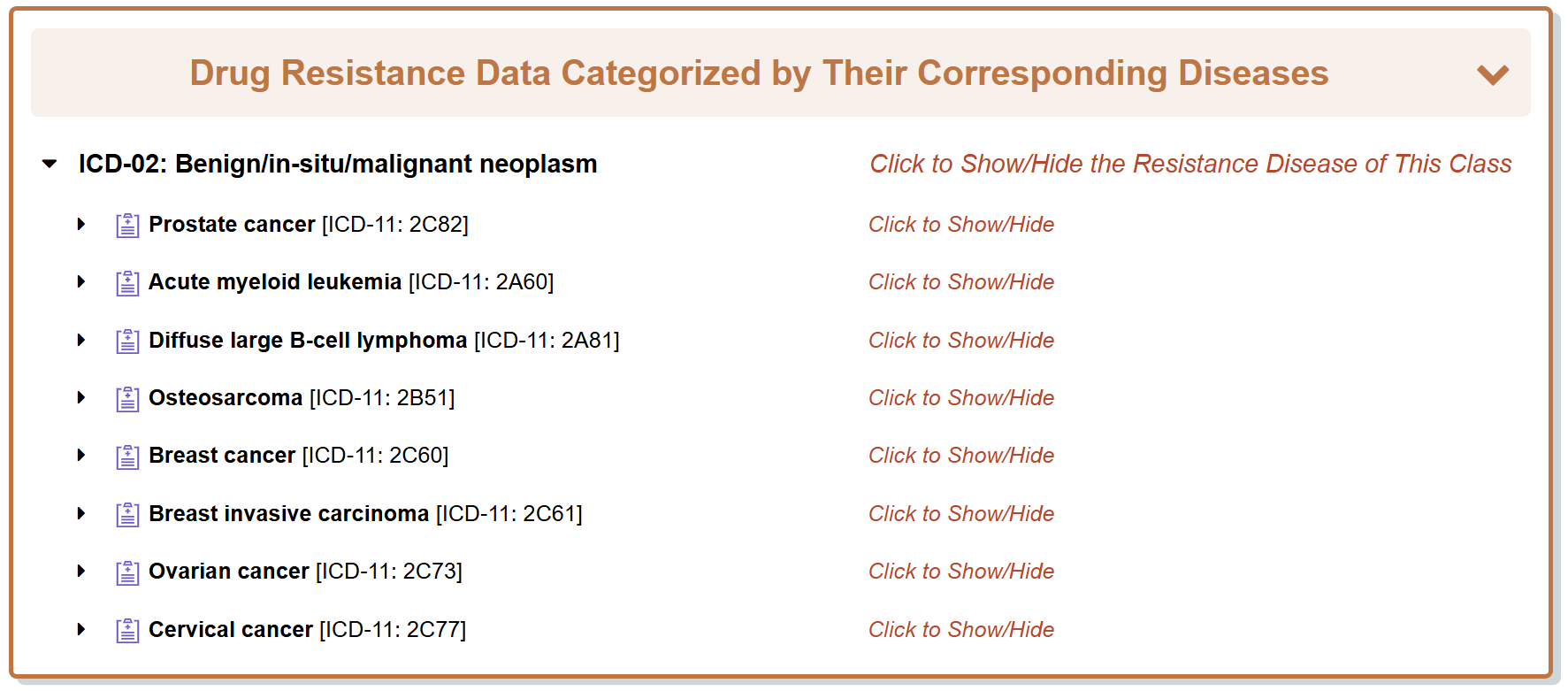
6.2. By clicking the “Disease Info” button, the detailed disease information page for particular “Representative Drug Response Disease” will be displayed.
Take Doxorubicin as an example, “General Information of the Disease” section displays the general information of Acute myeloid leukemia which the Doxorubicin responds to, including its Name, ICD and Resistance Map.
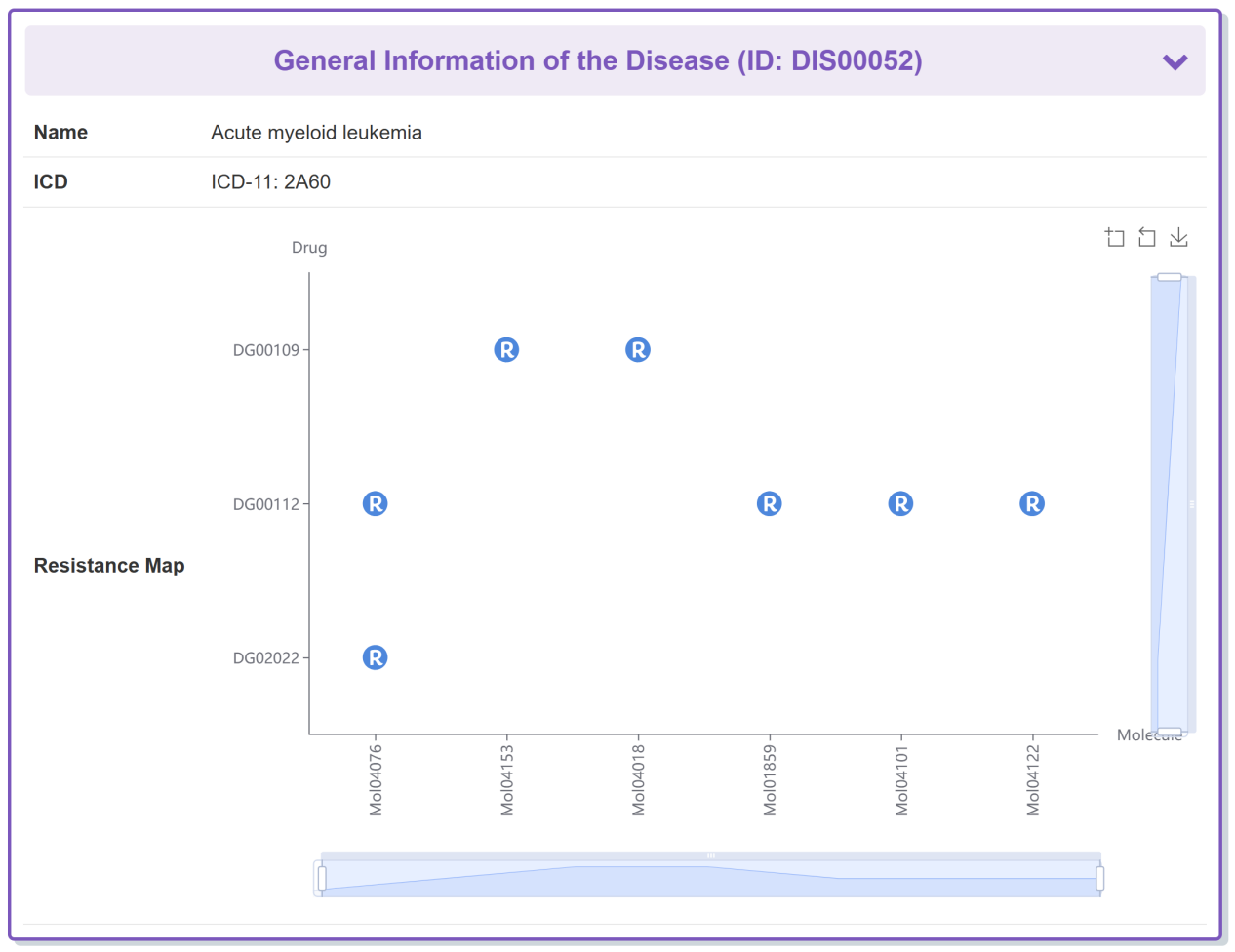
“Disease Metabolic Reprogramming of This Disease” section displays a single type of interactions between drug, disease and mechanism.
“Drug Resistance Data Categorized by Drug” section classifies all the drugs that respond to this disease.

6.3. By clicking the “Molecule Info” button, the detailed molecule information page for particular “Representative Drug Response Molecule” will be displayed.
Take Bafetinib as an example, “General Information of the Molecule” section displays the general information of Histone H3, Homo sapiens, which the Bafetinib responds to, including its Name, Synonyms, Molecule Type, 3D-Structure, Gene Name, Gene ID, Location, Sequence, Function, Uniprot ID, Ensembl ID, HGNC ID, PDB ID, Classification, Method and Resolution.
“Disease Metabolic Reprogramming of This Molecule” section displays a single type of interactions between drug, disease and mechanism.
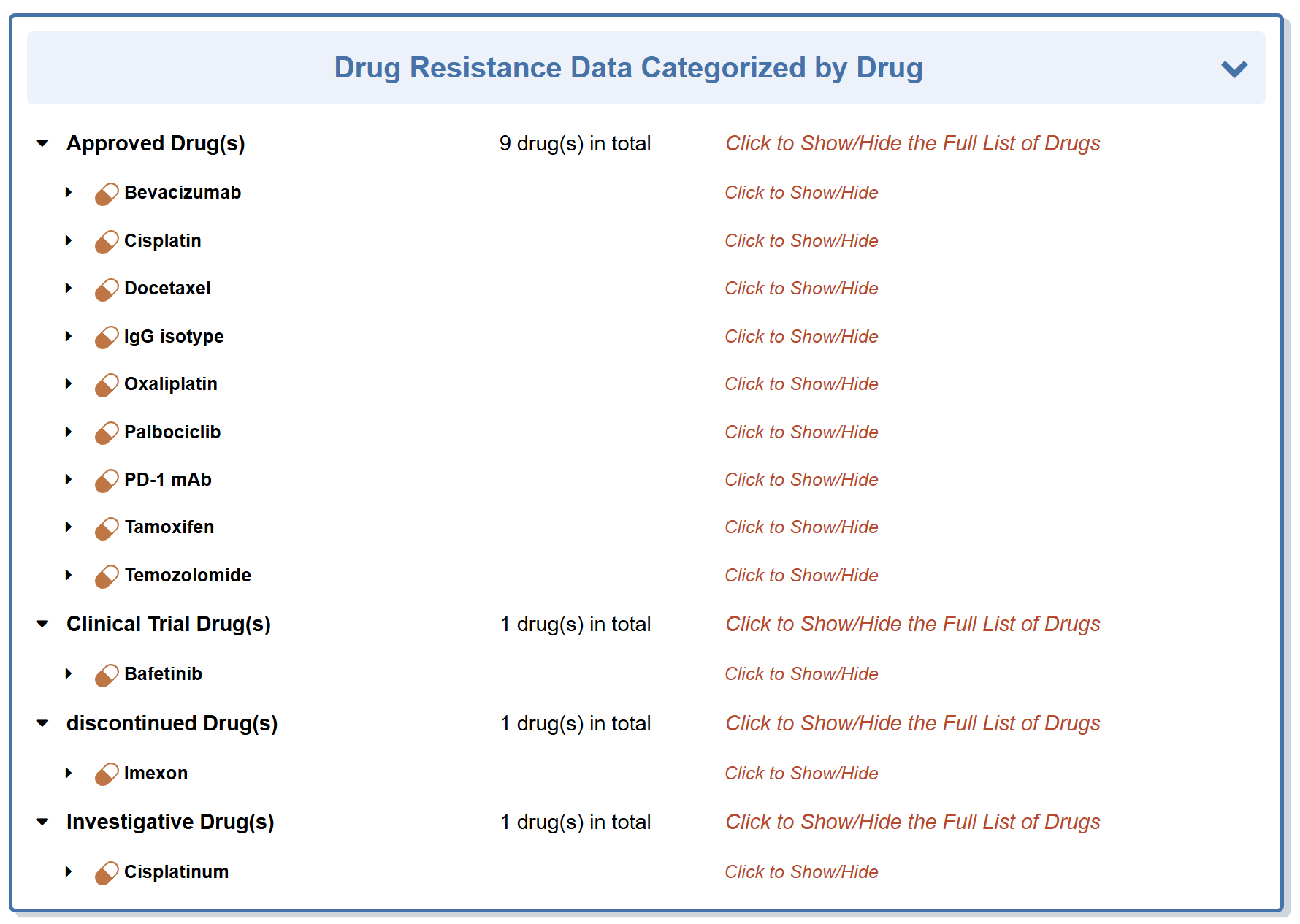
“Drug Resistance Data Categorized by Drug” section classifies all the drugs that are responded to this molecule.
In the field of “Search for drug”, user can find drug entries by searching drug name, disease name, molecule name, resistance mechanism name, and so on among the entire textual component of DRESIS. Query can be submitted by entering keywords into the main searching frame. The resulting webpage displays profiles of all the drugs directly related to the search term, including Drug structure, Drug name, Highest Status, Disease and Drug information link.
In order to facilitate a more customized input query, the wild characters of “*” and “?” are also supported.
(1) If search “Sunitinib”, find a single API entry which is named “Sunitinib”;
(2) If search: “Kidney cancer”, finds 12 entries with drug names;
(3) If search: “Sunitin*”, find a single entry which is named “Sunitinib”;
For example, if you want to know the detail information of DRUGRE for Sunitinib, you can search “Sunitinib” in the “Search for drug” field.

Search result of the “Sunitinib” shows the information of Drug ID, Drug Name, Drug Clinical Status, Drug Indication, Drug Target, Representative Drug Response Molecule, Representative Drug Response Disease and External buttons. The “Drug Info” button links to the detailed drug information page of Sunitinib. The “Molecule Info” button links to the detailed molecule information page of ARID1A. The “Disease Info” button links to the detailed disease information page of Acute myeloid leukemia.

If you want to know the detail information of DRUGRE indicating a specific disease, you can select “Disease Class” in the “Search for DRUGRE Entries by Disease Class” field, then select a drug to search for detail information. For example, if you want to know drug resistance information for Gemcitabine indicating gastric cancer, you can select “2B72: Gastric cancer” at “Step 1” and “Gemcitabine” at “Step 2”.

If you want to know the detail information of DRUGRE based on the resistance mechanism, you can select “Resistance Mechanism” in the “Search for DRUGRE Entries by Resistance Mechanism” field, then select a drug to search for detail information. For example, if you want to know drug resistance information for Brigatinib based on the resistance mechanism of ADTT (Aberration of the Drug's Therapeutic Target), you can select “ADTT: Aberration of the Drug's Therapeutic Target” at “Step 1” and “Brigatinib” in the “Step 2”

3.1. By clicking the “Drug Info” button, the detailed drug information page for particular drug will be displayed.
Take Sunitinib as an example, “General Information of the Drug” section displays the general information of Sunitinib including its Name, Synonyms, Indication, Structure, Resistant Disease, Target, Molecular Information and External Link(s).

Some drugs in DRESIS also have corresponding drug resistance verified in the in-vitro screen using cancer cell lines. Take Methylene Blue cation as an example, “Drug Resistance Verified in the In-Vitro Screen using Cancer Cell Lines” section displays the general cancer cell lines resistance or sensitivity information of Methylene Blue cation including its IC50, GI50, LC50 and Z-Score.
(1) The IC50 endpoint values are interpolated from the Percent of Treated cell growth as a fraction of Control cell growth.
(2) The GI50 and LC50 values are determined by simple interpolation of GIPRCNT values above and below 50 and -50 respectively, GIPRCNT is the percent of treated cell growth as a fraction of control cell growth corrected for the count of cells at the time of drug addiction in the assay. 100 is control growth, 0 is complete inhibition of growth (cytostasis), and -100 is complete cell kill.
(3) The Z-score describes the value's relationship to the mean of these data, which is from negative log10[GI50(molar)] data across NCI-60 for single drug and combo-score for paired drug. Generally, a lower score value indicates the drug is severely drug resistance.

“Type(s) of Resistant Mechanism of this Drug” section displays five types of interactions between drug, disease and mechanism.

“Comprehensive Drug Resistance Data Categorized by Disease” section displays all kinds of disease which confer resistance to Sunitinib.

3.2. By clicking the “Molecule Info” button, the detailed molecule information page for particular “Representative Drug Response Molecule” will be displayed.
Take Vemurafenib as an example, “General Information of the Molecule” section displays the general information of Induced myeloid leukemia cell differentiation protein Mcl-1 (MCL1) which the Vemurafenib responds to, including its Name, Synonyms, Molecule Type, Structure, Gene Name, Gene ID, Location, Sequence, Function, Uniprot ID, Ensembl ID and HGNC ID.

“Type(s) of Resistant Mechanism of this Molecule” section displays two types of interactions between drug, disease and mechanism.

“Comprehensive Drug Resistance Data Categorized by Drug” section classifies all the drugs that are responded to this molecule.

“Disease-Specific Expression Quantity of This Molecule” section displays the expression quantity of this molecule in those disease.

3.3. By clicking the “Disease Info” button, the detailed disease information page for particular “Representative Drug Response Disease” will be displayed.
Take Sunitinib as an example, “General Information of the Disease” section displays the general information of Kidney cancer which the Sunitinib responds to, including its Name, ICD and Resistance Map.

“Type(s) of Resistant Mechanism of this Disease” section displays six types of interactions between drug, disease and mechanism.

“Comprehensive Drug Resistance Data Categorized by Drug” section classifies all the drugs that respond to this disease.

In the field of “Search for Disease”, user can find disease entries by searching drug name, disease name, molecule name, resistance mechanism name and so on among the entire textual component of DRESIS. Query can be submitted by entering keywords into the main searching frame. The resulting webpage displays profiles of all the diseases directly related to the search term, including Disease name, Disease ICD, Disease class, Mechanism and Disease information link.
In order to facilitate a more customized input query, the wild characters of “*” and “?” are also supported.
(1) If search: “Liver cancer”, find a single API entry which is named “Liver cancer”;
(2) If search “Vemurafenib”, find 4 entries with diseases which related to Vemurafenib;
(3) If search: “Liver*”, finds 4 entries with disease names;
For example, if you want to know the detail information of DISERE for Liver cancer, you can search “Liver cancer” in the “Search for disease” field.

Search result of the “Liver cancer” shows the information of Disease ID, Disease Name, Disease ICD, Disease Class, Disease Specific Mechanism, Representative Disease Response Drug, Representative Disease Response Molecule and External buttons. The “Disease Info” button links to the detailed disease information page of Liver cancer. The “Mech Info” button links to the detailed mechanism information page of Liver cancer. The “Drug Info” button links to the detailed drug information page of Gemcitabine. The “Molecule Info” button links to the detailed molecule information page of PCBP2-OT1

If you want to know the detail information of DISERE belonging to a specific disease class, you can select “Disease Class” in the “Search for DISERE Entries by Disease Class” field, then select a disease to search for detail information. For example, if you want to know drug resistance information for Liver cancer belonging to Benign/in-situ/malignant neoplasm, you can select “Benign/in-situ/malignant neoplasm” at “Step 1” and “2C12: Liver cancer” at “Step 2”.

If you want to know the detail information of DISERE based on the resistance mechanism, you can select “Resistance Mechanism” in the “Search for DISERE Entries by Resistance Mechanism” field, then select a disease to search for detail information. For example, if you want to know drug resistance information for Ovarian cancer based on the resistance mechanism of DISM (Drug Inactivation by the Structure Modification), you can select “DISM: Drug Inactivation by the Structure Modification” at “Step 1” and “2C73: Ovarian cancer” in the “Step 2”.

4.1. By clicking the “Disease Info” button, the detailed disease information page for particular disease will be displayed.
Take Liver cancer as an example, “General Information of the Disease” section displays the general information of Liver cancer including its Name, ICD and Resistance Map.

“Type(s) of Resistant Mechanism of this Disease” section displays seven types of interactions between drug, disease and mechanism.

“Comprehensive Drug Resistance Data Categorized by Drug” section classifies all the drugs that respond to this disease.

4.2. By clicking the “Drug Info” button, the detailed drug information page for particular “Representative Drug Response Disease” will be displayed.
Take Liver cancer as an example, “General Information of the Drug” section displays the general information of Sorafenib which responds to Liver cancer, including its Name, Synonyms, Indication, Structure, Target, Molecular Information and External Link(s).

“Type(s) of Resistant Mechanism of this Drug” section displays seven types of interactions between drug, disease and mechanism.

“Comprehensive Drug Resistance Data Categorized by Disease” section displays all disease that confer resistance to Gemcitabine.

4.3. By clicking the “Molecule Info” button, the detailed molecule information page for particular “Representative Disease Response Molecule” will be displayed.
Take Kidney cancer as an example, “General Information of the Molecule” section displays the general information of Signal transducer activator transcription 3 (STAT3) which responds to Kidney cancer, including its Name, Synonyms, Molecule Type, Gene ID, Location, Ensembl ID and HGNC ID.

“Type(s) of Resistant Mechanism of this Molecule” section displays two types of interactions between drug, disease and mechanism.

“Comprehensive Drug Resistance Data Categorized by Drug” section classifies all the drugs that are responded to this molecule.

“Disease-Specific Expression Quantity of This Molecule” section displays the expression quantity of this molecule in those disease.

In the field of “Search for molecule”, user can find molecule entries by searching drug name, disease name, molecule name, resistance mechanism name and so on among the entire textual component of DRESIS. Query can be submitted by entering keywords into the main searching frame. The resulting webpage displays profiles of all the molecules directly related to the search term, including Molecule Name, Gene Name, Molecule Type and Molecule information link.
In order to facilitate a more customized input query, the wild characters of “*” and “?” are also supported.
(1) If search “PTEN”, find a single API entry which is named “Phosphatase and tensin homolog (PTEN)”
(2) If search: “Colorectal cancer”, finds 8 entries with molecules which related to colorectal cancer;
(3) If search: “ABCB*”, finds 13 entries with molecule names;
For example, if you want to know the detail information of MOLERE for PTEN, you can search “PTEN” in the “Search for molecule” field.

Search result of the “PTEN” shows the information of Molecule ID, Molecule Name, Gene Name, Gene ID, Molecule Type, Representative Molecule Response Drug, Representative Molecule Response Disease and External buttons. The “Molecule Info” button links to the detailed molecule information page of PTEN. The “Drug Info” button links to the detailed drug information page of Carmustine. The “Disease Info” button links to the detailed disease information page of Brain cancer.

If you want to know the detail information of MOLERE belonging to a specific molecule type, you can select “Molecule Type” in the “Search for MOLERE Entries by Molecular Name” field, then select a molecule to search for detail information. For example, if you want to know drug resistance information for HOX transcript antisense RNA belonging to LncRNA, you can select “LncRNA” at “Step 1” and “HOX transcript antisense RNA (HOTAIR)” at “Step 2”.

If you want to know the detail information of MOLERE based on the resistance mechanism, you can select “Resistance Mechanism” in the “Search for MOLERE Entries by Resistance Mechanism” field, then select a molecule to search for detail information. For example, if you want to know drug resistance information for B-lymphocyte antigen CD19 based on the resistance mechanism of ADTT (Aberration of the Drug's Therapeutic Target), you can select “ADTT: Aberration of the Drug's Therapeutic Target” at “Step 1” and “B-lymphocyte antigen CD19 (CD19)” in the “Step 2”.

If you want to know the detail information of MOLERE based on the disease, you can select “Disease Class” in the “Search for MOLERE Entries by Disease Name” field, then select a molecule to search for detail information. For example, if you want to know drug resistance information for PTEN which is related to the resistance of Liver cancer, you can select “2C12: Liver cancer” at “Step 1” and “Phosphatase and tensin homolog (PTEN)” in the “Step 2”.

5.1. By clicking the “Molecule Info” button, the detailed molecule information page for particular molecule will be displayed.
Take PTEN as an example, “General Information of the Molecule” section displays the general information of Liver cancer including its Name, Synonyms, Molecule Type, Gene Name, Gene ID, Location, Sequence, Function, Uniprot ID, Ensemble ID and HGNC ID.

“Type(s) of Resistant Mechanism of this Molecule” section displays two types of interactions between drug, disease and mechanism.

“Comprehensive Drug Resistance Data Categorized by Drug” section classifies all the drugs that are responded to this molecule.

“Disease-Specific Expression Quantity of This Molecule” section displays the expression quantity of this molecule in those disease

5.2. By clicking the “Drug Info” button, the detailed drug information page for particular “Representative Molecule Response Drug” will be displayed.
Take PTEN as an example, “General Information of the Drug” section displays the general information of Sunitinib which responds to PTEN, including its Name, Synonyms, Indication, Structure, Resistant Disease, Target, Molecular Information and External Link(s).

“Type(s) of Resistant Mechanism of this Drug” section displays five types of interactions between drug, disease and mechanism.

“Comprehensive Drug Resistance Data Categorized by Disease” section displays all disease that confer resistance to Tamoxifen.

5.3. By clicking the “Disease Info” button, the detailed disease information page for particular “Representative Molecule Response Disease” will be displayed.
Take PTEN as an example, “General Information of the Disease” section displays the general information of Brain cancer which PTEN related to, including its Name, ICD and Resistance Map.

“Type(s) of Resistant Mechanism of this Disease” section displays six types of interactions between drug, disease and mechanism.

“Comprehensive Drug Resistance Data Categorized by Drug” section classifies all the drugs that respond to this disease.


6.1. By clicking the “Drug structure similarity search” button, users can search for drugs either by structure similarity or by the SMILES.

In “Input a structure by JMSE editor”, draw the drug compound structure, click “submit” button for searching. In “Input a structure by SMILES”, input a SMILES of drug and click “submit” button for searching. “Drug structure similarity search” allows approximate searching.
6.2. By clicking the “Molecule sequence similarity search” button, users can search for molecule information by protein/ncRNA sequence in FASTA format

Input the protein sequence or RNA sequence in FASTA format and click “submit” button for searching. “Molecule sequence similarity search” allows approximate searching.
