Molecule Information
General Information of the Molecule (ID: Mol00667)
| Name |
Transcription factor A (TFAM)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
mtTFA; Mitochondrial transcription factor 1; MtTF1; Transcription factor 6; TCF-6; Transcription factor 6-like 2; TCF6; TCF6L2
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
TFAM
|
||||
| Gene ID | |||||
| Location |
chr10:58385345-58399220[+]
|
||||
| Sequence |
MAFLRSMWGVLSALGRSGAELCTGCGSRLRSPFSFVYLPRWFSSVLASCPKKPVSSYLRF
SKEQLPIFKAQNPDAKTTELIRRIAQRWRELPDSKKKIYQDAYRAEWQVYKEEISRFKEQ LTPSQIMSLEKEIMDKHLKRKAMTKKKELTLLGKPKRPRSAYNVYVAERFQEAKGDSPQE KLKTVKENWKNLSDSEKELYIQHAKEDETRYHNEMKSWEEQMIEVGRKDLLRRTIKKQRK YGAEEC Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Binds to the mitochondrial light strand promoter and functions in mitochondrial transcription regulation. Component of the mitochondrial transcription initiation complex, composed at least of TFB2M, TFAM and POLRMT that is required for basal transcription of mitochondrial DNA. In this complex, TFAM recruits POLRMT to a specific promoter whereas TFB2M induces structural changes in POLRMT to enable promoter opening and trapping of the DNA non-template strand. Required for accurate and efficient promoter recognition by the mitochondrial RNA polymerase. Promotes transcription initiation from the HSP1 and the light strand promoter by binding immediately upstream of transcriptional start sites. Is able to unwind DNA. Bends the mitochondrial light strand promoter DNA into a U-turn shape via its HMG boxes. Required for maintenance of normal levels of mitochondrial DNA. May play a role in organizing and compacting mitochondrial DNA.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
1 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [1] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 |
| MDA-MB-231/DDP cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTT assay; Annexin V-FITC apoptosis assay | |||
| Mechanism Description | miR199a-3p enhances breast cancer cell sensitivity to cisplatin by downregulating TFAM. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 2.83E-12; Fold-change: 1.55E-01; Z-score: 3.10E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.96E-04; Fold-change: 2.68E-01; Z-score: 4.51E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
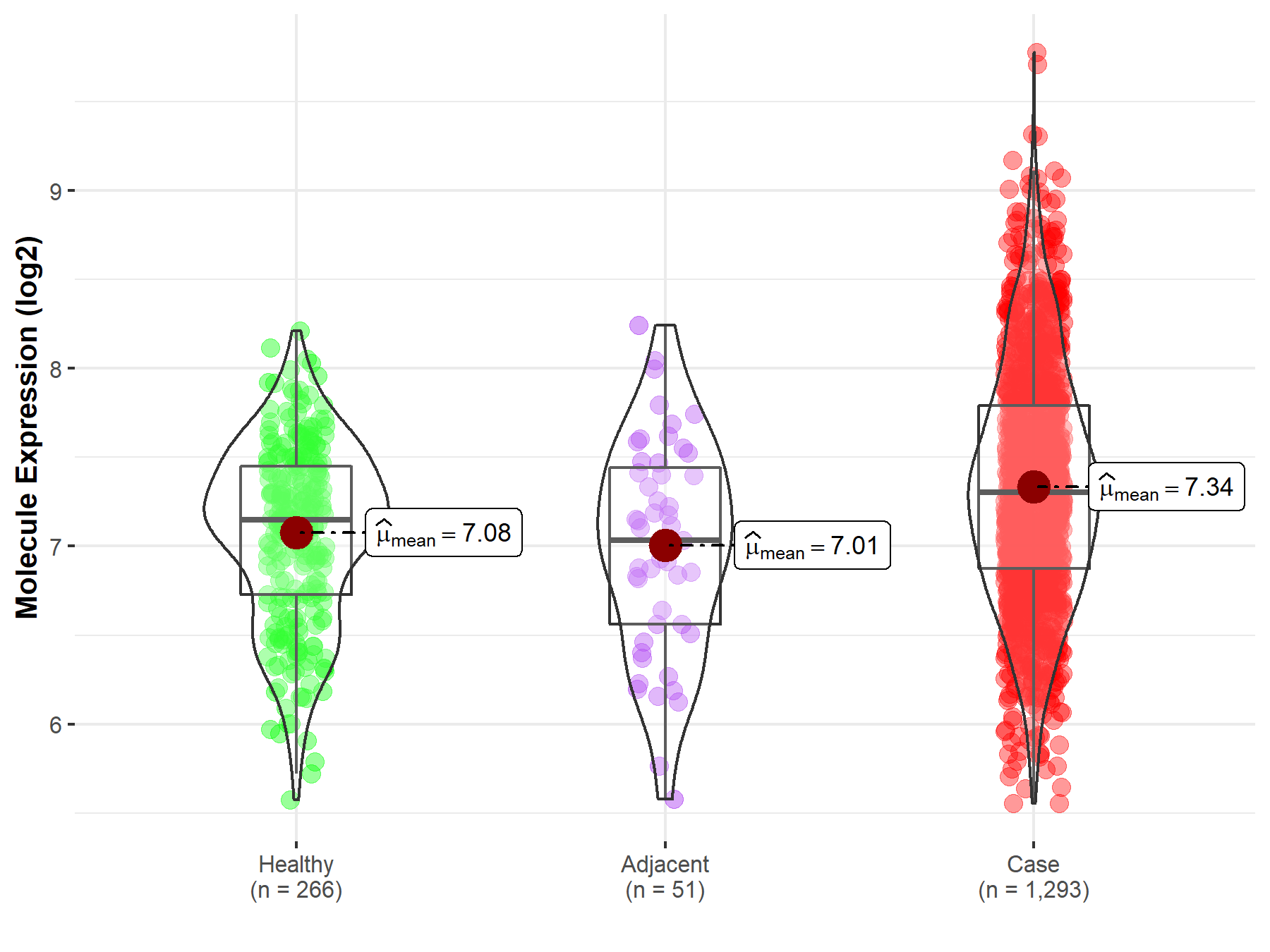
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

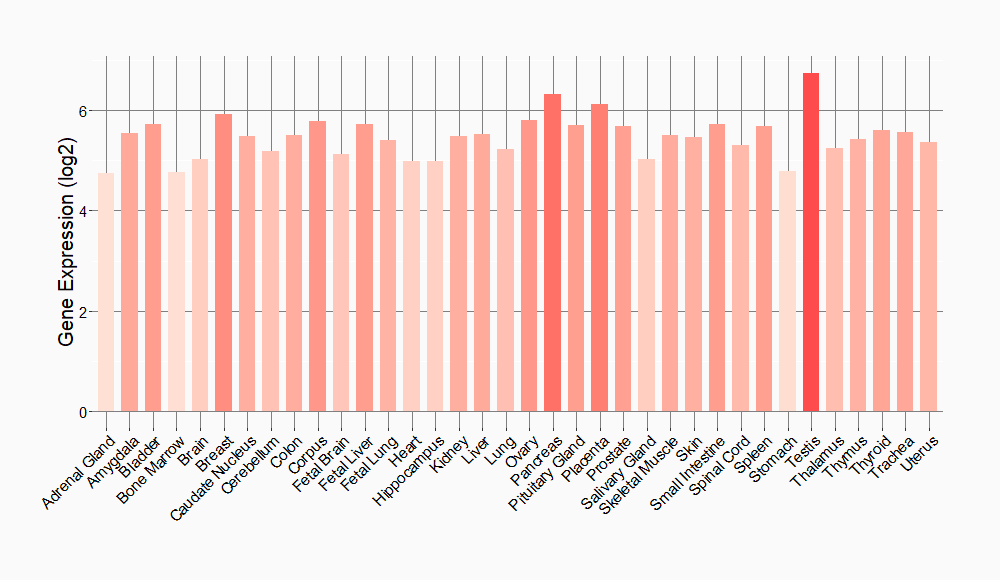
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
