Molecule Information
General Information of the Molecule (ID: Mol00531)
| Name |
Serine/threonine-protein kinase NLK (NLK)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
Nemo-like kinase; Protein LAK1; LAK1
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
NLK
|
||||
| Gene ID | |||||
| Location |
chr17:28041737-28196381[+]
|
||||
| Sequence |
MSLCGARANAKMMAAYNGGTSAAAAGHHHHHHHHLPHLPPPHLHHHHHPQHHLHPGSAAA
VHPVQQHTSSAAAAAAAAAAAAAMLNPGQQQPYFPSPAPGQAPGPAAAAPAQVQAAAAAT VKAHHHQHSHHPQQQLDIEPDRPIGYGAFGVVWSVTDPRDGKRVALKKMPNVFQNLVSCK RVFRELKMLCFFKHDNVLSALDILQPPHIDYFEEIYVVTELMQSDLHKIIVSPQPLSSDH VKVFLYQILRGLKYLHSAGILHRDIKPGNLLVNSNCVLKICDFGLARVEELDESRHMTQE VVTQYYRAPEILMGSRHYSNAIDIWSVGCIFAELLGRRILFQAQSPIQQLDLITDLLGTP SLEAMRTACEGAKAHILRGPHKQPSLPVLYTLSSQATHEAVHLLCRMLVFDPSKRISAKD ALAHPYLDEGRLRYHTCMCKCCFSTSTGRVYTSDFEPVTNPKFDDTFEKNLSSVRQVKEI IHQFILEQQKGNRVPLCINPQSAAFKSFISSTVAQPSEMPPSPLVWE Click to Show/Hide
|
||||
| Function |
Serine/threonine-protein kinase that regulates a number of transcription factors with key roles in cell fate determination. Positive effector of the non-canonical Wnt signaling pathway, acting downstream of WNT5A, MAP3K7/TAK1 and HIPK2. Negative regulator of the canonical Wnt/beta-catenin signaling pathway. Binds to and phosphorylates TCF7L2/TCF4 and LEF1, promoting the dissociation of the TCF7L2/LEF1/beta-catenin complex from DNA, as well as the ubiquitination and subsequent proteolysis of LEF1. Together these effects inhibit the transcriptional activation of canonical Wnt/beta-catenin target genes. Negative regulator of the Notch signaling pathway. Binds to and phosphorylates NOTCH1, thereby preventing the formation of a transcriptionally active ternary complex of NOTCH1, RBPJ/RBPSUH and MAML1. Negative regulator of the MYB family of transcription factors. Phosphorylation of MYB leads to its subsequent proteolysis while phosphorylation of MYBL1 and MYBL2 inhibits their interaction with the coactivator CREBBP. Other transcription factors may also be inhibited by direct phosphorylation of CREBBP itself. Acts downstream of IL6 and MAP3K7/TAK1 to phosphorylate STAT3, which is in turn required for activation of NLK by MAP3K7/TAK1. Upon IL1B stimulus, cooperates with ATF5 to activate the transactivation activity of C/EBP subfamily members. Phosphorylates ATF5 but also stabilizes ATF5 protein levels in a kinase-independent manner.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
1 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [1] | |||
| Sensitive Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Breast cancer [ICD-11: 2C60] | |||
| The Specified Disease | Breast cancer | |||
| The Studied Tissue | Breast tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.10E-37 Fold-change: 1.34E-01 Z-score: 1.40E+01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell viability | Inhibition | hsa05200 | |
| Wnt signaling pathway | Inhibition | hsa04310 | ||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| T47D cells | Breast | Homo sapiens (Human) | CVCL_0553 | |
| BT549 cells | Breast | Homo sapiens (Human) | CVCL_1092 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | LncRNA TUG1 sensitized triple negative breast cancer to cisplatin by upregulating NLk expression via sponging miR-197. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.10E-37; Fold-change: 6.76E-01; Z-score: 1.08E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 6.57E-09; Fold-change: 4.46E-01; Z-score: 7.06E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
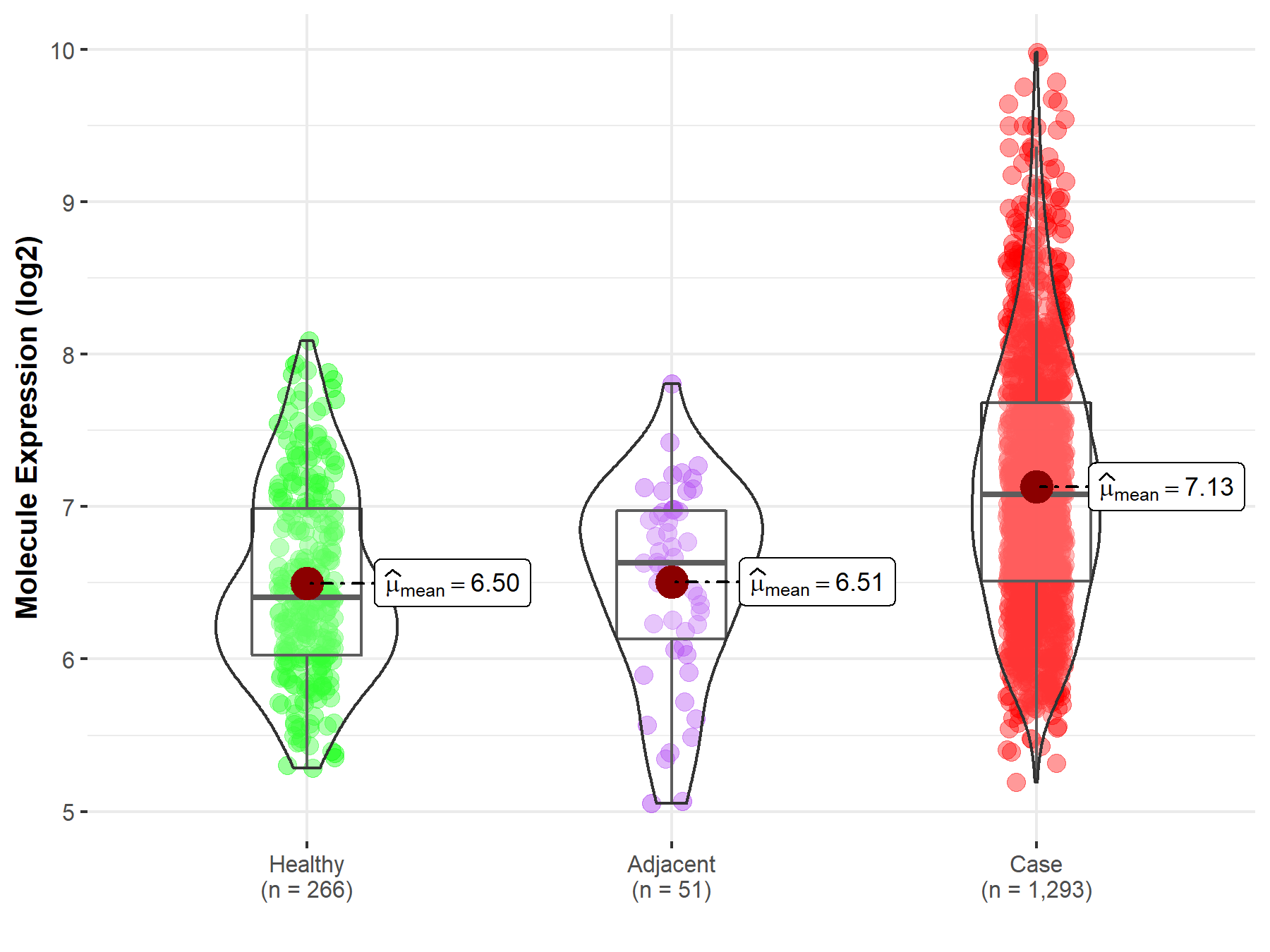
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

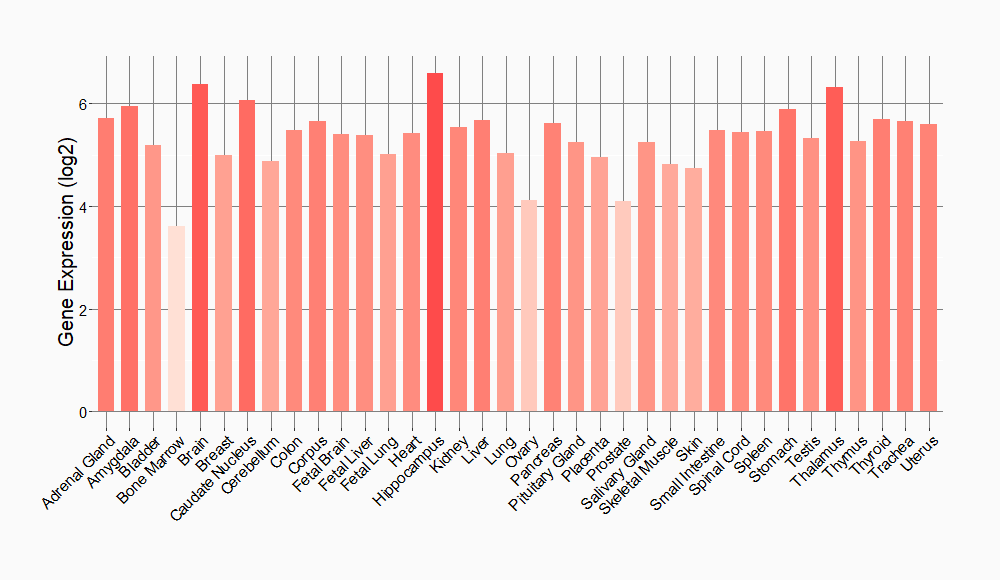
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
