Molecule Information
General Information of the Molecule (ID: Mol00118)
| Name |
Matrix metalloproteinase-9 (MMP9)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
MMP-9; 92 kDa gelatinase; 92 kDa type IV collagenase; Gelatinase B; GELB; CLG4B
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
MMP9
|
||||
| Gene ID | |||||
| Location |
chr20:46008908-46016561[+]
|
||||
| Sequence |
MSLWQPLVLVLLVLGCCFAAPRQRQSTLVLFPGDLRTNLTDRQLAEEYLYRYGYTRVAEM
RGESKSLGPALLLLQKQLSLPETGELDSATLKAMRTPRCGVPDLGRFQTFEGDLKWHHHN ITYWIQNYSEDLPRAVIDDAFARAFALWSAVTPLTFTRVYSRDADIVIQFGVAEHGDGYP FDGKDGLLAHAFPPGPGIQGDAHFDDDELWSLGKGVVVPTRFGNADGAACHFPFIFEGRS YSACTTDGRSDGLPWCSTTANYDTDDRFGFCPSERLYTQDGNADGKPCQFPFIFQGQSYS ACTTDGRSDGYRWCATTANYDRDKLFGFCPTRADSTVMGGNSAGELCVFPFTFLGKEYST CTSEGRGDGRLWCATTSNFDSDKKWGFCPDQGYSLFLVAAHEFGHALGLDHSSVPEALMY PMYRFTEGPPLHKDDVNGIRHLYGPRPEPEPRPPTTTTPQPTAPPTVCPTGPPTVHPSER PTAGPTGPPSAGPTGPPTAGPSTATTVPLSPVDDACNVNIFDAIAEIGNQLYLFKDGKYW RFSEGRGSRPQGPFLIADKWPALPRKLDSVFEERLSKKLFFFSGRQVWVYTGASVLGPRR LDKLGLGADVAQVTGALRSGRGKMLLFSGRRLWRFDVKAQMVDPRSASEVDRMFPGVPLD THDVFQYREKAYFCQDRFYWRVSSRSELNQVDQVGYVTYDILQCPED Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Matrix metalloproteinase that plays an essential role in local proteolysis of the extracellular matrix and in leukocyte migration. Could play a role in bone osteoclastic resorption. Cleaves KiSS1 at a Gly-|-Leu bond. Cleaves NINJ1 to generate the Secreted ninjurin-1 form. Cleaves type IV and type V collagen into large C-terminal three quarter fragments and shorter N-terminal one quarter fragments. Degrades fibronectin but not laminin or Pz-peptide.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Investigative Drug(s)
1 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [1] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Platinum | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Breast cancer [ICD-11: 2C60] | |||
| The Specified Disease | Breast cancer | |||
| The Studied Tissue | Breast tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.35E-110 Fold-change: 3.98E-01 Z-score: 2.85E+01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-231 cells | Breast | Homo sapiens (Human) | CVCL_0062 | |
| SkBR3 cells | Breast | Homo sapiens (Human) | CVCL_0033 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
qRT-PCR | |||
| Experiment for Drug Resistance |
Caspase 3/7 cleavage assays; Aldefluor assay; MTS assay; Flow cytometric analysis | |||
| Mechanism Description | Blocking the EZH2-interactiing domain of HOTAIR and disrupting the HOTAIR-EZH2 interaction resensitizes cancer cells to clinically relevant cytotoxic chemotherapies, reduces cell invasion and decreases NF-kB transcriptional activity and IL-6 and MMP-9 expression in vivo. Levels of genes like MMP9 shown to be up-regulated by HOTAIR. | |||
Approved Drug(s)
2 drug(s) in total
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Prostate cancer [ICD-11: 2C82.0] | [2] | |||
| Sensitive Disease | Prostate cancer [ICD-11: 2C82.0] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Prostate cancer [ICD-11: 2C82] | |||
| The Specified Disease | Prostate cancer | |||
| The Studied Tissue | Prostate | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.62E-01 Fold-change: -2.74E-02 Z-score: -4.42E-01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell invasion | Inhibition | hsa05200 | ||
| Cell migration | Inhibition | hsa04670 | ||
| Cell viability | Inhibition | hsa05200 | ||
| ERK signaling pathway | Inhibition | hsa04210 | ||
| Epithelial mesenchymal transition signaling pathway | Inhibition | hsa01521 | ||
| In Vitro Model | DU-145 cells | Prostate | Homo sapiens (Human) | CVCL_0105 |
| LNCaP cells | Prostate | Homo sapiens (Human) | CVCL_0395 | |
| PC3 cells | Prostate | Homo sapiens (Human) | CVCL_0035 | |
| VCaP cells | Prostate | Homo sapiens (Human) | CVCL_2235 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
CCK8 assay; Flow cytometry assay | |||
| Mechanism Description | UTMD mediated miR 205 transfection increased the expression of caspase 9, cleaved caspase 9, cytochrome c and E cadherin, and decreased the expression of MMP 9 and p ERk,inhibiting PCa cell proliferation, migration and invasion, and promoted apoptosis modulated by cisplatin. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Osteosarcoma [ICD-11: 2B51.0] | [3] | |||
| Sensitive Disease | Osteosarcoma [ICD-11: 2B51.0] | |||
| Sensitive Drug | Silibinin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Cell proliferation | Inhibition | hsa05200 | ||
| In Vitro Model | MG63 cells | Bone marrow | Homo sapiens (Human) | CVCL_0426 |
| Experiment for Molecule Alteration |
qPCR; Western blot analysis | |||
| Experiment for Drug Resistance |
TUNEL cell apoptosis assay; XTT cell proliferation assay; MTT cell proliferation assay; Colony formation assay | |||
| Mechanism Description | MMP-9 was targeted and negatively regulated by hsa-miR494 which inhibited cell proliferation and induced cell apoptosis. Silybin inhibited OS cell viability by altering the protein levels of beta-catenin and RUNX2, thus upregulating the level of hsa-miR494. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.35E-110; Fold-change: 2.48E+00; Z-score: 2.34E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 4.47E-08; Fold-change: 1.91E+00; Z-score: 1.20E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
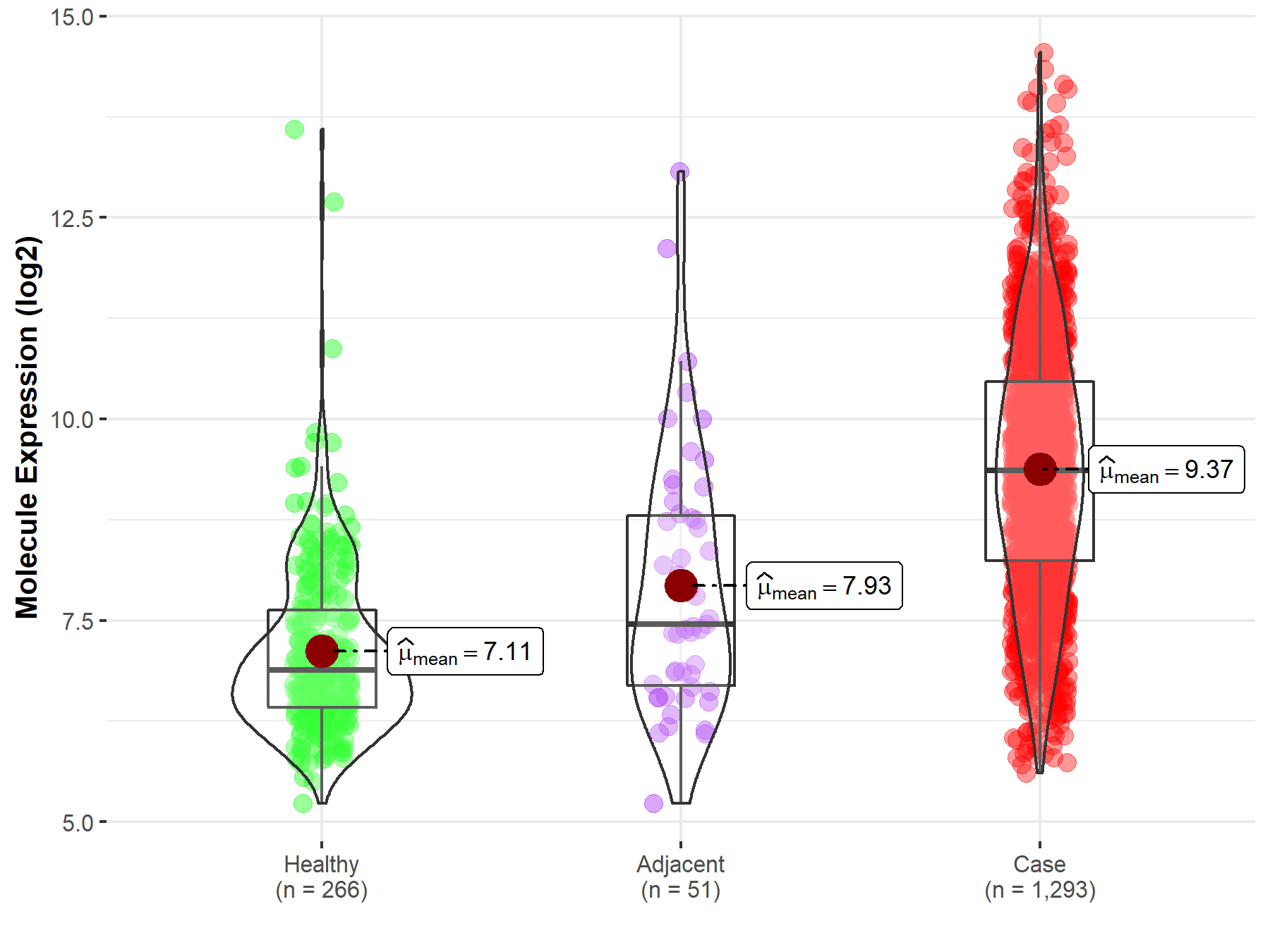
|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Prostate | |
| The Specified Disease | Prostate cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 6.62E-01; Fold-change: 5.13E-02; Z-score: 4.24E-02 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
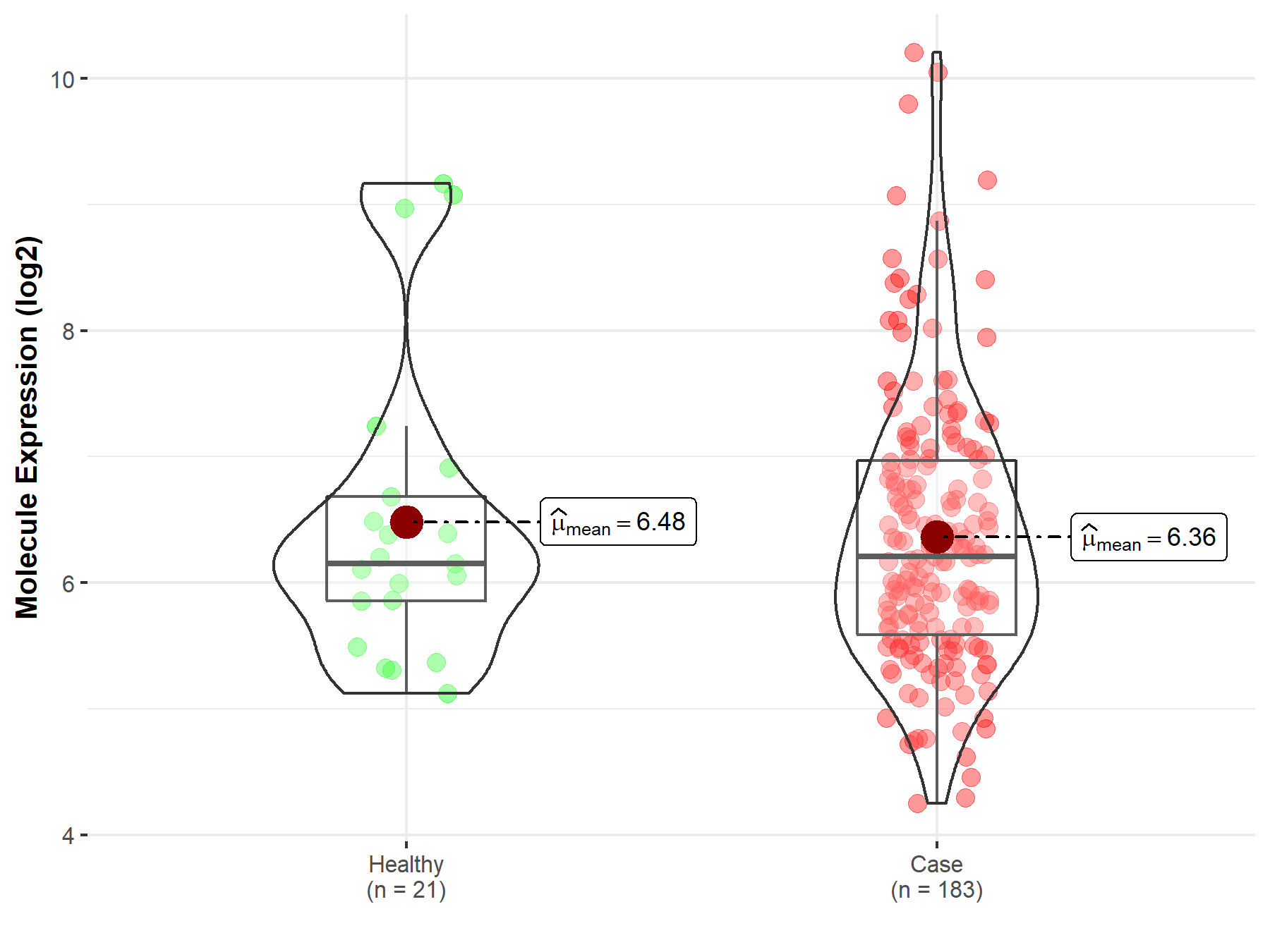
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

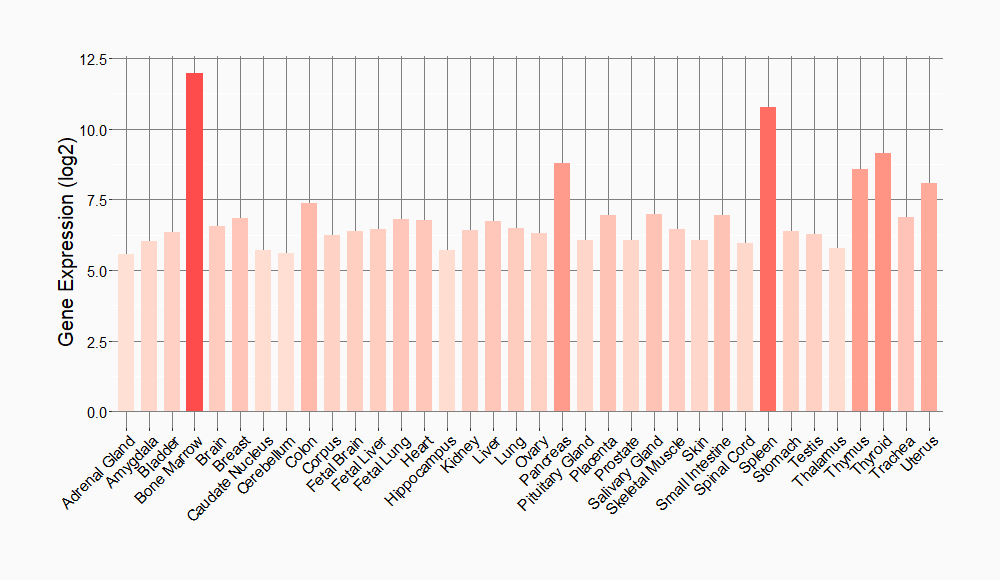
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
