Molecule Information
General Information of the Molecule (ID: Mol00018)
| Name |
Autophagy protein 5 (ATG5)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
APG5-like; Apoptosis-specific protein; APG5L; ASP
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
ATG5
|
||||
| Gene ID | |||||
| Location |
chr6:106045423-106325791[-]
|
||||
| Sequence |
MTDDKDVLRDVWFGRIPTCFTLYQDEITEREAEPYYLLLPRVSYLTLVTDKVKKHFQKVM
RQEDISEIWFEYEGTPLKWHYPIGLLFDLLASSSALPWNITVHFKSFPEKDLLHCPSKDA IEAHFMSCMKEADALKHKSQVINEMQKKDHKQLWMGLQNDRFDQFWAINRKLMEYPAEEN GFRYIPFRIYQTTTERPFIQKLFRPVAADGQLHTLGDLLKEVCPSAIDPEDGEKKNQVMI HGIEPMLETPLQWLSEHLSYPDNFLHISIIPQPTD Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
Involved in autophagic vesicle formation. Conjugation with ATG12, through a ubiquitin-like conjugating system involving ATG7 as an E1-like activating enzyme and ATG10 as an E2-like conjugating enzyme, is essential for its function. The ATG12-ATG5 conjugate acts as an E3-like enzyme which is required for lipidation of ATG8 family proteins and their association to the vesicle membranes. Involved in mitochondrial quality control after oxidative damage, and in subsequent cellular longevity. Plays a critical role in multiple aspects of lymphocyte development and is essential for both B and T lymphocyte survival and proliferation. Required for optimal processing and presentation of antigens for MHC II. Involved in the maintenance of axon morphology and membrane structures, as well as in normal adipocyte differentiation. Promotes primary ciliogenesis through removal of OFD1 from centriolar satellites and degradation of IFT20 via the autophagic pathway.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Approved Drug(s)
3 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [1] | |||
| Resistant Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Gastric cancer [ICD-11: 2B72] | |||
| The Specified Disease | Gastric cancer | |||
| The Studied Tissue | Gastric tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.87E-01 Fold-change: 4.38E-02 Z-score: 8.44E-01 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell autophagy | Activation | hsa04140 | |
| In Vitro Model | AGS cells | Gastric | Homo sapiens (Human) | CVCL_0139 |
| HGC27 cells | Gastric | Homo sapiens (Human) | CVCL_1279 | |
| Experiment for Molecule Alteration |
Western blot analysis; RT-qPCR | |||
| Experiment for Drug Resistance |
CCK8 assay | |||
| Mechanism Description | LncRNA MALAT1 potentiates autophagy associated cisplatin resistance by suppressing the microRNA 30b/autophagy related gene 5 axis in gastric cancer. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Gastric cancer [ICD-11: 2B72.1] | [3] | |||
| Sensitive Disease | Gastric cancer [ICD-11: 2B72.1] | |||
| Sensitive Drug | Cisplatin | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell proliferation | Inhibition | hsa05200 | |
| In Vitro Model | SGC7901/CDDP cells | Gastric | Homo sapiens (Human) | CVCL_0520 |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | miR-181a inhibited autophagy in cisplatin-resistant cell line SGC7901/CDDP. ATG5 was a potential target of miR-181a. miR-181a sensitized SGC7901/CDDP cells to cisplatin in vivo and in vitro. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Melanoma [ICD-11: 2C30.0] | [2] | |||
| Sensitive Disease | Melanoma [ICD-11: 2C30.0] | |||
| Sensitive Drug | Vemurafenib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Melanoma [ICD-11: 2C30] | |||
| The Specified Disease | Melanoma | |||
| The Studied Tissue | Skin | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.48E-01 Fold-change: -3.74E-02 Z-score: -1.49E+00 |
|||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | Cell autophagy | Inhibition | hsa04140 | |
| In Vitro Model | A375 cells | Skin | Homo sapiens (Human) | CVCL_0132 |
| A375-R cells | Skin | Homo sapiens (Human) | CVCL_6234 | |
| G-361 cells | Skin | Homo sapiens (Human) | CVCL_1220 | |
| G361/R cells | Skin | Homo sapiens (Human) | CVCL_IW13 | |
| MeWo cells | Skin | Homo sapiens (Human) | CVCL_0445 | |
| In Vivo Model | Nude mouse xenograft model | Mus musculus | ||
| Experiment for Molecule Alteration |
RT-PCR | |||
| Experiment for Drug Resistance |
MTT assay; Flow cytometric analysis | |||
| Mechanism Description | miR216b enhances the efficacy of vemurafenib by targeting Beclin-1, UVRAG and ATG5 in melanoma. miR216b suppresses autophagy in both BRAFi-sensitive and -resistant melanoma cells. | |||
| Drug Sensitivity Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Chronic myeloid leukemia [ICD-11: 2A20.0] | [4] | |||
| Sensitive Disease | Chronic myeloid leukemia [ICD-11: 2A20.0] | |||
| Sensitive Drug | Imatinib | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| Cell Pathway Regulation | Cell apoptosis | Activation | hsa04210 | |
| Mitochondrial signaling pathway | Activation | hsa04217 | ||
| In Vitro Model | K562 cells | Blood | Homo sapiens (Human) | CVCL_0004 |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
Flow cytometry assay | |||
| Mechanism Description | miR-30a mimic or knockdown of autophagy genes (ATGs) such as Beclin 1 and ATG5 by short hairpin RNA enhances imatinib-induced cytotoxicity and promotes mitochondria-dependent intrinsic apoptosis. In contrast, knockdown of miR-30a by antagomiR-30a increases the expression of Beclin 1 and ATG5, and inhibits imatinib-induced cytotoxicity. | |||
Investigative Drug(s)
1 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | [5] | |||
| Resistant Disease | Pancreatic ductal adenocarcinoma [ICD-11: 2C10.0] | |||
| Resistant Drug | 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone | |||
| Molecule Alteration | Expression | Down-regulation |
||
| Experimental Note | Revealed Based on the Cell Line Data | |||
| Cell Pathway Regulation | beta2AR-Akt feedback loop signalling pathway | Regulation | N.A. | |
| In Vitro Model | Panc1 cells | Pancreas | Homo sapiens (Human) | CVCL_0480 |
| Experiment for Molecule Alteration |
Western blot assay; qRT-PCR | |||
| Experiment for Drug Resistance |
Colony formation assay; Stem cell sphere formation assay | |||
| Mechanism Description | In this study, we found that NNK promoted stemness and gemcitabine resistance in pancreatic cancer cell lines. Moreover, NNK increased autophagy and elevated the expression levels of the autophagy-related markers autophagy-related gene 5 (ATG5), autophagy-related gene 7 (ATG7), and Beclin1. Furthermore, the results showed that NNK-promoted stemness and gemcitabine resistance was partially dependent on the role of NNK in cell autophagy, which is mediated by the beta2-adrenergic receptor (beta2AR)-Akt axis. Finally, we proved that NNK intervention could not only activate beta2AR, but also increase its expression, making beta2AR and Akt form a feedback loop. Overall, these findings show that the NNK-induced beta2AR-Akt feedback loop promotes stemness and gemcitabine resistance in pancreatic cancer cells. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Whole blood | |
| The Specified Disease | Myelofibrosis | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.88E-01; Fold-change: 2.10E-02; Z-score: 4.90E-02 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| The Studied Tissue | Whole blood | |
| The Specified Disease | Polycythemia vera | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 5.79E-01; Fold-change: -5.08E-02; Z-score: -1.24E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Gastric tissue | |
| The Specified Disease | Gastric cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 4.87E-01; Fold-change: 1.27E-01; Z-score: 2.60E-01 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 2.35E-02; Fold-change: 1.14E-01; Z-score: 3.94E-01 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Skin | |
| The Specified Disease | Melanoma | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 1.48E-01; Fold-change: -1.84E-01; Z-score: -2.75E-01 | |
|
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |

|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals

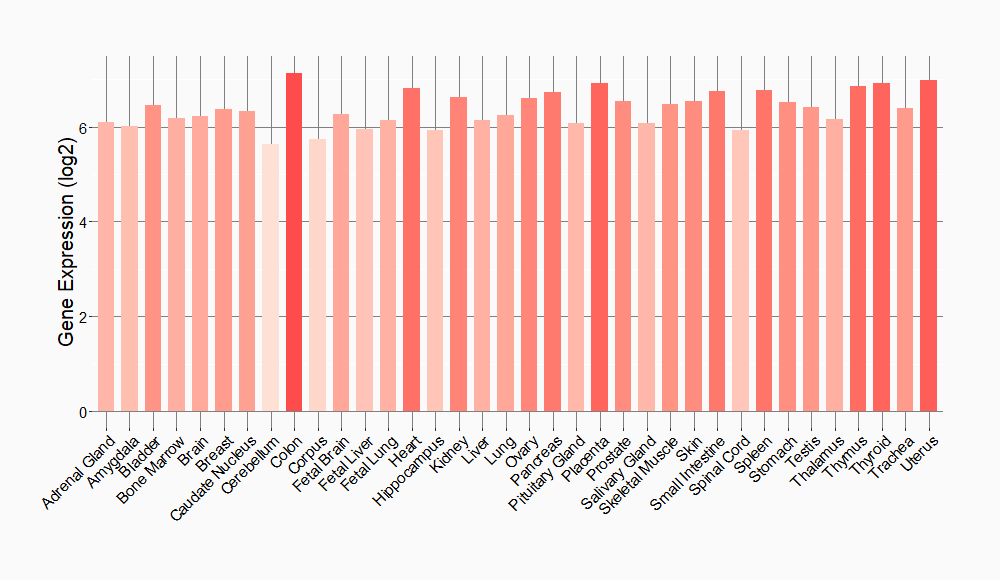
|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
