Molecule Information
General Information of the Molecule (ID: Mol01965)
| Name |
Ubiquitin protein ligase E3 component n-recognin 5 (UBR5)
,Homo sapiens
|
||||
|---|---|---|---|---|---|
| Synonyms |
UBR5; EDD; EDD1; HYD; KIAA0896
Click to Show/Hide
|
||||
| Molecule Type |
Protein
|
||||
| Gene Name |
UBR5
|
||||
| Gene ID | |||||
| Location |
chr8:102,252,273-102,412,759[-]
|
||||
| Sequence |
MTSIHFVVHPLPGTEDQLNDRLREVSEKLNKYNLNSHPPLNVLEQATIKQCVVGPNHAAF
LLEDGRVCRIGFSVQPDRLELGKPDNNDGSKLNSNSGAGRTSRPGRTSDSPWFLSGSETL GRLAGNTLGSRWSSGVGGSGGGSSGRSSAGARDSRRQTRVIRTGRDRGSGLLGSQPQPVI PASVIPEELISQAQVVLQGKSRSVIIRELQRTNLDVNLAVNNLLSRDDEDGDDGDDTASE SYLPGEDLMSLLDADIHSAHPSVIIDADAMFSEDISYFGYPSFRRSSLSRLGSSRVLLLP LERDSELLRERESVLRLRERRWLDGASFDNERGSTSKEGEPNLDKKNTPVQSPVSLGEDL QWWPDKDGTKFICIGALYSELLAVSSKGELYQWKWSESEPYRNAQNPSLHHPRATFLGLT NEKIVLLSANSIRATVATENNKVATWVDETLSSVASKLEHTAQTYSELQGERIVSLHCCA LYTCAQLENSLYWWGVVPFSQRKKMLEKARAKNKKPKSSAGISSMPNITVGTQVCLRNNP LYHAGAVAFSISAGIPKVGVLMESVWNMNDSCRFQLRSPESLKNMEKASKTTEAKPESKQ EPVKTEMGPPPSPASTCSDASSIASSASMPYKRRRSTPAPKEEEKVNEEQWSLREVVFVE DVKNVPVGKVLKVDGAYVAVKFPGTSSNTNCQNSSGPDADPSSLLQDCRLLRIDELQVVK TGGTPKVPDCFQRTPKKLCIPEKTEILAVNVDSKGVHAVLKTGNWVRYCIFDLATGKAEQ ENNFPTSSIAFLGQNERNVAIFTAGQESPIILRDGNGTIYPMAKDCMGGIRDPDWLDLPP ISSLGMGVHSLINLPANSTIKKKAAVIIMAVEKQTLMQHILRCDYEACRQYLMNLEQAVV LEQNLQMLQTFISHRCDGNRNILHACVSVCFPTSNKETKEEEEAERSERNTFAERLSAVE AIANAISVVSSNGPGNRAGSSSSRSLRLREMMRRSLRAAGLGRHEAGASSSDHQDPVSPP IAPPSWVPDPPAMDPDGDIDFILAPAVGSLTTAATGTGQGPSTSTIPGPSTEPSVVESKD RKANAHFILKLLCDSVVLQPYLRELLSAKDARGMTPFMSAVSGRAYPAAITILETAQKIA KAEISSSEKEEDVFMGMVCPSGTNPDDSPLYVLCCNDTCSFTWTGAEHINQDIFECRTCG LLESLCCCTECARVCHKGHDCKLKRTSPTAYCDCWEKCKCKTLIAGQKSARLDLLYRLLT ATNLVTLPNSRGEHLLLFLVQTVARQTVEHCQYRPPRIREDRNRKTASPEDSDMPDHDLE PPRFAQLALERVLQDWNALKSMIMFGSQENKDPLSASSRIGHLLPEEQVYLNQQSGTIRL DCFTHCLIVKCTADILLLDTLLGTLVKELQNKYTPGRREEAIAVTMRFLRSVARVFVILS VEMASSKKKNNFIPQPIGKCKRVFQALLPYAVEELCNVAESLIVPVRMGIARPTAPFTLA STSIDAMQGSEELFSVEPLPPRPSSDQSSSSSQSQSSYIIRNPQQRRISQSQPVRGRDEE QDDIVSADVEEVEVVEGVAGEEDHHDEQEEHGEENAEAEGQHDEHDEDGSDMELDLLAAA ETESDSESNHSNQDNASGRRSVVTAATAGSEAGASSVPAFFSEDDSQSNDSSDSDSSSSQ SDDIEQETFMLDEPLERTTNSSHANGAAQAPRSMQWAVRNTQHQRAASTAPSSTSTPAAS SAGLIYIDPSNLRRSGTISTSAAAAAAALEASNASSYLTSASSLARAYSIVIRQISDLMG LIPKYNHLVYSQIPAAVKLTYQDAVNLQNYVEEKLIPTWNWMVSIMDSTEAQLRYGSALA SAGDPGHPNHPLHASQNSARRERMTAREEASLRTLEGRRRATLLSARQGMMSARGDFLNY ALSLMRSHNDEHSDVLPVLDVCSLKHVAYVFQALIYWIKAMNQQTTLDTPQLERKRTREL LELGIDNEDSEHENDDDTNQSATLNDKDDDSLPAETGQNHPFFRRSDSMTFLGCIPPNPF EVPLAEAIPLADQPHLLQPNARKEDLFGRPSQGLYSSSASSGKCLMEVTVDRNCLEVLPT KMSYAANLKNVMNMQNRQKKEGEEQPVLPEETESSKPGPSAHDLAAQLKSSLLAEIGLTE SEGPPLTSFRPQCSFMGMVISHDMLLGRWRLSLELFGRVFMEDVGAEPGSILTELGGFEV KESKFRREMEKLRNQQSRDLSLEVDRDRDLLIQQTMRQLNNHFGRRCATTPMAVHRVKVT FKDEPGEGSGVARSFYTAIAQAFLSNEKLPNLECIQNANKGTHTSLMQRLRNRGERDRER EREREMRRSSGLRAGSRRDRDRDFRRQLSIDTRPFRPASEGNPSDDPEPLPAHRQALGER LYPRVQAMQPAFASKITGMLLELSPAQLLLLLASEDSLRARVDEAMELIIAHGRENGADS ILDLGLVDSSEKVQQENRKRHGSSRSVVDMDLDDTDDGDDNAPLFYQPGKRGFYTPRPGK NTEARLNCFRNIGRILGLCLLQNELCPITLNRHVIKVLLGRKVNWHDFAFFDPVMYESLR QLILASQSSDADAVFSAMDLAFAIDLCKEEGGGQVELIPNGVNIPVTPQNVYEYVRKYAE HRMLVVAEQPLHAMRKGLLDVLPKNSLEDLTAEDFRLLVNGCGEVNVQMLISFTSFNDES GENAEKLLQFKRWFWSIVEKMSMTERQDLVYFWTSSPSLPASEEGFQPMPSITIRPPDDQ HLPTANTCISRLYVPLYSSKQILKQKLLLAIKTKNFGFV Click to Show/Hide
|
||||
| 3D-structure |
|
||||
| Function |
E3 ubiquitin-protein ligase which is a component of the N-end rule pathway. Recognizes and binds to proteins bearing specific N-terminal residues that are destabilizing according to the N-end rule, leading to their ubiquitination and subsequent degradation (By similarity). Involved in maturation and/or transcriptional regulation of mRNA by activating CDK9 by polyubiquitination. May play a role in control of cell cycle progression. May have tumor suppressor function. Regulates DNA topoisomerase II binding protein (TopBP1) in the DNA damage response. Plays an essential role in extraembryonic development. Ubiquitinates acetylated PCK1. Also acts as a regulator of DNA damage response by acting as a suppressor of RNF168, an E3 ubiquitin-protein ligase that promotes accumulation of 'Lys-63'-linked histone H2A and H2AX at DNA damage sites, thereby acting as a guard against excessive spreading of ubiquitinated chromatin at damaged chromosomes.
Click to Show/Hide
|
||||
| Uniprot ID | |||||
| Ensembl ID | |||||
| HGNC ID | |||||
| Click to Show/Hide the Complete Species Lineage | |||||
Type(s) of Resistant Mechanism of This Molecule
Drug Resistance Data Categorized by Drug
Investigative Drug(s)
1 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [1] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | INK128 | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Differential expression of the molecule in resistant disease | ||||
| Classification of Disease | Breast cancer [ICD-11: 2C60] | |||
| The Specified Disease | Breast cancer | |||
| The Studied Tissue | Breast tissue | |||
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.41E-80 Fold-change: 1.02E-01 Z-score: 2.18E+01 |
|||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-436 cells | Breast | Homo sapiens (Human) | CVCL_0623 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | High nuclear EDD expression in a cohort of 151 women with serous ovarian carcinoma was associated with an increased risk of disease recurrence following first-line chemotherapy, and siRNA-knockdown of EDD gene expression partially restored cisplatin sensitivity in cisplatin-resistant ovarian cancer cells in vitro. Loss of EDD induced cell-cycle arrest at G1 through upregulation of tumour suppressor p53 and p21 proteins in osteosarcoma cells in vitro. | |||
Approved Drug(s)
2 drug(s) in total
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [1] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Cisplatin | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-436 cells | Breast | Homo sapiens (Human) | CVCL_0623 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | High nuclear EDD expression in a cohort of 151 women with serous ovarian carcinoma was associated with an increased risk of disease recurrence following first-line chemotherapy, and siRNA-knockdown of EDD gene expression partially restored cisplatin sensitivity in cisplatin-resistant ovarian cancer cells in vitro. Loss of EDD induced cell-cycle arrest at G1 through upregulation of tumour suppressor p53 and p21 proteins in osteosarcoma cells in vitro. | |||
| Drug Resistance Data Categorized by Their Corresponding Mechanisms | ||||
|
|
||||
| Disease Class: Breast cancer [ICD-11: 2C60.3] | [1] | |||
| Resistant Disease | Breast cancer [ICD-11: 2C60.3] | |||
| Resistant Drug | Sirolimus | |||
| Molecule Alteration | Expression | Up-regulation |
||
| Experimental Note | Identified from the Human Clinical Data | |||
| In Vitro Model | MCF-7 cells | Breast | Homo sapiens (Human) | CVCL_0031 |
| MDA-MB-436 cells | Breast | Homo sapiens (Human) | CVCL_0623 | |
| Experiment for Molecule Alteration |
Western blot analysis | |||
| Experiment for Drug Resistance |
MTS assay | |||
| Mechanism Description | High nuclear EDD expression in a cohort of 151 women with serous ovarian carcinoma was associated with an increased risk of disease recurrence following first-line chemotherapy, and siRNA-knockdown of EDD gene expression partially restored cisplatin sensitivity in cisplatin-resistant ovarian cancer cells in vitro. Loss of EDD induced cell-cycle arrest at G1 through upregulation of tumour suppressor p53 and p21 proteins in osteosarcoma cells in vitro. | |||
Disease- and Tissue-specific Abundances of This Molecule
ICD Disease Classification 02

| Differential expression of molecule in resistant diseases | ||
| The Studied Tissue | Breast tissue | |
| The Specified Disease | Breast cancer | |
| The Expression Level of Disease Section Compare with the Healthy Individual Tissue | p-value: 9.41E-80; Fold-change: 6.30E-01; Z-score: 1.72E+00 | |
| The Expression Level of Disease Section Compare with the Adjacent Tissue | p-value: 3.25E-09; Fold-change: 5.74E-01; Z-score: 1.09E+00 | |
|
Molecule expression in the normal tissue adjacent to the diseased tissue of patients
Molecule expression in the diseased tissue of patients
Molecule expression in the normal tissue of healthy individuals
|
||
| Disease-specific Molecule Abundances |
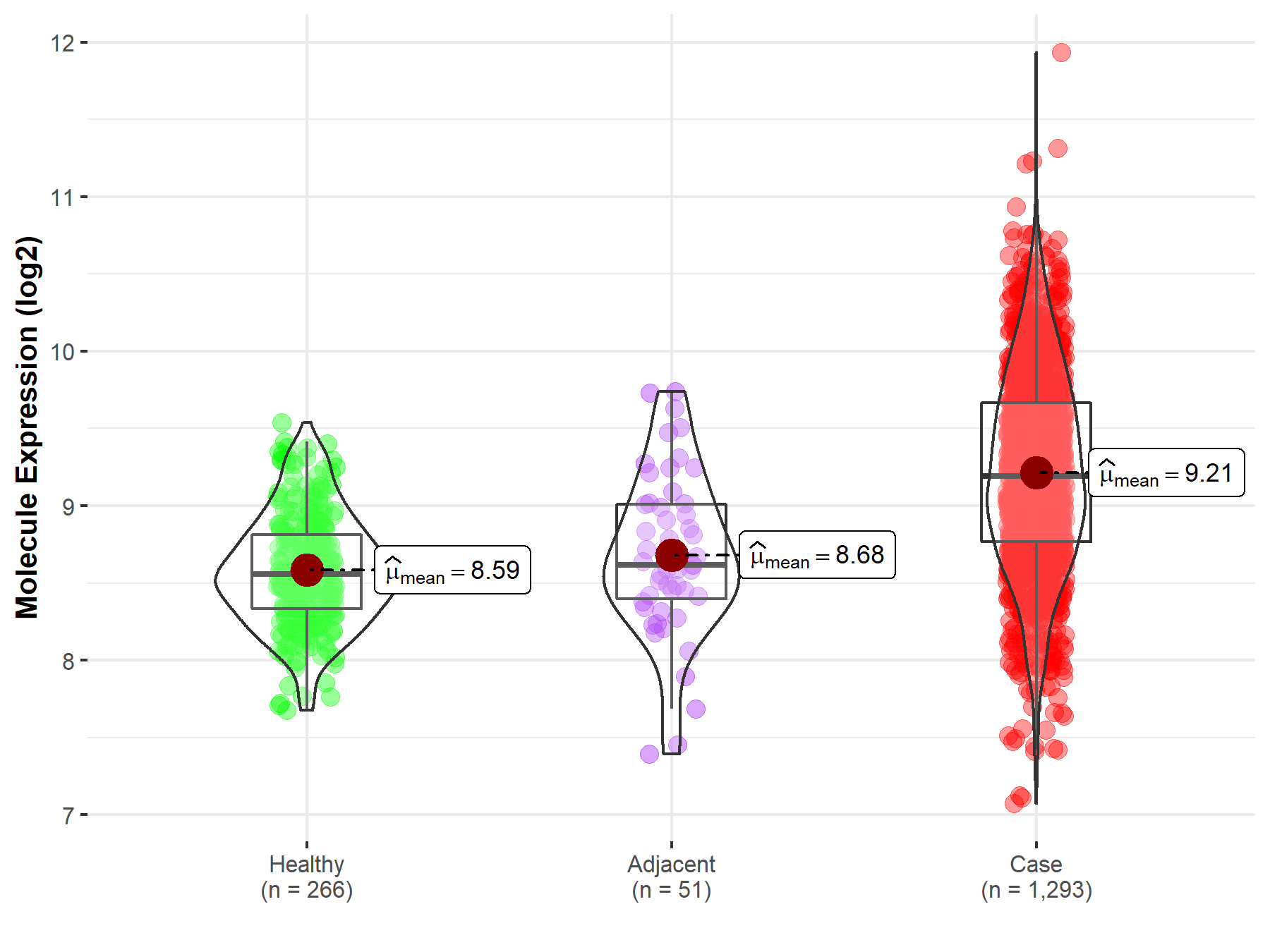
|
Click to View the Clearer Original Diagram |
Tissue-specific Molecule Abundances in Healthy Individuals


|
||
References
If you find any error in data or bug in web service, please kindly report it to Dr. Sun and Dr. Yu.
